Abstract
The genus Streptomyces is a unique subgroup of actinomycetes bacteria that are well-known as prolific producers of antibiotics and many other bioactive secondary metabolites. Various environmental and physiological signals affect the onset and level of production of each antibiotic. Here we highlight recent findings on the regulation of antibiotic biosynthesis in Streptomyces by signaling molecules, with special focus on autoregulators such as hormone-like signaling molecules and antibiotics themselves. Hormone-like signaling molecules are a group of small diffusible signaling molecules that interact with specific receptor proteins to initiate complex regulatory cascades of antibiotic biosynthesis. Antibiotics and their biosynthetic intermediates can also serve as autoregulators to fine-tune their own biosynthesis or cross-regulators of disparate biosynthetic pathways. Advances in understanding of signaling molecules-mediated regulation of antibiotic production in Streptomyces may aid the discovery of new signaling molecules and their use in eliciting silent antibiotic biosynthetic pathways in a wide range of actinomycetes.
Keywords: antibiotic biosynthesis, regulation, Streptomyces, hormone-like signaling molecule, antibiotic biosynthetic intermediate, elicitor
Introduction
The genus Streptomyces, a unique subgroup of actinomycetes bacteria, is best-known for their ability to produce an enormous variety of bioactive secondary metabolites including antibiotics. The onset and production level of each antibiotic is subject to complex control by various environmental and physiological signals (Liu et al., 2013; Niu et al., 2016). Several excellent and comprehensive reviews have focused on the roles of signaling molecules in the regulation of antibiotic production of Streptomyces species (Takano, 2006; Sidda and Corre, 2012; Niu et al., 2016; Okada and Seyedsayamdost, 2017). Hormone-like signaling molecules are a group of small diffusible signaling molecules that can elicit antibiotic production and/or induce morphological differentiation at nanomolar concentrations. Typically, a hormone-like signaling molecule binds to its specific receptor, and exerts regulatory function through regulators at different hierarchical levels, including global regulators, pleiotropic regulators, and cluster-situated regulators (CSRs) (Liu et al., 2013; Niu and Tan, 2015; Wei et al., 2018). The most-studied regulatory system is the A-factor cascade that involves the signaling molecule A-factor and its receptor ArpA necessary for streptomycin and grixazone production in Streptomyces griseus (Horinouchi, 2007; Horinouchi and Beppu, 2007). Aside from the hormone-like signaling molecules, accumulating evidence suggests that antibiotics and their biosynthetic intermediates can also serve as autoregulators to modulate their own biosynthesis and as cross-regulators of disparate biosynthetic pathways (Niu et al., 2016). This review is not intended to be comprehensive, but to highlight recent findings on the regulation of antibiotic biosynthesis in Streptomyces by small molecules including hormone-like signaling molecules and antibiotics/their biosynthetic intermediates. A better understanding of the regulatory cascade of antibiotic production by signaling molecules in Streptomyces may aid the discovery of new signaling molecules and speed up their application in eliciting silent antibiotic biosynthetic pathways in actinomycetes.
Representative Hormone-Like Signaling Molecules Controlling Antibiotic Biosynthesis
Hormone-like signaling molecules are diffusible small molecules that can elicit antibiotic production and/or induce morphological differentiation at nanomolar concentrations. They exert their regulatory effect via specific receptor proteins that usually belong to the TetR family of transcriptional regulators (Takano, 2006; Niu et al., 2016). The TetR family regulators are characterized by a N-terminal helix-turn-helix (HTH) DNA-binding motif and a C-terminal ligand regulatory domain. They are widely distributed among bacteria and regulate diverse cellular processes in bacteria, especially antibiotic biosynthesis in Streptomyces (Cuthbertson and Nodwell, 2013). In Streptomyces species, A-factor, the first signaling molecule to be discovered, induces streptomycin production and morphological differentiation through a regulatory cascade involving the receptor ArpA, the pleiotropic regulator AdpA, and the CSR activator StrR in S. griseus (Horinouchi, 2007; Horinouchi and Beppu, 2007). Inspired by studies on the A-factor regulatory cascade, many more hormone-like signaling molecules have been identified, and great efforts has been directed to understanding signaling molecules-mediated regulatory cascades of antibiotic biosynthesis in Streptomyces.
Five Major Classes of Hormone-Like Signaling Molecules
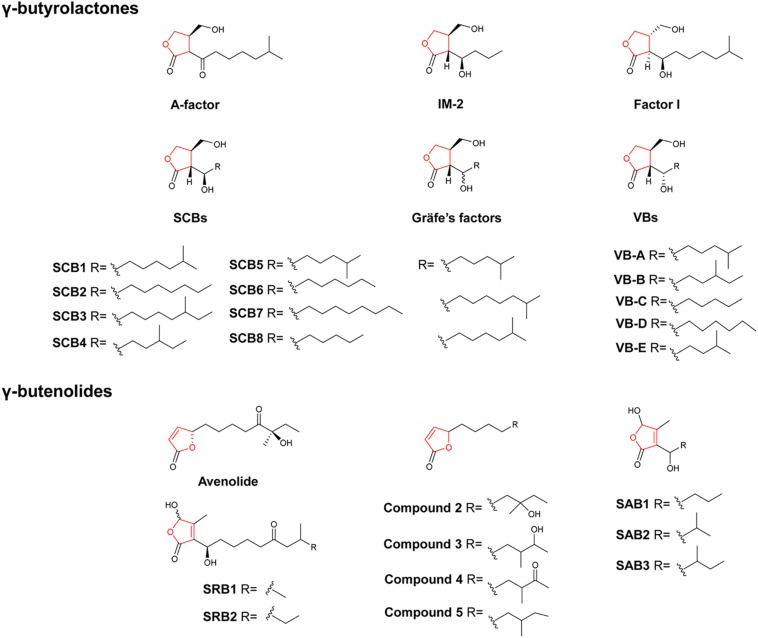
Similar to the well-documented N-acyl-homoserine lactones (AHLs) in Gram-negative bacteria, hormone-like signaling molecules in the Gram-positive Streptomyces bacteria are diffusible low molecular weight chemical substances that can elicit antibiotic production and/or induce morphological differentiation at nanomolar concentrations (Swift et al., 1994; Takano, 2006). To date, hormone-like signaling molecules identified from Streptomyces are classified into five major classes, including γ-butyrolactones (GBLs), furans, γ-butenolides as well as PI factor and N-methylphenylalanyl-dehydrobutyrine diketopiperazine (MDD) (Niu et al., 2016). GBLs, furans and γ-butenolides are based on five-membered heterocyclic rings containing four carbons and one oxygen, while PI factor and MDD have quite different structures (Recio et al., 2004; Matselyukh et al., 2015). As most recent investigations are largely on GBLs and γ-butenolides, we herein only summarize recent findings on these two important classes. GBLs, the largest group of these signaling molecules, share a characteristic 2,3-disubstituted GBL core skeleton but differ in the length, branching and stereochemistry of the acyl side chain (Takano, 2006). So far, a total of 19 GBLs have been identified in