Abstract
A recent hypothesis-free pathway-level analysis of genome-wide association study (GWAS) datasets suggested that the overall genetic variation measured by single nucleotide polymorphisms (SNPs) in the nucleotide excision repair (NER) pathway was associated with breast cancer (BC) risk, but no detailed SNP information was provided. To substantiate this finding, we performed a larger meta-analysis of 14 previously published GWAS datasets in the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) Study with 53107 European descent. Using a hypothesis-driven approach, we selected 138 candidate genes from the NER pathway using the “Molecular Signatures Database (MsigDB)” and “PathCards”. All SNPs were imputed using IMPUTE2 with the 1000 Genomes Project Phase 3. Logistic regression was used to estimate BC risk, and pooled ORs for each SNP were obtained from the meta-analysis using the false discovery rate (FDR) for multiple test correction. RegulomeDB, HaploReg, SNPinfo and expression quantitative trait loci (eQTL) analysis were used to assess the SNP functionality. We identified four independent SNPs associated with BC risk, BIVM-ERCC5 rs1323697_C (OR=1.06, 95% CI=1.03–1.10), GTF2H4 rs1264308_T (OR=0.93, 95% CI=0.89–0.97), COPS2 rs141308737_C deletion (OR=1.06, 95% CI=1.03–1.09) and ELL rs1469412_C (OR=0.93, 95% CI=0.90–0.96). Their combined genetic score was also associated with BC risk (OR=1.12, 95% CI=1.08–1.16, Ptrend<0.0001). The eQTL analysis revealed that BIVM-ERCC5 rs1323697 C and ELL rs1469412 C alleles were correlated with increased mRNA expression of their genes in 373 lymphoblastoid cell lines (P=0.022 and 2.67×10−22, respectively). These SNPs might have biological roles in the BC etiology, likely through modulating their corresponding gene expression.
Keywords: breast cancer susceptibility, single nucleotide polymorphism, DNA repair, expression quantitative trait loci analysis
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer and the leading cause of cancer deaths among women worldwide, with an estimated 1.7 million cases and 521 900 deaths in 2012, accounting for 25% of all cancer cases and 15% of all cancer deaths among women1. Despite the declining mortality rate due to early screenings and advanced medical therapies, the incidence rate of BC has remained steady over the past two decades in the US (https://seer.cancer.gov/statfacts/html/breast.html). Therefore, it is necessary to identify additional genetic factors that can be used for defining susceptible individuals at risk for BC.
Although the mechanisms of breast carcinogenesis are still not fully understood, a variety of risk factors have already been identified2–5. Some studies have shown that mammalian cells can convert estrogen into related compounds that not only generate free radicals capable of damaging DNA but also bind to DNA, causing the loss of a nucleotide base, a process known as depurination. The resulting mutations can convert a normal cell into a cancerous one6–8.
Another putative risk factor is smoking. Although there are no consistent results about the association between smoking and BC risk, there are carcinogens in tobacco smoke such as polycyclic aromatic hydrocarbons (PAH), aromatic amines, and nitrosoamines, and these carcinogens might cause DNA damage and adduct formation in mammary epithelial cells9, 10.
In addition, many epidemiologic studies reported a positive association between BC risk and alcohol consumption. Animal models of BC, although not entirely consistent, do provide the support for an enhancing action of ethanol on mammary carcinogenesis11. Overall, evidence from human studies, animal studies and cell culture experiments support some biologically plausible mechanisms, such as an increase in circulating estrogens and androgens, enhancement of mammary gland susceptibility to carcinogenesis, increased mammary carcinogen-induced DNA damage, and a greater potential for invasiveness of BC cells11, 12.
These mechanisms are all likely involved in DNA damage leading to the initiation of mutations and carcinogenesis, and thus the DNA repair system plays a critical role in protecting against mutations, maintaining genomic integrity and preventing carcinogenesis of the breasts13.
One of the DNA repair pathways is nucleotide excision repair (NER), a highly versatile and sophisticated DNA damage removal mechanism that counteracts the deleterious effects of a multitude of DNA lesions, including major types of damage induced by environmental mutagens and carcinogens. The most relevant lesions to be repaired by NER are cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts (6–4PPs) produced by the shortwave UV component of sunlight. In addition, numerous bulky chemical adducts are eliminated by this repair process as well14, 15. Given the importance of NER in the repair of UV-induced DNA damage, it seems that the NER pathway may not be relevant to BC risk, because there is no evidence that UV light may cause BC; however, it is likely that tobacco smoke may cause DNA damage in breast tissues.
A recent large study with pathway-level analysis using hierarchical modelling across five cancers, including 11 DRIVE (the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer) GWAS datasets of 33832 BC study subjects of European descent, did not find any specific risk-associated SNPs in genes involved in NER, but the limited study power did not allow the investigators to find an association with the overall genetic variation of the NER pathway16. Other prior studies also investigated associations between SNPs in DNA repair pathway genes and BC risk, but these studies had relatively small sample sizes without a focus on the NER pathway, although they have some notable findings, such as XRCC3 and ERCC4. 17–24.
Therefore, we hypothesize that genetic variants in the NER pathway genes are associated with BC risk. To assess the role of functional SNPs of the NER pathway genes in the BC etiology, we performed a much larger meta-analysis of 14 previously published DRIVE GWAS datasets with 53107 study subjects of European descent. In contrast to the previously published studies, the present analysis had a much larger sample size to focus on functional SNPs in the NER pathway genes. Hence, using a hypothesis-driven pathway-based approach with a much increased study power, we expected to identify some susceptibility loci in the NER pathway genes that have biologically relevant functions and thus play a role in the BC etiology.
