Abstract
Long noncoding RNAs (lncRNAs) are RNA molecules comprising more than 200 nucleotides, which are not translated into proteins. Many studies have shown that lncRNAs are involved in regulating a variety of biological processes, including immune, cancer, stress, development and differentiation at the transcriptional, epigenetic or post‐transcriptional levels. Here, we review the role of lncRNAs in the process of neurodevelopment, neural differentiation, synaptic function, and pathogenesis of Parkinson's disease (PD). These pathomechanisms include protein misfolding and aggregation, disordered protein degradation, mitochondrial dysfunction, oxidative stress, autophagy, apoptosis, and neuroinflammation. This information will provide the basis of lncRNA‐based disease diagnosis and drug treatment for PD.
Keywords: long noncoding RNAs, neural development, neural differentiation, Parkinson's disease, synapses
1. INTRODUCTION
Parkinson's disease (PD) is the second major neurodegenerative disease affecting the health of middle‐aged and elderly people.1 The main pathological feature of PD is the progressive loss of dopaminergic neurons in the substantia nigra of the brain, leading to a lack of dopamine in the striatum which subsequently causes motor dysfunction.1, 2, 3 The main PD‐associated motor symptoms include resting tremor, rigidity, posture instability and bradykinesia.4 Motor impairment can also be preceded by a range of nonmotor symptoms, including anxiety, dysosmia, autonomic nervous dysfunction, and emotional and memory disorders.5 With an increase in the society's aged population, the number of PD patients is rising year after year, which greatly affects the quality of life of the patients and their families, representing a heavy burden on the society. Therefore, more effective treatments for PD are urgently required.
The pathogenesis of PD is not yet completely clear, although genetic susceptibility and environmental factors are generally thought to play a role.6 The formation of lewy bodies (LBs) in the cytoplasm of the residual neurons and the accumulation of α‐synuclein have been implicated as major factors involved in the mechanism of PD.7 Multiple studies have shown that the degeneration and apoptosis of the dopaminergic neurons in the substantia nigra are closely related to an abnormal proteasome function, mitochondrial dysfunction, oxidative stress, neuroinflammation, and transcriptional signal changes.8 In recent years, in‐depth studies of various aspects of PD have gradually clarified the pathogenesis of this condition. Transcriptome studies of PD have revealed alterations in a variety of genes, pathways, and biological processes in the samples of brain tissue, blood, and cerebrospinal fluid.9 Studies of PD in brain tissues have shown changes in pathways involving biological processes such as synaptic transmission, dopamine metabolism, vesicular transport, neuroinflammation, mitochondrial function, protein degradation, oxidative stress, autophagy and apoptosis.10 Similarly, studies of PD patients' blood samples have shown alterations in biological pathways such as programmed cell death, immune function, inflammation, mitochondrial function, RNA processing and protein chaperones.11 It is worth mentioning that the consistency of changes in several pathways, such as inflammatory immunity, protein degradation and mitochondrial function, identified in the brain tissue and blood samples from PD patients suggests that the process of PD is comprehensive and systematic.9
The protein‐coding region accounts for only a small proportion (approximately 1.5%‐2%) of the human genome, while the remaining sequence has no protein‐coding capacity and is transcribed to form noncoding RNAs (ncRNAs). These molecules are considered to play an important role in normal physiological activities as well as the pathological mechanisms of different diseases.12 In recent years, the roles of ncRNAs in the process of neural development, regeneration and neurodegenerative diseases have become a research hotspot. Generally, long non‐coding RNA (lncRNA) is a type of ncRNA, which is longer than 200 nucleotides (nt),13 and is structurally similar to mRNA (capped, spliced, poly a tail), but some do not exhibits these characteristics.14 Long noncoding RNAs (lncRNAs) are transcribed by RNA polymerase II and III at several loci of the eukaryotic genome.14 Structurally, lncRNAs belong to regulatory ncRNAs, acting as decoys, signals, guides, scaffolds, and so on.14 They regulate the gene expression patterns by changing the accessibility of the DNA and the chromatin structure.15 Unlike protein‐coding mRNAs, lncRNAs perform unique functions by regulating various cell biological processes, such as cell transcription, histone modification, and DNA methylation.16 Accumulating evidence demonstrates the important roles of lncRNAs in many biological processes, affecting the pathogenesis of various neurodegenerative diseases including PD. Hence, this review aims to summarize the currently identified neural lncRNAs with a focus on the roles of lncRNAs in PD.
2. BIOLOGICAL CHARACTERISTICS AND FUNCTIONS OF THE LONG NONCODING RNAs
2.1. Biological characteristics of lncRNAs
LncRNAs are a class of ncRNAs that consist of more than 200 nt in length and represent one of the largest fraction in the genome.17 LncRNAs are usually 5'‐capped, spliced and 3'‐polyadenylated, and transcribed by RNA polymerase II.18 In many aspects, lncRNAs are similar to the mRNAs that encode proteins, but have lower transcription numbers, mostly existing in the nucleus, with poor interspecific sequence conservation.19 Based on the relative positions of the lncRNA’s coding sequences and protein‐coding genes, they are categorized as (Figure 1): (a) sense lncRNAs: lncRNA sequences overlapping with the protein‐coding genes; (b) antisense lncRNAs: lncRNA sequences overlapping with the antisense strands of protein‐coding genes; (c) bidirectional lncRNAs: lncRNA sequences transcribed from the divergent bidirectional promoters relative to the protein‐coding genes; (d) intronic lncRNAs: lncRNA sequences derived entirely from the introns of transcripts, which may be truly independent transcripts or processing products of the mRNA precursor; (e) intergenic lncRNAs: lncRNA sequences located between but not overlapping with the protein‐coding genes.
Figure 1.
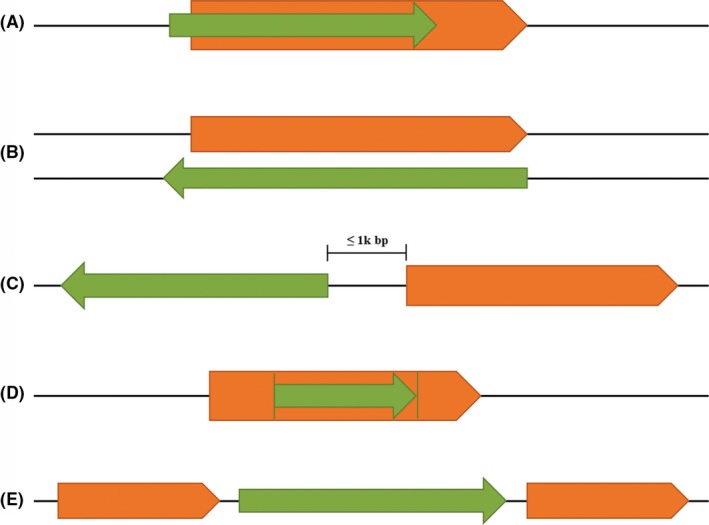
Classification of the lncRNAs on the basis of the relative positions of their coding sequences (in green) and the protein‐coding genes (in orange). Arrows indicate the direction of transcription. (A) Sense lncRNAs: lncRNA sequences overlapping with the protein‐coding genes; (B) antisense lncRNAs: lncRNA sequences overlapping with the antisense strands of the protein‐coding genes; (C) bidirectional lncRNAs: lncRNA sequences transcribed from the divergent bidirectional promoters relative to the protein‐coding genes; (D) intronic lncRNAs: lncRNA sequences derived entirely from introns of the transcripts, which may be truly independent transcripts or processing products of the precursor mRNA; (E) intergenic lncRNAs: lncRNA sequences located between but not overlapping the protein‐coding genes
Studies have shown that lncRNAs interact with DNA, RNA or protein molecules to regulate gene expression and exert effects through a variety of mechanisms. As regulators of gene expression, lncRNAs can be roughly divided into three categories according to the mode of regulation: transcriptional, epigenetic and post‐transcriptional (Figure 2). Firstly, lncRNAs can regulate gene expression levels by acting as a scaffold or bridge to regulate the related chromatin‐modifying enzymes during transcription. For example, snRNA 7SK forms scaffolds for the HEXIM1, HEXIM2, LARP7, and P‐TEFb protein complexes during transcription in the embryonic stem cells. The complexes inactivate P‐TEFb kinase, which affects the transcription of related genes.20 Secondly, lncRNAs affect epigenetic modifications by inducing DNA and histone methylation, histone acetylation and sumoylation. LncRNA HOTAIR at the HOXC site inhibits transcription of the HOXD by changing the methylation state of the chromatin.21 Thirdly, lncRNAs regulate post‐transcriptional gene expression through various mechanisms, including indirect regulation of miRNA or direct regulation of mRNA. Binding of the linc‐MD1 to its corresponding microRNAs (miRNAs), blocks the binding of its target genes to miRNAs and thereby prevents the transcriptional inhibition of the target genes.22 The complementary lncRNA and mRNA form the RNA double‐stranded bodies, which mask the key components of mRNA needed to bind the reaction factor, thus potentially affecting gene expression at the post‐transcriptional level via mechanisms such as mRNA precursor processing, splicing, transport, translation, and degradation.23
Figure 2.
