Abstract
Using the taxono-genomics concept, we describe here a strictly anaerobic Gram-positive bacillus. This strain was isolated from the stool sample of a 50-year-old healthy Bedouin woman. The 16S rRNA gene sequence analysis and the whole-genome sequencing showed that this isolate belonged to the genus Gordonibacter in the family Eggerthellaceae. Based on these criteria, we propose the creation of Gordonibacter massiliensis sp. nov., strain Marseille-P2775T (= CSUR P2775).
Keywords: Culturomics, Gordonibacter massiliensissp. nov., human gut, stool, taxono-genomics
Introduction
Members of the genus Gordonibacter are Gram-positive bacteria belonging to the recent family Eggerthellaceae [1]. These species are part of the human gut microbiota and have the capacity to metabolize polyphenols from diet into bioavailable metabolites known as urolithin [2,3]. During the last decades, culturomics studies have brought insight into the human microbiota, which has led to the discovery of previously uncultured bacteria [4,5]. Culturomics, including different culture conditions, is completed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) identification and sequencing of the 16S rRNA gene, in order to explore the microbial diversity of the human gut [6,7]. This new bacterial species was described using a combination of genotypic and phenotypic characteristics according to the previously reported taxono-genomics approach [8,9].
Herein, we give details of the isolation and taxono-genomics characters of strain Marseille-P2775T, which is the type strain of Gordonibacter massiliensis sp. nov.
Isolation and growth conditions
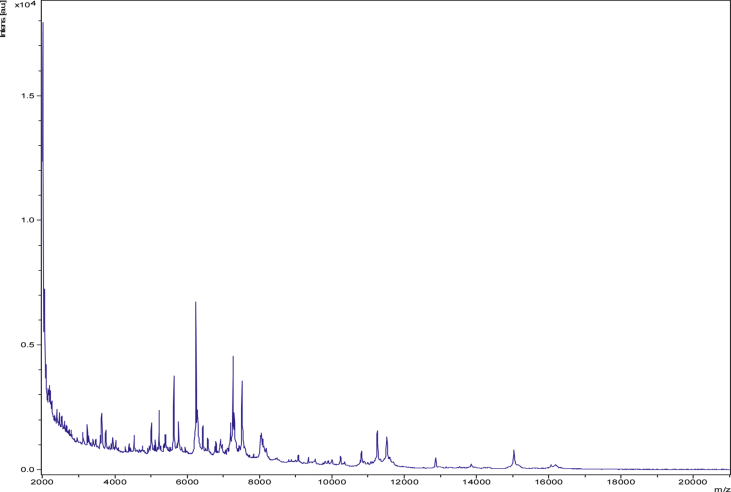
The strain was isolated in 2016 from the stool sample of a 50-year-old healthy Bedouin woman living in the Jazan region of Saudi Arabia. This study was performed in Franceafter approval from the ethics committee of the King Abdulaziz University (Saudi Arabia) and the local ethics committee of the IFR48 (Marseille, France) under numbers 014-CEGMR-2-ETH-P and 09-022, respectively. Isolation and growth conditions of strain were performed as previously described [10]. The initial growth of strain Marseille-P2775 was obtained after 2 days of incubation in a Colombia agar supplemented with 5% sheep's blood (COS, bioMérieux, Marcy l’Étoile, France) under strict anaerobic conditions at 37°C. Identification of this bacterial strain was attempted using MALDI-TOF mass spectrometry. The screening was performed on a Microflex LT spectrometer (Bruker Daltonics, Bremen, Germany) as previously reported [11]. The spectra obtained were saved into MALDI Biotyper 3.0 software (Bruker Daltonics) and analysed against the main spectra of the bacteria included in the local URMS database (https://www.mediterranee-infection.com/urms-data-base) (Fig. 1).
Fig. 1.
MALDI-TOF MS reference spectrum generated from the Biotyper 3.0 software. Spectra from 12 individual colonies were compared with the aim to obtain consensual spectrum.