Populations and Methods
Study populations
This meta-analysis included a sub-dataset of SNPs in the NER pathway genes from each of 14 previously published BC GWASs for a total of 28758 BC cases and 24349 controls of European ancestry from the DRIVE study (phs001265.v1.p1), which is different from the DRIVE-Genome-Wide Association meta-analysis (phs001263.v1.p1) previously used by others16 (Supplementary Table 1). The DRIVE study (phs001265.v1.p1), which included 17 GWASs, was one of the five projects funded in 2010 as part of the NCI’s Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative. For this meta-analysis, we excluded three studies including the “Women of African Ancestry Breast Cancer Study (WAABCS)”, which is a study of African ancestry, and “The Sister Study (SISTER)” and “The Two Sister Study (2 SISTER)”, which had a different study design that used cases’ sisters as the controls. These 14 GWAS studies consist of Breast Oncology Galicia Network (BREOGAN); Copenhagen General Population Study (CGPS); Cancer Prevention Study-II Nutrition Cohort (CPSII); European Prospective Investigation Into Cancer and Nutrition (EPIC); Melbourne Collaborative Cohort Study (MCCS); Multiethnic Cohort (MEC); Nashville Breast Health Study (NBHS); Nurses’ Health Study (NHS); Nurses’ Health Study 2 (NHS2); NCI Polish Breast Cancer Study (PBCS); The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO); Study of Epidemiology and Risk factors in Cancer Heredity (SEARCH); Swedish Mammography Cohort (SMC); and Women’s Health Initiative (WHI). The details of case and control recruitment and their characteristics are summarized in Supplementary Table 2. For all of the GWAS datasets, Illumina Infinium OncoArray-500k BeadChip genotyping platforms were used, and only two main de-identified variables (sex and age at interview) were available to us. For the cases, other three de-identified variables (age at diagnosis, estrogen receptor status, and histology type) were available. Each of the 14 studies was reviewed and approved by the corresponding Institutional Review Board and thus exempted by Duke Institutional Review Board.
Gene and SNP selection
Candidate genes in the NER pathway were selected according to the online datasets “Molecular Signatures Database v6.1 (MsigDB)” (http://software.broadinstitute.org/gsea/msigdb/search.jsp) and “PathCards” (http://pathcards.genecards.org/) using the key words “nucleotide excision repair”. In total, we selected 138 candidate genes from eight NER-related pathways after excluding duplicate genes, pseudo genes and withdrawn genes (LOC652672 and LOC652857) in the National Center for Biotechnology Information (NCBI). The detailed genes selection results are listed in Supplementary Table 3.
To avoid poor quality markers to be included in the imputation, we performed stringent quality control before imputation by including the following criteria: the minor allelic frequency (MAF) ≥1%, genotyping rate ≥95%, missing rate ≤90%, Hardy-Weinberg equilibrium (HWE) ≥1×10−6, All SNPs were flipped to forward strand and aligned with the reference genome data. Ambiguous SNPs with A-T or G-C alleles that are hard to determine the strand orientation by allele frequency were removed. According to the multi-population reference panels from the 1000 Genomes Project Phase 3, SNPs within the aforementioned 138 candidate genes and their ± 500 kb flanking regions were also extracted, and imputation for each study was performed using IMPUTE2 software 25. Imputed SNPs within 2-kb up- and down-stream of each gene’s region were extracted for further analysis. After imputation, SNPs that met the following quality control criteria were included in further analysis: imputation SNPs with information score ≥0.80 in IMPUTE2; a minor allele frequency (MAF) ≥5%; and a P value for the Hardy–Weinberg Equilibrium test ≥10−6. Due to differences between the 14 studies, 8433 to 9016 common SNPs remained in each study for further analysis. The final analysis included 7345 SNPs that were common to all 14 studies.
Functional analysis
To investigate the functions of candidate SNPs, we searched for functional annotation of the SNPs in three online functional prediction website: RegulomeDB (http://regulomedb.org/), HaploReg (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) and SNPinfo (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html). In addition, we performed the expression quantitative trait loci (eQTL) analysis by using data from multiple sources: lymphoblastoid cellline data of 373 subjects from the European Variation in Health and Disease Study (GEUVADIS) and the 1000 Genomes Project (phase I integrated release 3, March 2012)26. Furthermore, we used Genotype-Tissue Expression project (GTEx) results to obtain the corresponding mRNA expression in whole blood and breast tissues (https://gtexportal.org/home/)27.
Statistical analysis
For each study and the combined dataset, principal components (PCs) were calculated using the Genome-wide Complex Trait Analysis (GCTA) on the LD-pruned subset of the whole-genome-typed dataset28. The top 20 PCs were assessed for their associations with BC risk using univariate logistic regression analysis. Those PCs with significant associations in each study were included as covariates in further analyses of associations between SNPs and BC risk. For each SNP, we estimated odds ratios (ORs) and 95% confidence intervals (CIs) by unconditional logistic regression of case/control groups with adjustment for age and PCs. We performed the meta-analysis by using the inverse variance method to combine the results of the 14 studies. We defined heterogeneity as a Cochran’s Q test P ≤ 0.10 or I2 >50.0%. We used fixed-effects models, if no heterogeneity existed among the 14 studies, and random effects models were used, when heterogeneity existed. To assess the robustness of the results, we performed a sensitivity analysis by omitting each study one by one29. The false discovery rate (FDR) with a critical cut-off value of 0.05 using the linear step-up method of Benjamini and Hochberg was mainly used to correct for multiple comparisons to reduce the probability of false-positive findings30. To observe the combined effect of significant SNPs, we used the number of unfavorable genotypes (NUGs) of the significant SNPs as a genetic score to assess classification performance of the model. According to the frequency of each group and the effect values, we also dichotomized all the individuals into a low-risk group (0–2 NUGs) and a high-risk group (3–4 NUGs). In the eQTL analysis, we calculated the correlations between SNPs and specific mRNA expression levels by using a general linear regression model. Statistical analyses were performed using PLINK (version 1.9), SAS (version 9.3; SAS Institute, Cary, NC, USA) and R (version 3.0.2). The Manhattan plots and linkage disequilibrium (LD) plots were generated by Haploview v4.2, and regional association plots were constructed by LocusZoom (http://locuszoom.sph.umich.edu/locuszoom/).
Results
General characteristics of the study populations
The overall analysis included 28758 BC cases and 24349 controls from 14 studies (Supplementary Table 2). All subjects were women. The median age of controls was 60 years. The age distribution was statistically different between cases and controls (P<0.0001), with the control group being younger than the case group (≤ 60 years: 52.47% versus 49.73%). The proportion of estrogen receptor (ER) positive patients was 84%, and the proportion of patients with invasive tumors was 92% after deleting the missing data. The significant PCs among the first 20 in each study (Supplementary Table 4) were included in the analyses of associations between SNPs and BC risk. Therefore, age and PCs were adjusted for as possible confounders in the multivariate logistic regression analysis.
