Abstract
Fusarium oxysporum is a globally distributed soilborne fungal pathogen causing root rots, bulb rots, crown rots and vascular wilts on a range of horticultural plants. Pathogenic F. oxysporum isolates are highly host specific and are classified as formae speciales. Narcissus is an important ornamental crop and both the quality and yield of flowers and bulbs can be severely affected by a basal rot caused by F. oxysporum f. sp. narcissi (FON); 154 Fusarium isolates were obtained from different locations and Narcissus cultivars in the United Kingdom, representing a valuable resource. A subset of 30 F. oxysporum isolates were all found to be pathogenic and were therefore identified as FON. Molecular characterisation of isolates through sequencing of three housekeeping genes, suggested a monophyletic origin with little divergence. PCR detection of 14 Secreted in Xylem (SIX) genes, previously shown to be associated with pathogenicity in other F. oxysporum f. spp., revealed different complements of SIX7, SIX9, SIX10, SIX12 and SIX13 within FON isolates which may suggest a race structure. SIX gene sequences were unique to FON and SIX10 was present in all isolates, allowing for molecular identification of FON for the first time. The genome of a highly pathogenic isolate was sequenced and lineage specific (LS) regions identified which harboured putative effectors including the SIX genes. Real-time RT-PCR, showed that SIX genes and selected putative effectors were expressed in planta with many significantly upregulated during infection. This is the first study to characterise molecular variation in FON and provide an analysis of the FON genome. Identification of expressed genes potentially associated with virulence provides the basis for future functional studies and new targets for molecular diagnostics.
Keywords: Fusarium oxysporum f. sp. narcissi, Narcissus, daffodil, basal rot, pathogenicity, Secreted in Xylem
Introduction
Fusarium oxysporum is a globally important plant pathogen, causing root rots, bulb rots, crown rots and vascular wilts on a wide range of horticultural crops and ornamental plants including onion, lettuce, tomato, cucumber, pea, daffodil (Narcissus), carnation, lily, tulips and Gladiolus (Leslie and Summerell, 2006; Michielse and Rep, 2009). Pathogenic F. oxysporum isolates are highly host specific and are therefore classified as formae speciales (Leslie and Summerell, 2006; Michielse and Rep, 2009). There are now 106 well-characterised formae speciales, 37 with insufficient evidence to definitively classify and 58 plant species susceptible to F. oxysporum that do not have a defined forma specialis (Edel-Hermann and Lecomte, 2019). Recent research on F. oxysporum f. sp. lycopersici (FOL, infecting tomato) has led to the discovery that host specific virulence is associated with lineage specific (LS) chromosomes; for instance, it has been demonstrated that a non-pathogenic F. oxysporum isolate can acquire pathogenicity against tomato by transfer of an LS chromosome (Ma et al., 2010; Rep and Kistler, 2010; Ma et al., 2015). LS chromosomes have a low gene density (after transposons are taken into account) and are enriched for small secreted proteins (Ma et al., 2010; Schmidt et al., 2013; Armitage et al., 2018) which include those encoded by Secreted in Xylem (SIX) genes, 14 of which have been identified in FOL (Houterman et al., 2009; Schmidt et al., 2013). Functional studies have shown that SIX1, SIX3, SIX5 and SIX6 have a direct role in FOL virulence (Rep, 2005; Houterman et al., 2009; Takken and Rep, 2010; Gawehns et al., 2014; Ma et al., 2015); however, SIX genes show little or no homology to each other or any other genes of known function. Several studies have identified SIX gene homologues in a range of F. oxysporum f. spp. and their presence is very often associated with pathogenicity (Lievens et al., 2009; Fraser-Smith et al., 2014; Taylor et al., 2016; van Dam et al., 2016). In addition, members of nine transcription factor families on FOL LS regions (TF1-9) have roles in virulence through controlling effector gene expression (van der Does et al., 2016). In particular, research investigating F. oxysporum f. sp. phaseoli, causing wilt of common bean, revealed that Fusarium transcription factor (FTF1) from the TF1 gene family, plays a role in virulence through controlling the expression of at least two SIX genes (Nino-Sanchez et al., 2016).
Daffodil (Narcissus spp.) is one of the most widely cultivated ornamental bulb crops of temperate regions. The major production areas are the United Kingdom (3808 ha per annum), Netherlands (1756 ha) and United States (410 ha) although smaller areas are cultivated across the world (Hanks, 2013). In the United Kingdom, bulbs are particularly prone to infection by soil-borne pathogens due to the standard biennial growing system employed whereby after 2 years in the field, bulbs are lifted and used as a replanting stock (Hanks, 2013). One of the most economically damaging pathogens of Narcissus is F. oxysporum f. sp. narcissi (FON) Snyder and Hansen, which infects the roots or damaged basal plates resulting in soft and rotting bulbs (Linfield, 1994). Affected bulbs do not sprout, or produce short-lived shoots, or chlorotic, prematurely senescing foliage with few or no flowers. Infection can occur in the field or in storage (Hanks, 1996). Although FON infection is favoured by higher temperatures, a minimum temperature range of 8–10°C has been reported for germination of chlamydospores and growth of mycelium (Price, 1977) with infection optimum at 29°C (McClennan, 1952).
Control of FON is challenging, mainly due to the production of chlamydospores which can survive in the soil for many years. Under laboratory conditions, chlamydospores of a F. oxysporum isolate from muskmelon were shown to be viable for at least 17 years (McKeen and Wensley, 1961). In the United Kingdom, daffodil bulbs which are lifted from the soil are routinely subjected to hot water treatment (44.4°C for 3 h; Qiu et al., 1993; Hanks and Linfield, 1999) during which bulbs can become infected with FON (Hawker, 1935). Until the late 2000s, formalin was also added to the hot water tank to enhance nematode control and to kill FON chlamydospores (Linfield, 1994; Hanks and Linfield, 1999). Subsequently, formalin has been replaced by fungicides such as thiabendazole and chlorothalonil (Hanks, 1996). There is some evidence that dipping bulbs in a biological control agent such as Streptomyces, as part of an integrated control approach, can help reduce the incidence of basal rot but this is an area which requires more research (Hiltunen et al., 1995). Whilst some partial resistance to FON has been identified, particularly in material related to the cv. St. Keverne and also in some related wild germplasm (Nicholson et al., 1989; Bowes et al., 1992; Linfield, 1992), the genetics of this resistance have not been investigated. It has been proposed that the mechanism of resistance in St. Keverne is controlled by multiple factors, including the accumulation of antifungal compounds and cell wall modification (Nicholson et al., 1989).
Whilst differences in pathogenicity and morphology between FON isolates has been occasionally reported (Beale and Pitt, 1989; Linfield, 1994), these studies were conducted more than 20 years ago and as such may not represent the current diversity of FON isolates. No research has specifically characterised isolates based on DNA sequence or attempted to identify genes potentially associated with FON virulence. This study assembled a contemporary collection of FON isolates from different United Kingdom Narcissus cultivars and locations and characterised them through PCR and sequencing of housekeeping and putative effector genes. Genome sequencing of a single FON isolate was carried out to allow comparative analysis with FOL and F. oxysporum f. sp. cepae (FOC), which causes a basal rot of onion bulbs, including identification of putative LS effector candidates. The expression of putative FON effectors in planta was also examined using real-time RT-PCR.
Materials and Methods
Collection of Fusarium Isolates From Narcissus and Pathogenicity Testing
Narcissus bulb samples with symptoms of basal rot representing 30 cultivars were obtained from 39 different locations in the United Kingdom in 2012 (Supplementary Table S1). Five bulbs from each sample were split lengthways and isolations carried out from internal infected scales as previously described for onion (Taylor et al., 2016). Fungal colonies identified as Fusarium were assigned to one of eight groups based on their morphology (colour/growth habit) on 50% potato dextrose agar (PDA). Isolates were placed in medium- and long-term storage on PDA slopes (4°C) and as spore suspensions in potato dextrose broth containing 20% glycerol (−80°C), respectively. A subset of 30 Fusarium isolates from the different morphology groups were selected for further analysis based on their relative frequencies and geographic origin (Table 1).
TABLE 1.
Lesion size on Narcissus bulbs, culture morphology group and presence/absence of putative effector genes (SIX, BFJ63_5681, BFJ63_18062, BFJ63_18633) and Fusarium transcription factor (FTF) genes (as determined by PCR) for 30 FON isolates from different Narcissus cultivars and locations.
