Systems pharmacology integrates structural biological and pharmacological knowledge and experimental data, enabling dissection of organism and drug properties and providing excellent predictivity. The development of systems pharmacology models is a significant task requiring massive amounts of background information beyond individual trial data. The qualification of models needs repetitive demonstration of successful predictions. Open Systems Pharmacology is a community that develops, qualifies, and shares professional open source software tools and models in a collaborative open‐science way.
In recent years, systems pharmacology and its applications in pharmaceutical development have gained significant visibility and attention as an important pillar of model‐informed drug (discovery and) development.1 Physiologically‐based pharmacokinetics (PBPK) is just one of the many potential applications of systems pharmacology. Nevertheless, because of the generic relevance of pharmacokinetics in any pharmaceutical application, PBPK has seen the most rapid growth in terms of the number of organizations and scientists applying it and the published case studies. Regulatory use is documented in reviews of the European and US regulatory agencies2, 3 and has lately led both the US Food and Drug Administration (FDA)4 and European Medicines Agency5 to issue guidances for PBPK use. For some applications, PBPK has even become a recommended method (the FDA's Pharmaceutical Science and Clinical Pharmacology Advisory Committee majority vote in support of the use of PBPK modeling for pediatric drug development, March 2012). In many regards, the slow development of PBPK from the early concepts in the 1930s6 over many decades to its accelerated uptake in some regulatory applications2, 3, 7 within just a few years is a striking example of the catalytic effects of a diverse scientific community (see Important Milestones of PBPK Modeling Supplement).
Catalyzing Progress in Systems Pharmacology With an Open‐Science Approach
In light of the success seen with some applications of PBPK, it is important to find effective ways to further grow more applications of systems pharmacology (a much broader concept than mechanistic pharmacokinetics modeling alone) to a level where their full potential in supporting pharmaceutical sciences and innovation is exploited. The development of any systems pharmacology model is a significant task requiring large amounts of high‐quality background information. The qualification of models builds on the repetitive demonstration of successful predictions. Only in exceptional cases, individual researchers or organizations possess the resources and time for this undertaking. The integrative nature of system pharmacology, however, lends itself to an open‐science approach where information, models, and software are jointly developed.
To this end, building on 2 decades of experience with PBPK and system pharmacology modeling, we have established the Open Systems Pharmacology (OSP) community. OSP offers professional open access and open source software tools (PK‐Sim/MoBi) and models via a platform for collaborative development and qualification.
OSP already hosts several general systems pharmacology activities (for an example, see the OSP Links Supplement: Glucose‐Insulin Model , 8) addressing the broader scope of mechanistic disease and drug‐response modeling. A strong focus of the current OSP projects is on the demonstration of reliability and the characterization of predictive power of PBPK. The aims in this latter area are exemplary (and hopefully a convincing role model for other applications to follow) for the benefits of an open‐science approach. We intend to gain full confidence in PBPK as tool for (i) mechanistic investigations and (ii) simulations as a surrogate for clinical trial data. In particular, the latter will also require regulatory acceptance in compliance with applicable guidances. The “qualification for intended use” introduced in the European Medicines Agency guideline translates into the following two major challenges:
The provision of a sufficient package of successful prediction case studies
Full transparency of the approach, processes, tools, and models used
For any specific use such as first‐in‐man, drug–drug interactions, or kinetics in special populations, a series of overall successful prospective predictions is needed, and for none of the popular platforms has a general qualification scheme been established yet. Despite the expectations for model/platform qualifications as detailed in the guidances,4, 5 it is very likely that any of the individual stakeholders in the PBPK field (academics, software providers and consultancy firms, industry, regulatory agencies) will no longer be able to compile the evidence required for a convincing qualification of a given PBPK use in isolation. This is a simple fact even more so because the requirements cover the full scope from scientific content (e.g., clinical and preclinical data) to technical implementation (validation of computerized system) and documentation. Collaborative approaches are needed to compile the requested evidence in all these domains.
The Contribution of an Open Sciences Community
The OSP's vision is to provide robust and reliable, easy‐to‐use modeling and simulation tools, processes, and models for pharmaceutical and other life‐sciences applications. They shall be qualified and accepted by the scientific community representing academia, regulatory agencies, and industry and available and open to everyone (see OSP Links Supplement: Vision & Mission). The OSP community wants to provide a platform for joint development, review, and qualification; application of state‐of‐the‐art tools for PBPK and systems pharmacology modeling; and an open library of models for application as well as method and tool qualification processes.