Association analysis of single locus and BC risk
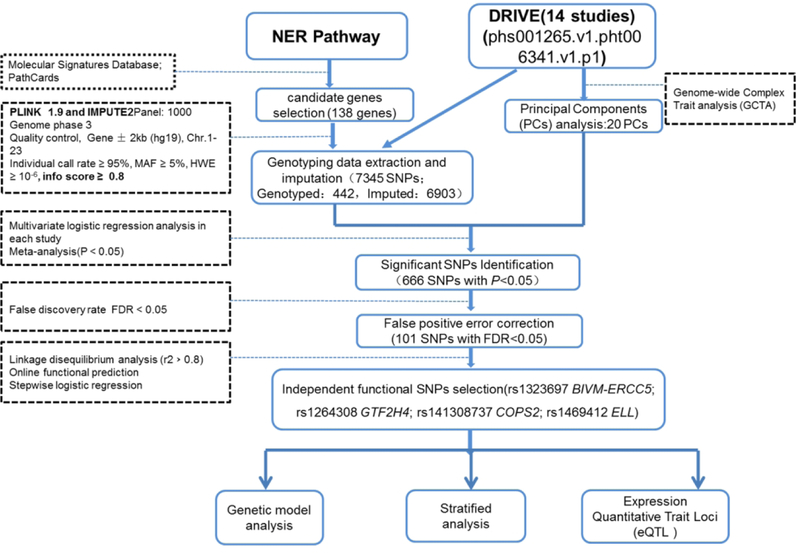
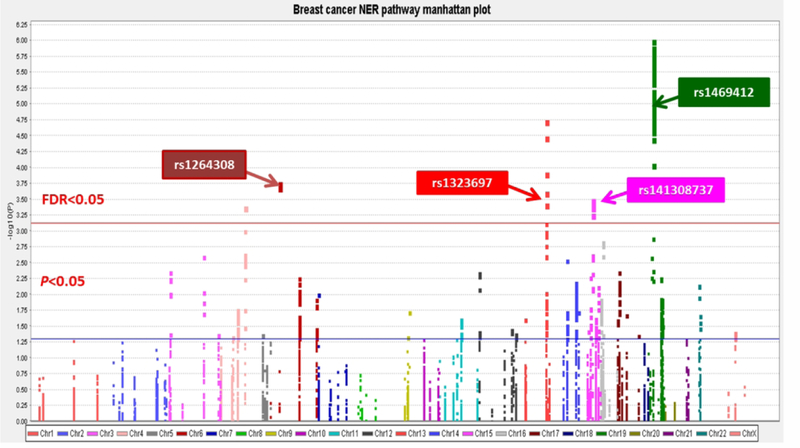
The workflow of the present study is shown in Figure 1. In total, we used 7345 SNPs that passed QC in the analysis, including 442 genotyped SNPs and 6903 imputed SNPs. Multivariate logistic regression and meta-analysis results showed that there were 666 SNPs significantly associated with BC risk (P<0.05), of which 101 SNPs remained significant after FDR correction < 0.05. The associations between SNPs of genes involved in the NER pathway and BC risk in the DRIVE study are shown in Figure 2. These 101 top SNPs were mapped to BIVM-ERCC5, GTF2H4, COPS2, ELL and COPS4 (Supplementary Table 5). COPS4 rs75870305 was deleted due to mapping or clustering errors on the NCBI website. Seven tagSNPs remained for additional analysis after removal of SNPs in high pairwise LD (Supplementary Figure 1). To search for functional SNPs, we used stringent criteria with RegulomeDB scores ≤ 3 and with eQTL evidence in breast tissues or blood cells (Supplementary Table 6 and Supplementary Table 7). As a result, BIVM-ERCC5 rs12870541 and ELL rs34664859 were left out, because of no functional annotation. Finally, the stepwise analysis kept four independent, potentially functional SNPs for further analysis (Table 1).
Figure 1.
The workflow of present study.
Figure 2.
Manhattan Plot of the 7345 SNPs of NER pathway Genes in the DRIVE. The x-axis represents each chromosome. The y-axis represents the association P values with breast cancer risk. Horizontal blue line means nominal P values of 0.05 and red line means FDR threshold 0.05.
Table 1.
Predictors of Risk Obtained from Stepwise Logistic Regression Analysis of Selected Variables in the DRIVE Study
| Variables | Category2 | Frequency3 | OR (95% CI) | P1 |
|---|---|---|---|---|
| Age | in years | 52994 | 1.003 (1.001–1.005) | 0.0003 |
| BIVM-ERCC5 rs1323697_C | GG/GC/CC | 35461/15712/1821 | 1.064 (1.031–1.097) | 0.0001 |
| GTF2H4 rs1264308_T | CC/CT/TT | 41872/10401/721 | 0.928 (0.893–0.964) | 0.0001 |
| COPS2 rs141308737_* | CC/C-/-- | 32971/17645/2378 | 1.062 (1.031–1.094) | <0.0001 |
| ELL rs1469412_C | TT/TC/CC | 33899/16933/2162 | 0.934 (0.906–0.962) | <0.0001 |
Abbreviations: DRIVE: Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; OR: odds ratio; CI: confidence interval.
Stepwise logistic regression analysis included age, PC1, PC3, PC4, PC5, PC6, PC10, PC16 and 5 SNPs (rs1323697, rs1264308, rs141308737, rs1469412 and rs4808136).
The most left-hand side “category” was used as the reference.
52994 subjects were included in the final stepwise analysis with deletion of the missing data.
This SNP is a base deletion.
Note: there were 20 PCs in the combined datasets as listed in Supplementary Table 4, of which seven remained significant and were adjusted in the final stepwise logistic regression analysis.
Supplementary Figure 2 presents the forest plots of the meta-analysis of the four independent SNPs. The results showed that SNP rs1323697 G>C and rs141308737 C>deletion were associated with a significantly increased risk of BC (OR = 1.06, 95% CI = 1.03–1.10, P = 2.66 × 10−4; OR = 1.06, 95% CI = 1.03–1.09, P = 3.60 × 10−4, respectively), while two other SNPs were associated with a significantly decreased BC risk (rs1264308 C>T: OR = 0.93, 95% CI = 0.89–0.97, P = 2.21 × 10–4; and rs1469412 T>C: OR = 0.93, 95% CI = 0.90–0.96, P = 3.09 × 10−6). There was no heterogeneity observed for the effect estimates of these four SNPs from the 14 GWASs. Dropping any one of the studies in the DRIVE study, did not change the pooled ORs and their 95% CIs (Supplementary Table 8). The association results from different genetic models for each SNP, including additive and dominant models, showed that all of the SNPs were significantly associated with BC risk in all of the genetic models (Table 2). Although the SNP rs4808801 (in the chromosome region 19p13.11 where ELL is located) has been previously reported by a GWAS31, the ELL rs1469412 that we identified in the present study has a moderate LD with rs4808801 (r2=0.471).