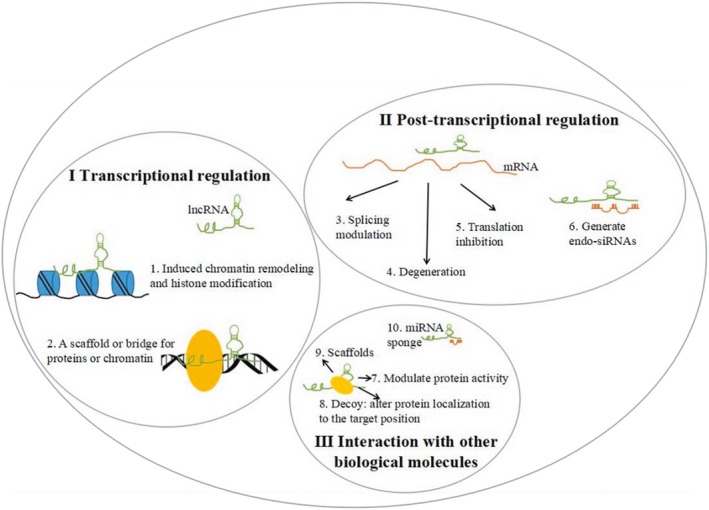
Functions of lncRNAs. I) Transcriptional regulation: lncRNAs can (1) induce chromatin remodeling and modification, and (2) act as a scaffold or bridge for proteins or chromatin. II) Post‐transcriptional regulation: lncRNAs can be combined with mRNA by base complementary pairs, to block the splice sites of the spliceosome, resulting in (3) alternatively spliced transcripts, (4) mRNA degeneration, (5) translation inhibition, or (6) the generation of endo‐siRNAs. III) Interaction with other biological molecules: lncRNAs can (7) bind to the specific protein partners to modulate protein activity, (8) act as a decoy to alter protein localization, (9) serve as scaffolds to allow the formation of larger RNA–protein complexes, or (10) interact with miRNA as a miRNA sponge
2.2. Biological functions of lncRNAs
Although studies on the functions of lncRNAs are still in the preliminary stage, these molecules are known to be involved in regulation of a variety of biological processes in the nucleus and cytoplasm, including immune, cancer, stress, development and differentiation. Studies have shown that lncRNAs are involved in many aspects of immune responses and play important roles in the differentiation and activation of immune cells, including T cells, B cells, macrophages and NK cells. The lncRNA NeST expressed in Th1 CD4+ T cells, CD8+ T cells and NK cells, regulates the production of IFN‐gamma to control the microbial susceptibility of mice.24 LncRNAs are closely related to the occurrence, development and migration of cancer. Furthermore, as a relatively large and heterogeneous group of ncRNAs, lncRNAs have been identified as suitable biomarkers for cancer diagnosis and prognosis. Increased lncRNA PVT1 expression is significantly correlated with positive lymph node metastasis, positive distant metastasis, advanced tumor‐lymph node metastasis stage and poor differentiation.25 In terms of functional enrichment, lncRNAs in lung cancer with significantly changed expression levels were associated with more comprehensive damage than mRNAs, suggesting that lncRNAs represent valuable markers for the development of effective diagnosis and development of prevention strategies.26
LncRNAs are closely related to the occurrence and development of stress conditions. LncRNAs respond to stress to maintain cellular homeostasis by enabling signals associated with the process of cell survival or apoptosis. Studies have shown that lncRNAs can be used as alternative indicators of chemical stress responses in the human induced pluripotent stem cells.27 LncRNAs undergo expression changes to mediate cell defense during the course of genotoxic induction of human glioma cell death.28 LncRNAs play an important role during embryogenesis and organ differentiation, affecting not only the intracellular signal transduction pathways, but also those involved in the development of organisms. For example, the lateral mesodermal specific lncRNA Fendrr is required for the normal development of the mouse hearts and body wall.29 Studies have confirmed that lncRNAs function as active regulators of cell pluripotency and lineage differentiation.
Recently, studies on the underlying mechanism of lncRNAs in different biological funtions are constantly emerging. LncRNAs play a regulatory role in the process of disease apoptosis. LncRNA FTX is notably reduced in the hippocampus of status epilepticus (SE) rat models.30 Overexpression of lncRNA FTX can inhibit apoptosis of the hippocampus neuron in the SE rat models by targeting the 3′‐UTR of miR‐21‐5p to inhibit the miR‐21‐5p/SOX7 axis.30 Studies have shown that lncRNA can help to determine the identity code of cells. Antisense lncRNA transcription can drive stochastic activation of Protocadherin (Pcdh) α promoter choice that generates cell‐surface identity code in functional neurons participating in the neural circuit assembly.31 Pcdhα choice involves the activation of an antisense promoter located in the first exon.31 Antisense lncRNA transcribes from the antisense promoter and extends through the sense promoter, regulating DNA demethylation at the CTCF sites that can bind to both promoters, looping cohesin‐mediated DNA with distal enhancer (HS5‐1), locking the Pcdhα gene in the transcriptional state.31 LncRNA is also essential to maintain the stability of the genome, such as the NORAD lncRNA, a specific lncRNA localized to the nucleus upon activation of the replication stress or DNA damage. NORAD interacts with RBMX which is the component of the DNA‐damage, controls the ability of RBMX to assemble a ribonucleoprotein complex, referred as the NORAD‐activated ribonucleoprotein complex 1 (NARC1), involved in maintaining the stability of the genome.32 More lncRNAs await more extensive and in‐depth studies.
Further, the relationship between the structure and function of lncRNAs has yet to be clarified. This information is critical for the development of new strategies and methods for the treatment of human diseases.
3. LONG NONCODING RNAs IN NEURODEVELOPMENT, NEURAL DIFFERENTIATION AND SYNAPSES
3.1. LncRNAs in neural development and differentiation
Brain development and neurogenesis are complex and precisely regulated processes, in which the dynamic and spatiotemporal expression of specific lncRNAs plays an important role. High‐throughput studies have shown that lncRNAs are spatially and temporally expressed in different brain regions and at different ages. Whole‐transcriptome sequencing data showed different expression patterns of 66 lncRNAs in six adjacent layers of the somatosensory cortex of mice, and the enrichment of these 66 lncRNAs was related to their activity.33 In a RNA‐seq study of the temporal cortical region of the neocortices surgically resected from 36 human donors of various ages, different patterns of lncRNA expression were detected in samples ranging from infancy to adulthood.34 Thus, it can be speculated that these changes are related to maturation and aging in this region of the brain.34
Neurogenesis, which is strictly regulated during the process of brain development, occurs in the subgranular zone (SGZ) of the hippocampus dentate gyrus (DG) and the subventricular zone (SVZ) lined by the lateral ventricle (LV). It is a dynamic process in which the neural stem cells (NSCs) and the neural progenitor cells develop into neurons. The newly generated granulosa cells in the DG area eventually integrate into the local neural network, while neuroblasts formed in the LV area differentiate into neurons and migrate to the olfactory bulb through the rostral migration stream.35 Stratified cluster correlation analysis showed differential expression of lncRNAs in the three brain regions (SVZ, OB and DG), with higher levels of expression in a time‐specific manner compared with mRNAs.36 LncRNA Pnky is enriched in the nucleus and transcribed from the developmental transcription factor.37 The absence of the cortical Pnky in the developmental cortex leads to the production of the projection neuron from NSCs in a cell‐autonomous mode, changing the cortical stratification after birth to transcriptionally regulate the process of neural development.37 As a specialized part of the central nervous system (CNS), the retina is an easy organ to study in vivo, and some lncRNAs are shown to be related to the process of development of retinal. For example, lncRNA Vax2os1 is specifically expressed in the ventral retina of mice, where it regulates cell cycle progression of mouse retinal photoreceptor progenitor cells.38
The dynamic expression of lncRNAs in the CNS reveals their potential role in determining the neural cell fate. The differentiation of NSCs into different types of nerve cells (including neurons and glial cells) is the result of many complex and comprehensive interactions between lncRNAs and other factors. Neurons, the most important structures in the CNS are divided according to their function as motor and sensory neurons. It is known that the number of GABAergic interneurons in the hippocampus and DG of Evf2 mutant mice decreased in the early postnatal period.39 Although the number of GABAergic interneurons returned to normal in the adult hippocampus, their synaptic inhibition was reduced.39 Another study showed that the long intergenic noncoding (linc)RNA TUNA promotes neural differentiation.40 LncRNAs also regulate the differentiation of NSCs into glial cells. For example, the expression levels of lncRNA Dlx1as and Six3os affect the differentiation direction of NSCs, with high expression directing the neuronal pathway, while low expression leads to the generation of astrocytes.36 Nkx2.2AS and NEAT1 have been shown to play regulatory roles in the process of differentiation of NSCs into oligodendrocytes.41, 42 Neurodegenerative diseases (including PD) are intimately related to the process of neural development and differentiation. Recent advances in research and the continuous discovery of the functional lncRNAs have provided a more comprehensive understanding of the regulation of neural development and differentiation by lncRNAs and the association with the neurodegenerative diseases, in addition to highlighting potential new strategies for the treatment of diseases.