This scope translates into the following three tasks (Figure 1 a):
Provision of fully transparent (open source) high‐quality software, models, and data sets free of license costs (open access)
Organization of a community that jointly develops scientific concepts, applications, and best practices and invests in training and education of its members and other interested parties (open science)
Development and certification of best practices, qualification packages for specific PBPK and systems pharmacology use, and facilitation of scientific advice
Figure 1.
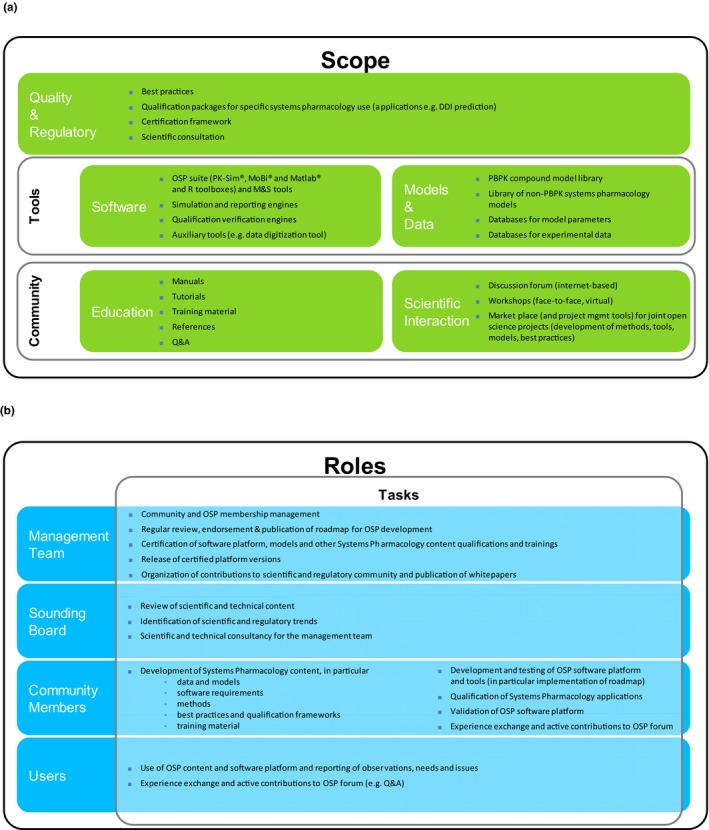
Scope and organization of the OPS community. (a) The scope of the OSP community—open science in a community that organizes development of tools and ensures and certifies quality. (b) Four different roles in the OSP community can be distinguished: users of OSP content, actively contributing community members, members of the sounding board, and the management team. DDI, drug–drug interactions; mgmt., management; M&S, modeling and simulation; OSP, Open Systems Pharmacology; PBPK, physiologically‐based pharmacokinetics.
OSP Tools—Software, Models, and Data
Open systems pharmacology uses GitHub (GitHub, San Francisco, CA, USA) as its collaboration and exchange platform. From the release of version 7.0 in early 2017 onward, already eight releases of the formerly commercial OSP suite comprising PK‐Sim (PBPK) and MoBi (general systems pharmacology modeling) have been published as open‐source software (latest release 8.0; see OSP Links Supplement: Releases; GPLv2 license, see OSP Links Supplement: License).
The OSP suite provides functionality for the full range of PBPK applications from physico‐chemistry‐based prediction models for all relevant (pre)clinical species to elaborated simulations for drug–drug interactions and special populations. The suite comprises functionalities for parameter identification, model qualification and automated reporting as well as interfaces to R (The R Project for Statistical Computing, http://www.R-project.org) and Matlab (The Mathworks, Natick, MA, USA) (Figure 2 a; see OSP Links Supplement: OSP Suite).
Figure 2.