| Isolate | Morphology | Location | Cultivar | Lesion area (cm2) | SIX7 | SIX9 | SIX10 | SIX12 | SIX13 | 18062 | 5681 | 18633 | FTF1 | FTF2 |
| FON139 | 4 | Falmouth, Cornwall | Scrumpy | 8.28 | + | + | + | + | – | + | + | + | A | + |
| FON81 | 4 | Truro, Cornwall | Salome | 7.49 | – | – | + | – | – | + | + | + | – | + |
| FON63 | 5 | Truro, Cornwall | Magnificence | 7.37 | + | + | + | + | + | + | + | + | B | + |
| FON115 | 4 | Falmouth, Cornwall | Pinza | 7.22 | + | + | + | + | – | + | + | + | – | + |
| FON46 | 4 | Truro, Cornwall | Salome | 7.05 | – | – | + | – | – | + | + | + | – | + |
| FON7 | 7 | Norfolk | White Lion | 6.84 | + | + | + | + | – | + | + | + | A | + |
| FON42 | 1 | Holt, Norfolk | Golden Ducat | 6.81 | + | + | + | + | – | + | + | + | A | + |
| FON75 | 4 | Norwich, Norfolk | Pheasant’s Eye | 6.79 | + | + | + | + | – | + | + | + | – | + |
| FON3 | 4 | Norfolk | White Lion | 6.71 | – | – | + | – | – | + | + | + | – | + |
| FON89 | 4 | Cornwall | Jedna | 6.54 | + | + | + | + | – | + | + | + | B | + |
| FON97 | 2 | Cornwall | Hollywood | 6.42 | + | + | + | + | – | + | + | + | A | + |
| FON122 | 7 | Falmouth, Cornwall | Unique | 6.4 | + | + | + | + | – | + | + | + | A | + |
| FON24 | 3 | Boston, Lincs | Carlton | 6.36 | + | + | + | + | – | + | + | + | B | + |
| FON29 | 8 | Norfolk | Fortune | 6.18 | + | + | + | + | – | + | + | + | A | + |
| FON11 | 7 | E Cornwall | Carlton | 6.12 | + | + | + | + | – | + | + | + | A | + |
| FON38 | 2 | Spalding, Lincs | Carlton | 5.86 | + | + | + | + | – | + | + | + | B | + |
| FON34 | 4 | Holt, Norfolk | Sempre Avanti | 5.78 | + | + | + | + | – | + | + | + | A | + |
| FON94 | 7 | Cornwall | Hollywood | 5.53 | + | + | + | + | – | + | + | + | A | + |
| FON 87 | 6 | Cornwall | Golden Ducat | 5.36 | + | + | + | + | – | + | + | + | B | + |
| FON118 | 1 | Falmouth, Cornwall | Orkney | 4.59 | + | + | + | + | – | + | + | + | A | + |
| FON77 | 1 | Norwich, Norfolk | Pheasant’s Eye | 4.44 | – | + | + | – | – | + | + | + | B | + |
| FON55 | 7 | Penzance | White Lion | 4.44 | + | + | + | + | – | + | + | + | B | + |
| FON58 | 2 | Spalding, Lincs | Great Leap | 4.15 | + | + | + | + | – | + | + | + | A | + |
| FON141 | 1 | Falmouth, Cornwall | Mithrel | 4.02 | + | + | + | + | – | + | + | + | B | + |
| FON129 | 6 | Falmouth, Cornwall | Whiskey Galore | 4.02 | + | + | + | + | – | + | + | + | A | + |
| FON19 | 1 | Spalding, Lincs | Quirinus | 3.64 | + | + | + | + | – | + | + | + | B | + |
| FON15 | 4 | Spalding, Lincs | St Keverne | 3.53 | + | + | + | + | – | + | + | + | A | + |
| FON152 | 4 | Moulton, Lincs | Carlton | 3.36 | + | + | + | + | – | + | + | + | – | + |
| FON133 | 7 | Falmouth, Cornwall | Hampton Court | 3.3 | + | + | + | + | – | + | + | + | A | + |
| FON68 | 7 | Spalding, Lincs | Spellbinder | 3.24 | + | + | + | + | – | + | + | + | B | + |
Isolates were tested for pathogenicity on Narcissus bulbs of the susceptible cultivar “Carlton.” Agar plugs (5 mm diameter) from actively growing colonies of each isolate were used to inoculate five replicate “bulb units” prepared by peeling away the outer scales (Supplementary Figure S1). Sterile agar plugs were used as a control and the non-pathogenic isolate Fo47 (Aimé et al., 2013) was also included. Inoculated bulb units were placed on damp tissue in a sealed container and incubated at 25°C. Lesions were photographed at 7, 12, 15 and 20 days after inoculation and the area of each lesion calculated at each time point using ImageJ software (Schneider et al., 2012). The experiment was repeated once. The significance of differences in lesion sizes at 20 dpi was assessed by ANOVA using Genstat version 18 (VSN International).
Molecular Characterisation of Fusarium Isolates From Narcissus
DNA was extracted from freeze-dried mycelium of each isolate using a DNeasy Mini Kit (Qiagen) and PCR amplification carried out for three housekeeping genes using previously published primers and cycling conditions (Taylor et al., 2016; Supplementary Table S2); translation elongation factor 1 alpha (EF-α, exTEF-F/FUexTEF-R primers), β-tubulin (TUB2, T1/T22 primers) and RNA polymerase II second largest subunit (RPB2, 7cF/11aR primers). PCR products were purified using a QIAquick PCR Purification Kit (Qiagen) and then sequenced using the forward primer (Eurofins-GATC). Fusarium isolates were identified to species level based on EF-α sequence using BLAST searches (Boratyn et al., 2013). Sequences of all three genes were concatenated and aligned (CLUSTALW method) using MEGA version 7 (Kumar et al., 2016) and a maximum likelihood tree constructed using the gamma distributed Kimura two-parameter model (Kimura, 1980). Sequences from selected F. oxysporum f. spp. were also included for comparison and the tree rooted using Fusarium proliferatum.
Identification and Sequencing of SIX Genes, Transcription Factors and Putative Novel Effectors
The 30 Fusarium isolates from Narcissus were screened for the presence of 14 SIX genes by PCR using previously published primers and thermocycling conditions designed for both FOC (Taylor et al., 2016) and FOL (Lievens et al., 2009). Where PCR amplicons were obtained, this was confirmed by an additional (conventional) PCR using published primers designed for real-time PCR (Supplementary Table S2, data not shown). Primers were also designed to amplify FTF1 and FTF2, based on the genome sequence of isolate FON139 (described below); FON FTF1-F (5′-GGGTTGAATCTCACGTATCCTGC-3′)/FON FTF1-R (5′-TCCATTCGAGCCCTGCCCAAAG-3′) and FON FTF2-F (5′-CGGTCAAGCAATTCGCATGGC-3′/FON FTF2-R (5′-GTTCTCTGTCTTGTGGACGTCG-3′). Primers were used at a final concentration of 0.5 μM each in a 20 μl reaction following the same conditions outlined for SIX gene detection in FOC (Taylor et al., 2016) but with an annealing temperature of 65°C for FTF1 and 63°C for FTF2. PCR amplicons for FTF1 were purified and sequenced as described previously and sequences aligned (CLUSTALW method) and compared using MEGA version 7 (Kumar et al., 2016). Finally, all 30 FON isolates were screened for the presence of three other putative effectors identified by the presence of a mimp motif from genome analysis (Table 3, described below), BFJ63_18062 (FON18062 F: 5′-ATGGCGAACTGGTCTTGGCTCC-3′/FON18062 R” 5′-CTTGAGCACCCCACGGAACACT-3′); BFJ63_5681 (FON 5681 F: 5′-CCGTGTTCCTTGCTTTTACTGCCG-3′/FON5681 R: 5′-TGCAGCCGCCATCCTTGTAGTAC-3′) and BFJ63_18633 (FON18633 F: 5′-CCAGGCTTTTCCTCGAACCGCA-3′/FON 18633 R: 5′-TCGACGCTGTAGTCCGCAAAGAG-3′), following the same reaction set-up and cycling conditions outlined for SIX gene detection in FOC (Taylor et al., 2016) but with an annealing temperature of 65°C.
TABLE 3.
Details of SIX gene homologs and an additional 27 genes in the FON genome located within 2 kb of mimps and encoding predicted secreted proteins (Sec + mimp).
| FON gene ID | FON contig | Sec. + mimp | Summarised effector function | Orthogroup | Orthogroup contents |
| BFJ63_17906 | Contig_728 (LS) | No | SIX7 homolog | Transmembrane domain | Orthogroup13453 | FON(1):FOL(0):FOC(1):FO(0) |
| BFJ63_19066 | Contig_1485 (LS) | Yes | SIX9 homolog | EffP | Orthogroup5087 | FON(1):FOL(2):FOC(2):FO(0) |
| BFJ63_17905 | Contig_728 (LS) | Yes | SIX10 homolog | EffP | Orthogroup6913 | FON(2):FOL(1):FOC(1):FO(0) |
| BFJ63_17909 | Contig_728 (LS) | No | SIX12 homolog | Orthogroup13592 | FON(1):FOL(0):FOC(1):FO(0) |
| BFJ63_3347 | Contig_16 (C) | Yes | CAZY | IPR017853;Glycoside hydrolase superfamily | Orthogroup6617 | FON(1):FOL(1):FOC(1):FO(1) |
| BFJ63_5681 | Contig_32 (C) | Yes | EffP | IPR002532;Hantavirus glycoprotein G2 | Orthogroup11209 | FON(1):FOL(1):FOC(1):FO(1) |
| BFJ63_17239 | Contig_492 (LS) | Yes | IPR031348;Fungal N-terminal domain of STAND protein | Orthogroup4168 | FON(2):FOL(0):FOC(2):FO(1) |
| BFJ63_17356 | Contig_522 (LS) | Yes | IPR005152;Lipase, secreted | Orthogroup1532 | FON(5):FOL(1):FOC(2):FO(1) |
| BFJ63_17513 | Contig_579 (LS) | Yes | EffP | Orthogroup13095 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_17710 | Contig_648 (LS) | Yes | PF12697;Alpha/beta hydrolase family | Orthogroup3054 | FON(3):FOL(1):FOC(1):FO(1) |
| BFJ63_17810 | Contig_686 (LS) | Yes | CAZY | IPR011050;Pectin lyase fold/virulence factor | Orthogroup299 | FON(8):FOL(6):FOC(4):FO(5) |
| BFJ63_18064 | Contig_796 (LS) | Yes | EffP | Singleton | – |
| BFJ63_18062 | Contig_796 (LS) | Yes | EffP | Orthogroup13571 | FON(1):FOL(0):FOC(1):FO(0) |
| BFJ63_18183 | Contig_848 (LS) | Yes | EffP | Orthogroup13372 | FON(1):FOL(0):FOC(1):FO(0) |
| BFJ63_18181 | Contig_848 (LS) | Yes | EffP | Orthogroup13378 | FON(1):FOL(0):FOC(1):FO(0) |
| BFJ63_18368 | Contig_947 (LS) | Yes | CAZY | IPR011050;Pectin lyase fold/virulence factor | Orthogroup2534 | FON(2):FOL(1):FOC(3):FO(1) |
| BFJ63_18450 | Contig_995 (LS) | Yes | IPR001563;Peptidase S10, serine carboxypeptidase | Orthogroup1545 | FON(3):FOL(2):FOC(2):FO(2) |
| BFJ63_18534 | Contig_1045 (LS) | Yes | EffP | IPR032710;NTF2-like domain | Orthogroup13078 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_18557 | Contig_1059 (LS) | Yes | EffP | Orthogroup13112 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_18633 | Contig_1107 (LS) | Yes | EffP | Singleton | – |
| BFJ63_18752 | Contig_1194 (LS) | Yes | IPR001506;Peptidase M12A, astacin | Orthogroup12558 | FON(1):FOL(0):FOC(2):FO(0) |
| BFJ63_18778 | Contig_1221 (LS) | Yes | – | Singleton | – |
| BFJ63_18926 | Contig_1352 (LS) | Yes | EffP | Orthogroup13360 | FON(1):FOL(0):FOC(1):FO(0) |
| BFJ63_18986 | Contig_1408 (LS) | Yes | EffP | G3DSA:2.10.80.10 | Orthogroup4560 | FON(4):FOL(0):FOC(1):FO(0) |
| BFJ63_18985 | Contig_1408 (LS) | Yes | EffP | Orthogroup13095 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_19093 | Contig_1508 (LS) | Yes | EffP | Orthogroup13113 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_19121 | Contig_1536 (LS) | Yes | EffP | Singleton | – |
| BFJ63_19127 | Contig_1542 (LS) | Yes | EffP | G3DSA:2.10.80.10; | Orthogroup4560 | FON(4):FOL(0):FOC(1):FO(0) |
| BFJ63_19143 | Contig_1559 (LS) | Yes | EffP | Orthogroup13098 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_19153 | Contig_1568 (LS) | Yes | EffP | Orthogroup13114 | FON(2):FOL(0):FOC(0):FO(0) |
| BFJ63_19631 | Contig_2189 (LS) | Yes | – | Orthogroup13584 | FON(1):FOL(0):FOC(1):FO(0) |
Genomic location within the FON assembly is shown including whether the contig was identified as core (C) or lineage specific (LS). Summaries of Interproscan annotations are provided along with indication of whether the gene is identified as encoding a protein with an effector-like structure (EffP) or carbohydrate active enzyme (CAZY). The orthology group is shown for each gene as well as the number of proteins assigned to that orthogroup from each of the genomes (FON, FOC, FOL, FO) used in the analysis. Genes analysed for their expression in planta are in bold.