3.2. Roles of lncRNAs in synapses
Synapse formation, neurite growth and synaptic plasticity involve complex signal transduction pathways and are tightly regulated. Nerve cells in the brain develop axons and dendrites to form synapses and functional links. The number and strength of these synaptic connections are altered in response to an increase or decrease in nerve activity.43 Synaptic plasticity is the biological basis of the cellular level of synaptic development, transmission and morphological plasticity. LncRNAs play a role in these biological processes through different mechanisms, including RNA interference‐mediated inhibition of mRNA expression, ribosomal RNA modification, selective splicing, and regulatory mechanisms mediated by miRNA‐mRNA interactions.43, 44, 45, 46, 47, 48, 49, 50, 51, 52
Among the lncRNAs, lncRNA BC1/BC200 and Malat1 have been extensively investigated. LncRNA BC1 and lncRNA BC200, expressed in rodents and primates respectively, are essential for maintaining the process of synaptic plasticity, inhibiting local translation and regulating protein synthesis within the synapse.43, 44, 45 The destruction of lncRNA BC1 and lncRNA BC200 in synapses results in abnormal neuronal activity and multiple behavioral deficits.43, 44, 45 For example, decreased BC1 expression increases the lateral branch synapses that deliver glutamatergic transmission.44 Mice without BC1 show increased intrasynaptic glutamate receptors and greater excitatory postsynaptic current in the cortex, leading to excessive excitability of neurons, and resulting in anxiety, twitching and abnormal exploration behaviors.44, 45 Malat1, which is highly expressed in neurons and located in nuclear speckles, may maintain the survival and synaptic formation of neurocytoma cells by activating the ERK/MAPK signaling pathway.46 Malat1‐deficient neuroblastoma cells affect the expression of genes related not only to nuclear processes, but also to dendritic development and synaptic function.46 In cultured hippocampal neurons, Malat1 interacts with the serine/arginine‐rich family of the splicing proteins to regulate synapse formation.47 Overexpression of Malat1 increases synaptic density, while its knockdown has the opposite effect.47 However, adult Malat1 knockout mice showed no changes in either the behavior or the brain tissue morphology.48
In addition, numerous lncRNAs have been found to play regulatory roles in synapses. The nucleolar enriched lncRNA LoNA is negatively regulated by neuronal activation and memory, inhibiting the formation of ribosome‐assembled rRNA and synaptic transport in the hippocampus.49 Brain‐enriched lncRNA, Paupar interacts with the nearby Pax6 to regulate the transcription of many distal genes involved in synaptic plasticity, such as Camk2a, Syn3, and Ank3.50 LncRNA Ube3a1 is selectively cleaved by E3 ubiquitin ligase Ube3a to inhibit the process of dendritic growth by inhibiting miR‐134 and promoting spinal maturation of hippocampal neurons.51 Antisense lncRNAs are also involved in the regulation of many important proteins related to the processes of neuron differentiation and synaptic growth, such as BDNF, GDNF, and EPHB2.52 Inhibiting lncRNA BDNF‐AS, which is the antisense transcript of BDNF, improves BDNF expression and promotes neuron survival and neurite growth.52
LncRNAs are exclusively expressed across specific regions of the brain in certain developmental time points to perform specific functions (Table 1).53 For example, there are 806 of 5195 lncRNAs that are differentially expressed between the cortical pyramidal and neural cells, performing their functions of maintenance and identification of the neural cell type.54 A study on the spatiotemporal transcriptome of embryonic neurogenesis in Drosophila melanogaster, that absolutely requires accurate regulation of gene in space and time during early nervous system development, reveals a highly complex dynamic and spatiotemporal regulation of lncRNAs.55 LncRNA Riken‐201 and Riken‐203 can regulate Sox6 by sequestering both miR‐96 and miR‐467a‐3p to modulate neural development.56 A number of lncRNAs that determine the pluripotency or neural cell fate in neurogenesis are shown in Table 1. LncRNA NTAB expressed in the developing and adult rat brain is involved in the regulation of neuronal processes.57 LncRNA Rmst, exclusively expressed in the brain is implicated in the differentiation of neural cells, supporting the role of RMST in neurogenesis, binding to SOX2 and modulating a number of neurogenic genes including NEUROG2, ASCL1, HEY2, and DLX1.58 Moreover, absence of Rmst is known to promote the process of neural differentiation.58 LncKdm2b activates Kdm2b in cis to regulate the process of cortical neuronal differentiation.59 Recently, many studies have reported that lncRNAs and miRNAs are involved in different biological processes including neurodevelopment, neural differentiation and maturation, associating with the development of neurodegenerative diseases (such as AD, Alzheimer's disease and PD), particularly gene expression, introduced as possible biomarkers for diagnosis and prognosis.60 In addition, lncRNAs can also play important roles in neurological impairment. For example, repression of the lncRNA MEG3 restores nerve growth in rat models of cerebral ischemia‐reperfusion injury,61 while lncRNA H19 regulates the p53/Notch1 pathway to inhibit neurogenesis in ischemic stroke.62 A summary of regulated lncRNAs, their corresponding functions and mechanisms in neurodevelopment is shown in Table 1.36, 37, 38, 39, 40, 41, 42, 50, 56, 58, 59, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72
Table 1.
A summary of regulated lncRNAs, their corresponding functions and mechanisms in neurodevelopment
| Long noncoding RNAs | Regions | Mechanisms | Functions | References |
|---|---|---|---|---|
| Dlx1as, Six3os | NSCs | Recruits EZH2 and EYA | Glial‐neuronal lineage specification | 36 |
| Pnky | Nucleus of NSCs | Interacts with Pou3f2 | Neurogenesis and neural proliferation | 37 |
| Vax2os1 | Retina | Cell cycle regulator | Cell cycle progression of mouse retinal photoreceptor progenitor cells | 38 |
| Evf2 | Hippocampus and dentate gyrus | Recruits DLX and MECP2 | Differentiation of GABAergic neurons | 39 |
| TUNA | NSCs | Recruits NCL, PTBP1, and hnRNP‐K | Neural differentiation | 40 |
| Nkx2.2AS | Nucleus of NSCs | — | Oligodendrocyte lineage differentiation | 41 |
| NEAT1 | Nucleus of NSCs | Paraspeckle integrity | Oligodendrocyte lineage differentiation | 42 |
| Paupar | Glioma | Interacts with PAX6 and localize to SOX2, NANOG and HES1 | Neuroblastoma differentiation | 50 |
| LncRNA Riken‐201 and Riken‐203 | Neural differentiation of mouse embryonic stem cells | Regulate Sox6 by sequestering miR‐96 and miR‐467a‐3p | Neural development and differentiation | 56 |
| Rmst | NSCs | Recruits SOX2 | Neural differentiation | 58 |
| LncKdm2b | Cortical projection neurons | Activates Kdm2b in cis | Cortical neuronal differentiation | 59 |
| LncND | Radial glia cells of the ventricular and subventricular zones of developing brain | Targets Notch signaling by binding and release of miR‐143‐3p | Neural development and differentiation | 63 |
| LncR492 | Murine embryonic stem cells | Interacts with the mRNA binding protein HuR and facilitates inhibitory function by activating Wnt signaling | Inhibits neural differentiation of murine embryonic stem cells | 64 |
| Sox2ot | Pluripotent stem cells and NSCs | Regulates Sox2 by an allele‐specific mechanism | Neural development | 65 |
| LncRNA‐1604 | Cytoplasm | Regulates lncRNA‐1604/miR‐200c/ZEB axis | Neural differentiation | 66 |
| LincRNA1230 | Embryonic stem (ES) cells | Modulation of bivalent modifications at the promoters of early neural genes; interacts with Wdr5 and inhibits the enrichment of H3K4me3 modifications | Neural differentiation | 67 |
| Dali | Glioma | Interacts with POU3F and DNMT1 | Neuroblastoma differentiation | 68 |
| Brn1b | Neocortex | Regulates BRN1 protein | Migration and differentiation of cortex | 69 |
| HAR1F | Cortex | Upregulates reelin | Neural development | 70 |
| Pou3f2 | NSCs | — | Neural stem cell proliferation | 71 |
| HOTAIR | Glioma | Recruits EZH2 | Neural differentiation/neuroblastoma differentiation | 72 |
4. ROLES OF LONG NONCODING RNAs IN PD
Accumulating evidence shows that lncRNAs play an important role in PDs (Table 2). Changes in lncRNAs may be the key factors in the pathogenesis of diseases and represent disease‐specific markers. Studies have shown that lncRNAs are closely related to PD. Compared to the normal tissues, significant changes in the expression of 87 lncRNAs were identified in the substantia nigra of PD patients.73 Among these, lncRNA AL049437 and lncRNA AK021630 showed the most significant changes.73 Based on experimental verification, it is speculated that lncRNA AL049437 and lncRNA AK021630 function to promote and inhibit the occurrence of PD, respectively.73 Interestingly, abnormal lncRNA expression was detected not only in brain tissues, but also in the peripheral blood and cerebrospinal fluid of PD patients. Amongst the greater than 6000 lncRNAs detected in the peripheral white blood cells of PD patients, 13 lncRNAs showed decreased expression and selective PD‐induced changes, and four lncRNAs were reversed after deep brain stimulation treatment.74 Comparisons of the peripheral white blood cells and brain tissue samples of the PD patients and healthy individuals reveal common differences in lncRNAs, with the same expression trends.74 Studies have shown that there is decreased expression of the neurodegenerative gene‐related lncRNAs in PD patients; six in the substantia nigra and three in the cerebellum.75 Furthermore, all disorders of lncRNAs were detected in the peripheral blood mononuclear cells, with four in the cerebrospinal fluid‐derived exosomes.75 Thus, lncRNAs are implicated as potential diagnostic markers or novel therapeutic targets in PD.