Strain identification
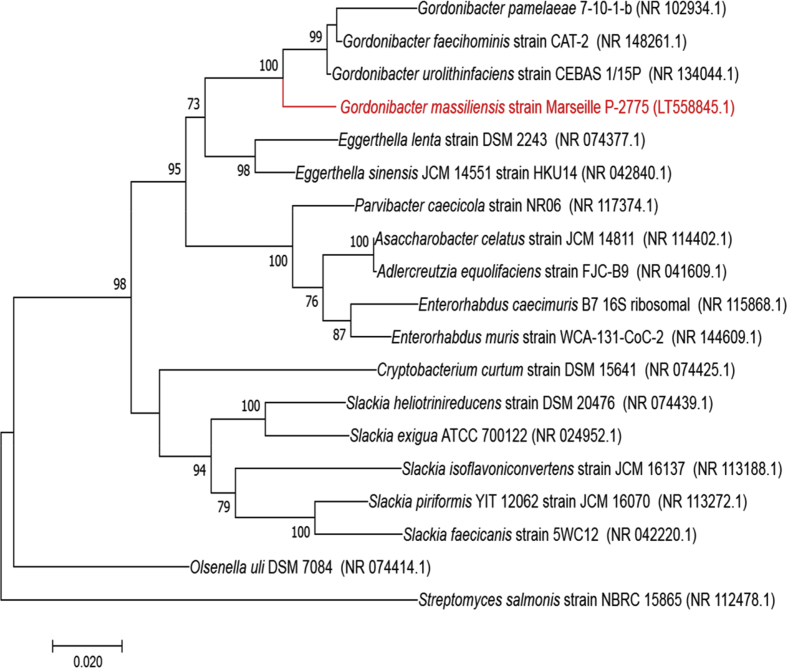
To classify the strain Marseille-P2775, its 16S rRNA gene was amplified using the fD1 and rP2 primer pair (Eurogentec, Angers, France) and sequenced using the Big Dye® Terminator v1.1 Cycle Sequencing Kit and 3500xLGenetic Analyser capillary sequencer (Thermofisher, Saint-Aubin, France) [12,13] as described previously. The 16S rRNA nucleotide sequences were assembled and corrected using CodonCode Aligner software (http://www.codoncode.com). The 16S rRNA gene sequence analysis of strain Marseille-P2775 showed 97.19% identity with Gordonibacter urolithinfaciens strain CEBAS 1/15P (GenBank accession number: NR134044), the phylogenetically closest species with a standing in nomenclature (Fig. 2). This value was <98.7% of similarity, the threshold above which a strain is considered a new species [14].
Fig. 2.
Phylogenetic tree showing the position of Gordonibacter massiliensis strain Marseille-P2775T and other phylogenetically close neighbours. The respective GenBank accession numbers for 16S rRNA genes are indicated in parenthesis. Sequences were aligned using Muscle v7.0.26 with default parameters and phylogenetic inferences were obtained using the maximum likelihood method with MEGA 7 software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 1000 times to generate a majority consensus tree. Only bootstrap values > 70% were retained. The scale bar indicates a 5% nucleotide sequence divergence.
Phenotypic characteristics
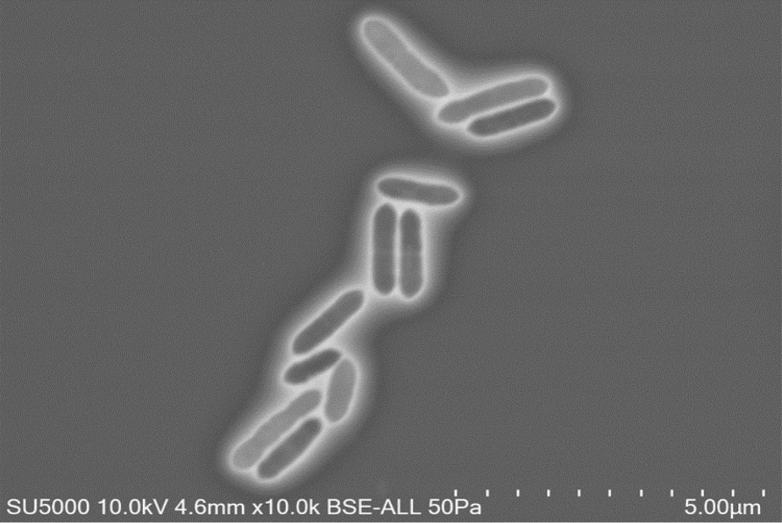
Growing on Columbia blood agar, colonies of strain Marseille-P2775 appeared beige with a mean diameter of 1 mm. Bacterial cells were Gram-positive, strictly anaerobic short-rod bacilli with a mean length of 1.2 μm and 0.5 μm in diameter (Fig. 3). Strain Marseille-P2775 was motile, non-haemolytic and non-spore-forming. It presented catalase-positive and oxidase-negative activities. Carbohydrate metabolism and enzymatic characteristics of the strain Marseille-P2775 were tested under strict anaerobic conditions at 37°C using API 50 CH and ZYM, respectively (Table 1). A comparative study of the differential characteristics of this strain with other closely related species is shown in Table 2. Cellular fatty acid methyl ester (FAME) analysis was performed as previously described [15,16]. The major fatty acids were 9-octadecenoic acid (41%) and hexadecanoic acid (24%). Several branched structures were also described with lower abundances (Table 3).