Expression of FON SIX Genes and Other Putative Effectors in planta
Dried scales were removed from Narcissus bulbs (cv. Carlton) and inner scales separated to obtain 1–2 mm thick sections. These were then cut into 2 × 2 cm squares and surface sterilised by immersing in 70% ethanol for 2 min before rinsing twice in sterile distilled water. Scale sections were inoculated by placing a 5 mm agar plug removed from the edge of an actively growing colony of FON63 in the centre, mycelium side down. Scales were placed on moist chromatography paper (Whatman 3MM, Fisher Scientific, United Kingdom) in clear plastic boxes with lids (three per box) and incubated at 25°C in the dark. Four replicate samples of infected scales were taken each day from 0 (pre-inoculation) to 6 dpi by removing a 10 mm section around the inoculation point with a cork borer. Agar plugs were then removed and the infected Narcissus tissue samples flash frozen in liquid N before storing at −80°C. Mock inoculations were set up using plugs of PDA. RNA was extracted using the Spectrum plant total RNA extraction kit (Sigma, United Kingdom) following the manufacturers guidelines for a starchy plant storage organ. DNA was removed using DNase I (Sigma, United Kingdom) and first strand cDNA synthesised from 300 ng of RNA using Superscript II reverse transcriptase (Life Technologies). The expression of five SIX genes (SIX7, SIX9, SIX10, SIX12, SIX13), three other putative effectors (BFJ63_5681, BFJ63_18062, BFJ63_18633; Table 3) and two housekeeping genes (EF1α and TUB2) was then determined using previously published primers and cycling conditions (Taylor et al., 2016) with the exception of SIX13 where new primers were designed (5′-ACAGCACGGGACAGCTTACA-3′/5′-CGTCAGAGGGGTAGCCACAT-3′, annealing temperature 60 °C). Primers for the three putative effectors were as described in the previous section. Quantitative PCR was carried out using a Roche Lightcycler 480 and expression determined using a standard curve method and normalised to the geometric mean of EF1α and TUB2. The statistical significance of differences in relative expression compared to that at 1 dpi was determined using paired, two-tailed t-tests.
FON Genome Sequencing, Assembly, Gene Prediction and Orthology Analysis
DNA was extracted from freeze-dried mycelium of FON139 using the GenElute Plant Genomic DNA Miniprep Kit (Sigma) and genomic libraries prepared using the TruSeq DNA Sample Prep Kit (Illumina), following the manufacturer’s Low Sample Protocol. Libraries were sequenced using Illumina Miseq v3 2 × 300 bp PE (MS-102-3003). De novo assembly was performed using Spades v.3.5.0 (Nurk et al., 2013) with assembly quality statistics summarised using Quast (Gurevich et al., 2013). Single copy core Sodariomycete genes were identified using BUSCO v3 and used to assess assembly completeness (Simão et al., 2015). RepeatModeler, RepeatMasker and transposonPSI were used to identify repetitive and low complexity regions1,2. Gene prediction was performed on softmasked genomes using Braker1 v.2 (Hoff et al., 2016, fungal flag), a pipeline for automated training and gene prediction of AUGUSTUS 3.1 (Stanke and Morgenstern, 2005). Additional gene models were called in intergenic regions using CodingQuarry v.2 (Testa et al., 2015, pathogen flag). RNAseq data generated from FOC and the non-pathogenic Fo47 (FO, Armitage et al., 2018) were aligned to the FON genome using STAR (Dobin et al., 2013), and used in the training of Braker and CodingQuarry gene models for FON.
Orthology was identified between predicted FON proteins and those in the publicly available genomes for the non-pathogenic F. oxysporum isolate Fo47, FOC isolate Fus2 and FOL isolate 4287 (Ma et al., 2010; Armitage et al., 2018). OrthoMCL v.2.0.9 (Li et al., 2003) was run with an inflation value of 5 on the combined set of predicted proteins. Venn diagrams visualising genes common between proteomes were plotted using the R package VennDiagram (Chen and Boutros, 2011). Genes in orthogroups containing a single gene from both FON and FOL were used to assess synteny between genomes. Genes in a 1:1 relationship were used visualise synteny between FON contigs and FOL chromosomes with plots generated with Circos v.0.69 (Krzywinski et al., 2009).
Draft functional annotations were determined for gene models using InterProScan-5.18-57.0 (Jones et al., 2014) and through identifying homology between predicted proteins and those contained in the July 2016 release of the SwissProt database (Bairoch and Apweiler, 2000) using BLASTP (E-value > 1 × 10–100). Putative secreted proteins were identified through prediction of signal peptides using SignalP v.4.1 and removing those predicted to contain transmembrane domains using TMHMM v.2.0 (Krogh et al., 2001; Petersen et al., 2011). EffectorP v1.0 was used to screen secreted proteins for characteristics of length, net charge and amino acid content typical of fungal effectors (Sperschneider et al., 2016). Secreted proteins were also screened for carbohydrate active enzymes using HMM models from the dbCAN database (Huang et al., 2018) and HMMER3 (Mistry et al., 2013).
The mimp family of miniature inverted-repeat transposable elements has previously been associated with putative effector genes (Schmidt et al., 2013; van Dam and Rep, 2017; Armitage et al., 2018). Genes were identified within 2 kb of a mimp sequence in the assembled and reference genomes using the consensus sequence for the mimp 3′ inverted repeat (Schmidt et al., 2013) using Perl regular expressions/CAGTGGG..GCAA[TA]AA/ and /TT[TA]TTGC..CCCACTG/.
Searches for homologs of known FTFs were performed using tBLASTx against the FON genome. Searches were performed for SGE1, a core region transcription factor involved in pathogenicity response, and a further nine transcription factor families (TF1-9), with genes predominantly found on F. oxysporum LS regions (Nino-Sanchez et al., 2016; van der Does et al., 2016; Armitage et al., 2018). Sequences used were those detailed in Nino-Sanchez et al. (2016) and Armitage et al. (2018). Genes intersecting the locations of these blast hits were identified using Bedtools v.2.2.6 (Quinlan and Hall, 2010). Coding sequence of these genes was extracted and aligned in Geneious 10.0.33 using the MAFFT algorithm, along with query sequences. A cladogram was generated for TF1 family homologs of FTF1 and FTF2 genes in Geneious using a Neighbour joining approach and the consensus tree displayed using the R package GGtree v1.12.4 (Yu et al., 2017).
Results
Fusarium Isolates From Narcissus Vary in Morphology and Virulence
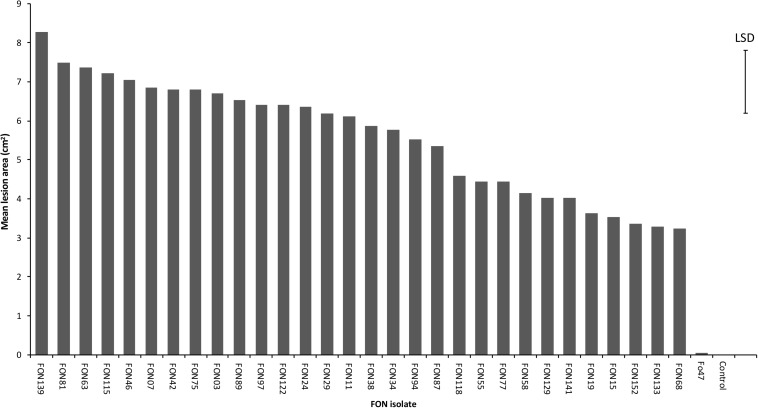
A total of 154 Fusarium isolates were obtained from Narcissus bulb samples and classified into eight morphology groups ranging in colour from purple/pink to pale orange/white (Supplementary Table S1 and Supplementary Figure S2). The number of isolates in each group (Table 1) ranged from four in group 3 (purple/red colonies) to 50 in group 4 (white/peach colonies) and this latter group was by far the most common (32.5% of total isolates). Culture morphology was not related to isolate origin and generally, isolates from a single location or Narcissus variety comprised of more than one morphology group. The 30 Fusarium isolates selected for further analysis comprised group 1, five isolates; group 2, three isolates; group 3, one isolate; group 4, 10 isolates; group 5, one isolate, group 6, two isolates; group 7, seven isolates; group 8, one isolate (Table 1). All isolates were pathogenic on Narcissus cv. Carlton but significant differences in virulence were observed (P < 0.001, Figure 1 and Table 1) with mean lesion sizes ranging from 3.2 cm2 (FON68) to 8.3 cm2 (FON139). No lesions were observed on mock inoculated (control) bulbs. The mean lesion size caused by the known non-pathogenic isolate Fo47 was 0.05 cm2, which was not significantly different from the control treatment (Figure 1).
FIGURE 1.
Virulence of 30 Fusarium oxysporum isolates against Narcissus (cv. Carlton) as measured by mean lesion area at 20 dpi (cm2). The error bar indicates the maximum LSD (5% level) following ANOVA. Standard error values ranged from 0.20 to 0.87 with a mean of 0.55.