streptomycetes, including the A-factor from S. griseus, eight butanolides (SCB1-8) from Streptomyces coelicolor, five virginiae butanolides (VBs A–E) from Streptomyces virginiae, IM-2 from Streptomyces lavendulae, factor 1 from Streptomyces viridochromogenes, and three Gräfe’s factors from Streptomyces bikinensis and Streptomyces cyaneofuscatus (Figure 1; Niu et al., 2016; Sidda et al., 2016). To gain insight to the distribution of GBLs, an A-factor-deficient mutant of S. griseus was used as an indicator strain to screen against 203 actinomycete strains. Thirty of the strains tested showed distinct A-factor activity, indicating that A-factor was widely distributed among actinomycetes (Hara and Beppu, 1982). Earlier studies identified the butenolide synthase AfsA as a key enzyme for A-factor biosynthesis in S. griseus, and the AfsA homolog ScbA was required for the biosynthesis of the three SCBs in S. coelicolor (Takano et al., 2001; Hsiao et al., 2007). Among the eight butanolides identified in the model organism S. coelicolor, SCB1-3 were identified in the 2000s (Takano et al., 2000; Hsiao et al., 2009). Recently, the engineered strain S. coelicolor M1152 was found to overproduce GBLs SCB1–3 as well as five novel GBLs designated as SCB4–8 (Sidda et al., 2016). The S. coelicolor M1152 is a derivative of S. coelicolor M145 lacking four biosynthetic gene clusters (BGCs) for the biosynthesis of actinorhodin (ACT), undecylprodigiosin (RED), calcium-dependent antibiotic (CDA) and coelimycin. It should be noted that the deletion of the coelimycin gene cluster included scbR2, which encodes a repressor of SCBs biosynthetic gene scbA, and thereby contributed to the overproduction of SCBs in S. coelicolor M1152 (Gomez-Escribano and Bibb, 2012; Sidda et al., 2016). In a BLAST survey, we found one to three afsA-like genes in each of nine Streptomyces genomes (Niu et al., 2016). In another study, BLAST search for homologs of AfsA from S. griseus within the actinobacteria genomes available from public databases showed that AfsA-like proteins are present in most actinomycetes (Ahmed et al., 2017). The widespread distribution of GBLs was further confirmed by using VB- and IM-2-deficient strains as indicators to examine VB and IM-2 distribution. A total of 10 strains of the 40 Streptomyces and 11 endophytic actinomycetes tested produce IM-2, and the same number of strains produced VB active compounds (Thao et al., 2017). These studies reinforce the notion that GBLs are the most common hormone-like signaling molecules among actinomycetes. More efforts should be directed to identifying chemical structures of these signaling molecules.
FIGURE 1.
Chemical structures of representative γ-butyrolactones and γ-butenolides. Compounds 2–5: the four γ-butenolides from Streptomyces albus J1074.
Representative γ-butenolides include two butenolides (SRB1 and SRB2) from Streptomyces rochei (Arakawa et al., 2012; Arakawa, 2014), the avenolide from Streptomyces avermitilis (Kitani et al., 2011), four butenolides from Streptomyces albus J1074 (Nguyen et al., 2018), and three butenolides (SAB1-3) from Streptomyces ansochromogenes (Wang et al., 2018; Figure 1). Of special note is the avenolide that is necessary for triggering avermectin production in S. avermitilis (Kitani et al., 2011). The gene cluster responsible for avenolide biosynthesis consists of three genes (avaR1-3) that encode GBL receptor homologs, and two genes (aco and cyp17) that encode an acyl-CoA oxidase and a cytochrome P450 hydroxylase, respectively. Studies suggested that both the acyl-CoA oxidase Aco and the cytochrome P450 hydroxylase Cyp17 were required for the enzymatic production of avenolide. Furthermore, Aco/Cyp17 homologs were also found in the genomes of Streptomyces fradiae, Streptomyces ghanaensis, Streptomyces griseoauranticus, and S. albus J1074, suggesting that they have the capacity to produce avenolide or avenolide-like autoregulators (Kitani et al., 2011; Ahmed et al., 2017). Recently, Luzhetskyy and colleagues identified an avenolide-like compound 4 in a genetically modified strain of S. albus 1074 (Ahmed et al., 2017). In another study, Nihira and colleagues investigated the distribution of avenolide by using a S. avermitilis aco disruptant as a biosensor in a collection of 40 Streptomyces and 11 endophytic actinomycetes. They showed that 12 of the 51 actinomycetes strains exhibited avenolide activity with S. albus J1074 showing the highest activity (Thao et al., 2017). Metabolite profiling of a disruptant of the S. albus aco gene led to the identification of the compound 4 along with three other butenolides (compounds 2, 3, and 5). The four avenolide-like compounds showed different levels of avenolide activity in stimulating avermectin production in S. avermitilis (Nguyen et al., 2018). It is interesting to note that the four compounds have been isolated previously from marine-derived Streptomyces strains, though there are no reports on their connection with antibiotic biosynthesis in the native producers (Cho et al., 2001; Strand et al., 2014; Viegelmann et al., 2014; Igarashi et al., 2015). In a recent study, Tan and colleagues identified a putative BGC for autoregulator biosynthesis in S. ansochromogenes (nikkomycin producer). Within the gene cluster, sabA encodes an AfsA-like enzyme, whereas sabP and sabD encode phosphatase and dehydrogenase enzymes, respectively. Heterologous expression of sabAPD in E. coli and Streptomyces led to the identification of three novel butenolides (SAB1, 2, and 3) (Wang et al., 2018). Since there are only limited numbers of natural GBLs and γ-butenolides identified in Streptomyces, these studies provide effective strategies for the discovery of new signaling molecules, which are normally produced in very small quantities. It is noteworthy that several butenolides have also been identified in Streptomyces species (Wang T. et al., 2014; de Oliveira et al., 2019), especially from marine-derived Streptomyces (Cho et al., 2001; Strand et al., 2014; Viegelmann et al., 2014; Igarashi et al., 2015). The roles of these butenolides in their native producers await further investigation.