Table 2.
Associations genotypes of the Four Independent SNPs and risk of BC in the DRIVE Study
| Genotype | Univariate analysis |
Multivariate analysis& |
||||
|---|---|---|---|---|---|---|
| NControl/NCase | OR (95% CI) | P | NControl/NCase | OR (95% CI) | P | |
| BIVM-ERCC5 rs1323697 G>C@ | ||||||
| GC | 16495/19029 | ref. | 16432/19029 | ref. | ||
| GC | 7079/8666 | 1.06 (1.02–1.10) | 0.0020 | 7049/8666 | 1.05 (1.01–1.09) | 0.0071 |
| CC | 766/1057 | 1.20 (1.09–1.32) | 0.0002 | 764/1057 | 1.17 (1.07–1.29) | 0.0010 |
| Trend test | <0.0001 | <0.0001 | ||||
| GC+CC | 7845/9723 | 1.07 (1.04–1.11) | 0.0001 | 7813/9723 | 1.07 (1.03–1.11) | 0.0007 |
| GTF2H4 rs1264308 C>T$ | ||||||
| CC | 19106/22861 | ref. | 19028/22861 | ref. | ||
| CT | 4880/5539 | 0.95 (0.91–0.99) | 0.0161 | 4863/5539 | 0.94 (0.90–0.98) | 0.0032 |
| TT | 363/358 | 0.82 (0.71–0.95) | 0.0098 | 363/358 | 0.80 (0.69–0.93) | 0.0029 |
| Trend test | 0.0011 | <0.0001 | ||||
| CT+TT | 5243/5897 | 0.94 (0.90–0.98) | 0.0037 | 5226/5897 | 0.93 (0.89–0.97) | 0.0005 |
| CC# | 19106/22861 | 1.06 (1.02–1.11) | 0.0037 | 19028/22861 | 1.08 (1.03–1.13) | 0.0005 |
| COPS2 rs141308737 C>_* | ||||||
| CC | 15327/17705 | ref. | 15278/17705 | ref. | ||
| C- | 7994/9693 | 1.05 (1.01–1.09) | 0.0096 | 7957/9693 | 1.05 (1.02–1.09) | 0.0064 |
| -- | 1028/1360 | 1.15 (1.05–1.25) | 0.0016 | 1019/1360 | 1.15 (1.06–1.26) | 0.0008 |
| Trend test | 0.0002 | <0.0001 | ||||
| C-/-- | 9022/11053 | 1.06 (1.02–1.10) | 0.0011 | 8976/11053 | 1.06 (1.03–1.10) | 0.0006 |
| ELL rs1469412 T>C$@ | ||||||
| TT | 15316/18646 | ref. | 15263/18646 | ref. | ||
| TC | 8003/8973 | 0.92 (0.89–0.96) | <0.0001 | 7965/8973 | 0.92 (0.89–0.95) | <0.0001 |
| CC | 1030/1136 | 0.91 (0.83–0.99) | 0.0259 | 1026/1136 | 0.91 (0.83–0.99) | 0.0328 |
| Trend test | <0.0001 | <0.0001 | ||||
| TC+CC | 9033/10109 | 0.92 (0.89–0.95) | <0.0001 | 8991/10109 | 0.92 (0.89–0.95) | <0.0001 |
| TT# | 15316/18646 | 1.09 (1.05–1.13) | <0.0001 | 15263/18646 | 1.09 (1.05–1.13) | <0.0001 |
Abbreviations: SNP: single nucleotide polymorphism; BC: breast cancer; DRIVE: Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; OR: odd ratio; CI: confidence interval; NUGs: number of unfavorable genotypes.
Adjusted for age, PC1, PC3, PC4, PC5, PC6, PC10 and PC16.
rs1323697 has 9 controls and 6 cases missing; rs1469412 has 3 cases missing.
For consistent with the risk SNPs, we transfer the protected SNPs into risk ones.
This SNP is a base deletion.
Risk genotypes were rs1323697 GC+CC, rs1264308 CC, rs141308737 C-/-- and rs1469412 TT.
Association analysis of the combined score of SNPs and BC risk
The effect size (beta) values of each SNPs are very similar, from 0.056 to 0.074, so we did not consider the weight of each SNP in our analysis of the combined risk genotypes. Using a dominant model, we combined risk genotypes of rs1323697 GC+CC, rs141308737 C-/--, rs1264308 CC and rs1469412 TT into a genetic score as the number of unfavourable genotypes (NUGs). The trend test indicated a significant association between an increased NUGs and an increased risk of BC (P<0.0001, Table 3). Stratified analyses were performed to assess subgroups defined by age, ER status and invasiveness. We found that the risk associated with NUGs was more evident in the younger group (OR = 1.14, 95% CI = 1.08–1.20, P < 0.0001, Supplementary Table 9), but no heterogeneity or interaction were observed between these strata (P = 0.227 and 0.274, respectively, Supplementary Table 9). Subgroups analysis (ER status and histological type) also showed similar results by age among patients with ER+ and invasive tumors (Supplementary Table 9). Additionally, we found no significant differences between ER+ and ER− patients (P = 0.990) or between invasive and in situ carcinomas (P = 0.945).
Table 3.
Combined Risk Genotypes of the Four Validated SNPs and Risk of BC in the DRIVE Study
| Genotype | Univariate analysis |
Multivariate analysis2 |
||||
|---|---|---|---|---|---|---|
| NControl/NCase | OR (95% CI) | P | NControl/NCase | OR (95% CI) | P | |
| NUG1 | ||||||
| 0 | 854/877 | ref. | 852/877 | ref. | ||
| 1 | 5268/5769 | 1.07 (0.96–1.18) | 0.2139 | 5252/5769 | 1.07 (0.97–1.19) | 0.1805 |
| 2 | 10067/11776 | 1.14 (1.03–1.26) | 0.0091 | 10024/11776 | 1.15 (1.04–1.27) | 0.0057 |
| 3 | 6734/8365 | 1.21 (1.10–1.34) | 0.0002 | 6705/8365 | 1.22 (1.10–1.35) | <0.0001 |
| 4 | 1417/1962 | 1.35 (1.20–1.52) | <0.0001 | 1412/1962 | 1.36 (1.21–1.53) | <0.0001 |
| Trend | <0.0001 | <0.0001 | ||||
| 0–2 | 16189/18422 | ref. | 16128/18422 | ref. | ||
| 3–4 | 8151/10327 | 1.11 (1.07–1.15) | <0.0001 | 8117/10327 | 1.12 (1.08–1.16) | <0.0001 |
Abbreviations: SNPs: single nucleotide polymorphisms; BC: breast cancer; DRIVE: Discovery, Biology, and Risk of Inherited Variants in Breast Cancer; NUG: number of unfavorable genotypes; OR: odd ratio; CI: confidence interval.