FON Isolates From Narcissus Are From a Single Lineage
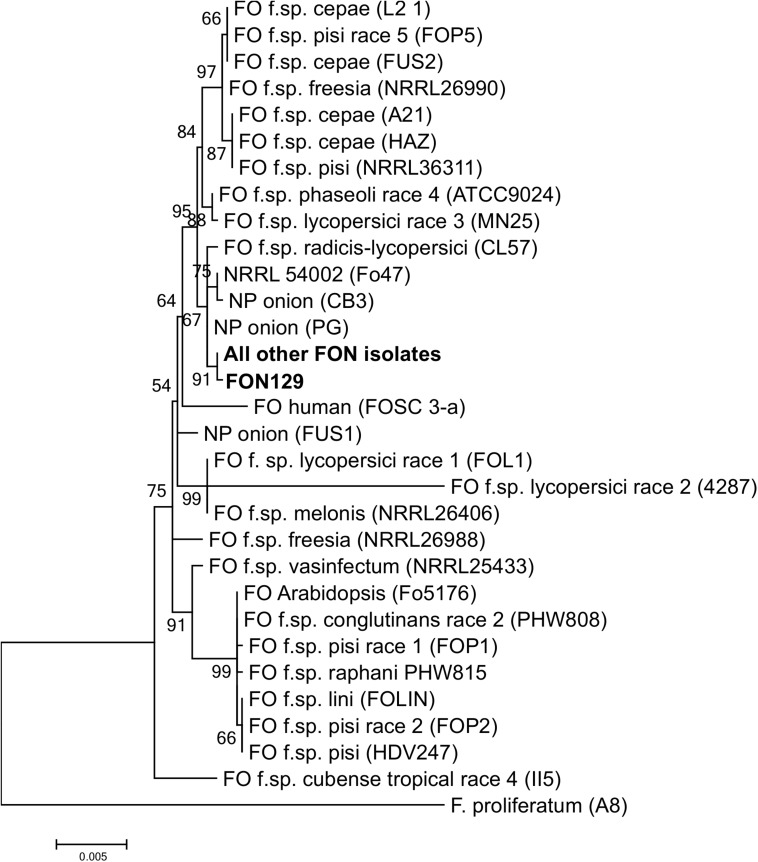
BLAST analysis of EF1-α sequences identified all 30 selected Fusarium isolates as F. oxysporum and as they were all pathogenic on Narcissus they were designated as FON. Despite the wide variation in morphology, the concatenated sequences of three housekeeping genes (EF1-α, TUB2 and RPBII), totalling 2698 bp, revealed that all 30 FON isolates were identical with the exception of isolate FON129 which showed a single base change (T to A) in the TUB2 region resulting in a single amino acid change from phenylalanine to tyrosine. The ML tree showed that all FON isolates clustered in a single clade that was separated from other F. oxysporum f. spp. suggesting they were from a single lineage (Figure 2).
FIGURE 2.
Maximum likelihood tree showing relationships between Fusarium oxysporum isolates from Narcissus and other hosts based on a concatenated alignment of β-tubulin, translation elongation factor 1-α and RNA polymerase II second largest subunit. Numbers represent bootstrap (1000 replicates) and the scale bar indicates 0.005 substitutions per site. An isolate of F. proliferatum was used to root the tree. Additional sequences were obtained from Taylor et al. (2016). NP, non-pathogenic.
SIX and FTF Gene Complements Differ Within FON
The 30 FON isolates contained different complements of SIX genes as determined by PCR (Table 1). Three isolates contained SIX10 only, one isolate SIX9 and SIX10, 25 isolates SIX7, SIX9, SIX10 and SIX12, with a single isolate (FON63) containing SIX7, SIX9, SIX10, SIX12 and SIX13. The sequences of the SIX gene amplicons were identical for all FON isolates. There were high levels of similarity between FON and FOL SIX genes with at least 84% protein identity and 93% nucleotide identity (Supplementary Table S3). In the case of SIX9, no amplification was observed for FON using the FOL primers, but products were obtained with the FOC primers; the FON SIX9 gene was 99% identical to FOC SIX9. Despite the high level of sequence homology, SIX gene sequences were unique to FON following BLAST searches. The three putative effector genes identified the genome analysis (BFJ63_5681, BFJ63_18062, BFJ63_18633) were all present in each of the 30 FON isolates.
All 30 FON isolates contained the FTF2 gene while 24 isolates contained FTF1 where two different sequence types (A, B) were identified (Table 1). Based on percentage amino acid identity, FON FTF1a and FTF1b were closely related to the corresponding FTF1a and FTF1b genes of F. oxysporum f. sp. melonis (Nino-Sanchez et al., 2016). Analysis of the FON139 assembled genome confirmed the presence of a single copy of both FTF1 and FTF2 in this isolate (described below).
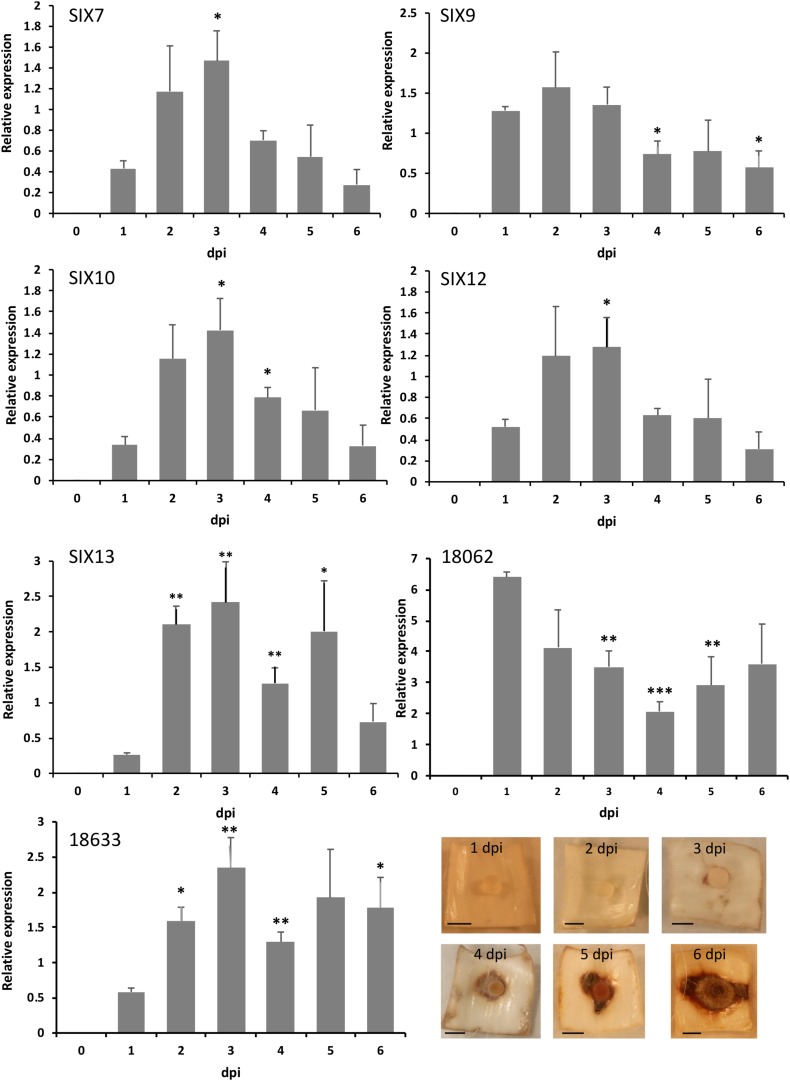
FON SIX Genes and Other Putative Effectors Are Expressed in planta
Isolate FON63 was chosen for expression analyses as it was highly virulent and contained the maximum complement of five SIX genes. All SIX genes tested were expressed in planta and significant upregulation was observed for SIX7, SIX10, SIX12 and SIX13 (Figure 3). No upregulation was observed for SIX9, but a high level of expression was seen at 1 dpi, possibly suggesting that this gene is upregulated at an earlier time. SIX gene expression peaked at 3 dpi when disease symptoms were just starting to develop in the scale tissue, after which expression declined. As for SIX9, the putative effector gene BFJ63_18062, showed a very high expression level at 1 dpi while BFJ63_18633 showed a similar expression pattern to the SIX genes with significant upregulation and peak of expression at 3 dpi. The putative effector BFJ63_5681 was expressed at levels that were too low for accurate analysis (data not shown).
FIGURE 3.
Quantitative expression of SIX genes and two other putative effectors in planta following inoculation of Narcissus (cv. Carlton) scales with isolate FON63. Expression was calculated relative to the geometric mean of translation elongation factor 1-α and β-tubulin. Error bars show the standard error of the mean of four biological replicates; dpi, days post-inoculation. Asterisks indicate expression levels significantly different from 1 dpi based on two-tailed T-tests (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). Images show the progression of disease symptoms at the time-points used for RNA extractions; scale bars are 0.5 cm.
FON Genome Sequencing, Assembly and Gene Prediction
Illumina sequencing of FON139 led to generation of 40-fold coverage of the genome, which was assembled to 57.6 Mb in 4358 contigs (>500 bp), with an N50 metric of 159 kb (Table 2). Despite fragmentation in the assembly, homologs were detected for 3684 of 3725 (98.9%) core Sodariomycete genes, comparable to results from previous F. oxysporum sequencing projects (Table 2). Repetitive and low complexity regions within the assembly were identified, masking 5.7 Mb (9.88 %) of the genome, similar to the 5.6 Mb (10.54%) masked previously for FOC (Armitage et al., 2018).
TABLE 2.
Genomic statistics for FON139 and reference genomes for FOL, FOC and FO.
| Organism | FON | FOL | FOC | FO |
| Isolate | 139 | 4287 | Fus2 | fo47 |
| (A) Assembly stats: | ||||
| Assembly size (Mb) | 57.5 | 61.5 | 53.4 | 49.7 |
| Contigs | 4349 | 15 +73∗ | 34 | 124 |
| Largest contig (kb) | 1164 | 6855 | 6434 | 6199 |
| N50 (kb) | 181 | 4590 | 414 | 3844 |
| Kb Repeatmasked | 5690 | 10,092 | 5632 | 2819 |
| % Repeatmasked | 9.89 | 16.42 | 10.54 | 5.68 |
| Sodariomycete genes (BUSCO) | 3684 | 3599 | 3687 | 3687 |
| % Sodariomycete genes (BUSCO) | 98.9 | 96.6 | 99.0 | 99.0 |
| (B) Gene models: | ||||
| Total genes | 20493 | 20925 | 18855 | 18191 |
| Total proteins | 20701 | 27347 | 19371 | 24818 |
| Braker transcripts | 19059 | – | – | – |
| CodingQuarry transcripts | 1642 | – | – | – |
| Sodariomycete genes (BUSCO) | 3666 | 3577 | 3668 | 3687 |
| % Sodariomycete genes (BUSCO) | 98.4 | 96.0 | 98.5 | 99.0 |
| Secreted genes | 1560 | 1493 | 1449 | 1409 |
| (C) Effector-associated mimp motif: | ||||
| Mimps in genome | 207 | 158 | 153 | 25 |
| Genes in 2 kb of mimp | 119 | 108 | 155 | 24 |
| Secreted genes in 2 kb of mimp | 29 | 22 | 31 | 3 |
| (D) Effector candidates: | ||||
| Secreted and effector-like structure | 407 | 351 | 355 | 291 |
| Secreted CAZYmes | 399 | 386 | 386 | 382 |
Summarised assembly (A) and gene model (B) statistics are shown, as well as results of searches for pathogenicity-associated miniature impala (mimp) sequences (C) and identification of effector candidates (D). Gene models predicted in this study show values predicted by Braker and additional genes predicted by CodingQuarry. ∗FOL chromosomes + non-scaffolded contigs.
Gene prediction performed on the softmasked genome resulted in 20,493 genes, with 19,059 predicted from Braker and a further 1642 additional proteins predicted by CodingQuary in intergenic regions (Table 2). The predicted proteome was found to contain homologs to 3666 of 3725 (98.4%) core Sodariomycete genes, indicating that gene prediction led to the annotation of the majority of FON gene models. The assembly and gene models were deposited at GenBank as a Whole Genome Shotgun project under the accession PRJNA338236. The version described in this paper is the first version, MQTW00000000.