Receptors of Hormone-Like Signaling Molecules
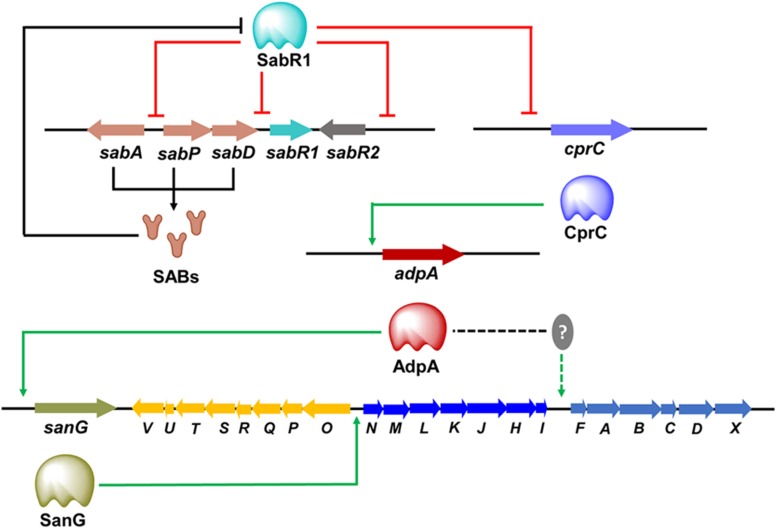
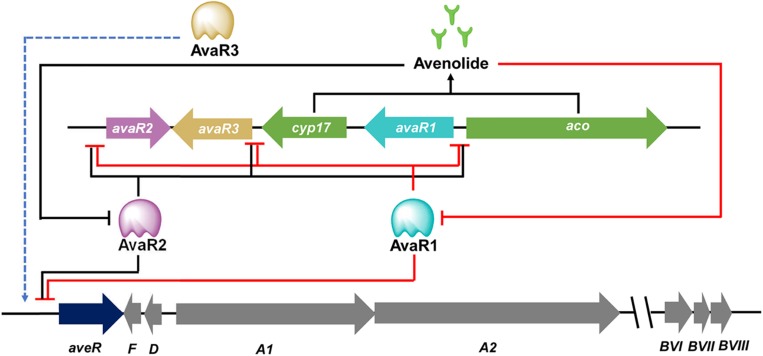
The transmission of chemical signals starts with the binding of an autoregulator to its specific receptor protein, that usually belongs to the TetR family of transcriptional regulators. It is not uncommon that many Streptomyces genomes contain multiple genes for ArpA-like GBL receptors. Examples include ScbR, ScbR2, CprA and CprB in S. coelicolor, JadR2 and JadR3 in Streptomyces venezuelae, and AvaR1, AvaR2, and AvaR3 in S. avermitilis, SabR1 and SabR2 in S. ansochromogenes. It is noteworthy that most genes encoding these GBL receptors are closely linked both to each other and to the afsA-like genes, except that cprA and cprB are not linked to an afsA-like gene and therefore referred to as “orphan GBL receptor” genes (Onaka et al., 1998). To date, only a few identified receptors have been shown to interact with endogenous GBL molecules. In the model organism S. coelicolor, ScbR was characterized as a genuine receptor of SCBs (Takano et al., 2005; Gomez-Escribano et al., 2012). ScbR binds the scbR-scbA intergenic region to repress its own expression, while interacting with ScbA to activate scbA expression in response to SCBs (Takano et al., 2001). Another known target of ScbR is kasO, the CSR activator of the coelimycin biosynthetic pathway (Takano et al., 2005). In S. venezuelae, JadR3 was identified as a genuine receptor of SVB1, a GBL identical in structure to SCB3 of S. coelicolor. The SVB1 receptor JadR3 has dual activation and repression effects on jadomycin biosynthesis (Zou et al., 2014; Niu et al., 2016). In S. ansochromogenes, SabR1, the cognate receptor of SABs, was shown to repress the expression of sabA, sabR1, sabR2 and cprC by binding directly to their promoter regions. Interestingly, CprC, a homolog of CprA and CprB in S. coelicolor, was identified as an activator of the pleiotropic regulatory gene adpA, which in turn stimulated nikkomycin biosynthesis via activating the transcriptional initiation of the CSR activator gene sanG and biosynthetic genes of the sanF-X operon (Figure 2; Pan et al., 2009; Wang et al., 2018). Of the three GBL receptors in S. avermitilis, AvaR1 acts as an avenolide receptor to control avenolide and avermectin production by directly repressing the transcription of aco, aveR, its own gene and the other two GBL receptor homologous genes (avaR2 and avaR3) (Figure 3; Zhu et al., 2017). ScbR2, JadR2, AvaR2 and SabR2 were identified as pseudo GBL receptors (Xu et al., 2010; Zhu et al., 2016; Wang et al., 2018), and the roles of these pseudo GBL receptors in the regulation of antibiotic biosynthesis will be discussed in a subsequent section. AvaR3 contains an extra 75-amino acid stretch that is not present in typical GBL receptors, and promotes avermectin production through a yet-unknown regulatory mechanism (Miyamoto et al., 2011). It is interesting to note that AvaR3 may represent a new subgroup of GBL receptors, and therefore further studies are needed to reveal its role in avermectin production.
FIGURE 2.
Cascade regulation of nikkomycin biosynthesis. The SABs synthesized by SabA, SabP, and SabD exert regulatory functions via the receptor SabR1. SabR1 represses the transcription of cprC and other target genes (sabA, sabR1, sabR2) by binding directly to their promoter regions. Binding of SABs to SabR1 causes the dissociation of SabR1 from cprC promoter, and thereby releases its repression on the transcription of cprC, which in turn activates adpA transcription to stimulate nikkomycin production. AdpA activates nikkomycin biosynthesis via activating the transcriptional initiation of sanG. SanG promotes nikkomycin biosynthesis through directly binding to the bidirectional sanN–sanO promoter region and activating the transcription of the biosynthetic genes. AdpA can also activate the transcription of sanF-X through an unknown mediator.
FIGURE 3.
Cascade regulation of avermectin biosynthesis. The complex regulation of avermectin production involves three GBL receptors. The avenolide receptor AvaR1 inhibits avenolide and avermectin production by directly repressing the transcription of aco, aveR, avaR1, avaR2, and avaR3. The pseudo GBL receptor AveR2 also acts as a repressor of avenolide and avermectin biosynthesis by binding to the same targets as AveR1. Another GBL receptor AvaR3 promotes avermectin production through an unknown regulatory mechanism. The DNA binding activities of AvaR1 and AvaR2 are modulated by avenolide, which are the enzymatic product of Aco and Cyp17.
Antibiotics as Regulatory Ligands Regulating Antibiotic Biosynthesis
Other than the hormone-like signaling molecules described above, antibiotics and their biosynthetic intermediates can also serve as regulatory ligands to regulate their own biosynthesis or cross-regulators of disparate biosynthetic pathways. In jadomycin biosynthesis, jadomycin and its biosynthetic intermediates actively participate in both feedback and feedforward cascade control of their own production in S. venezuelae. Such interplay makes jadomycin biosynthesis an illuminating model for regulatory studies of antibiotic production mediated by antibiotics and their biosynthetic intermediates (Niu et al., 2016). Aside from roles in regulating their own biosynthesis, antibiotics can also serve as autoregulators to control disparate antibiotic biosynthetic pathways. Typically, antibiotics exert their regulatory effect through CSRs that are associated with BGCs or pleiotropic regulators situated elsewhere in the genome (Table 1).
TABLE 1.
Representative regulators of antibiotic biosynthesis with antibiotics and/or their biosynthetic intermediates as regulatory ligands.
| Regulators | Ligands | Description | References |
| JadR1 | JdB, JdA, DHU, DHR | TetR family, activator of jadomycin biosynthesis; repressor of chloramphenicol biosynthesis | Wang et al., 2009 |
| RedZ | RED | TetR family, activator of RED biosynthesis | Wang et al., 2009 |
| JadR∗ | DHU, DHR, JdA, JdB | TetR family, repressor of jadomycin biosynthesis | Zhang et al., 2013 |
| ChlF1 | CHL, DM-CHL, deschloro-CHL | TetR family, repressor of chlorothricin biosynthesis | Li et al., 2016 |
| CalR3 | calcimycin, cezomycin | TetR family, repressor of calcimycin biosynthesis | Gou et al., 2017 |
| NosP | NOS, NOS-AC | SARP family, activator of nosiheptide production | Li et al., 2018 |
| RifQ | rifamycin B | TetR family, repressor of rifamycin biosynthesis | Lei et al., 2018 |
| PhlH | 2,4-DAPG, MAPG | TetR family, repressor of 2,4-diacetylphloroglucinol biosynthesis | Yan et al., 2017 |
| ScbR2 | ACT, RED | TetR family, repressor of coelimycin biosynthesis | Xu et al., 2010 |
| JadR2 | JdB, Cm | TetR family, repressor of jadomycin biosynthesis; activator of chloramphenicol | Xu et al., 2010 |
JdB, jadomycin B; JdA, jadomycin A; DHU, 2,3-dehydro-UWM6; DHR, dehydrorabelomycin; RED, undecylprodigiosin; CHL, chlorothricin; DM-CHL, demethylsalicycloyl chlorothricin; deschloro-CHL, deschloro-chlorothricin; NOS, nosiheptide; NOS-AC, the intermediate of NOS; 2,4-DAPG, 2,4-diacetylphloroglucinol; MAPG, monoacetylphloroglucinol; ACT, actinorhodin; Cm, chloramphenicol.