Risk genotypes were rs1323697 GC+CC, rs1264308 CC, rs141308737 C-/-- and rs1469412 TT.
Multivariate logistic regression analyses were adjusted for age and PCs.
In silico functional validation
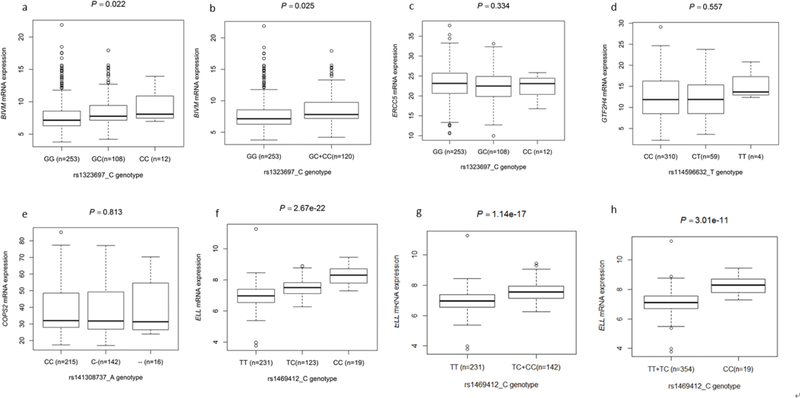
The in silico eQTL analysis among 373 European descendants with both SNP genotype and mRNA expression data showed that BIVM-ERCC5 rs1323697 C allele demonstrated a significant association with increased mRNA expression levels of BIVM in both additive (P = 0.022) and dominant models (P = 0.025) (Figure 3a and 4b). The ELL rs1469412 C allele also demonstrated a significant association with increased mRNA expression levels of ELL in all genetic models (Figure 3f, 4g and 4h: P = 2.67E-22, 1.14E-17 and 3.01E-11, respectively). However, no significant associations between the other two SNPs and corresponding mRNA expression levels were found (Figure 3d and 4e). In addition, GTF2H4 rs114596632, the same SNP with rs1264308, has been reported significantly associated with a decreased mRNA expression levels in 270 lymphoblastoid cell lines from HapMap32.
Figure 3.
The Correlations between identified putative functional SNPs and corresponding gene’s mRNA expression in the 1000 Genome Project. rs1323697 (a, additive model, P = 0.022; b, dominant model, P = 0.025; c, additive model, P = 0.334), rs114596632, which is the same position of rs1264308, (d, additive model, P = 0.557), rs141308737 (e, additive model, P = 0.813), rs1469412 (f, additive model, P = 2.67e-22; g, dominant model, P = 1.14e-17; h, recessive model, P = 3.01e-11).
To further examine the correlation between the significant SNPs and mRNA expression levels, we searched GTEx as well and found that BIVM-ERCC5 rs1323697, GTF2H4 rs1264308 and ELL rs1469412 were correlated with their specific mRNA expression levels in the whole blood cells (P =0.003, 0.032 and <0.0001, respectively), but COPS2 rs141308737 was unrelated to its gene expression levels. In addition, COPS2 rs141308737 (P =0.026) and BIVM-ERCC5 rs1323697 (P =0.001) had a positive correlation with their gene specific mRNA expression levels in breast tissues (Supplementary Table 10).
Discussion
To determine whether genetic variants in the NER pathway genes contribute to BC susceptibility, we performed association analyses between 7345 SNPs in 138 genes and BC risk with a large sample size of 28758 cases and 24349 controls. As a result, we identified four novel susceptibility variants, BIVM-ERCC5 rs1323697 at 13q33.1, GTF2H4 rs1264308 at 6p21.33, COPS2 rs141308737 at 15q21.2, and ELL rs1469412 at 19p13.11. In addition, the eQTL analysis results revealed that BIVM-ERCC5 rs1323697 C allele was associated with an increased mRNA expression levels, as was the ELL rs1469412 C allele. These results indicate that these two SNPs might influence mRNA expression levels and thus the functions of the genes, a possible mechanism underlying the observed associations. These findings suggest that variants in the NER pathway genes play an important role in the development of BC possibly by influencing mRNA expression.
The NER pathway is a mechanism that recognizes and repairs bulky DNA damage caused by chemical compounds, environmental carcinogens, and exposure to UV-light. The repair of damaged DNA involves at least 30 polypeptides within two different sub-pathways of NER known as transcription-coupled repair (TC-NER) and global genome repair (GG-NER)33. The TCR and GGR processes are different in terms of damage recognition: RNA polymerase II (RNAP II) is needed in TC-NER, while XPC-hHR23B complexes together with XPE complex are needed in GG-NER. In general, genes of GG-NER have been associated with cancer predisposition15. However, the present study indicated some genes of TC-NER also might be involved in BC susceptibility, such as ELL, COPS2 and GTF2H4, but their exact mechanisms involved in the BC etiology need to be further studied.
BIVM-ERCC5 rs1323697 is located on 13q33.1, which has not been reported by any of the GWASs included in the present analysis. Based on the NCBI website (https://www.ncbi.nlm.nih.gov/gene/100533467), this locus represents naturally occurring read-through transcription between the neighbouring BIVM (basic, immunoglobulin-like variable motif containing) and ERCC5 (excision repair cross-complementing rodent repair deficiency, complementation group 5) genes on chromosome 13. The read-through transcript encodes a fusion protein that shares sequence identity with the products of each individual gene (Supplementary Figure 3). Because the present study mainly indicated that rs1323697 was correlated with BIVM gene expression levels, the discussion will focus on the function of BIVM only. Previous studies have shown that BIVM possesses virtually no sequence similar to any currently described protein, making the prediction of a function challenging34. It is highly likely that BIVM is essential for some aspect of basic cellular functioning and is expressed in a nearubiquitous manner34. The presence of a CpG island at the 5’ end of BIVM and its wide tissue distribution suggest that it may function as a housekeeping gene34, 35. While others think it is likely that the immunoglobulin-like motif in BIVM may have functions similar to an immunoglobulin, but this remains to be experimentally confirmed36. Furthermore, we found that SNP rs1323697 is located at the LUN-1 motif, as shown by the position weight matrix (PWM) based Sequence Logo (Supplementary Figure 4 and Supplementary Table 7).