Table 2.
Long noncoding RNAs in Parkinson's disease
| Long noncoding RNAs | Regions | Functions | References |
|---|---|---|---|
| LncRNA NEAT1 | MPTP mouse model and MPP+‐induced SH‐SY5Y | Regulating neuronal injury by targeting miR‐124; promoting autophagy through stabilizing PINK1 protein | 78, 100 |
| LncRNA HAGLROS | MPTP mouse model and MPP+‐induced SH‐SY5Y | Regulating apoptosis and autophagy via regulating miR‐100/ATG10 axis and PI3K/Akt/mTOR pathway activation | 79 |
| LncRNA HOTAIR | Midbrain tissue of MPTP induced PD mice and SH‐SY5Y cells exposed to MPP+ | Promoting PD by upregulating LRRK2 expression; targeting miR‐126‐5p to promote PD through RAB3IP | 83, 112 |
| LncRNA SNHG1 | SH‐SY5Y cells | Promoting α‐synuclein aggregation and toxicity by targeting miR‐15b‐5p to activate SIAH1 | 84 |
| LncRNA MALAT1 | MPTP mouse model and MPP+‐induced SH‐SY5Y; MN9D cells | α‐synuclein protein expression; regulating cell apoptosis by directly targeting LRRK2 through lncRNA MALAT1/miR‐205‐5p axis | 85, 98 |
| LncRNA UCA1 | Substantia nigra striatum of 6‐OHDA PD model; MPTP mouse model and MPP+‐induced SH‐SY5Y | The damage of dopaminergic neurons, oxidative stress and inflammation by the PI3K/Akt signaling pathway; promoting PD development by upregulating SNCA | 94, 99 |
| LncRNA‐p21 | SH‐SY5Y cells | Regulating MPP+‐induced neuronal injury by targeting miR‐625 and derepressing TRPM2 | 95 |
| Nrf2‐related lncRNAs | Substantia nigra of paraquat and MPTP induced mouse model | Oxidative stress | 96 |
| LncRNA AS Uchl1 | Dopaminergic cells' differentiation in vitro and in neurochemical models of PD | Expression is under the regulation of Nurr1 | 110 |
| PINK1‐AS, UCHL1‐AS, BCYRN1, SOX2‐OT, ANRIL and HAR1A | PD in the Hungarian population | Interfering with the binding affinity of transcription factor HNF4A, potentially resulting in abnormal expression of target genes, such as BCYRN1 | 111 |
| Sox2OT | Anti‐NGF AD11 transgenic mouse | Regulating cotranscribed Sox2 gene expression to down neurogenesis | 113 |
Both animal and cell models are commonly used in PD research. The 6‐hydroxydopamine (6‐OHDA) rat model of PD simulates the pathological features of the disease, characterized by the loss of numerous dopaminergic neurons in substantia nigra and decreased levels of dopamine in the striatum. Transcriptome sequencing of the striatum tissue revealed 512 differentially expressed lncRNAs associated with the biological processes of innate immune response, GTPase activity, and 2′‐5′‐oligoadenylate synthase activity, and with a possible regulatory role in the pathogenesis of PD.76 Abnormal expression of approximately 756 lncRNAs was detected in the substantia nigra pars compacta (SNpc) of the presymptomatic mice overexpressing human A30P*A53T α‐synuclein.77 The mRNAs coexpressed with lncRNAs are mainly concentrated in the axon‐guided signaling pathway.77 The function of these abnormally expressed lncRNAs may provide new targets for the treatment of early PD.77 SH‐SY5Y cells are treated with methyl‐4‐phenylpyridinium (MPP+) to generate the cellular model of PD which has been extensively used to study the role of lncRNAs in cell injury.78, 79 Although animal and cell models cannot fully replicate the pathological changes occurring in human PD, these experimental tools provide important insights into the mechanism of the PD pathogenesis and the role of lncRNAs in the process, thus offering new insights into strategies for the diagnosis and treatment of PD.
4.1. LncRNAs in the ubiquitin‐proteasome system, protein decomposition, protein misfolding and aggregation
The ubiquitin‐proteasome system (UPS) is a nonlysosomal pathway of protein degradation, which removes mutant, damaged and aberrant proteins in the cells, regulates cell cycle DNA damage and repairs apoptosis. UPS dysfunction results in abnormal accumulation of protein within the cell and plays an important role in the pathogenesis of PD, acting synergistically with mitochondrial dysfunction, oxidative stress and other factors.80, 81 Ubiquitin carboxy‐terminal hydrolase L1 (Uchl1) is a gene involved in the UPS of PD.82 Antisense Uchl1 (UCHL1‐AS1), a new functional class of lncRNAs, increases synthesis of the UCHL1 protein.82 UCHL1‐AS1 contains 5′ overlapping sequence of UCHL1 mRNA and an embedded inverted SINEB2 element, which is necessary for protein translation.82 LncRNA UCHL1‐AS1 directly affects the translation of the UCHL1 protein, leading to disruption of the UPS.80
Lewy body formation is a typical pathological change associated with PD. α‐synuclein, which is the main component of LBs, is related to dopamine synthesis, neuronal remodeling, signal transduction, synaptic vesicle transport and is degraded by UPS. The abnormal folding, aggregation and diffusion of α‐synuclein under the influence of various factors play an important role in the pathogenesis of PD. LncRNA HOTAIR has been shown to affect the progression of PD.83 A recent study showed that HOTAIR regulates miR‐126‐5p and RAB3IP in a ceRNA‐dependent manner to promote progression of PD.83 In vivo knockout of HOTAIR reduces the number of α‐synuclein‐positive cells and reduces apoptosis of dopaminergic neurons.83 LncRNA SNHG1 combines directly with miR‐15‐5p to inhibit its expression, thereby promoting the α‐synuclein polymerization and α‐synuclein‐induced apoptosis, in a process that is implicated in the mechanism of LB formation and its role in neuron loss in PD.84 In the 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine (MPTP) mouse model of PD, β‐asarone increases the number of tyrosine hydroxylase‐positive cells and simultaneously reduces the expression of lncRNA MALAT1 and α‐synuclein, thereby playing a neuroprotective function.85 Microarray expression analysis showed that during the inhibition of α‐synuclein toxicity by the commonly used traditional Chinese medicine Acanthopanax senticosus (AS), 341 lncRNAs were differentially expressed under the stimulation of α‐synuclein.86 Among these, 29 lncRNAs were involved in the AS mediated inhibition of the α‐synuclein neurotoxicity mechanism, and 19 were found to be potentially related to the α‐synuclein neurotoxicity.86 Elucidation of the lncRNA‐mediated regulation of the α‐synuclein associated mechanisms (including abnormal modification of α‐synuclein after translation, aggregation of α‐synuclein, toxic effects, and degradation) is important for clarification of the pathogenesis of PD and the discovery of new therapeutic targets and methods.