Antibiotics and Their Biosynthetic Intermediates as Autoregulators of Their Own Biosynthesis
The first line of antibiotics as autoregulators came from studies on the roles of two atypical response regulators (ARRs) in antibiotic biosynthesis. The CSR activator JadR1, an OmpR-type ARR of S. venezuelae, activates the expression of jadomycin B (JdB) biosynthetic genes in the presence of a low concentration of JdB and its homologs, but high JdB concentrations cause the dissociation of JadR1 from its target promoters. Thus, JdB interacts with JadR1 directly in a dose-dependent manner to control JdB production in a feedback regulatory mechanism (Wang et al., 2009). Similarly, a NarL-type ARR RedZ was shown to promote RED biosynthesis in S. coelicolor by activating the CSR activator gene redD, and the DNA-binding activity of RedZ to its target gene was released by adding RED (Wang et al., 2009). Other than the end-product, studies also suggest that biosynthetic intermediates of antibiotics can also serve as autoregulatory molecules to modulate their own biosynthesis. For example, the TetR family regulator JadR∗ can respond to early jadomycin intermediates [2,3-dehydro-UWM6 (DHU) and dehydrorabelomycin (DHR)] and release its repression on jadY transcription to ensure the timely supply of cofactors for JadG to convert DHR to the late intermediate jadomycin A (JdA) in jadomycin biosynthesis (Zhang et al., 2013).
Inspired by studies on the regulation of jadomycin biosynthesis in S. venezuelae, more antibiotics and their biosynthetic intermediates have been found to serve as autoregulatory molecules to modulate their own biosynthesis in many other streptomycetes. For illustration purposes, we herein only highlight several recent findings. For example, ChlF1, a TetR family regulator involved in chlorothricin biosynthesis of Streptomyces antibioticus, was found to repress the transcription of chlF1, chlG (encoding a major facilitator superfamily transporter), and chlK (encoding a type II thioesterase) but activates chlJ (encoding acyl-CoA carboxyl transferase) by binding to their promoter regions. However, ChlF1 was disassociated from its target promoters in the presence of chlorothricin and its biosynthetic intermediates (demethyl salicyloyl chlorothricin and deschloro-chlorothricin), which directly interacted with the regulator in a concentration-dependent manner (Li et al., 2016). In another study, CalR3, a TetR family regulator involved in calcimycin biosynthesis of Streptomyces chartreusis NRRL 3882, was found to repress the transcription of calR3 and its adjacent calT (encoding a putative transmembrane efflux pump protein of the MMPL family) by binding to their promoter regions. Similarly, both calcimycin and its biosynthetic intermediate (cezomycin) can act as ligands to dissociate CalR3 from its target promoters in a concentration-dependent manner (Gou et al., 2017; Wu et al., 2018). Other than TetR family regulators, Streptomyces antibiotic regulatory proteins (SARPs) family regulator was also found to regulate antibiotic biosynthesis in response to antibiotics and their biosynthetic intermediates. For example, NosP was found to regulate nosiheptide production in response to both the end-product nosiheptide (NOS) and biosynthetic intermediate (NOS-AC) (Li et al., 2018). It should be noted that such findings are not limited to the genus Streptomyces. One example comes from the actinomycete Amycolatopsis mediterranei, in which rifamycin B (the end-product of rifamycin biosynthesis) serves as an extracellular signaling molecule to regulate rifamycin export in a feedback mechanism (Lei et al., 2018). Another study showed that 2,4-diacetylphloroglucinol and its biosynthetic intermediate could serve as autoregulatory ligands to regulate its own biosynthesis in Pseudomonas fluorescens (Yan et al., 2017). These studies suggested that antibiotics and their biosynthetic intermediates could function as autoregulators to regulate their own biosynthesis, mainly through the modulation of the binding activity of CSRs to their target genes within their cognate gene clusters.
Antibiotics as Cross-Regulators of the Biosynthesis of Other Antibiotics
Earlier studies suggest that CSRs of one biosynthetic pathway can also control the biosynthesis of disparate antibiotic biosynthetic pathways. For example, RedZ, CSR of the red gene cluster, modulates the production of RED as well as that of ACT and CDA (Huang et al., 2005). In another study, the candicidin CSR FscRI was found to control the biosynthesis of candicidin as well as antimycin, the product of a disparate BGC in Streptomyces albidoflavus S4 (previously known as Streptomyces albus S4) (McLean et al., 2016; Li et al., 2019). Similar observations were also made with coordinated production of geldanamycin and elaiophylin by GdmRIII in Streptomyces autolyticus CGMCC0516 (Jiang et al., 2017), production of cephamycin C and clavulanic acid by CcaR in Streptomyces clavuligerus (Santamarta et al., 2002), and production of jadomycin and chloramphenicol by JadR1 in S. venezuelae (Wang et al., 2009). Recent years see growing evidence that antibiotics can also act as autoregulators regulating other biosynthetic pathways. The first examples to be described involve pseudo GBL receptors. In S. coelicolor, ScbR2, represses the same target, kasO, as the SCBs-interactive ScbR. Although scbR2 is linked to the gene cluster for coelimycin biosynthesis, ScbR2 also responds to the endogenous antibiotics ACT and RED, and thereby regulates the production of these antibiotics (Xu et al., 2010). In S. venezuelae, JadR2, a close homolog of ScbR2, modulates JdB biosynthesis via direct repression of jadR1. This repression is sensitive to the jadomycin end products. Likewise, although jadR2 is linked to the jad gene cluster for jadomycin biosynthesis, JadR2 could also bind to the endogenous antibiotic chloramphenicol, the product of the distant cml biosynthetic gene cluster (Xu et al., 2010). These studies suggest that CSRs of one biosynthetic pathway can also control disparate antibiotic biosynthetic pathways in response to different antibiotics. The pseudo GBL receptor AvaR2 of S. avermitilis was as an important repressor of avermectin and avenolide biosynthesis. It directly repressed transcription of aveR, aco, its own gene and the other two GBL receptor homologous genes (avaR1 and avaR3) (Figure 3; Zhu et al., 2016). Interestingly, DNA-binding activity of AvaR2 can be modulated by endogenous avenolide in a concentration-dependent manner, indicating that avenolide serves as ligands for both the genuine GBL receptor AvaR1 and the pseudo GBL receptor AvaR2. Furthermore, DNA-binding activity of AvaR2 can also be modulated by exogenous antibiotics JdB and aminoglycoside antibiotics such as apramycin, hygromycin B, kanamycin and streptomycin (Zhu et al., 2016). It would be interesting to examine if JdB from S. venezuelae can serve as signals to modulate avermectin production in S. avermitilis under physiological conditions.