GTF2H4, known as a general transcription factor IIH subunit 4, encodes a subunit of transcription factor IIH (TFIIH), a helicase that is responsible for unwinding DNA structure, allowing repair of the damaged DNA, and it is involved in both NER process and transcription control interacting with variable factors important in carcinogenesis37. The TFIIH complex has both ATPase and helicase activities and opens DNA at sites of DNA distorting damage, and the TFIIH4 subunit may regulate the ATPase activity of the TFIIH subunit (XPB, a protein coded by ERCC3)38. Previous studies have found that some GTF2H4 SNPs were significantly associated with lung cancer risk and survival, multiple sclerosis risk and cervical cancer32, 39–41, but there is no report on the associations between GTF2H4 SNPs and BC risk to date. There is an interesting finding that BC and lung cancer risk was associated with the same SNP, GTF2H4 rs126430832. GWAS catalog results indicated that some of the adjacent genes shared the same location 6p21.33, including ABCF1, PPP1P18 and LOC105375013, have a high LD with rs1264308 (Supplementary Table 11). As an intron SNP, GTF2H4 rs1264308 may have an effect on the disease by changing motif FOXJ3 (Supplementary Figure 4) or by mechanisms of interacting with other genes above-mentioned. However, none of the other genes have a known function in NER.
In the present study, COPS2 rs141308737 has no functional clues from mRNA expression levels. However, a study showed that over-expression of COPS2 was linked to chromosome instability42. Functional prediction software shows that rs141308737 is located at the ER motif (Supplementary Figure 4) and can bind to the CJUN protein (Supplementary Table 7). It has been reported that endogenous c-Jun plays a key role in ErbB2-induced migration and invasion of mammary epithelial cells and mediates the expansion of a self-renewing population of mammary tumor stem cells via the production of CCL5 and SCF to enhance BC tumor invasiveness43.
As for ELL rs1469412, although some SNPs in this region have been reported by GWASs, it is necessary to include this SNP, because it was only in moderate-to-low LD with other reported SNPs (Supplementary Table 11) and lack of functional analysis in the previously published study. ELL is known as an elongation factor for RNA polymerase II, which is an important gene in the TC-NER sub-pathway. One study reported that ELL encoded an elongation factor that could increase the catalytic rate of RNA polymerase II transcription by suppressing transient pausing by polymerase at multiple sites along the DNA44. Another study showed that ELL was a key regulator of transcriptional elongation, suggesting that, as an E3 ubiquitin ligase for c-Myc and a potential tumor suppressor, ELL may function as a partner of steroid receptors, hypoxiainducible factor 1-alpha (HIF-1α), E2F1 and the TFIIH complex, modulating their binding partner’s activity45. The present study showed that the ELL rs1469412 C allele was associated with an increase in mRNA expression levels, exerting a protective effect on BC risk. However, further studies are needed to investigate biological mechanisms underlying the observed associations between ELL rs1469412 and BC risk.
It should also be mentioned that the present study has some limitations. Firstly, due to the limited access to phenotypes of the published GWAS datasets with many PCs included in the analysis, we could not adjust for some known risk factors, such as smoking, menstrual, reproductive and lactational history46, and the findings need to be verified in other BC studies with more detailed information about the known risk factors. Secondly, we did not have access to the target tissues collected by the participating GWAS studies, and we only did in silico analysis using published data for the functional prediction of the identified SNPs. Therefore, the biological mechanisms by which the four SNPs of the genes may influence BC risk remain unclear. Thirdly, the study populations were of non-Hispanic whites, and thus the findings may not generalizable to other ethnic groups, and thus additional studies in other ethnic groups are warranted.
In conclusion, this large-scale meta-analysis of 14 published GWASs among 53107 subjects of European descent identified four novel BC susceptibility loci in the NER pathway genes (i.e., BIVM-ERCC5 rs1323697, GTF2H4 rs1264308, COPS2 rs141308737 and ELL rs1469412) and also provided some evidence for their functional relevance. Further studies on the exact biological mechanisms and functional analysis of these SNPs in the BC etiology are needed.
Supplementary Material
Novelty and Impact:.
Mechanisms of some risk factors of breast cancer are likely involved in DNA damage. As one of DNA repair pathways, the nucleotide excision repair (NER) pathway plays a critical role in prevention of breast cancer (BC). In the present study, we identified four independent SNPs (BIVM-ERCC5 rs1323697_C, GTF2H4 rs1264308_T, COPS2 rs141308737_C deletion and ELL rs1469412_C) associated with BC risk. BIVM-ERCC5 rs1323697 C and ELL rs1469412 C alleles were correlated with increased mRNA expression of their genes.
Acknowledgement
DRIVE: OncoArray genotyping and phenotype data harmonization for the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE; dbGaP#: phs001265.v1.p1) breastcancer case control samples was supported by X01 HG007491 and U19 CA148065 and by Cancer Research UK (C1287/A16563). Genotyping was conducted by the Centre for Inherited Disease Research (CIDR), Centre for Cancer Genetic Epidemiology, University of Cambridge, and the National Cancer Institute. The following studies contributed germline DNA from breast cancer cases and controls: the Two Sister Study (2SISTER), Breast Oncology Galicia Network (BREOGAN), Copenhagen General Population Study (CGPS), Cancer Prevention Study 2 (CPSII), The European Prospective Investigation into Cancer and Nutrition (EPIC), Melbourne Collaborative Cohort Study (MCCS), Multi-ethnic Cohort (MEC), Nashville Breast Health Study (NBHS), Nurses’ Health Study (NHS), Nurses’ Health Study 2 (NHS2), Polish Breast Cancer Study (PBCS), Prostate Lung Colorectal and Ovarian Cancer Screening Trial (PLCO), Studies of Epidemiology and Risk Factors in Cancer Heredity (SEARCH), The Sister Study (SISTER), Swedish Mammographic Cohort (SMC), Women of African Ancestry Breast Cancer Study (WAABCS), Women’s Health Initiative (WHI).