4.2. LncRNAs in mitochondrial dysfunction, oxidative stress, autophagy and apoptosis
Mitochondrial dysfunction and oxidative stress can damage cells and lead to PD.87, 88 Mitochondrial electron transfer is a major source of intracellular free radicals. Reduced mitochondrial complex I activity promotes free radical formation and enhances the susceptibility of tissues to oxidative stress, resulting in damage to cellular DNA, lipids and proteins.89 Furthermore, studies have shown that the neurotoxicity associated with PD is due to inhibition of respiratory chain complex Ι activity, leading to dysfunction of ATP synthesis, cell degeneration and death.89 Oxidative stress occurs when the oxidation and antioxidant systems are unbalanced, resulting in cell destruction caused by changes in membrane fluidity and the lipid peroxidation chain reaction. Under normal circumstances, multiple antioxidant systems exist in cells to clear reactive oxygen species (ROS).87 The main free radical scavenging system in the brain is the glutathione system.87 PD patients show high oxidative stress in the brain, while dopaminergic neurons lack the ability to control oxidative stress.87 6‐OHDA can produce iron‐mediated free radicals and inhibit the activity of mitochondrial complex Ι.90 MPTP also blocks mitochondrial electron transfer by inhibiting mitochondrial complex Ι.91
Studies have reported that mice lacking the lncRNA Norad exhibit genomic instability and mitochondrial dysfunction, leading to a multi‐system degenerative phenotype similar to premature aging.92 This suggests that mitochondrial‐related lncRNAs may play important roles in the pathophysiology of neurodegenerative diseases.92 Recently, it has been shown that the lncRNA NEAT1, which is overexpressed in the substantia nigra of PD, plays a neuroprotective role against drug‐induced oxidative stress.93 In the 6‐OHDA rat model of PD, downregulation of lncRNA UCA1 inhibits the PI3K/Akt signaling pathway, with a resultant reduction in the damage of the dopaminergic neurons, as well as the oxidative stress and inflammatory response associated with PD.94 In the MPP+‐induced SH‐SY5Y cell model of PD, lncRNA‐p21 knockdown weakens the cytotoxicity and apoptosis induced by MPP+ and reduces the production of ROS, oxidative stress and neuroinflammation.95 Nrf2 is an important cell signaling molecule that protects against oxidative stress. During oxidative stress, Nrf2 dissociates and translocates from the cytoplasm to the nucleus, upregulates the expression of the antioxidant protein genes, and enhances the ability of cells to survive from oxidative stress. Using microarray technology, Nrf2‐related lncRNAs have been shown to be present in the substantia nigra of the paraquat and MPTP mouse models of PD.96
Protein degradation mediated by autophagy is a process in which the proteins are engulfed within vesicles which fuse with the lysosomes to degrade proteins. Autophagy consists of three pathways: endocytosis, macrovesicle autophagy and chaperone mediated autophagy. Mitochondrial autophagy is a process of selective clearance of excessive or damaged mitochondria. Various neurodegenerative diseases are associated with abnormalities in this process.97 Enhanced autophagy can effectively counter PD. In SH‐SY5Y cell (MPP+‐induced cells) and mouse (MPTP‐induced) models of PD, lncRNA HAGLROS regulates the apoptosis and autophagy processes by regulating the miR‐100/ATG10 axis to activate the PI3K/Akt/mTOR pathway.79 Another study showed that the lncRNA NEAT1 promotes the MPTP‐induced autophagy in PD by inhibiting the degradation of PINK1 protein.78
The process of apoptosis is closely related to mitochondrial dysfunction and oxidative stress and plays an important role in the death of the dopaminergic neurons in PD.98 Mitochondria is the site where the process of apoptosis takes place as it houses many apoptotic factors, such as caspases and cytochrome C. Oxidative stress results in the generation of ROS, which in turn act as second messengers to regulate the mitochondrial permeability transition pore and increase the mitochondrial membrane permeability. This leads to the release of cytochrome C and apoptotic inducers into the cytoplasm, which bind with the caspase‐9 precursor to form apoptotic corpuscular bodies. Under pathological conditions, prolonged increase in the levels of ROS leads to opening of the mitochondrial permeability transition pore, resulting in a series of reactions, including uncoupling of the mitochondrial electron transport chain from oxidative phosphorylation, reduced ATP synthesis, loss of mitochondrial membrane potential, Ca2+ outflow, glutathione reduction, increased levels of intracellular ROS, and release of cytochrome C and apoptosis‐inducing factors, which eventually initiate the cell apoptosis process. LncRNA MALAT1 regulates the MPP+‐induced apoptosis of the MN9D cells by inhibiting miR‐205‐5p targeting LRRK2.98 Knockdown of lncRNA‐UCA1 inhibits caspase‐3 activity, thereby reducing MPP+‐induced SH‐SY5Y cell apoptosis.99 LncRNA NEAT1 regulates MPP+‐induced SH‐SY5Y cell apoptosis by targeting miR‐124.100
4.3. LncRNAs and inflammation
Neuroinflammation is an important factor in PD. The inflammatory cytokines secreted by the glial cells participate in neuroinflammatory responses to induce apoptosis of the dopaminergic neurons. Microglia belong to the mononuclear phagocyte lineage and are the primary innate immune cells of the CNS.101 When the neural microenvironment is unbalanced, microglial cells are activated and release a variety of biological active molecules, including inflammatory cytokines, chemokines and neuromodulins, which cause chronic and persistent inflammatory reactions and promote the injury and death of dopaminergic neurons resultant in the pathological process that leads to PD. The astrocytes are the supporting cells which provide nutrients to neurons, but can also cause neuroinflammation. Long‐term dysfunction of astrocytes causes lack of neuronutrients and increased secretion of inflammatory cytokines, affecting the integrity of the blood‐brain barrier and synaptic plasticity, which accelerates the aging and degeneration of dopaminergic neurons. Microarray technology used to detect the PD‐associated differentially expressed genes and lncRNAs in the human blood revealed that the downregulated differentially expressed genes in the regulatory network were enriched in the immune response.102 In the 6‐OHDA rat PD model, downregulating lncRNA UCA1 inhibits the PI3K/Akt signaling pathway, alleviating the damage of the dopaminergic neuron, oxidative stress and inflammatory response associated with PD.94 Study has shown that in the SH‐SY5Y of AD cell model, lncRNA RP11‐543N12.1 targets miR‐324‐3p to modify microglia‐induced cell apoptosis of SH‐SY5Y.101 In the hippocampus tissues of AD rats, lncRNA MEG3 alleviates neuronal damage and inhibits the activation of astrocytes by inactivating the PI3K/Akt signaling pathway.103 In addition, lncRNA TUG1 regulates the process of microglial polarization and production of inflammatory cytokines after oxygen‐glucose deprivation through sponging miR‐145a‐5p, providing therapeutic hope from the inflammatory injury.104
4.4. Other functions of lncRNAs in PD
During the process of dopamine metabolism, dopamine oxidation intermediates bind to α‐synuclein, which inhibits dopamine activity and selectively aggregates α‐synuclein. This imbalance in dopamine anabolism leads to a vicious cycle of increased free dopamine associated cytotoxicity, and the obstruction of the transport of the dopamine vesicles. LncRNAs are found in the nucleus and cytoplasm of dopaminergic neurons.105 Knockout of the lncRNA Gomafu/MIAT/Rncr2 enhances the effect of methamphetamine, which increases the release of dopamine into the nucleus accumbens.106 Neurotrophic factors, including BDNF, GDNF, and NT‐3, can support the survival of neurons, induce the growth of nerve processes and maintain the functions of neurons. These neurotrophic factors, which play a neuroprotective role in providing nutrients and promoting the regeneration and repair of neurons play an important role in the pathogenesis and treatment of PD. Many lncRNAs have been shown to promote or inhibit the expression of these neurotrophic factors, thereby affecting neurons and the intracerebral microenvironment.107, 108 In addition, many lncRNAs act on PD‐related genes. For example, the lncRNA NEAT1 promotes the MPTP‐induced autophagy in PD by inhibiting degradation of the PINK1 protein.78 LncRNAs are also associated with PD‐related genes such as LRRK2, DJ‐1, Uchl1, and Nurr1.98, 109, 110 In addition, the roles of lncRNAs in neurodevelopment, neural differentiation and synapses may also affect the pathogenesis and treatment strategies of PD.