Signaling Molecules in the Improvement of Antibiotic Titers and the Construction of Genetic Circuits
As mentioned above, signaling molecules serve as elicitors for antibiotic production in Streptomyces. It is reasonable that signaling molecules can be used to improve antibiotic production through exogenous addition. However, due to the fact that natural GBLs are produced in very small quantities, it is impractical to collect sufficient amount of GBLs for the improvement of antibiotic production. Synthetic GBLs and their analogs are then used for this purpose. Early studies showed that addition of chemically synthesized VB-C enhanced virginiamycin production in Streptomyces virginiae (Yang et al., 1995, 1996). Similarly, addition of chemically synthesized SCB1 promoted ACT production in S. coelicolor. Synthetic GBL analogs are also an alternative (Yang et al., 2009). In two recent studies, the addition of synthetic 1,4-butyrolactone enhanced validamycin A production in Streptomyces hygroscopicus 5008 and bitespiramycin production in Streptomyces spiramyceticus WS1-195, respectively (Tan et al., 2013; Gao et al., 2019). These studies showed that exogenous addition of signaling molecules can be used as an effective strategy to increase metabolic titers of antibiotics. However, this method has limited application due to the fact that there are only a few natural and synthetic analogs available. Therefore, it is necessary to expand the reservoir of both natural and synthetic signaling molecules. Furthermore, there is a growing interest in the construction of GBL-based genetic circuits in heterologous systems, which has been summarized by Takano and colleagues in 2015 (Biarnes-Carrera et al., 2015). Recently, orthogonal regulatory circuits based on the S. coelicolor GBL system have been constructed in E. coli (Biarnes-Carrera et al., 2018), reinforcing the promising applications of GBL systems from Streptomyces as a regulatory tool for synthetic biology in heterologous systems.
Signaling Molecules in the Discovery of Novel Natural Products
High-throughput DNA sequencing technologies have resulted in an explosion of microbial genome sequences. Genome sequencing of multiple well-known actinomycetes has revealed that they harbor a great number of BGCs that are predicted to direct the biosynthesis of diverse natural products (Nett et al., 2009). It has become clear that these enormous reservoir of uncharacterized BGCs can serve as an important source of novel bioactive compounds. However, most of these BGCs are not expressed efficiently or not at all under routine laboratory culturing conditions (Mao et al., 2018). Though the reason for the silence of these gene clusters is complicated and remains obscure, a lack of specific signaling molecules may be one of the contributing factors (Liu et al., 2013). Interestingly, antibiotics can be used as chemical elicitors to trigger the expression of these cryptic BGCs and expand the chemical diversity of natural products.
Antibiotics as Chemical Elicitors for the Discovery of Novel Natural Products
In recent years, growing evidence suggests that antibiotics, at sub-inhibitory concentrations, can potentiate antibiotic production in multiple Streptomyces species. For example, lincomycin at a sub-inhibitory concentration resulted in an elevated expression of the CSR activator gene actII-ORF4, and therefore increased ACT overproduction in S. coelicolor (Imai et al., 2015). A recent study suggests that lincomycin produced profound changes in gene expression profiles of S. coelicolor (Ishizuka et al., 2018). In another example, a sub-inhibitory concentration of JdB (the angucycline from S. venezuelae) induced early RED production and premature differentiation (formation of sporulating aerial mycelium) in S. coelicolor. Other angucyclines were also found to elicit similar phenotypes (Wang W. et al., 2014). Examples also include perturbation of antibiotics production by ribosome-targeting antibiotics (thiostrepton, spectinomycin, and chloramphenicol) in multiple Streptomyces species (Tanaka et al., 2017; Wang et al., 2017). Furthermore, antibiotics can also be used as chemical elicitors for the discovery of novel natural products. For example, S. lividans 1326 grown in the presence of lincomycin at a sub-inhibitory concentration produced abundant antibacterial compounds that were absent in cells grown in lincomycin-free medium. Some of these antibacterial compounds were revealed as novel congeners of CDA (Imai et al., 2015). In another case, polyether antibiotics, including promomycin and closely related salinomycin, monensin, and nigercin, at sub−inhibitory concentrations elicited antibiotic production in multiple Streptomyces strains (Amano et al., 2010, 2011). These studies suggest that antibiotics have the potential to serve as chemical elicitors for the discovery of novel bioactive natural products.
High-Throughput Elicitor Screening for the Discovery of Novel Natural Products
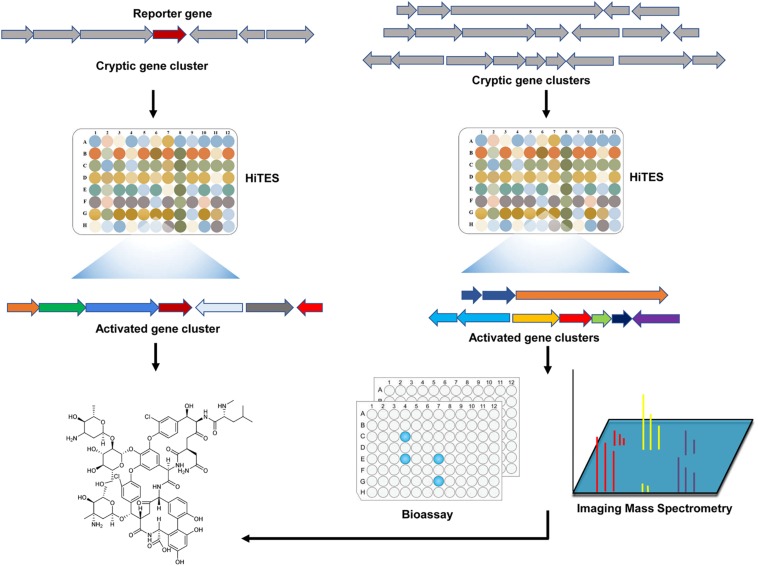
It is well-known that the cryptic gene clusters represent a treasure trove of new natural products, and various strategies and methods has been devised for the activation of these gene clusters (Liu et al., 2013; Niu et al., 2016; Niu, 2018; Niu and Li, 2019). Of special note is a chemogenetic method referred to as high-throughput elicitor screening approach (HiTES) (Figure 4). In this approach, a reporter gene is inserted into the BGC of interest to allow a rapid read-out for its expression. The resulting reporter strain is then screened against small molecule libraries to identify candidate elicitors (Seyedsayamdost, 2014). This approach was initially applied to activate two cryptic gene clusters in the Gram-negative bacterium Burkholderia thailandensis, leading to the identification of a new malleilactone analog. Interestingly, almost all elicitors identified from a library of 800 compounds were antibiotics (Seyedsayamdost, 2014). Furthermore, Seyedsayamdost and colleagues have employed this approach to identify 14 novel specialized products in S. albus J1074 (Xu et al., 2017). HiTES with S. albus J1074 using a library of ∼500 natural products identified two classical antibiotics (ivermectin and etoposide) as the best elicitors of a cryptic non-ribosomal peptide synthetase (NRPS) gene cluster (Xu et al., 2017). A major limitation of this method is that genetic manipulations and/or molecular biology approaches are required for the insertion of the reporter gene. To circumvent this problem, HiTES is then combined with bioactivity assays or imaging mass spectrometry (IMS) to identify novel antibiotics. In these approaches, a wild-type microorganism is subjected to HiTES. The resulting induced cultures are then screened directly for biological activity or subject to IMS (Figure 4). Application of these modified methods have uncovered two novel specialized products in Streptomyces hiroshimensis (Moon et al., 2019b), and many other novel specialized metabolites in diverse microorganisms (Moon et al., 2019a; Xu et al., 2019). These studies suggest that HiTES represents a promising new avenue for the discovery of novel bioactive natural products in a high-throughput manner.