GTEx: The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: GTEx Analysis V7 (dbGaP Accession phs000424.v7.p2).
Grant support
Jie Ge was supported by the Qiqihar Medical University’s Support Grant (QY2016B-13). Qingyi Wei was in part supported by the Duke Cancer Institute’s P30 Cancer Center Support Grant (NIH CA014236).
Abbreviations:
- BC
breast cancer
- NER
nucleotide excision repair
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association study
- DRIVE
Discovery, Biology, and Risk of Inherited Variants in Breast Cancer
- BREOGAN
Breast Oncology Galicia Network
- CGPS
Copenhagen General Population Study
- CPSII
Cancer Prevention Study-II Nutrition Cohort
- EPIC
European Prospective Investigation Into Cancer and Nutrition
- MCCS
Melbourne Collaborative Cohort Study
- MEC
Multiethnic Cohort
- NBHS
Nashville Breast Health Study
- NHS
Nurses’ Health Study
- NHS2
Nurses’ Health Study 2
- PBCS
NCI Polish Breast Cancer Study
- PLCO
The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial
- SEARCH
Study of Epidemiology and Risk factors in Cancer Heredity
- SMC
Swedish Mammography Cohort
- WHI
Women’s Health Initiative
- MAF
minor allele frequency
- eQTL
expression quantitative trait loci
- PCs
principal components
- OR
odds ratio
- CI
confidence interval
- FDR
false discovery rate
- NUGs
number of unfavorable genotypes
- LD
linkage disequilibrium
Footnotes
Conflict of interest
The authors state no conflict of interest.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancerstatistics, 2012. CA: a cancer journal for clinicians 2015;65: 87–108. [DOI] [PubMed] [Google Scholar]
- 2.Maas P, Barrdahl M, Joshi AD, Auer PL, Gaudet MM, Milne RL, Schumacher FR, Anderson WF, Check D, Chattopadhyay S, Baglietto L, Berg CD, et al. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA oncology 2016;2: 1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjerkaas E, Parajuli R, Weiderpass E, Engeland A, Maskarinec G, Selmer R, Gram IT.Smoking duration before first childbirth: an emerging risk factor for breast cancer? Results from 302,865 Norwegian women. Cancer causes & control : CCC 2013;24: 1347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP.Risk Factors and Preventions of Breast Cancer. International journal of biological sciences 2017;13: 1387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight JA, Fan J, Malone KE, John EM, Lynch CF, Langballe R, Bernstein L, Shore RE, Brooks JD, Reiner AS, Woods M, Liang X, et al. Alcohol consumption and cigarette smoking in combination: A predictor of contralateral breast cancer risk in the WECARE study. International journal of cancer 2017;141: 916–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller K. Estrogen and DNA damage: the silent source of breast cancer? Journal of the National Cancer Institute 2003;95: 100–2. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, Eccles D, Evans DG, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nature genetics 2009;41: 585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. The New England journal of medicine 2006;354: 270–82. [DOI] [PubMed] [Google Scholar]
- 9.Kawai M, Malone KE, Tang MT, Li CI. Active smoking and the risk of estrogenreceptor-positive and triple-negative breast cancer among women ages 20 to 44 years. Cancer 2014;120: 1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catsburg C, Kirsh VA, Soskolne CL, Kreiger N, Rohan TE. Active cigarette smokingand the risk of breast cancer: a cohort study. Cancer epidemiology 2014;38: 376–81. [DOI] [PubMed] [Google Scholar]
- 11.Singletary KW, Gapstur SM. Alcohol and breast cancer: review of epidemiologic andexperimental evidence and potential mechanisms. Jama 2001;286: 2143–51. [DOI] [PubMed] [Google Scholar]
- 12.Ellingjord-Dale M, Vos L, Hjerkind KV, Hjartaker A, Russnes HG, Tretli S, Hofvind S, Dos-Santos-Silva I, Ursin G. Alcohol, Physical Activity, Smoking, and Breast Cancer Subtypes in a Large, Nested Case-Control Study from the Norwegian Breast Cancer Screening Program. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2017;26: 1736–44. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy DO, Agrawal M, Shen J, Terry MB, Zhang FF, Senie RT, Motykiewicz G,Santella RM. DNA repair capacity of lymphoblastoid cell lines from sisters discordant for breast cancer. Journal of the National Cancer Institute 2005;97: 127–32. [DOI] [PubMed] [Google Scholar]
- 14.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotideexcision repair. Genes & development 1999;13: 768–85. [DOI] [PubMed] [Google Scholar]
- 15.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotideexcision repair and its roles in cancer and ageing. Nature reviews Molecular cell biology 2014;15: 465–81. [DOI] [PubMed] [Google Scholar]
- 16.Scarbrough PM, Weber RP, Iversen ES, Brhane Y, Amos CI, Kraft P, Hung RJ, Sellers TA, Witte JS, Pharoah P, Henderson BE, Gruber SB, et al. A Cross-Cancer Genetic Association Analysis of the DNA Repair and DNA Damage Signaling Pathways for Lung, Ovary, Prostate, Breast, and Colorectal Cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2016;25: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang LL, Chen FY, Wang H, Hu XL, Dai X, Mao J, Shen ZT, Wu YH, Wang SM, Hai J, Yan GJ, Li H, et al. Haplotype analysis of eight genes of the monoubiquitinated FANCD2-DNA damage-repair pathway in breast cancer patients. Cancer epidemiology 2013;37: 311–7. [DOI] [PubMed] [Google Scholar]
- 18.Sapkota Y, Mackey JR, Lai R, Franco-Villalobos C, Lupichuk S, Robson PJ, Kopciuk K, Cass CE, Yasui Y, Damaraju S. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PloS one 2014;8: e64896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva SN, Tomar M, Paulo C, Gomes BC, Azevedo AP, Teixeira V, Pina JE, Rueff J,Gaspar JF. Breast cancer risk and common single nucleotide polymorphisms in homologous recombination DNA repair pathway genes XRCC2, XRCC3, NBS1 and RAD51. Cancer epidemiology 2010;34: 85–92. [DOI] [PubMed] [Google Scholar]