Fig. 3.
Scanning electron micrograph of Gordonibacter massiliensis strain Marseille-P2775T obtained from TM4000 microscope. Scale bar and acquisition settings are shown on the picture.
Table 1.
Biochemical tests performed on strain Marseille-P2775 using API strips 50 CH and ZYM
| API 50 CH |
API ZYM |
||||
|---|---|---|---|---|---|
| Tests | Results | Tests | Results | Tests | Results |
| Control | − | Esculin ferric citrate | + | Control | − |
| Glycerol | + | Salicin | + | Alkaline phosphatase | − |
| Erythritol | − | d-cellobiose | − | Esterase (C4) | + |
| d-arabinose | − | d-maltose | + | Esterase lipase (C8) | + |
| l-arabinose | + | d-lactose | + | Lipase (C14) | − |
| d-ribose | − | d-melibiose | − | Leucine arylamidase | + |
| d-xylose | + | d-saccharose | + | Valine arylamidase | − |
| l-xylose | − | d-trehalose | + | Cystine arylamidase | + |
| d-adonitol | − | Inulin | − | Trypsin | − |
| Methyl β-d-xylopyranoside | − | d-melezitose | + | α-chymotrypsin | − |
| d-galactose | + | d-raffinose | − | Acid phosphatase | + |
| d-glucose | + | Amidon | − | Naphthol-AS-BI-phosphohydrolase | + |
| d-fructose | + | Glycogen | − | α-galactosidase | − |
| d-mannose | − | Xylitol | − | β-galactosidase | + |
| l-sorbose | − | Gentiobiose | + | β-glucuronidase | − |
| l-rhamnose | − | d-tyranose | + | α-glucosidase | − |
| Dulcitol | − | d-lyxose | − | β-glucosidase | − |
| Inositol | − | d-tagatose | − | N-acetyl-β-glucosaminidase | + |
| d-mannitol | + | d-fucose | − | α-mannosidase | − |
| d-sorbitol | + | l-fucose | − | α-fucosidase | − |
| Methyl α-d-mannopyranoside | − | d-arabitol | − | ||
| Methyl α-d-glucopyranoside | − | l-arabitol | − | ||
| N-acetyl-glucosamine | + | Potassium gluconate | − | ||
| Amygdalin | − | Potassium 2-ketogluconate | − | ||
| Arbutin | − | Potassium 5-ketogluconate | − | ||
Table 2.
Differential characteristics of Gordonibacter massiliensis strain Marseille-P2775, Gordonibacter urolithinfaciens, Gordonibacter pamelaeae, Eggerthella lenta and Eggerthella sinensis
| Properties | Gordonibacter massiliensis | Gordonibacter urolithinfaciens | Gordonibacter pamelaeae | Eggerthella lenta | Eggerthella sinensis |
|---|---|---|---|---|---|
| Cell diameter (μm) | 0.5 | 0.4–0.6 | 0.5–0.6 | 0.5 | 0.5 |
| Oxygen requirement | Anaerobic | Anaerobic | Anaerobic | Anaerobic | Anaerobic |
| Shape | Coccobacilli | Coccobacilli | Coccobacilli | Coccobacilli | Coccobacilli |
| Gram stain | + | + | + | + | + |
| Motility | + | + | + | − | − |
| Sporulation | − | − | − | − | − |
| Production of: | |||||
| Alkaline phosphatase | − | − | − | − | − |
| Catalase | + | + | + | Variable | + |
| Oxidase | − | NA | NA | NA | + |
| β-galactosidase | + | + | NA | − | − |
| N-acetyl-glucosamine | + | NA | NA | − | − |
| Acid from: | |||||
| l-arabinose | + | − | − | − | − |
| Mannose | − | − | − | + | − |
| d-fucose | − | + | NA | NA | NA |
| l-fucose | − | + | − | − | + |
| d-glucose | + | − | − | − | − |
| Trehalose | − | − | − | + | − |
| d-fructose | + | + | − | NA | NA |
| G + C content (%) | 65.1 | 66.4 | 66.4 | 62 | 65.6 |
| Habitat | Human gut | Human gut | Human gut | Human gut | Human gut |
+, positive result; −, negative result; NA, data not available.
Table 3.