Orthology Analysis and Identification of Core and Unplaced Regions of the FON Genome
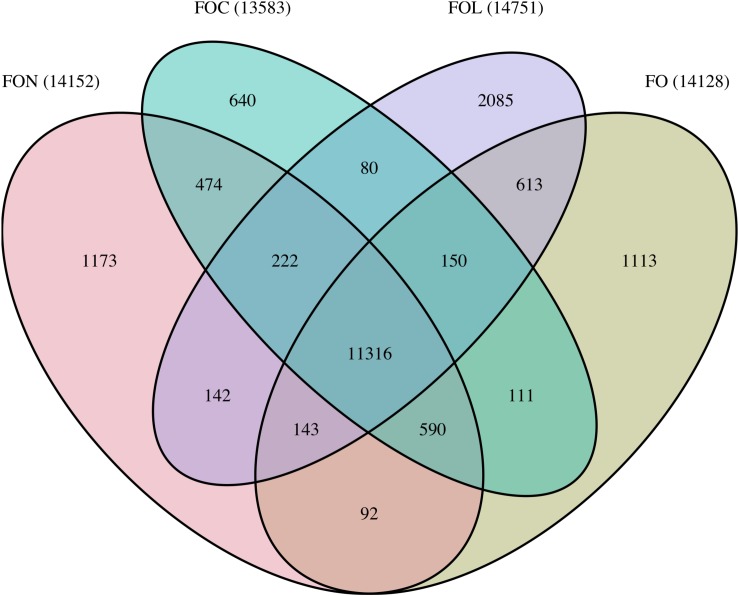
Orthology analysis clustered 92,237 proteins from FO, FON, FOC and FOL proteomes into 18,944 orthologue groups (Figure 4). Of these, 11,316 orthogroups were common to all isolates, containing 77,393 proteins including 17,147 proteins from FON. The remaining 3554 FON proteins were shared between a subset of isolates or unique to FON. Orthologous genes were used to explore macrosynteny and conservation of effectors, as below.
FIGURE 4.
Summary of shared and unique orthogroups between FON, FOC, FOL and FO genomes. Bracketed numbers indicate the total number of orthogroups for each genome.
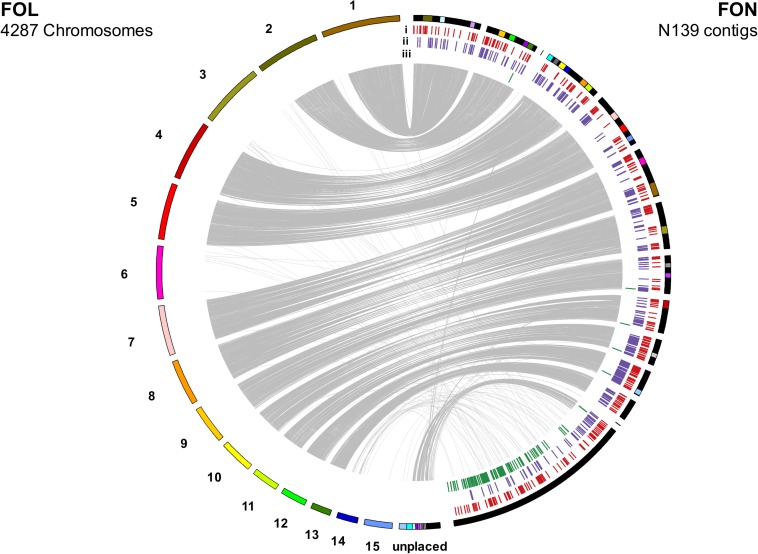
Macrosynteny between FON and FOL assemblies was assessed using 7369 proteins in a 1:1 relationship between FOL and FON. The size of the FON core genome was consistent with that found in other F. oxysporum genomes with 273 contigs, equivalent to 42.7 Mb which showed macrosynteny with the 11 FOL core chromosomes (Figure 5). Remaining contigs comprised a total size of 14.9 Mb, representing the putative LS portion of the FON genome.
FIGURE 5.
Synteny between FOL and FON genomes as determined by linking single copy orthologous genes. The 14.9 Mb of FON contigs not showing synteny with FON chromosomes are grouped following syntenous contigs. Annotation tracks under FON contigs show distribution of: (i) secreted genes with an effector-like structure, (ii) secreted carbohydrate active enzymes and (iii) mimps throughout the genome.
SIX Genes Are Located Within Putative LS Regions of the FON Genome
BLAST searches identified single homologs of FOL SIX7, 9, 10, 12 in putative LS regions of the FON genome. FON homologs of SIX7, 10 and 12 were located on a single 7594 bp contig (contig_728; Table 3) whereas SIX9 was located on a 2809 bp contig (contig_1485; Table 3). The FON SIX9 homolog was a member of the same orthogroup as the SIX9 homologs from both FOC and FOL. FON SIX7 and SIX12 genes were assigned to orthogroups containing their corresponding SIX7 and SIX12 genes from FOC, but not FOL, reflecting the divergence between these rapidly evolving genes. Similarly, SIX10 did not cluster with any other genes. The presence of FON SIX gene homologs in LS regions but lack of synteny with FOL and FOC indicates a common origin for these regions but divergence in gene content.
Mimps Are Expanded in the FON Genome and Allow Prediction of Novel Effector Candidates
The FON genome was found to contain over 30% more mimps than FOC or FOL genomes, with 207 mimp sequences identified (Table 2). These mimp sequences were primarily located in putative LS regions of the FON genome, with only six on contigs that showed synteny to FOL chromosomes, four on FON_contig_172 that showed synteny to an unplaced FOL contig DS231739.1 and the remaining 197 on unplaced FON contigs. Despite the increased numbers of mimps in FON compared with FOC, only 119 genes were within 2 kb (a similar number to FOC) and of these, only 29 encoded secreted proteins (Table 2).
Putative effectors and pathogenicity factors were identified in the FON genome using EffectorP, CAZY and AntiSMASH (Table 2), with particular focus on genes located on LS contigs and the 29 genes encoding secreted proteins within 2 kb of a mimp (Table 3). These 29 genes included homologs of SIX9 and SIX10, but not SIX7 and SIX12 (not predicted to be secreted) and included 19 that encoded proteins with a predicted effector-like structure and three encoding secreted CAZYmes. Of the remaining seven genes, annotations included one STAND domain, one alpha/beta hydrolase, one lipase, two peptidases, and two proteins with no recognisable domains (Table 3). Two of the 29 effector candidates were located on FON contigs corresponding to core FOL chromosomes 10 and 11 (BFJ63_3347, BFJ63_5681), with both being members of orthogroups containing a single gene from all isolates, including the non-pathogenic FO. None of the other genes were in orthogroups with a 1:1 relationship between FON and FO, indicating that these represent genes on LS regions.
Identification of Transcription Factors in the FON Genome
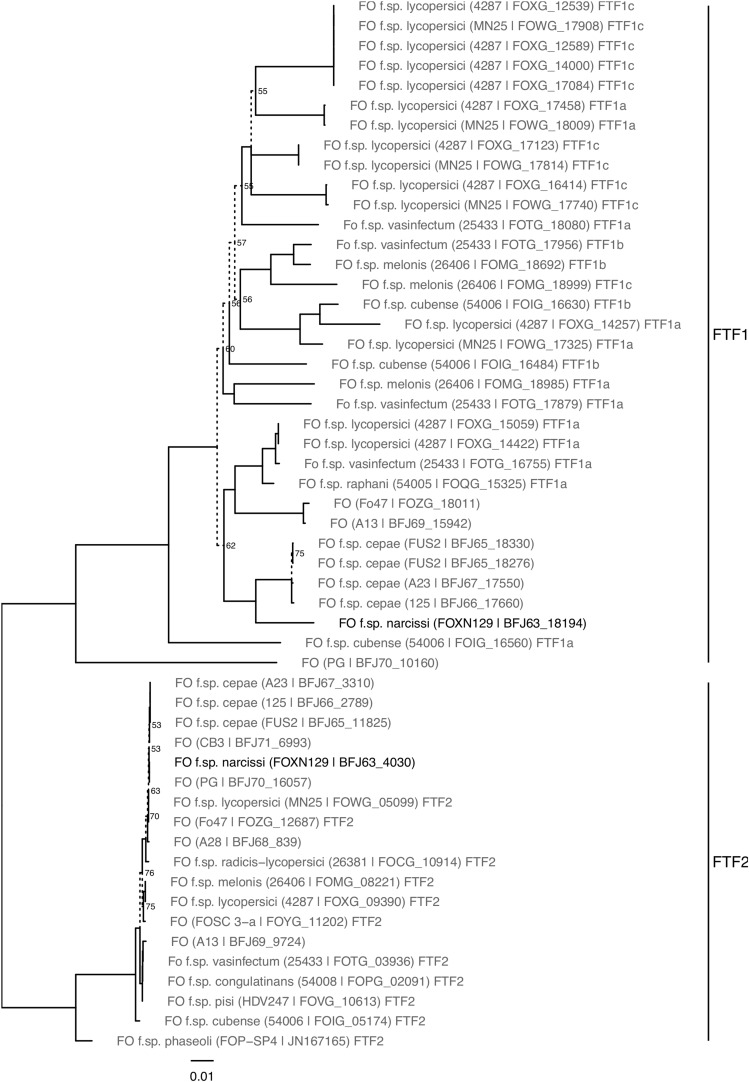
Pathogenicity factors within F. oxysporum are regulated by genes present on both core and LS regions (van der Does et al., 2016). Orthogroups were identified containing the FOL core genome transcription factor SGE1 and genes from nine additional FOL transcription factor families (TF1–9), including those from FOL LS regions (Table 4). SGE1 had a single FON homolog within the core region of the genome identified as syntenous to FOL chromosome 9. The TF1 family (including FTF1 and FTF2) was identified in FON, with all previously identified FOL and FOC FTF1 and FTF2 genes present in a single orthogroup (Orthogroup 472) including two genes from FON. This confirmed the PCR results. Phylogenetic analysis of this orthogroup identified one of these genes as FTF2 and the other as an FTF1 homolog distinct from previously sequenced FTF1 genes (Figure 6). These genes were located on a contig showing synteny to FOL chromosome 9 and a putative LS contig, respectively. Of the eight other transcription factor gene families, seven had homologs in FON, with no homologs found to TF3 genes. In each case, a single homolog was located within contigs syntenous to FOL core chromosomes. In addition to these core genome homologs, additional homologs were identified to TF4, TF6, TF7, TF8 and TF9 genes, all of which were located in putative LS FON contigs (Table 4).