FIGURE 4.
A general workflow of HiTES for the discovery of novel antibiotics. Initially, HiTES uses a reporter gene to allow a rapid read-out for the expression of the target cyptic gene cluster. The resulting reporter strain is then subjected to HiTES screening for the identification of novel antibiotics. Alternatively, microbial genomes encoding numerous cryptic gene clusters are directly subjected to HiTES screening. Cryptic gene clusters activated by small molecule elicitors are then subjected to high throughput bioassays for novel bioactive compounds. They can also be subjected to Imaging Mass Spectrometry to detect the natural products in a rapid and untargeted fashion. HiTES: high-throughput elicitor screening.
Conclusion and Perspectives
Over the past several decades, significant advances have been made in understanding the regulation of antibiotic biosynthesis in Streptomyces. The emerging picture shows a complex interplay of various signals and regulatory proteins. It is interesting to note that both hormone-like signaling molecules and antibiotics/their biosynthetic intermediates actively participate in the regulation of antibiotic production. These small molecule regulators exert their function through the modulation of the binding activity of different regulators to their target genes. Further studies are needed to reveal the complex interplay of autoregulators and multiple GBL receptor homologs in antibiotic biosynthesis of different Streptomyces. However, the number of hormone-like signaling molecules identified from Streptomyces is still limited, most likely due to the fact that they are produced in very small quantities. BLAST searches for homologs of AfsA from S. griseus within the actinobacteria genomes available from public databases indicated that AfsA-like proteins are present in most actinomycetes (Niu et al., 2016; Ahmed et al., 2017). Similar observations have been made with a BLAST search for homologs of Aco and Cyp17 proteins from S. avermitilis (Ahmed et al., 2017). It is strongly believed that GBLs and γ-butenolide autoregulators are widely distributed among actinomycetes. It is therefore necessary to identify more hormone-like signaling molecules. Currently, several approaches have been used to discover new signaling molecules in Streptomyces, including deletion of gene encoding repressor of signaling molecule biosynthetic gene (Sidda et al., 2016), bioassay using disruptants of known signaling molecule biosynthetic genes as indicator strains (Thao et al., 2017; Nguyen et al., 2018), and heterologous expression of putative signaling molecule biosynthetic genes in model microorganisms (Wang et al., 2018). These approaches will speed up the process of identifying new signaling molecules, and their signal transduction pathways. For autoregulators of antibiotics and their biosynthetic intermediates, more efforts should be geared toward understanding their effect on the fine-tuning regulation of antibiotic biosynthesis under physiological conditions. A better understanding of the complex regulatory pathways of signaling molecules in Streptomyces may be used to increase the metabolic titer of industrially and medically important antibiotics as well as activate silent antibiotic BGCs for the discovery of novel bioactive natural products in a wide range of actinomycetes.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all members of our lab for their helpful comments and discussion during the preparation of this manuscript. We also thank the two reviewers for their helpful comments. We sincerely apologize to those authors whose work we could not cite due to space limitations.
Footnotes
Funding. This work was supported in part by the National Natural Science Foundation of China (31870061), grants from the Chongqing Science and Technology Commission (cstccxljrc201904, cstc2018jcyjAX0066, and cstc2017jcyjAX0467), a start-up grant from the Southwest University (SWU117015), and the Fundamental Research Funds for the Central Universities (SWU119034).
References
- Ahmed Y., Rebets Y., Tokovenko B., Brotz E., Luzhetskyy A. (2017). Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci. Rep. 7:9784. 10.1038/s41598-017-10316-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano S., Morota T., Kano Y. K., Narita H., Hashidzume T., Yamamoto S., et al. (2010). Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot. 63 486–491. 10.1038/ja.2010.68 [DOI] [PubMed] [Google Scholar]
- Amano S. I., Sakurai T., Endo K., Takano H., Beppu T., Furihata K., et al. (2011). A cryptic antibiotic triggered by monensin. J. Antibiot. 64:703. 10.1038/ja.2011.69 [DOI] [PubMed] [Google Scholar]
- Arakawa K. (2014). Genetic and biochemical analysis of the antibiotic biosynthetic gene clusters on the Streptomyces linear plasmid. Biosci. Biotechnol. Biochem. 78 183–189. 10.1080/09168451.2014.882761 [DOI] [PubMed] [Google Scholar]
- Arakawa K., Tsuda N., Taniguchi A., Kinashi H. (2012). The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. Chembiochem 13 1447–1457. 10.1002/cbic.201200149 [DOI] [PubMed] [Google Scholar]
- Biarnes-Carrera M., Breitling R., Takano E. (2015). Butyrolactone signalling circuits for synthetic biology. Curr. Opin. Chem. Biol. 28 91–98. 10.1016/j.cbpa.2015.06.024 [DOI] [PubMed] [Google Scholar]
- Biarnes-Carrera M., Lee C. K., Nihira T., Breitling R., Takano E. (2018). Orthogonal regulatory circuits for Escherichia coli based on the (-butyrolactone system of Streptomyces coelicolor. ACS Synth. Biol. 7 1043–1055. 10.1021/acssynbio.7b00425 [DOI] [PubMed] [Google Scholar]
- Cho K. W., Lee H. S., Rho J. R., Kim T. S., Mo S. J., Shin J. (2001). New lactone-containing metabolites from a marine-derived bacterium of the genus Streptomyces. J. Nat. Prod. 64 664–667. 10.1021/np000599g [DOI] [PubMed] [Google Scholar]
- Cuthbertson L., Nodwell J. R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77 440–475. 10.1128/MMBR.00018-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira J. A. M., Williams D. E., Andersen R. J., Sarragiotto M. H., Baldoqui D. C. (2019). Mycenolide A, new butenolide from a marine sediment-derived bacterium Streptomyces sp. 4054. Nat. Prod. Res. 10.1080/14786419.2019.1597351 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Gao X., Wang Y., Chu J. (2019). A preliminary study on the impact of exogenous A-Factor analogue 1,4-butyrolactone on stimulating bitespiramycin biosynthesis. Bioprocess Biosyst. Eng. 42 1903–1913. 10.1007/s00449-019-02184-9 [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Bibb M. J. (2012). Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol. 517 279–300. 10.1016/B978-0-12-404634-4.00014-0 [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Song L., Fox D. J., Yeo V., Bibb M. J., Challis G. L. (2012). Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 3 2716–2720. 10.1039/C2SC20410J [DOI] [Google Scholar]
- Gou L., Han T., Wang X., Ge J., Liu W., Hu F., et al. (2017). A novel TetR family transcriptional regulator, CalR3, negatively controls calcimycin biosynthesis in Streptomyces chartreusis NRRL 3882. Front. Microbiol. 8:2371. 10.3389/fmicb.2017.02371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara O., Beppu T. (1982). Mutants blocked in streptomycin production in Streptomyces griseus - the role of A-factor. J. Antibiot. 35 349–358. 10.7164/antibiotics.35.349 [DOI] [PubMed] [Google Scholar]