Cellular fatty acid composition (%) of strain Marseille-P2775T compared with other Gordonibacter species
| Fatty acids | Name | Gordonibacter massiliensis | Gordonibacter pamelaeae | Gordonibacter faecihominis |
|---|---|---|---|---|
| 12:00 | Dodecanoic acid | TR | 2.70 | ND |
| 13:00 | Tridecanoic acid | TR | 4.20 | ND |
| 13:0 anteiso | 10-methyl-Dodecanoic acid | 1.0 | TR | ND |
| 13:0 iso | 11-methyl-Dodecanoic acid | 2.3 | 1.09 | ND |
| 14:00 | Tetradecanoic acid | 8.5 | 8.71 | 8.9 |
| 14:0 iso | 12-methyl-Tridecanoic acid | 2.6 | 6.78 | 8.4 |
| 15:00 | Pentadecanoic acid | 1.0 | TR | ND |
| 15:0 anteiso | 12-methyl-tetradecanoic acid | 6.6 | 19.79 | 8.4 |
| 15:0 iso | 13-methyl-tetradecanoic acid | 1.9 | 2.96 | 4.3 |
| 16:00 | Hexadecanoic acid | 23.9 | 2.42 | 10.5 |
| 16:1n7 | 9-Hexadecenoic acid | TR | 1.69 | 3.6 |
| 17:00 | Heptadecanoic acid | TR | 0.68 | ND |
| 17:01 | Heptadecenoic acid | TR | ND | ND |
| 18:00 | Octadecanoic acid | 7.2 | TR | TR |
| 18:1n9 | 9-Octadecenoic acid | 40.8 | 3.65 | ND |
| 18:2n6 | 9,12-Octadecadienoic acid | 1.5 | TR | ND |
TR, trace amounts <1%; ND, not detected.
Genome sequencing
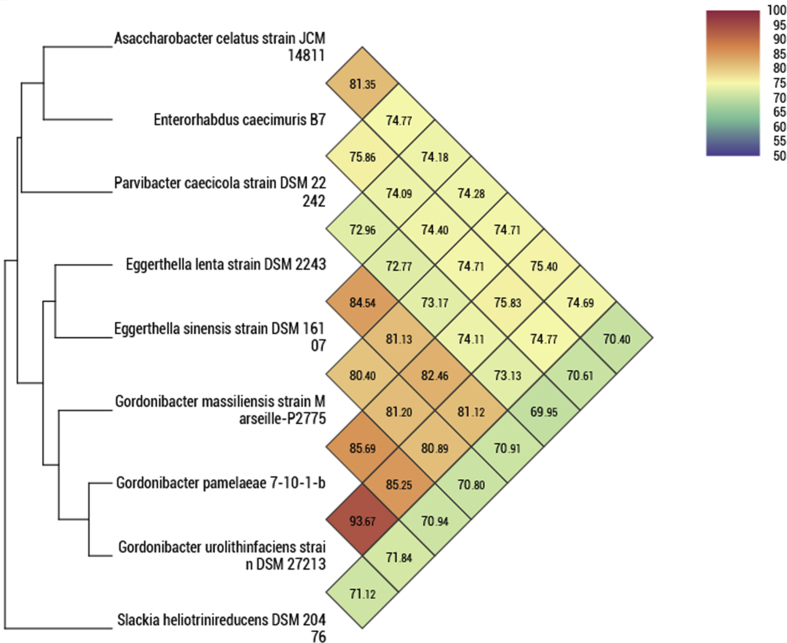
The DNA genomic extraction was performed using the EZ1 biorobot and the EZ1 DNA tissue kit. Genomic DNA (gDNA) was quantified by a Qubit assay. The sequencing was performed using MiSeq technology (Illumina, San Diego, CA, USA) with the Paired-End (Illumina). The assembly was performed with Spades software [17]. The reads with low quality were trimmed using Trimmomatic software [18]. GapCloser software [19] was used to reduce the assembly gap. Scaffolds < 800 bp and scaffolds with a depth value < 25% of the mean depth were removed. The total length of the G. massiliensis genome is 3.9 megabases encompassing 1 contig with a G + C content of 65.1 mol%. The predicted genes analysis reported 3248 genes. The degree of genomic similarity of strain Marseille-P2775 with closely related species was estimated using the OrthoANI software [20]. OrthoANI values ranged from 69.95% between Parvibacter caecicola strain DSM 22242 and Slackia heliotrinireducens strain DSM 20476 to 93.67% between Gordonibacter urolithinfaciens strain DSM 27213 and Gordonibacter pamelaeae DSM 19378. When G. massiliensis strain Marseille-P2775T was compared with these closely related species, we found values ranging from 70.94% with Slackia heliotrinireducens strain DSM 20476 to 85.69% with G. pamelaeae strain DSM 19378 (Fig. 4).