TABLE 4.
Members of SGE and TF1-9 transcription factor families in FON.
| Transcription factor | Function | Orthogroup | Orthogroup contents |
FON genes in orthogroup |
||
| FON gene ID | FON contig | FOL chromosome | ||||
| SGE | Orthogroup10386 | FON(1):FOL(1):FOC(1):FO(1) | BFJ63_6144 | Contig_35 | 9 | |
| TF1 (FTF2) | Zn(2)-C6 fungal transcription factor | Orthogroup472 | FON(2):FOL(11):FOC(3):FO(2) | BFJ63_4030 | Contig_20 | 9 |
| TF1 (FTF1) | BFJ63_18194 | Contig_853 | – | |||
| TF2 | Forkhead transcription factor | Orthogroup4647 | FON(1):FOL(2):FOC(1):FO(1) | BFJ63_4125 | Contig_21 | 1 |
| TF4 | Zn finger containing protein | Orthogroup1633 | FON(2):FOL(3):FOC(2):FO(2) | BFJ63_8640 | Contig_61 | – |
| BFJ63_17657 | Contig_629 | – | ||||
| TF5 | Basic-leucine zipper (bZIP) transcription factor | Orthogroup4960 | FON(1):FOL(2):FOC(1):FO(1) | BFJ63_10518 | Contig_87 | 8 |
| TF6 | Zn finger containing protein | Orthogroup2432 | FON(3):FOL(2):FOC(1):FO(1) | BFJ63_8562 | Contig_60 | 5 |
| BFJ63_16235 | Contig_305 | – | ||||
| BFJ63_16773 | Contig_388 | – | ||||
| TF7 | Zn finger containing protein | Orthogroup413 | FON(2):FOL(12):FOC(1):FO(5) | BFJ63_13207 | Contig_140 | 9 |
| BFJ63_16716 | Contig_379 | – | ||||
| TF8 | Zn(2)-C6 fungal transcription factor | Orthogroup174 | FON(5):FOL(21):FOC(2):FO(3) | BFJ63_7317 | Contig_46 | 7 |
| BFJ63_16714 | Contig_379 | – | ||||
| BFJ63_17643 | Contig_624 | – | ||||
| BFJ63_19029 | Contig_1453 | – | ||||
| BFJ63_19368 | Contig_1830 | – | ||||
| TF9 | Basic-leucine zipper (bZIP) transcription factor | Orthogroup457 | FON(5):FOL(11):FOC(2):FO(1) | BFJ63_8227 | Contig_56 | 5 |
| BFJ63_16715 | Contig_379 | – | ||||
| BFJ63_18199 | Contig_857 | – | ||||
| BFJ63_18750 | Contig_1192 | – | ||||
| BFJ63_18830 | Contig_1255 | – | ||||
The contig location in the FON genome is shown for each gene, as well as the corresponding location in the FOL genome, allowing identification of LS genes within these families.
FIGURE 6.
Neighbour joining phylogeny of FTF gene sequences from FON and reference sequences. Branches with bootstrap support under 80% from 1000 replicates are marked with dashed lines and labelled with % support values.
Discussion
Fusarium was consistently isolated from Narcissus bulbs samples with typical basal rot symptoms obtained from different locations and cultivars in the United Kingdom and the collection of over 150 isolates represents a valuable resource. The 30 isolates selected on the basis of culture morphology and origin were all identified as F. oxysporum and as they were all pathogenic on Narcissus bulbs were designated as FON. This confirms previous research that identified FON as the main causal agent for Narcissus basal rot in the United Kingdom (Price, 1975; Beale and Pitt, 1989). While the FON isolates showed considerable variation in morphology and virulence as reported by Linfield (1994) (a common feature of F. oxysporum in other pathosystems, Gordon and Martyn, 1997), there was no variation within the three housekeeping genes except a single base change in TUB2 for isolate FON129. Mutations in this gene have been associated with resistance to benzimidazole fungicides in many fungi including F. oxysporum where the same substitution but in a different position has been reported (Chen et al., 2014; Petkar et al., 2017). As the fungicide thiabendazole is extensively used to control FON (Hanks, 2013), it is possible that this mutation in FON129 confers resistance to benzimidazole fungicides but this has yet to be tested. The concatenated phylogenetic tree grouped all FON isolates in a single clade, suggesting a single evolutionary lineage. Similarly, a monophyletic origin has also been reported for FOC and F. oxysporum f. sp. ciceris (Demers et al., 2014; Taylor et al., 2016), which is in contrast to FOL (Biju et al., 2017), F. oxysporum f. sp. apii (Epstein et al., 2017) and F. oxysporum f. sp. cubense (Czislowski et al., 2018) which are polyphyletic. In this latter case, effector genes allowing F. oxysporum f. spp. to infect specific hosts are thought to have been gained by horizontal gene transfer (van Dam et al., 2016).
Despite an apparent monophyletic origin for FON, significant variation was observed in the SIX gene complement. This is highly unlikely to be due to primer mismatches as these primers are effective on a range of F. oxysporum f. spp. (Lievens et al., 2009; Taylor et al., 2016) and genome analysis of isolate FON139 revealed 95–100% primer matches. Two FON isolates contained SIX10 only with the majority of isolates containing SIX7, SIX9, SIX10 and SIX12. SIX genes are consistently associated with pathogenicity of other F. oxysporum f. spp., (Edel-Hermann and Lecomte, 2019) and several studies have shown that they are secreted into the xylem sap during infection (Houterman et al., 2007; Ligat et al., 2011; Schmidt et al., 2013; de Sain and Rep, 2015). FON SIX9 and SIX10 as well as a range of putative novel effectors were predicted to be secreted. Whilst FON SIX7 and SIX12 were not predicted to be secreted, FOL SIX12 lacks a signal peptide yet was isolated from the xylem sap of infected tomato plants (Schmidt et al., 2013) so must be secreted by a different mechanism. Functional studies have shown that knocking out certain SIX genes can lead to a reduction in virulence against tomato (Rep, 2005; Houterman et al., 2009; Gawehns et al., 2014; Ma et al., 2015). Similarly, studies on F. oxysporum infecting banana and cabbage have also shown that knocking out SIX genes directly impacts on virulence, confirming the importance of these genes across multiple F. oxysporum f. spp. (Li et al., 2016; Niu et al., 2016; Widinugraheni et al., 2018). Therefore, it is highly likely that SIX genes are involved in pathogenicity against Narcissus though gene knockouts would be required to definitively prove this. In FOL, at least four SIX genes (SIX 1, 3, 4 and 5) are recognised by host resistance genes (Rep et al., 2004; Houterman et al., 2008, 2009; Takken and Rep, 2010; Ma et al., 2015) and in the future, it may be possible to identifiy resistance genes in Narcissus which recognise FON SIX genes.
Previously, we identified a single FOC isolate which exhibited reduced virulence and contained only two of the seven FOC SIX genes (Taylor et al., 2016), suggesting that multiple SIX genes contribute to full virulence. In FON, the second most pathogenic isolate (FON81) only contained SIX10 while conversely, the least pathogenic isolate (FON68) contained four SIX genes. This suggests that there may be considerable functional redundancy in known SIX genes in FON. This is not unprecedented as FOL race 2 emerged through loss of SIX4 in order to avoid detection by a corresponding resistance gene introgressed into tomato (Houterman et al., 2008; Takken and Rep, 2010). New races can also emerge through mutation of a SIX gene, as for FOL race 3 where a single base change in the SIX3 gene resulted in resistance breakdown through avoidance of detection by another resistance gene (Houterman et al., 2009). Similarly, races of F. oxysporum f. sp. cubense can be separated by differences in SIX gene complements and sequence variations (Meldrum et al., 2012; Fraser-Smith et al., 2014). Past studies have identified cv. St. Keverne as resistant to basal rot (Nicholson et al., 1989; Bowes et al., 1992) while subsequent anecdotal evidence has indicated resistance breakdown (Hanks, 1996), supporting the theory that different races of FON may have evolved. However, the genetic basis for FON resistance in Narcissus has not been determined and further work is required to assess whether there is variation in response of Narcissus cultivars from different genetic backgrounds to isolates of FON with differing SIX gene profiles. FON15 was isolated from an infected St. Keverne bulb but showed no evidence of effector gene mutations although a whole genome sequence of this isolate would be required to definitively address this question.
All five SIX genes identified in FON63, as well as two putative novel effectors, were expressed in planta and significant upregulation was observed for SIX7, 10, 12 and 13. The upregulation of FON SIX genes coupled with the very high level of SIX gene sequence conservation between FOL and FON strongly suggests a conserved function, supporting the hypothesis that these genes are effectors in FON. As noted above, FON isolates containing only SIX10 were still highly virulent. Therefore, it is likely that other putative effectors, as identified in the FON genome analysis, are involved in pathogenicity and that there is redundancy in SIX gene function. Genome analysis identified 119 FON genes within 2 kb of a mimp sequence of which 29 encoded secreted proteins, representing further effector candidates. Mimp sequences have been shown to be present in the core genome of other F. oxysporum f. sp. and present in F. proliferatum and Fusarium hostae indicating that their presence is not unique to LS regions (van Dam and Rep, 2017; Armitage et al., 2019). The lack of orthologous genes in the non-pathogenic FO isolate fo47 gene models supports the LS status of 19 of these genes. The LS gene BFJ63_18633 (and potentially the LS gene BFJ63_18062) showed significant upregulation over time in planta, strongly suggesting a role for these genes in virulence on Narcissus. Lack of quantifiable expression for BFJ63_5681, located in the core region of the FON genome, questions whether effector candidates in the vicinity of mimps have the same association with pathogenicity when located in the core genome. Whilst this study focuses on the role of effector genes in pathogenicity, it should be noted that other mechanisms of pathogenicity may also be important. For example, production of toxins such as Fusaric acid has been shown to directly impact pathogenicity in F. oxysporum (López-Díaz et al., 2018).
The molecular characterisation of FON presented here provides potential targets for distinguishing this pathogen from other F. oxysporum f. spp. which has previously been very challenging due to the polyphyletic nature of the F. oxysporum complex (O’Donnell et al., 1998; Baayen et al., 2000). Previous work indicated that F. oxysporum f. spp. narcissi and dianthi could not be separated based on SIX gene complement/sequence (Taylor et al., 2016) but we have subsequently discovered that this historic isolate from the Warwick collection was mislabelled as it is actually pathogenic on Narcissus (Taylor et al., unpublished) and hence is now identified as FON. Whilst EF1-α sequences could be used as a preliminary tool to identify FON, sequence differences from other F. oxysporum f. spp. are not sufficient for molecular diagnostics. FON SIX10 is one potential target for a diagnostic locus but has a high level of sequence identity with FOC SIX10 (96% nucleotide identity). The newly identified putative effectors from the FON genome analysis therefore may represent more suitable diagnostic targets.