5. CONCLUSION AND FUTURE PERSPECTIVES
In summary, many studies have shown that lncRNAs have widespread tissue expression with numerous biological functions. However, the mechanisms by which lncRNAs function in complex physiological and pathological conditions such as PD have not yet been fully elucidated. However, it is known that a variety of triggering factors work together leading to the protein misfolding and aggregation, dysregulated protein degradation, mitochondrial dysfunction, oxidative stress, autophagy, apoptosis, and neuroinflammation (Figure 3) that ultimately account for the pathological manifestations and clinical symptoms of PD. The spatiotemporal expression of specific lncRNAs play a role in the pathological processes that lead to PD. In‐depth studies of the functions of the lncRNAs in PD will provide significant information that can be used to develop early and effective diagnostic methods and therapeutic targets. Although the regulatory function of the lncRNAs has been demonstrated in the cell and animal models (Table 2),78, 79, 83, 84, 85, 94, 95, 96, 98, 99, 100, 110, 111, 112, 113 this is only a snapshot of the dynamic changes that occur with the progression of the disease. Due to the complexity of diseases, different pathological processes are usually accompanied by changes in multiple lncRNAs. Therefore, the selection of the most promising and critical lncRNAs for further investigation remains a significant challenge. Because PD involves a complex interaction of genes and their regulatory factors, with the lncRNAs mediating complex coexpression regulatory networks, the diagnostic value of a single biological marker is limited in the diagnosis and treatment of PD. Therefore, the combined application of multiple biological markers to improve the reliability and effectiveness of diagnosis and treatment is now a focus of research. In conclusion, developing new strategies using lncRNAs for disease diagnosis and drug treatment holds promise towards tackling the pathogenesis of PD.
Figure 3.
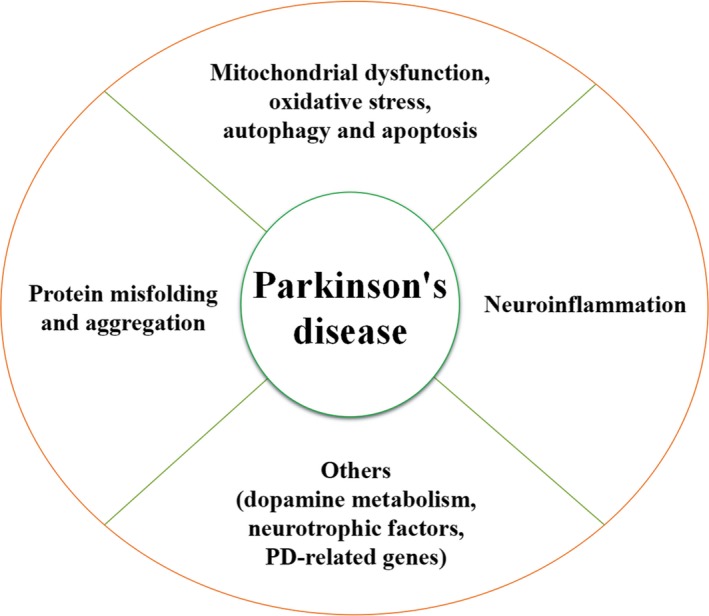
Regulatory role of lncRNAs in Parkinson's disease. A variety of lncRNAs are associated with different mechanisms like protein misfolding and aggregation, dysregulated protein degradation, mitochondrial dysfunction, oxidative stress, autophagy, apoptosis, neuroinflammation, and other pathomechanisms (including dopamine metabolism, neurotrophic factors, and PD‐related genes) that ultimately account for the pathological manifestations and clinical symptoms of PD
CONFLICT OF INTEREST
None.
ACKNOWLEDGMENTS
The work was supported by grants from the Beijing Natural Science Foundation (5171001) and CAMS Innovation Fund for Medical Sciences (CIFMS) (2017‐I2M‐2‐005, 2016‐I2M‐2‐006).
Lyu Y, Bai L, Qin C. Long noncoding RNAs in neurodevelopment and Parkinson’s disease. Animal Model Exp Med. 2019;2:239–251. 10.1002/ame2.12093
Contributor Information
Lin Bai, Email: bailin49@163.com.
Chuan Qin, Email: qinchuan@pumc.edu.cn.
REFERENCES
- 1. Tang Y, Meng LI, Wan C‐M, et al. Identifying the presence of Parkinson's disease using low‐frequency fluctuations in BOLD signals. Neurosci Lett. 2017;645:1‐6. [DOI] [PubMed] [Google Scholar]
- 2. Shao Y‐M, Ma X, Paira P, et al. Discovery of indolylpiperazinylpyrimidines with dual‐target profiles at adenosine A2A and dopamine D2 receptors for Parkinson's disease treatment. PLoS ONE. 2018;13(1):e188212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chung Y‐C, Ko H‐W, Bok E‐G, et al. The role of neuroinflammation on the pathogenesis of Parkinson's disease. BMB Rep. 2010;43(4):225‐232. [DOI] [PubMed] [Google Scholar]
- 4. Devos D, Moreau C, Dujardin K, Cabantchik I, Defebvre L, Bordet R. New pharmacological options for treating advanced Parkinson's disease. Clin Ther. 2013;35(10):1640‐1652. [DOI] [PubMed] [Google Scholar]
- 5. Rial D, Castro AA, Machado N, et al. Behavioral phenotyping of Parkin‐deficient mice: looking for early preclinical features of Parkinson's disease. PLoS ONE. 2014;9(12):e114216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou T, Kim TW, Chong CN, et al. A hPSC‐based platform to discover gene‐environment interactions that impact human beta‐cell and dopamine neuron survival. Nat Commun. 2018;9(1):4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stojkovska I, Krainc D, Mazzulli JR. Molecular mechanisms of alpha‐synuclein and GBA1 in Parkinson's disease. Cell Tissue Res. 2018;373(1):51‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Korkmaz OT, Tuncel N. Advantages of vasoactive intestinal peptide for the future treatment of Parkinson's disease. Curr Pharm Des. 2018;24(39):4693‐4701. [DOI] [PubMed] [Google Scholar]
- 9. Borrageiro G, Haylett W, Seedat S, Kuivaniemi H, Bardien S. A review of genome‐wide transcriptomics studies in Parkinson's disease. Eur J Neurosci. 2018;47(1):1‐16. [DOI] [PubMed] [Google Scholar]
- 10. Nair VD, Ge Y. Alterations of miRNAs reveal a dysregulated molecular regulatory network in Parkinson's disease striatum. Neurosci Lett. 2016;629:99‐104. [DOI] [PubMed] [Google Scholar]
- 11. Karlsson MK, Sharma P, Aasly J, et al. Found in transcription: accurate Parkinson's disease classification in peripheral blood. J Parkinsons Dis. 2013;3(1):19‐29. [DOI] [PubMed] [Google Scholar]
- 12. Khawar MB, Mehmood R, Abbasi MH, Sheikh N. Multifactorial role of long non‐coding RNAs (LncRNAs) in hematopoiesis. Front Biosci. 2018;10:119‐126. [DOI] [PubMed] [Google Scholar]
- 13. Zucchelli S, Fedele S, Vatta P, et al. Antisense transcription in loci associated to hereditary neurodegenerative diseases. Mol Neurobiol. 2019;56(8):5392‐5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non‐coding RNA: classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82‐92. [DOI] [PubMed] [Google Scholar]
- 15. Wang XQ, Crutchley JL, Dostie J. Shaping the genome with non‐coding RNAs. Curr Genomics. 2011;12(5):307‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Q, Dong C, Cui J, Wang Y, Hong X. Over‐expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up‐regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37(1):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484‐1488. [DOI] [PubMed] [Google Scholar]
- 18. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47‐62. [DOI] [PubMed] [Google Scholar]
- 19. Kour S, Rath PC. Long noncoding RNAs in aging and age‐related diseases. Ageing Res Rev. 2016;26:1‐21. [DOI] [PubMed] [Google Scholar]
- 20. Castelo‐Branco G, Amaral PP, Engström PG, et al. The non‐coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013;14(9):R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sørensen KP, Thomassen M, Tan Q, et al. Long non‐coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor‐positive primary breast cancer. Breast Cancer Res Treat. 2013;142(3):529‐536. [DOI] [PubMed] [Google Scholar]
- 22. Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adelman K, Egan E. Non‐coding RNA: More uses for genomic junk. Nature. 2017;543(7644):183‐185. [DOI] [PubMed] [Google Scholar]
- 24. Gomez JA, Wapinski OL, Yang YW, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon‐gamma locus. Cell. 2013;152(4):743‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu D, Luo P, Wang Q, Ye Y, Wang B. lncRNA PVT1 in cancer: A review and meta‐analysis. Clin Chim Acta. 2017;474:1‐7. [DOI] [PubMed] [Google Scholar]
- 26. Chen R, Li W‐X, Sun Y, et al. Comprehensive analysis of lncRNA and mRNA expression profiles in lung cancer. Clin Lab. 2017;63(2):313‐320. [DOI] [PubMed] [Google Scholar]
- 27. Tani H, Onuma Y, Ito Y, Torimura M. Long non‐coding RNAs as surrogate indicators for chemical stress responses in human‐induced pluripotent stem cells. PLoS ONE. 2014;9(8):e106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Q, Sun S, Yu W, et al. Altered expression of long non‐coding RNAs during genotoxic stress‐induced cell death in human glioma cells. J Neurooncol. 2015;122(2):283‐292. [DOI] [PubMed] [Google Scholar]
- 29. Grote P, Wittler L, Hendrix D, et al. The tissue‐specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X, Giri V, Cui Y, Yin M, Xian Z, Li J. LncRNA FTX inhibits hippocampal neuron apoptosis by regulating miR‐21‐5p/SOX7 axis in a rat model of temporal lobe epilepsy. Biochem Biophys Res Commun. 2019;512(1):79‐86. [DOI] [PubMed] [Google Scholar]