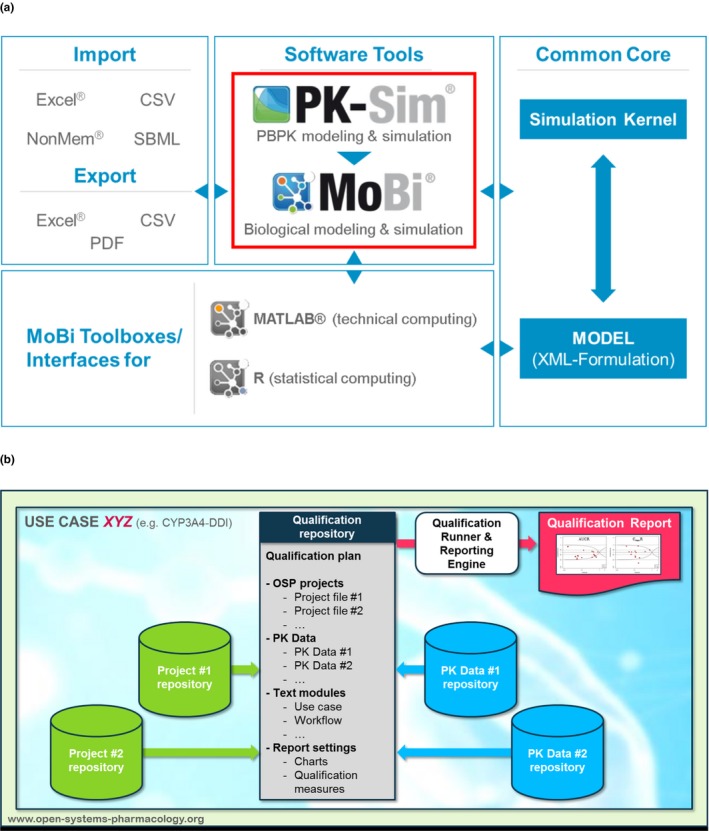
Architecture of the OSP suite and model qualification workflows. (a) The OSP suite consists of the graphical user interfaces PK‐Sim and MoBi, toolboxes for Matlab and R, and builds on a single computational simulation kernel. Import/export functionality for different data formats and model languages are built in. (b) Automated qualification support is a built‐in functionality of the OSP suite (see OSP Links Supplement: Qualification Workflow). A qualification repository consists of model project files, experimental data, and a formal qualification plan describing the simulations, visualizations, and reporting to be automatically generated by a qualification engine. Several qualification repositories for drug–drug interactions, and pediatric applications have already been published on GitHub (for an example, see OSP Links Supplement: Pediatric‐CYP2C8‐Qualification). CYP2C8, Cytochrome P4502C8; DDI, drug–drug interactions; OSP, Open Systems Pharmacology; PK, pharmacokinetics.
The continuous development of the software suite is run on GitHub and can be monitored by everyone in real time (see OSP Links Supplement: Continuous Development). Nightly builds of prerelease versions are available for all interested parties as is the source code.
For sharing of data and PBPK and systems pharmacology models, dedicated repositories exist. The current model library includes PBPK models of individual drugs in particular drug–drug interaction victims and perpetrators and sophisticated systems pharmacology models. An expression database for proteins is also part of the OSP suite.
OSP Community—Education and Scientific Interaction
Education and training of competent PBPK and systems pharmacology users is an important goal of the OSP community. A diverse group of knowledgeable users is the source of innovative ideas and new applications and workflows and creates a motivating environment. A series of videos and tutorials are already shared on GitHub that covers a broad range of topics from introductory lessons (e.g., understanding fundamental concepts of PBPK) to advanced applications (e.g., compliance with guidelines for model development and qualification; see OSP Links Supplement: Tutorials). Training curricula and teaching materials for use in graduate programs, industry, and regulatory environments are planned. For community members interested in providing training, the platform offers a natural “market place” for interaction with interested trainees.
A less formal method of education and support is provided by the discussion forum. It supports OSP users in learning from and providing support to each other. It also serves as a match‐making environment for formative assessment of ideas, models, and so on.
When groups of community members intend to jointly develop new data sets, models, or tools, the GitHub platform also provides project management functionality facilitating collaborative work.
OSP has been selected by several publicly funded projects as the provider of modeling and simulation tools and the technical platform for sharing of project results. Funding agencies of these projects include the German Ministry for Education and Research (BMBF), European Union, Bill & Melinda Gates Foundation9, and US FDA (see Acknowledgments).
OSP Quality and Certification
Because an important goal of OSP is regulatory‐accepted qualification of PBPK and systems‐pharmacology‐based approaches, a strong emphasis of the OSP activities is on qualification efforts and the development of a framework for certification. The OSP community establishes best practices and fully transparent processes for qualification and software validation that will enable it to certify compliance with regulatory standards as a learned society (see Figure 2 b, automated qualification as an example). The OSP management team is responsible for the development and maintenance of the OSP roadmap and the formal release of certified OSP suite versions, best practices, and the coordination of all community activities.