- Horinouchi S. (2007). Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 71 283–299. 10.1271/bbb.60627 [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. (2007). Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 83 277–295. 10.2183/pjab/83.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao N. H., Nakayama S., Merlo M. E., de Vries M., Bunet R., Kitani S., et al. (2009). Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem. Biol. 16 951–960. 10.1016/j.chembiol.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Hsiao N. H., Soding J., Linke D., Lange C., Hertweck C., Wohlleben W., et al. (2007). ScbA from Streptomyces coelicolor A3(2) has homology to fatty acid synthases and is able to synthesize (-butyrolactones. Microbiology 153 1394–1404. 10.1099/mic.0.2006/004432-0 [DOI] [PubMed] [Google Scholar]
- Huang J., Shi J., Molle V., Sohlberg B., Weaver D., Bibb M. J., et al. (2005). Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58 1276–1287. 10.1111/j.1365-2958.2005.04879.x [DOI] [PubMed] [Google Scholar]
- Igarashi Y., Ikeda M., Miyanaga S., Kasai H., Shizuri Y., Matsuura N. (2015). Two butenolides with PPAR(agonistic activity from a marine-derived Streptomyces. J. Antibiot. 68 345–347. 10.1038/ja.2014.151 [DOI] [PubMed] [Google Scholar]
- Imai Y., Sato S., Tanaka Y., Ochi K., Hosaka T. (2015). Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl. Environ. Microbiol. 81 3869–3879. 10.1128/aem.04214-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka M., Imai Y., Mukai K., Shimono K., Hamauzu R., Ochi K., et al. (2018). A possible mechanism for lincomycin induction of secondary metabolism in Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek 111 705–716. 10.1007/s10482-018-1021-0 [DOI] [PubMed] [Google Scholar]
- Jiang M., Yin M., Wu S., Han X., Ji K., Wen M., et al. (2017). GdmRIII, a TetR family transcriptional regulator, controls geldanamycin and elaiophylin biosynthesis in Streptomyces autolyticus CGMCC0516. Sci. Rep. 7:4803. 10.1038/s41598-017-05073-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani S., Miyamoto K. T., Takamatsu S., Herawati E., Iguchi H., Nishitomi K., et al. (2011). Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 108 16410–16415. 10.1073/pnas.1113908108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C., Wang J., Liu Y., Liu X., Zhao G., Wang J. (2018). A feedback regulatory model for RifQ-mediated repression of rifamycin export in Amycolatopsis mediterranei. Microb. Cell Fact. 17:14. 10.1186/s12934-018-0863-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Y., Niu G., Guo H., Qiu Y., Lin Z., et al. (2018). NosP-regulated nosiheptide production responds to both peptidyl and small-molecule ligands derived from the precursor peptide. Cell Chem. Biol. 25 e144. 10.1016/j.chembiol.2017.10.012 [DOI] [PubMed] [Google Scholar]
- Li Y., Li J., Tian Z., Xu Y., Zhang J., Liu W., et al. (2016). Coordinative modulation of chlorothricin biosynthesis by binding of the glycosylated intermediates and end product to a responsive regulator ChlF1. J. Biol. Chem. 291 5406–5417. 10.1074/jbc.M115.695874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Pinto-Tomas A. A., Rong X., Cheng K., Liu M., Huang Y. (2019). Population genomics insights into adaptive evolution and ecological differentiation in Streptomycetes. Appl. Environ. Microbiol. 85:e2555-18. 10.1128/AEM.02555-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Chater K. F., Chandra G., Niu G., Tan H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77 112–143. 10.1128/MMBR.00054-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D., Okada B. K., Wu Y., Xu F., Seyedsayamdost M. R. (2018). Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 45 156–163. 10.1016/j.mib.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matselyukh B., Mohammadipanah F., Laatsch H., Rohr J., Efremenkova O., Khilya V. (2015). N-methylphenylalanyl-dehydrobutyrine diketopiperazine, an A-factor mimic that restores antibiotic biosynthesis and morphogenesis in Streptomyces globisporus 1912-B2 and Streptomyces griseus 1439. J. Antibiot. 68 9–14. 10.1038/ja.2014.86 [DOI] [PubMed] [Google Scholar]
- McLean T. C., Hoskisson P. A., Seipke R. F. (2016). Coordinate regulation of antimycin and candicidin biosynthesis. mSphere 1 1–35. 10.1128/mSphere.00305-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K. T., Kitani S., Komatsu M., Ikeda H., Nihira T. (2011). The autoregulator receptor homologue AvaR3 plays a regulatory role in antibiotic production, mycelial aggregation and colony development of Streptomyces avermitilis. Microbiology 157 2266–2275. 10.1099/mic.0.048371-0 [DOI] [PubMed] [Google Scholar]
- Moon K., Xu F., Seyedsayamdost M. R. (2019a). Cebulantin, a cryptic lanthipeptide antibiotic uncovered using bioactivity-coupled HiTES. Angew. Chem. Int. Ed Engl. 58 5973–5977. 10.1002/anie.201901342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K., Xu F., Zhang C., Seyedsayamdost M. R. (2019b). Bioactivity-HiTES unveils cryptic antibiotics encoded in actinomycete bacteria. ACS Chem. Biol. 14 767–774. 10.1021/acschembio.9b00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M., Ikeda H., Moore B. S. (2009). Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26 1362–1384. 10.1039/b817069j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. B., Kitani S., Shimma S., Nihira T. (2018). Butenolides from Streptomyces albus J1074 act as external signals to stimulate avermectin production in Streptomyces avermitilis. Appl. Environ. Microbiol. 84:e2791-17. 10.1128/aem.02791-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G. (2018). Genomics-driven natural product discovery in actinomycetes. Trends Biotechnol. 36 238–241. 10.1016/j.tibtech.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Niu G., Chater K. F., Tian Y., Zhang J., Tan H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40 554–573. 10.1093/femsre/fuw012 [DOI] [PubMed] [Google Scholar]
- Niu G., Li W. (2019). Next-generation drug discovery to combat antimicrobial resistance. Trends Biochem. Sci. 44 961–972. 10.1016/j.tibs.2019.05.005 [DOI] [PubMed] [Google Scholar]
- Niu G., Tan H. (2015). Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends Microbiol. 23 110–119. 10.1016/j.tim.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Okada B. K., Seyedsayamdost M. R. (2017). Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 41 19–33. 10.1093/femsre/fuw035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H., Nakagawa T., Horinouchi S. (1998). Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28 743–753. 10.1046/j.1365-2958.1998.00832.x [DOI] [PubMed] [Google Scholar]
- Pan Y., Liu G., Yang H., Tian Y., Tan H. (2009). The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72 710–723. 10.1111/j.1365-2958.2009.06681.x [DOI] [PubMed] [Google Scholar]
- Recio E., Colinas A., Rumbero A., Aparicio J. F., Martin J. F. (2004). PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 279 41586–41593. 10.1074/jbc.M402340200 [DOI] [PubMed] [Google Scholar]