Genome sequencing at 40× coverage led to a FON assembly containing over 98% of expected Sodariomycete gene models. Despite fragmentation, synteny was identified between FON contigs and FOL chromosomes, allowing identification of genes in both core and LS regions of the genome. In total, putative LS regions represented 14.9 Mb, falling within the 4–19 Mb range of LS regions identified in other F. oxysporum f. spp. (Schmidt et al., 2016; van Dam et al., 2016; Williams et al., 2016). Transposons have been shown to play an important role in adaptation of fungal pathogens, driving genomic rearrangement (Faino et al., 2016). The genome of FON139 possessed 30% more mimps than identified in FOL or FOC, with the majority of these located in unplaced FON contigs. This number may reflect a conservative estimate of mimps in this genome as assemblies generated from Illumina sequence data have been shown to underestimate mimps in comparison to PacBio sequence data (van Dam and Rep, 2017). Despite this, FON carried similar numbers of mimp-associated secreted proteins to FOC and FOL. All SIX gene homologs were located within 2 kb of a mimp. However, mimp sequences are not determinants of gene regulation (Schmidt et al., 2013) and may play other roles in pathogen adaptation and evolution. Other miniature inverted-repeat transposable elements have been shown to be mobilised by full-length class II transposable elements (Feschotte et al., 2002; Dufresne et al., 2007; Bergemann et al., 2008) and impala sequences have been shown to be active in some F. oxysporum strains (Hua-Van et al., 2001). As there is diversity in effector gene complements between FON isolates and mimps are abundant, FON may be an important pathogen for studying their role in genomic rearrangement within and between dynamic LS regions. Pertinent questions include whether mimps are still active in FON, whether mimps are driving race evolution and whether LS regions are undergoing more reshuffling in FON.
Homologs to genes in transcription factor families known to regulate expression of virulence genes in F. oxysporum f. spp. were also identified in FON. FON139 carried a single homolog on the core genome to SGE and each of TF1-9 except TF3, congruent with the original description of these genes in FOL (van der Does et al., 2016). Additional genes in TF families were identified in unplaced FON regions likely represent LS transcription factors. TF gene families in FON showed a different pattern of expansion from those observed in FOL; TF2 and TF5 had an additional copy in FOL LS regions, but there were no additional copies in FON. In contrast, TF6 had a single copy in a FOL LS region, while FON contained two additional copies in unplaced contigs. Expansion of these TF1-9 families onto LS regions has been hypothesised to confer selective advantage through dosage effect, with more copies of TF1 being associated with greater virulence in F. oxysporum f. sp. phaseoli (de Vega-Bartol et al., 2010; van der Does et al., 2016). As such, genes regulated by these TFs may be of differing importance in FOL and FON host infection. Occurrence of the TF1 LS gene FTF1 was determined within the 30 FON isolates, leading to the identification of two distinct FTF1 haplotypes. Genes from this family have been shown to regulate SIX gene expression and contribute to virulence (Nino-Sanchez et al., 2016). This may also be the case in FON, although further work is required to understand how many genes are present in this family in different FON isolates. It may be that where a negative PCR was observed (6 out of 30), these isolates have an alternate, divergent copy of FTF1. The FON FTF2 homologue was present in all isolates and has been shown to exist in a wide range of ascomycetes (Nino-Sanchez et al., 2016).
This is the first study to characterise molecular variation in FON and provide an analysis of the FON genome. Identification of expressed genes potentially associated with virulence provides the basis for future functional studies and new targets for molecular diagnostics.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher. GenBank Accession Numbers: MQTW01000000 – FON139 genome assembly; MN078955 – FON EF1α sequence; MN078956 – FON RPB2 sequence; MN078957 – FON129 TUB2 sequence; MN078958 – FON TUB2 sequence; MN078959 – FON FTF1a sequence; MN078960 – FON FTF1b sequence; MN078961 – FON SIX7 sequence; MN078962 – FON SIX9 sequence; MN078963 – FON SIX10 sequence; MN078964 – FON SIX12 sequence; MN078965 – FON63 SIX13 sequence.
Author Contributions
AT planned and carried out the experiments, analysed the data, created figures, and drafted and edited the manuscript. AA carried out the FON genome analysis and all bioinformatics, created figures, and drafted and edited the manuscript. CH and MH carried out the experiments. AJ carried out the experiments and edited the manuscript. RH obtained funding, planned experiments, and edited the manuscript. JC obtained funding, planned experiments, and wrote and edited the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Agriculture and Horticulture Development Board and the Biotechnology and Biological Sciences Research Council for financial support.
Funding. This work was funded by the Biotechnology and Biological Sciences Research Council (Projects BB K020870 and BB K020730) and the Agriculture and Horticulture Development Board (Projects BOF 74 and FV POBOF 452).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02905/full#supplementary-material
References
- Aimé S., Alabouvette C., Steinberg C., Olivain C. (2013). The endophytic strain Fusarium oxysporum Fo47: a good candidate for priming the defense responses in tomato roots. Mol. Plant Microbe Interact. 26 918–926. 10.1094/MPMI-12-12-0290-R [DOI] [PubMed] [Google Scholar]
- Armitage A. D., Taylor A., Hulin M. T., Jackson A. C., Harrison R. J., Clarkson J. P. (2019). Draft genome sequence of an onion basal rot isolate of Fusarium proliferatum. Microbiol. Resour. Announc. 8:e1385-18. 10.1128/MRA.01385-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage A. D., Taylor A., Sobczyk M. K., Baxter L., Greenfield B. P. J., Bates H. J., et al. (2018). Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae. Sci. Rep. 8:13530. 10.1038/s41598-018-30335-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baayen R. P., O’Donnell K., Bonants P. J., Cigelnik E., Kroon L. P., Roebroeck E. J., et al. (2000). Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology 90 891–900. 10.1094/PHYTO.2000.90.8.891 [DOI] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28 45–48. 10.1093/nar/28.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale R. E., Pitt D. (1989). Variation in morphology and pathogenicity of Fusarium oxysporum f. sp. narcissi. Mycol. Res. 93 369–374. 10.1016/s0953-7562(89)80164-9 [DOI] [Google Scholar]
- Bergemann M., Lespinet O., M’barek S. B., Daboussi M. J., Dufresne M. (2008). Genome-wide analysis of the Fusarium oxysporum mimp family of MITEs and mobilization of both native and de novo created mimps. J. Mol. Evol. 67 631–642. 10.1007/s00239-008-9164-7 [DOI] [PubMed] [Google Scholar]
- Biju V. C., Fokkens L., Houterman P. M., Rep M., Cornelissen B. J. C. (2017). Multiple evolutionary trajectories have led to the emergence of races in Fusarium oxysporum f. sp. lycopersici. Appl. Environ. Microbiol. 83:e2548-16. 10.1128/AEM.02548-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn G. M., Camacho C., Cooper P. S., Coulouris G., Fong A., Ma N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41 W29–W33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes S. A., Edmondson R. N., Linfield C. A., Langton F. A. (1992). Screening immature bulbs of daffodil (Narcissus L.) crosses for resistance to basal rot disease caused by Fusarium oxysporum f. sp. narcissi. Euphytica 63 199–206. [Google Scholar]
- Chen H., Boutros P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Gao T., Liang S., Liu K., Zhou M., Chen C. (2014). Molecular mechanism of resistance of Fusarium fujikuroi to benzimidazole fungicides. FEMS Microbiol. Lett. 357 77–84. 10.1111/1574-6968.12504 [DOI] [PubMed] [Google Scholar]
- Czislowski E., Fraser-Smith S., Zander M., O’Neill W. T., Meldrum R. A., Tran-Nguyen L. T. T., et al. (2018). Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant Pathol. 19 1155–1171. 10.1111/mpp.12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sain M., Rep M. (2015). The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 16 23970–23993. 10.3390/ijms161023970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vega-Bartol J. J., Martín-Dominguez R., Ramos B., García-Sánchez M.-A., Díaz-Mínguez J. M. (2010). New virulence groups in Fusarium oxysporum f. sp. phaseoli: the xxpression of the gene coding for the transcription factor ftf1 correlates with virulence. Phytopathology 101 470–479. 10.1094/PHYTO-09-10-0252 [DOI] [PubMed] [Google Scholar]
- Demers J. E., Garzón C. D., Jiménez-Gasco M. (2014). Striking genetic similarity between races of Fusarium oxysporum f. sp. ciceris confirms a monophyletic origin and clonal evolution of the chickpea vascular wilt pathogen. Eur. J. Plant Pathol. 139 309–324. 10.1007/s10658-014-0387-8 [DOI] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne M., Hua-Van A., El Wahab H. A., Ben M’barek S., Vasnier C., Teysset L., et al. (2007). Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics 175 441–452. 10.1534/genetics.106.064360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel-Hermann V., Lecomte C. (2019). Current status of Fusarium oxysporum formae speciales and races. Phytopathology 109 512–530. 10.1094/PHYTO-08-18-0320-RVW [DOI] [PubMed] [Google Scholar]
- Epstein L., Kaur S., Chang P. L., Carrasquilla-Garcia N., Lyu G., Cook D. R., et al. (2017). Races of the celery pathogen Fusarium oxysporum f. sp. apii are polyphyletic. Phytopathology 107 463–473. 10.1094/PHYTO-04-16-0174-R [DOI] [PubMed] [Google Scholar]
- Faino L., Seidl M. F., Shi-Kunne X., Pauper M., Van Den Berg G. C. M., Wittenberg A. H. J., et al. (2016). Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 26 1091–1100. 10.1101/gr.204974.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Zhang X., Wessler S., Sue Wessler D. (2002). “Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons,” in Mobile DNA II, eds Craig N. L., Lambowitz A., Gragie R., Gellert M. (Washington, DC: ASM Press; ), 1147–1155. [Google Scholar]
- Fraser-Smith S., Czislowski E., Meldrum R. A., Zander M., O’Neill W., Balali G. R., et al. (2014). Sequence variation in the putative effector gene SIX8 facilitates molecular differentiation of Fusarium oxysporum f. sp. cubense. Plant Pathol. 63 1044–1052. [Google Scholar]
- Gawehns F., Houterman P. M., Ichou F. A., Michielse C. B., Hijdra M., Cornelissen B. J., et al. (2014). The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microbe Interact. 27 336–348. 10.1094/MPMI-11-13-0330-R [DOI] [PubMed] [Google Scholar]
- Gordon T. R., Martyn R. D. (1997). The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35 111–128. 10.1146/annurev.phyto.35.1.111 [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks G. R. (1996). Control of Fusarium oxysporum f.sp. narcissi, the cause of narcissus basal rot, with thiabendazole and other fungicides. Crop Prot. 15 549–558. 10.1016/0261-2194(96)00023-3 [DOI] [Google Scholar]