- 31. Canzio D, Nwakeze CL, Horta A, et al. Antisense lncRNA transcription mediates DNA demethylation to drive stochastic Protocadherin alpha promoter choice. Cell. 2019;177(3):639‐653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munschauer M, Nguyen CT, Sirokman K, et al. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561(7721):132‐136. [DOI] [PubMed] [Google Scholar]
- 33. Belgard T, Marques A, Oliver P, et al. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605‐616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lipovich L, Tarca AL, Cai J, et al. Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb Cortex. 2014;24(6):1451‐1459. [DOI] [PubMed] [Google Scholar]
- 35. Ayana R, Singh S, Pati S. Decoding crucial lncRNAs implicated in neurogenesis and neurological disorders. Stem Cells Dev. 2017;26(8):541‐553. [DOI] [PubMed] [Google Scholar]
- 36. Ramos AD, Diaz A, Nellore A, et al. Integration of genome‐wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12(5):616‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersen RE, Hong SJ, Lim JJ, et al. The long noncoding RNA Pnky is a trans‐acting regulator of cortical development in vivo. Dev Cell. 2019;49(4):632‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18(1):111‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bond AM, VanGompel MJW, Sametsky EA, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12(8):1020‐1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin N, Chang K‐Y, Li Z, et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53(6):1005‐1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tochitani S, Hayashizaki Y. Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem Biophys Res Commun. 2008;372(4):691‐696. [DOI] [PubMed] [Google Scholar]
- 42. Mercer TR, Qureshi IA, Gokhan S, et al. Long noncoding RNAs in neuronal‐glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Briz V, Restivo L, Pasciuto E, et al. The non‐coding RNA BC1 regulates experience‐dependent structural plasticity and learning. Nat Commun. 2017;8(1):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chung A, Dahan N, Alarcon JM, Fenton AA. Effects of regulatory BC1 RNA deletion on synaptic plasticity, learning, and memory. Learn Mem. 2017;24(12):646‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lewejohann L, Skryabin BV, Sachser N, et al. Role of a neuronal small non‐messenger RNA: behavioural alterations in BC1 RNA‐deleted mice. Behav Brain Res. 2004;154(1):273‐289. [DOI] [PubMed] [Google Scholar]
- 46. Chen L, Feng P, Zhu X, He S, Duan J, Zhou D. Long non‐coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J Cell Mol Med. 2016;20(11):2102‐2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear‐retained non‐coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082‐3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakagawa S, Ip JY, Shioi G, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487‐1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li D, Zhang J, Wang M, et al. Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat Commun. 2018;9(1):1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vance KW, Sansom SN, Lee S, et al. The long non‐coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33(4):296‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Valluy J, Bicker S, Aksoy‐Aksel A, et al. A coding‐independent function of an alternative Ube3a transcript during neuronal development. Nat Neurosci. 2015;18(5):666‐673. [DOI] [PubMed] [Google Scholar]
- 52. Modarresi F, Faghihi MA, Lopez‐Toledano MA, et al. Inhibition of natural antisense transcripts in vivo results in gene‐specific transcriptional upregulation. Nat Biotechnol. 2012;30(5):453‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hosseini E, Bagheri‐Hosseinabadi Z, De Toma I, Jafarisani M, Sadeghi I. The importance of long non‐coding RNAs in neuropsychiatric disorders. Mol Aspects Med. 2019;70:127‐140. [DOI] [PubMed] [Google Scholar]
- 54. Molyneaux BJ, Goff LA, Brettler AC, et al. DeCoN: genome‐wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85(2):275‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCorkindale AL, Wahle P, Werner S, et al. A gene expression atlas of embryonic neurogenesis in Drosophila reveals complex spatiotemporal regulation of lncRNAs. Development. 2019;146(6): dev175265 https://doi.org10.1242/dev.175265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang L, Xue Z, Yan J, Wang J, Liu Q, Jiang H. LncRNA Riken‐201 and Riken‐203 modulates neural development by regulating the Sox6 through sequestering miRNAs. Cell Prolif. 2019;52(3):e12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. French PJ, Bliss TV, O'Connor V. Ntab, a novel non‐coding RNA abundantly expressed in rat brain. Neuroscience. 2001;108(2):207‐215. [DOI] [PubMed] [Google Scholar]
- 58. Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51(3):349‐359. [DOI] [PubMed] [Google Scholar]
- 59. Li W, Shen W, Zhang B, et al. Long non-coding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b. Protein Cell. 2019. 10.1007/s13238-019-0650-z. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maniati MS, Maniati M, Yousefi T, Ahmadi‐Ahangar A, Tehrani SS. New insights into the role of microRNAs and long noncoding RNAs in most common neurodegenerative diseases. J Cell Biochem. 2019;120(6):8908‐8918. [DOI] [PubMed] [Google Scholar]
- 61. You D, You H. Repression of long non‐coding RNA MEG3 restores nerve growth and alleviates neurological impairment after cerebral ischemia‐reperfusion injury in a rat model. Biomed Pharmacother. 2019;111:1447‐1457. [DOI] [PubMed] [Google Scholar]
- 62. Wang J, Cao B, Zhao H, et al. Long noncoding RNA H19 prevents neurogenesis in ischemic stroke through p53/Notch1 pathway. Brain Res Bull. 2019;150:111‐117. [DOI] [PubMed] [Google Scholar]
- 63. Rani N, Nowakowski T, Zhou H, et al. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron. 2016;90(6):1174‐1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Winzi M, Casas Vila N, Paszkowski‐Rogacz M, et al. The long noncoding RNA lncR492 inhibits neural differentiation of murine embryonic stem cells. PLoS ONE. 2018;13(1):e191682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Messemaker TC, van Leeuwen SM, van den Berg PR, et al. Allele‐specific repression of Sox2 through the long non‐coding RNA Sox2ot. Sci Rep. 2018;8(1):386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Weng R, Lu C, Liu X, et al. Long noncoding RNA‐1604 orchestrates neural differentiation through the miR‐200c/ZEB axis. Stem Cells. 2018;36(3):325‐336. [DOI] [PubMed] [Google Scholar]
- 67. Wang C, Li G, Wu Y, Xi J, Kang J. LincRNA1230 inhibits the differentiation of mouse ES cells towards neural progenitors. Sci China Life Sci. 2016;59(5):443‐454. [DOI] [PubMed] [Google Scholar]
- 68. Chalei V, Sansom SN, Kong L, et al. The long non‐coding RNA Dali is an epigenetic regulator of neural differentiation. Elife. 2014;3:e4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sauvageau M, Goff LA, Lodato S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pollard KS, Salama SR, Lambert N, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443(7108):167‐172. [DOI] [PubMed] [Google Scholar]
- 71. Lin M, Pedrosa E, Shah A, et al. RNA‐Seq of human neurons derived from iPS cells reveals candidate long non‐coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE. 2011;6(9):e23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang K, Sun X, Zhou X, et al. Long non‐coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ni Y, Huang H, Chen Y, Cao M, Zhou H, Zhang Y. Investigation of long non‐coding RNA expression profiles in the substantia nigra of Parkinson's disease. Cell Mol Neurobiol. 2017;37(2):329‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Soreq L, Salomonis N, Guffanti A, Bergman H, Israel Z, Soreq H. Whole transcriptome RNA sequencing data from blood leukocytes derived from Parkinson's disease patients prior to and following deep brain stimulation treatment. Genom Data. 2015;3:57‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Elkouris M, Kouroupi G, Vourvoukelis A, et al. Long non‐coding RNAs associated with neurodegeneration‐linked genes are reduced in Parkinson's disease patients. Front Cell Neurosci. 2019;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li J, Sun Y, Chen J. Transcriptome sequencing in a 6‐hydroxydopamine rat model of Parkinson's disease. Genes Genet Syst. 2019;94(2):61‐69. [DOI] [PubMed] [Google Scholar]