Community Organization
Figure 1 b sketches the constitution of the OSP community where three types of active roles are distinguished. The OSP management team is a project group of (Arbeitsgemeinschaft für Angewandte Humanpharmakologie (AGAH) e.V.) the association for applied human pharmacology (https://www.agah.eu). It is responsible for regular review, endorsement, and publication of the OSP development roadmap. The management team provides certification of the software platform, models, and other content such parameter databases and qualification processes. It releases certified platform versions. A sounding board supports the management team in scientific and technological matters. Community members are all active contributors to the development of OSP content. The OSP community also invites any interested party to use the provided content (software, models, etc.) because it benefits from widespread use and feedback provided via the OSP forum and other communication channels. This includes input and feedback from users and developers of commercial platforms.
Conclusions
During the past decade, systems pharmacology and its PBPK applications have seen a rapid development from dominant academic use to widespread applications with regulatory relevance and high ethical and commercial impact. Although the concepts and tools have reached a considerable maturity, the next level of development, namely, the demonstration of regulatory acceptable qualification is a major challenge. To be successful, the PBPK and systems pharmacology community needs to collaborate. With OSP, we intend to lower hurdles and facilitate progress. OSP offers a framework for collaboration and open‐access, open‐source tools. The community organization aims to combine open science development with processes for review and certification from a learned society. We invite you to join us on http://www.open-systems-pharmacology.org.
Conflict of interest
All authors use Open Systems Pharmacology software, tools, or models in their professional roles.
Supporting information
S1. Important Milestones of PBPK Modeling. PBPK, physiologically‐based pharmacokinetics.
S2. OSP Links. OSP, Open Systems Pharmacology.
Acknowledgments
Funds for the creation of the article were provided by authors' organizations. On behalf of the community, the authors wish to thank US Food & Drug Administration (FDA) and European Union and German Ministry for Education and Research (BMBF) for providing funding for the further development of OSP. FDA: PBPK and Population Modeling Seamlessly Linked to Clinical Trial Simulation in an Open‐Source Software Platform (Agreement RGF011519‐A); European Union: Horizon 2020 Research and Innovation Programme–U‐PGx consortium (Grant 668353), BMBF: OSMOSES consortium (Grant 031L0161A), nanoCELL consortium (Grant 03XP0196C), VISION consortium (Grant 031L0153A).
References
- 1. Workgroup, E.M. et al Good practices in model‐informed drug discovery and development: practice, application, and documentation. CPT Pharmacometrics Syst. Pharmacol. 5, 93–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wagner, C. et al Application of physiologically based pharmacokinetic (PBPK) modeling to support dose selection: report of an FDA Public Workshop on PBPK. CPT Pharmacometrics Syst. Pharmacol. 4, 226–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shepard, T. , Scott, G. , Cole, S. , Nordmark, A. & Bouzom, F. Physiologically based models in regulatory submissions: output from the ABPI/MHRA forum on physiologically based modeling and simulation. CPT Pharmacometrics Syst. Pharmacol. 4, 221–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. US Food and Drug Administration . Physiologically Based Pharmacokinetic Analyses — Format and Content Guidance for Industry. <https://www.fda.gov/ucm/groups/fdagov-public/@fdagov-drugs-gen/documents/document/ucm531207.pdf>. Accessed October 28, 2019.
- 5. European Medicines Agency . Guideline on the reporting of physiologically based pharmacokinetic (PBPK) modelling and simulation. <https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-reporting-physiologically-based-pharmacokinetic-pbpk-modelling-simulation_en.pdf>. Accessed October 28, 2019.
- 6. Teorell, T. Kinetics of distribution of substances administered to the body, I: The extravascular modes of administration. Arch. Int. Pharmacodyn. Ther. 57, 205–225 (1937). [Google Scholar]
- 7. Leong, R. et al Regulatory experience with physiologically based pharmacokinetic modeling for pediatric drug trials. Clin. Pharmacol. Ther. 91, 926–931 (2012). [DOI] [PubMed] [Google Scholar]
- 8. Schaller, S. et al A generic integrated physiologically based whole‐body model of the glucose‐insulin‐glucagon regulatory system. CPT Pharmacometrics Syst. Pharmacol. 2, e65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lesko, L.J. et al Establishing a multidisciplinary framework to study drug‐drug interactions of hormonal contraceptives: an invitation to collaborate. CPT Pharmacometrics Syst. Pharmacol. 7, 706–708 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Important Milestones of PBPK Modeling. PBPK, physiologically‐based pharmacokinetics.
S2. OSP Links. OSP, Open Systems Pharmacology.