- 20.Sehl ME, Langer LR, Papp JC, Kwan L, Seldon JL, Arellano G, Reiss J, Reed EF,Dandekar S, Korin Y, Sinsheimer JS, Zhang ZF, et al. Associations between single nucleotide polymorphisms in double-stranded DNA repair pathway genes and familial breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2009;15: 2192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monsees GM, Kraft P, Chanock SJ, Hunter DJ, Han J. Comprehensive screen ofgenetic variation in DNA repair pathway genes and postmenopausal breast cancer risk. Breast cancer research and treatment 2011;125: 207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C, Srihari S, Lal S, Gautier B, Simpson PT, Khanna KK, Ragan MA, Le Cao KA.Personalised pathway analysis reveals association between DNA repair pathway dysregulation and chromosomal instability in sporadic breast cancer. Molecular oncology 2016;10: 179–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Haiman C, Niu T, Guo Q, Cox DG, Willett WC, Hankinson SE, Hunter DJ.Genetic variation in DNA repair pathway genes and premenopausal breast cancer risk. Breast cancer research and treatment 2009;115: 613–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haiman CA, Hsu C, de Bakker PI, Frasco M, Sheng X, Van Den Berg D, Casagrande JT, Kolonel LN, Le Marchand L, Hankinson SE, Han J, Dunning AM, et al. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Human molecular genetics 2008;17: 825–34. [DOI] [PubMed] [Google Scholar]
- 25.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 2011;1: 457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Liu Z, Wang Y, Stinchcombe TE, Owzar K, Han Y, Hung RJ, Brhane Y,McLaughlin J, Brennan P, Bickeboller H, Rosenberger A, et al. Functional variants in DCAF4 associated with lung cancer risk in European populations. Carcinogenesis 2017;38: 541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson G. Human genetics. GTEx detects genetic effects. Science 2015;348: 640–1. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. Genome-wide complex trait analysis(GCTA): methods, data analyses, and interpretations. Methods in molecular biology 2013;1019: 215–36. [DOI] [PubMed] [Google Scholar]
- 29.Teasdale N, Elhussein A, Butcher F, Piernas C, Cowburn G, Hartmann-Boyce J,Saksena R, Scarborough P. Systematic review and meta-analysis of remotely delivered interventions using self-monitoring or tailored feedback to change dietary behavior. The American journal of clinical nutrition 2018;107: 247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met 1995;57: 289–300. [Google Scholar]
- 31.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmi2dt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nature genetics 2013;45: 353–61, 61e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, Liu H, Liu Z, Yi X, Bickeboller H, Hung RJ, Brennan P, Landi MT, Caporaso N, Christiani DC, Doherty JA, Team TR, et al. Genetic variant in DNA repair gene GTF2H4 is associated with lung cancer risk: a large-scale analysis of six published GWAS datasets in the TRICL consortium. Carcinogenesis 2016;37: 888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadkarni A, Burns JA, Gandolfi A, Chowdhury MA, Cartularo L, Berens C, Geacintov NE, Scicchitano DA. Nucleotide Excision Repair and Transcription-coupled DNA Repair Abrogate the Impact of DNA Damage on Transcription. The Journal of biological chemistry 2016;291: 848–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoder JA, Hawke NA, Eason DD, Mueller MG, Davids BJ, Gillin FD, Litman GW.BIVM, a novel gene widely distributed among deuterostomes, shares a core sequence with an unusual gene in Giardia lamblia. Genomics 2002;79: 750–5. [DOI] [PubMed] [Google Scholar]
- 35.Mitra PS, Ghosh S, Zang S, Sonneborn D, Hertz-Picciotto I, Trnovec T, Palkovicova L, Sovcikova E, Ghimbovschi S, Hoffman EP, Dutta SK. Analysis of the toxicogenomic effects of exposure to persistent organic pollutants (POPs) in Slovakian girls: correlations between gene expression and disease risk. Environment international 2012;39: 188–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferraren DO, Liu C, Badner JA, Corona W, Rezvani A, Monje VD, Gershon ES, Bonner TI, Detera-Wadleigh SD. Linkage disequilibrium analysis in the LOC93081-KDELC1- BIVM region on 13q in bipolar disorder. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics 2005;133B: 12–7. [DOI] [PubMed] [Google Scholar]
- 37.Gervais V, Lamour V, Jawhari A, Frindel F, Wasielewski E, Dubaele S, Egly JM,Thierry JC, Kieffer B, Poterszman A. TFIIH contains a PH domain involved in DNA nucleotide excision repair. Nature structural & molecular biology 2004;11: 616–22. [DOI] [PubMed] [Google Scholar]
- 38.Oksenych V, Coin F. The long unwinding road: XPB and XPD helicases in damaged DNA opening. Cell cycle 2010;9: 90–6. [DOI] [PubMed] [Google Scholar]
- 39.Buch SC, Diergaarde B, Nukui T, Day RS, Siegfried JM, Romkes M, Weissfeld JL.Genetic variability in DNA repair and cell cycle control pathway genes and risk of smokingrelated lung cancer. Molecular carcinogenesis 2012;51 Suppl 1: E11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briggs FB, Goldstein BA, McCauley JL, Zuvich RL, De Jager PL, Rioux JD, Ivinson AJ, Compston A, Hafler DA, Hauser SL, Oksenberg JR, Sawcer SJ, et al. Variation within DNA repair pathway genes and risk of multiple sclerosis. American journal of epidemiology 2010;172: 217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SS, Gonzalez P, Yu K, Porras C, Li Q, Safaeian M, Rodriguez AC, Sherman ME, Bratti C, Schiffman M, Wacholder S, Burk RD, et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PloS one 2010;5: e8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wicker CA, Izumi T. Analysis of RNA expression of normal and cancer tissuesreveals high correlation of COP9 gene expression with respiratory chain complex components. BMC genomics 2016;17: 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiao X, Katiyar S, Willmarth NE, Liu M, Ma X, Flomenberg N, Lisanti MP, Pestell RG. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. The Journal of biological chemistry 2010;285: 8218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNApolymerase II elongation factor encoded by the human ELL gene. Science 1996;271: 1873–6. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Zhou C, Ji W, Mei Z, Hu B, Zhang W, Zhang D, Wang J, Liu X, Ouyang G,Zhou J, Xiao W. ELL targets c-Myc for proteasomal degradation and suppresses tumour growth. Nature communications 2016;7: 11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persson I. Estrogens in the causation of breast, endometrial and ovarian cancers evidence and hypotheses from epidemiological findings. The Journal of steroid biochemistry and molecular biology 2000;74: 357–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.