- 77. Jiao F, Wang Q, Zhang P, Bu L, Yan J, Tian B. Expression signatures of long non‐coding RNA in the substantia nigra of pre‐symptomatic mouse model of Parkinson's disease. Behav Brain Res. 2017;331:123‐130. [DOI] [PubMed] [Google Scholar]
- 78. Yan W, Chen ZY, Chen JQ, Chen HM. LncRNA NEAT1 promotes autophagy in MPTP‐induced Parkinson's disease through stabilizing PINK1 protein. Biochem Biophys Res Commun. 2018;496(4):1019‐1024. [DOI] [PubMed] [Google Scholar]
- 79. Peng T, Liu X, Wang J, et al. Long noncoding RNA HAGLROS regulates apoptosis and autophagy in Parkinson's disease via regulating miR‐100/ATG10 axis and PI3K/Akt/mTOR pathway activation. Artif Cells Nanomed Biotechnol. 2019;47(1):2764‐2774. [DOI] [PubMed] [Google Scholar]
- 80. Riva P, Ratti A, Venturin M. The long non‐coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res. 2016;13(11):1219‐1231. [DOI] [PubMed] [Google Scholar]
- 81. Kudriaeva AA, Belogurov AA. Proteasome: A nanomachinery of creative destruction. Biochemistry (Mosc). 2019;84(Suppl. 1):S159‐S192. [DOI] [PubMed] [Google Scholar]
- 82. Carrieri C, Cimatti L, Biagioli M, et al. Long non‐coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454‐457. [DOI] [PubMed] [Google Scholar]
- 83. Lin Q, Hou S, Dai Y, Jiang N, Lin Y. LncRNA HOTAIR targets miR‐126‐5p to promote the progression of Parkinson's disease through RAB3IP. Biol Chem. 2019;400(9):1217‐1228. [DOI] [PubMed] [Google Scholar]
- 84. Chen Y, Lian YJ, Ma YQ, Wu CJ, Zheng YK, Xie NC. LncRNA SNHG1 promotes alpha‐synuclein aggregation and toxicity by targeting miR‐15b‐5p to activate SIAH1 in human neuroblastoma SH‐SY5Y cells. Neurotoxicology. 2018;68:212‐221. [DOI] [PubMed] [Google Scholar]
- 85. Zhang QS, Wang ZH, Zhang JL, Duan YL, Li GF, Zheng DL. Beta‐asarone protects against MPTP‐induced Parkinson's disease via regulating long non‐coding RNA MALAT1 and inhibiting alpha‐synuclein protein expression. Biomed Pharmacother. 2016;83:153‐159. [DOI] [PubMed] [Google Scholar]
- 86. Li XZ, Zhang SN, Lu F, Liu SM. Microarray expression analysis for the paradoxical roles of Acanthopanax senticosus harms in treating alpha‐synucleinopathies. Phytother Res. 2016;30(2):243‐252. [DOI] [PubMed] [Google Scholar]
- 87. Jenner P. Oxidative stress in Parkinson's disease. Ann Neurol. 2003;53(Suppl. 3):S26‐S36, S36–S38. [DOI] [PubMed] [Google Scholar]
- 88. Greenamyre JT, Sherer TB, Betarbet R, Panov AV. Complex I and Parkinson's disease. IUBMB Life. 2001;52(3–5):135‐141. [DOI] [PubMed] [Google Scholar]
- 89. Brenner‐Lavie H, Klein E, Zuk R, Gazawi H, Ljubuncic P, Ben‐Shachar D. Dopamine modulates mitochondrial function in viable SH‐SY5Y cells possibly via its interaction with complex I: relevance to dopamine pathology in schizophrenia. Biochim Biophys Acta. 2008;1777(2):173‐185. [DOI] [PubMed] [Google Scholar]
- 90. Xu H, Liu X, Xia J, et al. Activation of NMDA receptors mediated iron accumulation via modulating iron transporters in Parkinson's disease. Faseb J. 2018;32:6100–6111. [DOI] [PubMed] [Google Scholar]
- 91. Li T, Zhang W, Kang X, et al. Salidroside protects dopaminergic neurons by regulating the mitochondrial MEF2D‐ND6 pathway in the MPTP/MPP(+) ‐induced model of Parkinson's disease. J Neurochem. 2019. [DOI] [PubMed] [Google Scholar]
- 92. Kopp F, Elguindy MM, Yalvac ME, et al. PUMILIO hyperactivity drives premature aging of Norad‐deficient mice. Elife. 2019;8:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Simchovitz A, Hanan M, Niederhoffer N, et al. NEAT1 is overexpressed in Parkinson's disease substantia nigra and confers drug‐inducible neuroprotection from oxidative stress. FASEB J. 2019;33(10):11223‐11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cai L, Tu LI, Li T, et al. Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson's disease through the inhibition of the PI3K/Akt signaling pathway. Int Immunopharmacol. 2019;75:105734. [DOI] [PubMed] [Google Scholar]
- 95. Ding XM, Zhao LJ, Qiao HY, Wu SL, Wang XH. Long non‐coding RNA‐p21 regulates MPP(+)‐induced neuronal injury by targeting miR‐625 and derepressing TRPM2 in SH‐SY5Y cells. Chem Biol Interact. 2019;307:73‐81. [DOI] [PubMed] [Google Scholar]
- 96. Wang L, Yang H, Wang Q, et al. Paraquat and MPTP induce alteration in the expression profile of long noncoding RNAs in the substantia nigra of mice: Role of the transcription factor Nrf2. Toxicol Lett. 2018;291:11‐28. [DOI] [PubMed] [Google Scholar]
- 97. Roberts RF, Tang MY, Fon EA, Durcan TM. Defending the mitochondria: the pathways of mitophagy and mitochondrial‐derived vesicles. Int J Biochem Cell Biol. 2016;79:427‐436. [DOI] [PubMed] [Google Scholar]
- 98. Chen Q, Huang X, Li R. lncRNA MALAT1/miR‐205‐5p axis regulates MPP(+)‐induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am J Transl Res. 2018;10(2):563‐572. [PMC free article] [PubMed] [Google Scholar]
- 99. Lu M, Sun WL, Shen J, et al. LncRNA‐UCA1 promotes PD development by upregulating SNCA. Eur Rev Med Pharmacol Sci. 2018;22(22):7908‐7915. [DOI] [PubMed] [Google Scholar]
- 100. Xie SP, Zhou F, Li J, Duan SJ. NEAT1 regulates MPP(+)‐induced neuronal injury by targeting miR‐124 in neuroblastoma cells. Neurosci Lett. 2019;708:134340. [DOI] [PubMed] [Google Scholar]
- 101. Cai M, Wang YW, Xu SH, et al. Regulatory effects of the long noncoding RNA RP11543N12.1 and microRNA3243p axis on the neuronal apoptosis induced by the inflammatory reactions of microglia. Int J Mol Med. 2018;42(3):1741‐1755. [DOI] [PubMed] [Google Scholar]
- 102. Chi LM, Wang LP, Jiao D. Identification of differentially expressed genes and long noncoding RNAs associated with Parkinson's disease. Parkinsons Dis. 2019;2019:6078251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yi J, Chen B, Yao X, Lei Y, Ou F, Huang F. Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer's disease through inactivating the PI3K/Akt signaling pathway. J Cell Biochem. 2019;120(10):18053‐18065. [DOI] [PubMed] [Google Scholar]
- 104. Wang H, Liao S, Li H, Chen Y, Yu J. Long non‐coding RNA TUG1 Sponges Mir‐145a‐5p to regulate microglial polarization after oxygen‐glucose deprivation. Front Mol Neurosci. 2019;12:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bannon MJ, Savonen CL, Jia H, et al. Identification of long noncoding RNAs dysregulated in the midbrain of human cocaine abusers. J Neurochem. 2015;135(1):50‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ip JY, Sone M, Nashiki C, et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci Rep. 2016;6:27204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bohnsack JP, Teppen T, Kyzar EJ, Dzitoyeva S, Pandey SC. The lncRNA BDNF‐AS is an epigenetic regulator in the human amygdala in early onset alcohol use disorders. Transl Psychiatry. 2019;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li E‐Y, Zhao P‐J, Jian J, et al. LncRNA MIAT overexpression reduced neuron apoptosis in a neonatal rat model of hypoxic‐ischemic injury through miR‐211/GDNF. Cell Cycle. 2019;18(2):156‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zucchelli S, Fasolo F, Russo R, et al. SINEUPs are modular antisense long non‐coding RNAs that increase synthesis of target proteins in cells. Front Cell Neurosci. 2015;9:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Carrieri C, Forrest ARR, Santoro C, et al. Expression analysis of the long non‐coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells' differentiation in vitro and in neurochemical models of Parkinson's disease. Front Cell Neurosci. 2015;9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Márki S, Göblös A, Szlávicz E, et al. The rs13388259 intergenic polymorphism in the genomic context of the BCYRN1 gene is associated with Parkinson's disease in the Hungarian population. Parkinsons Dis. 2018;2018:9351598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang S, Zhang X, Guo Y, Rong H, Liu T. The long noncoding RNA HOTAIR promotes Parkinson's disease by upregulating LRRK2 expression. Oncotarget. 2017;8(15):24449‐24456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Arisi I, D'Onofrio M, Brandi R, et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer's disease: mining of microarray data by logic classification and feature selection. J Alzheimers Dis. 2011;24(4):721‐738. [DOI] [PubMed] [Google Scholar]