- Hanks G. R. (2013). Narcissus Manual. Kenilworth: AHDB Horticulture. [Google Scholar]
- Hanks G. R., Linfield C. A. (1999). Evaluation of a peroxyacetic acid disinfectant in hot-water treatment for the control of basal rot (Fusarium oxysporum f. sp. narcissi) and stem nematode (Ditylenchus dipsaci) in Narcissus. J. Phytopathol. 147 271–279. 10.1111/j.1439-0434.1999.tb03830.x [DOI] [Google Scholar]
- Hawker L. E. (1935). Further experiments on the Fusarium bulb rot of Narcissus. Ann. Appl. Biol. 22 684–708. 10.1111/j.1744-7348.1935.tb07176.x 19279798 [DOI] [Google Scholar]
- Hiltunen L. H., Linfield C. A., White J. G. (1995). The potential for the biological control of basal rot of Narcissus by Streptomyces sp. Crop Prot. 14 539–542. 10.1016/0261-2194(95)00068-2 [DOI] [Google Scholar]
- Hoff K. J., Lange S., Lomsadze A., Borodovsky M., Stanke M. (2016). BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32 767–769. 10.1093/bioinformatics/btv661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman P. M., Cornelissen B. J. C., Rep M. (2008). Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4:e1000061. 10.1371/journal.ppat.1000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houterman P. M., Ma L., Van Ooijen G., De Vroomen M. J., Cornelissen B. J. C., Takken F. L. W., et al. (2009). The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 58 970–978. 10.1111/j.1365-313X.2009.03838.x [DOI] [PubMed] [Google Scholar]
- Houterman P. M., Speijer D., Dekker H. L., De Koster C. G., Cornelissen B. J. C., Rep M. (2007). The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 8 215–221. 10.1111/j.1364-3703.2007.00384.x [DOI] [PubMed] [Google Scholar]
- Huang L., Zhang H., Wu P., Entwistle S., Li X., Yohe T., et al. (2018). dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 46 D516–D521. 10.1093/nar/gkx894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A., Pamphile J. A., Langin T., Daboussi M. J. (2001). Transposition of autonomous and engineered impala transposons in Fusarium oxysporum and a related species. Mol. Gen. Genet. 264 724–731. 10.1007/s004380000395 [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., Mcanulla C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. 10.1007/bf01731581 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie J. F., Summerell B. A. (2006). The Fusarium Laboratory Manual. Oxford, UK: Blackwell Publishing. [Google Scholar]
- Li E., Wang G., Xiao J., Ling J., Yang Y., Xie B. (2016). A SIX1 homolog in Fusarium oxysporum f. sp. conglutinans is required for full virulence on cabbage. PLoS One 11:e0152273. 10.1371/journal.pone.0152273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J., Roos D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens B., Houterman P. M., Rep M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300 201–215. 10.1111/j.1574-6968.2009.01783.x [DOI] [PubMed] [Google Scholar]
- Ligat L., Lauber E., Albenne C., San Clemente H., Valot B., Zivy M., et al. (2011). Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11 1798–1813. 10.1002/pmic.201000781 [DOI] [PubMed] [Google Scholar]
- Linfield C. A. (1992). Wild Narcissus species as a source of resistance to Fusarium oxysporum f.sp. narcissi. Ann. Appl. Biol. 121 175–181. 10.1111/j.1744-7348.1992.tb03998.x [DOI] [Google Scholar]
- Linfield C. A. (1994). An Investigation into the Detection, Identification, Differentiation and Pathogenicity of Fusarium Oxysporum f. sp. Narcissi. Ph.D thesis, University of Reading, Reading. [Google Scholar]
- López-Díaz C., Rahjoo V., Sulyok M., Ghionna V., Martín-Vicente A., Capilla J., et al. (2018). Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 19 440–453. 10.1111/mpp.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Houterman P. M., Gawehns F., Cao L., Sillo F., Richter H., et al. (2015). The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytol. 208 507–518. 10.1111/nph.13455 [DOI] [PubMed] [Google Scholar]
- Ma L.-J., van der Does H. C., Borkovich K. A., Coleman J. J., Daboussi M.-J., Di Pietro A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464 367–373. 10.1038/nature08850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClennan W. D. (1952). Effect of temperature on the severity of Fusarium basal rot in Narcissus. Phytopathology 42 407–412. [Google Scholar]
- McKeen C. D., Wensley R. N. (1961). Longevity of Fusarium oxysporum in soil tube culture. Science 134 1528–1529. 10.1126/science.134.3489.1528 [DOI] [PubMed] [Google Scholar]
- Meldrum R. A., Fraser-Smith S., Tran-Nguyen L. T. T., Daly A. M., Aitken E. A. B. (2012). Presence of putative pathogenicity genes in isolates of Fusarium oxysporum f. sp. cubense from Australia. Australas. Plant Pathol. 41 551–557. 10.1007/s13313-012-0122-x [DOI] [Google Scholar]
- Michielse C. B., Rep M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10 311–324. 10.1111/j.1364-3703.2009.00538.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Finn R. D., Eddy S. R., Bateman A., Punta M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. 10.1093/nar/gkt263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P., Skidmore D. I., Ingram D. S. (1989). Resistance of narcissus to infection by Fusarium oxysporum f.sp. narcissi. Mycol. Res. 93 363–368. 10.1016/s0953-7562(89)80163-7 [DOI] [Google Scholar]
- Nino-Sanchez J., Casado-Del Castillo V., Tello V., de Vega-Bartol J. J., Ramos B., Sukno S. A., et al. (2016). The FTF gene family regulates virulence and expression of SIX effectors in Fusarium oxysporum. Mol. Plant Pathol. 17 1124–1139. 10.1111/mpp.12373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X., Zhao X., Ling K. S., Levi A., Sun Y., Fan M. (2016). The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp. niveum - watermelon pathosystem. Sci. Rep. 6:28146. 10.1038/srep28146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Bankevich A., Antipov D., Gurevich A., Korobeynikov A., Lapidus A., et al. (2013). Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. Berlin: Springer, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K., Kistler H. C., Cigelnik E., Ploetz R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. 95 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., Von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Petkar A., Langston D. B., Buck J. W., Stevenson K. L., Ji P. (2017). Sensitivity of Fusarium oxysporum f. sp. niveum to prothioconazole and thiophanate-methyl and gene mutation conferring resistance to thiophanate-methyl. Plant Dis. 101 366–371. 10.1094/PDIS-09-16-1236-RE [DOI] [PubMed] [Google Scholar]
- Price D. (1975). Pathogenicity of Fusarium oxysporum found on narcissus bulbs and in soil. Trans. Br. Mycol. Soc. 64 137–142. 10.1016/s0007-1536(75)80084-2 [DOI] [Google Scholar]
- Price D. (1977). Effects of temperature and inoculum concentration on infection of narcissus bulbs by Fusarium oxysporum f.sp. narcissi. Ann. Appl. Biol. 86 433–436. 10.1111/j.1744-7348.1977.tb01860.x [DOI] [Google Scholar]
- Qiu J., Westerdahl B. B., Giraud D., Anderson C. A. (1993). Evaluation of hot water treatments for management of Ditylenchus dipsaci and fungi in daffodil bulbs. J. Nematol. 25 686–694. [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M. (2005). Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253 19–27. 10.1016/j.femsle.2005.09.014 [DOI] [PubMed] [Google Scholar]
- Rep M., Kistler H. C. (2010). The genomic organization of plant pathogenicity in Fusarium species. Curr. Opin. Plant Biol. 13 420–426. 10.1016/j.pbi.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Rep M., van der Does H. C., Meijer M., Van Wijk R., Houterman P. M., Dekker H. L., et al. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53 1373–1383. 10.1111/j.1365-2958.2004.04177.x [DOI] [PubMed] [Google Scholar]
- Schmidt S. M., Houterman P. M., Schreiver I., Ma L., Amyotte S., Chellappan B., et al. (2013). MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics 14:119. 10.1186/1471-2164-14-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S. M., Lukasiewicz J., Farrer R., van Dam P., Bertoldo C., Rep M. (2016). Comparative genomics of Fusarium oxysporum f. sp. melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol. 209 307–318. 10.1111/nph.13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Sperschneider J., Gardiner D. M., Dodds P. N., Tini F., Covarelli L., Singh K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210 743–761. 10.1111/nph.13794 [DOI] [PubMed] [Google Scholar]
- Stanke M., Morgenstern B. (2005). AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33 W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F., Rep M. (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 11 309–314. 10.1111/j.1364-3703.2009.00605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A., Vagany V., Jackson A. C., Harrison R. J., Rainoni A., Clarkson J. P. (2016). Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Mol. Plant Pathol. 17 1032–1047. 10.1111/mpp.12346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A. C., Hane J. K., Ellwood S. R., Oliver R. P. (2015). CodingQuarry: highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genomics 16:170. 10.1186/s12864-015-1344-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam P., Fokkens L., Schmidt S. M., Linmans J. H., Kistler H. C., Ma L. J., et al. (2016). Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 18 4087–4102. 10.1111/1462-2920.13445 [DOI] [PubMed] [Google Scholar]
- van Dam P., Rep M. (2017). The distribution of miniature impala elements and SIX genes in the Fusarium genus is suggestive of horizontal gene transfer. J. Mol. Evol. 85 14–25. 10.1007/s00239-017-9801-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does H. C., Fokkens L., Yang A., Schmidt S. M., Langereis L., Lukasiewicz J. M., et al. (2016). Transcription factors encoded on core and accessory chromosomes of Fusarium oxysporum induce expression of effector genes. PLoS Genetics 12:e1006401. 10.1371/journal.pgen.1006401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widinugraheni S., Niño-Sánchez J., van der Does H. C., van Dam P., García-Bastidas F. A., Subandiyah S., et al. (2018). A SIX1 homolog in Fusarium oxysporum f.sp. cubense tropical race 4 contributes to virulence towards cavendish banana. PLoS One 13:e0205896. 10.1371/journal.pone.0205896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. H., Sharma M., Thatcher L. F., Azam S., Hane J. K., Sperschneider J., et al. (2016). Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17:191. 10.1186/s12864-016-2486-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y. (2017). ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8 28–36. 10.1111/2041-210x.12628 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher. GenBank Accession Numbers: MQTW01000000 – FON139 genome assembly; MN078955 – FON EF1α sequence; MN078956 – FON RPB2 sequence; MN078957 – FON129 TUB2 sequence; MN078958 – FON TUB2 sequence; MN078959 – FON FTF1a sequence; MN078960 – FON FTF1b sequence; MN078961 – FON SIX7 sequence; MN078962 – FON SIX9 sequence; MN078963 – FON SIX10 sequence; MN078964 – FON SIX12 sequence; MN078965 – FON63 SIX13 sequence.