- Santamarta I., Rodriguez-Garcia A., Perez-Redondo R., Martin J. F., Liras P. (2002). CcaR is an autoregulatory protein that binds to the ccaR and cefD-cmcI promoters of the cephamycin C-clavulanic acid cluster in Streptomyces clavuligerus. J. Bacteriol. 184 3106–3113. 10.1128/jb.184.11.3106-3113.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R. (2014). High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. U.S.A. 111 7266–7271. 10.1073/pnas.1400019111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidda J. D., Corre C. (2012). (-butyrolactone and furan signaling systems in Streptomyces. Methods Enzymol. 517 71–87. 10.1016/B978-0-12-404634-4.00004-8 [DOI] [PubMed] [Google Scholar]
- Sidda J. D., Poon V., Song L., Wang W., Yang K., Corre C. (2016). Overproduction and identification of butyrolactones SCB1-8 in the antibiotic production superhost Streptomyces M1152. Org. Biomol. Chem. 14 6390–6393. 10.1039/c6ob00840b [DOI] [PubMed] [Google Scholar]
- Strand M., Carlsson M., Uvell H., Islam K., Edlund K., Cullman I., et al. (2014). Isolation and characterization of anti-adenoviral secondary metabolites from marine actinobacteria. Mar. Drugs 12 799–821. 10.3390/md12020799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S., Bainton N. J., Winson M. K. (1994). Gram-negative bacterial communication by N-acyl homoserine lactones: a universal language? Trends Microbiol. 2 193–198. 10.1016/0966-842x(94)90110-q [DOI] [PubMed] [Google Scholar]
- Takano E. (2006). Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9 287–294. 10.1016/j.mib.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Takano E., Chakraburtty R., Nihira T., Yamada Y., Bibb M. J. (2001). A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41 1015–1028. 10.1046/j.1365-2958.2001.02562.x [DOI] [PubMed] [Google Scholar]
- Takano E., Kinoshita H., Mersinias V., Bucca G., Hotchkiss G., Nihira T., et al. (2005). A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56 465–479. 10.1111/j.1365-2958.2005.04543.x [DOI] [PubMed] [Google Scholar]
- Takano E., Nihira T., Hara Y., Jones J. J., Gershater C. J., Yamada Y., et al. (2000). Purification and structural determination of SCB1, a (-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275 11010–11016. 10.1074/jbc.275.15.11010 [DOI] [PubMed] [Google Scholar]
- Tan G. Y., Bai L., Zhong J. J. (2013). Exogenous 1,4-butyrolactone stimulates A-factor-like cascade and validamycin biosynthesis in Streptomyces hygroscopicus 5008. Biotechnol. Bioeng. 110 2984–2993. 10.1002/bit.24965 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Izawa M., Hiraga Y., Misaki Y., Watanabe T., Ochi K. (2017). Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs. Appl. Microbiol. Biotechnol. 101 4417–4431. 10.1007/s00253-017-8216-6 [DOI] [PubMed] [Google Scholar]
- Thao N. B., Kitani S., Nitta H., Tomioka T., Nihira T. (2017). Discovering potential Streptomyces hormone producers by using disruptants of essential biosynthetic genes as indicator strains. J. Antibiot. 70 1004–1008. 10.1038/ja.2017.85 [DOI] [PubMed] [Google Scholar]
- Viegelmann C., Margassery L. M., Kennedy J., Zhang T., O’Brien C., O’Gara F., et al. (2014). Metabolomic profiling and genomic study of a marine sponge-associated Streptomyces sp. Mar. Drugs 12 3323–3351. 10.3390/md12063323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhao G., Ding X. (2017). Morphology engineering of Streptomyces coelicolor M145 by sub-inhibitory concentrations of antibiotics. Sci. Rep. 7:13226. 10.1038/s41598-017-13493-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., et al. (2009). Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U.S.A. 106 8617–8622. 10.1073/pnas.0900592106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Jiang Y., Ma K. X., Li Y. Q., Huang R., Xie X. S., et al. (2014). Two new butenolides produced by an actinomycete Streptomyces sp. Chem. Biodivers. 11 929–933. 10.1002/cbdv.201300321 [DOI] [PubMed] [Google Scholar]
- Wang W., Ji J., Li X., Wang J., Li S., Pan G., et al. (2014). Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 111 5688–5693. 10.1073/pnas.1324253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhang J., Liu X., Li D., Li Y., Tian Y., et al. (2018). Identification of a butenolide signaling system that regulates nikkomycin biosynthesis in Streptomyces. J. Biol. Chem. 293 20029–20040. 10.1074/jbc.RA118.005667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., He L., Niu G. (2018). Regulation of antibiotic biosynthesis in actinomycetes: perspectives and challenges. Synth. Syst. Biotechnol. 3 229–235. 10.1016/j.synbio.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Liang J., Wang J., Liang W. J., Gou L., Wu Q., et al. (2018). Cezomycin is activated by CalC to its ester form for further biosynthesis steps in the production of calcimycin in Streptomyces chartreusis NRRL 3882. Appl. Environ. Microbiol. 84:e586-18. 10.1128/aem.00586-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Nazari B., Moon K., Bushin L. B., Seyedsayamdost M. R. (2017). Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 139 9203–9212. 10.1021/jacs.7b02716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Wu Y., Zhang C., Davis K. M., Moon K., Bushin L. B., et al. (2019). A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 15 161–168. 10.1038/s41589-018-0193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Wang J., Wang L., Tian X., Yang H., Fan K., et al. (2010). “Pseudo” (-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285 27440–27448. 10.1074/jbc.M110.143081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Yang R., Zhao R. X., Han J. T., Jia W. J., Li D. Y., et al. (2017). Transcriptional regulator PhlH modulates 2,4-diacetylphloroglucinol biosynthesis in response to the biosynthetic intermediate and end product. Appl. Environ. Microbiol. 83:e1419-17. 10.1128/AEM.01419-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. H., Kim T. W., Park S. H., Lee K., Park H. Y., Song E., et al. (2009). Cell-free Escherichia coli-based system to screen for quorum-sensing molecules interacting with quorum receptor proteins of Streptomyces coelicolor. Appl. Environ. Microbiol. 75 6367–6372. 10.1128/AEM.00019-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. K., Morikawa M., Shimizu H., Shioya S., Suga K., Nihira T., et al. (1996). Maximum virginiamycin production by optimization of cultivation conditions in batch culture with autoregulator addition. Biotechnol. Bioeng. 49 437–444. [DOI] [PubMed] [Google Scholar]
- Yang Y. K., Shimizu H., Shioya S., Suga K., Nihira T., Yamada Y. (1995). Optimum autoregulator addition strategy for maximum virginiamycin production in batch culture of Streptomyces virginiae. Biotechnol. Bioeng. 46 437–442. 10.1002/bit.260460507 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pan G., Zou Z., Fan K., Yang K., Tan H. (2013). JadR∗-mediated feed-forward regulation of cofactor supply in jadomycin biosynthesis. Mol. Microbiol. 90 884–897. 10.1111/mmi.12406 [DOI] [PubMed] [Google Scholar]
- Zhu J., Chen Z., Li J., Wen Y. (2017). AvaR1, a butenolide-type autoregulator receptor in Streptomyces avermitilis, directly represses avenolide and avermectin biosynthesis and multiple physiological responses. Front. Microbiol. 8:2577. 10.3389/fmicb.2017.02577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Sun D., Liu W., Chen Z., Li J., Wen Y. (2016). AvaR2, a pseudo (-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 102 562–578. 10.1111/mmi.13479 [DOI] [PubMed] [Google Scholar]
- Zou Z., Du D., Zhang Y., Zhang J., Niu G., Tan H. (2014). A (-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol. Microbiol. 94 490–505. 10.1111/mmi.12752 [DOI] [PubMed] [Google Scholar]