Fig. 4.
Heatmap generated with OrthoANI values calculated using the OAT software between Gordonibacter massiliensis strain Marseille-P2775T and other closely related species with standing in nomenclature.
Description of Gordonibacter massiliensis sp. nov.
Gordonibacter massiliensis (mas.si.li.en'sis. L. fem. adj., from massiliensis of Massilia, the Latin name of Marseille where the strain was first cultivated) is a Gram-positive, motile, non-spore-forming and obligate anaerobic coccobacillus. Bacterial cells had a mean diameter of 0.5 μm. Colonies appear beige on blood agar after 48 h of incubation at 37°C in an anaerobic environment. Major cellular fatty acids were 9-octadecenoic acid (41%) and hexadecanoic acid (24%). Catalase is positive but oxidase is negative. Utilization of l-arabinose and d-glucose distinguishes strain Marseille-P2775 from among the closest bacterial species. Also mannose, raffinose, fucose and β-glucosidase are not produced. The strain Marseille-P2775 is isolated from the stool sample of healthy woman living in Saudi Arabia. The G + C content of the genomic DNA is 65.1 mol%.
Conclusion
Based on phenotypic, genomic and phylogenetic analyses, we formally propose the creation of Gordonibacter massiliensis sp. nov., represented here by the strain Marseille-2775. This strain was isolated from a stool sample of a 50-year-old healthy Bedouin woman living in the Jazan region of Saudi Arabia.
Nucleotide sequence accession numbers
The 16S rRNA and genome sequences were deposited in GenBank under accession numbers LT558845 and LT827128, respectively.
Deposit in culture collections
Strain Marseille-P2775T was deposited in the Collection de Souches de l’Unité des Rickettsies under the following number: CSUR P2775.
Conflicts of interest
None to declare.
Funding sources
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the programme Investissements d'avenir, reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
Acknowledgements
The authors thank Catherine Robert for sequencing the genome and Aurelia Caputo for submitting the genomic sequence to GenBank.
References
- 1.Gupta R.S., Chen W.J., Adeolu M., Chai Y. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int J Syst Evol Microbiol. 2013;63:3379–3397. doi: 10.1099/ijs.0.048371-0. [DOI] [PubMed] [Google Scholar]
- 2.Selma M.V., Tomás-Barberán F.A., Beltrán D., García-Villalba R., Espín J.C. Gordonibacter urolithinfaciens sp. nov., a urolithin-producing bacterium isolated from the human gut. Int J Syst Evol Microbiol. 2014;64:2346–2352. doi: 10.1099/ijs.0.055095-0. [DOI] [PubMed] [Google Scholar]
- 3.Selma M.V., Romo-Vaquero M., García-Villalba R., González-Sarrías A., Tomás-Barberán F.A., Espín J.C. The human gut microbial ecology associated with overweight and obesity determines ellagic acid metabolism. Food Funct. 2016;7:1769–1774. doi: 10.1039/c5fo01100k. [DOI] [PubMed] [Google Scholar]
- 4.Lagier J.-C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Lagier J.-C., Dubourg G., Million M., Cadoret F., Bilen M., Fenollar F. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 6.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 7.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramasamy D., Mishra A.K., Lagier J.-C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 9.Fournier P.E., Lagier J.-C., Dubourg G., Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015;36:73–78. doi: 10.1016/j.anaerobe.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Ngom I.I., Mailhe M., Ricaboni D., Vitton V., Benezech A., Khelaifia S. Noncontiguous finished genome sequence and description of Mediterranea massiliensis gen. nov., sp. nov., a new member of the Bacteroidaceae family isolated from human colon. New Microbe New Infect. 2017;21:105–116. doi: 10.1016/j.nmni.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fall B., Lo C.I., Samb-Ba B., Perrot N., Diawara S., Gueye M.W. The ongoing revolution of MALDI-TOF mass spectrometry for microbiology reaches tropical Africa. Am J Trop Med Hyg. 2015;92:641–647. doi: 10.4269/ajtmh.14-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morel A.-S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.-P. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis. 2015;34:561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 13.Lo C.I., Padhamanabhan R., Fall B., Sambe-Ba B., Mediannikov O., Nguyen T.T. Noncontiguous finished genome sequence and description of Necropsobacter massiliensis sp. nov. New Microbe New Infect. 2015;8:41–50. doi: 10.1016/j.nmni.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier-Kolthoff J.P., Klenk H.P., Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 15.Sasser M. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME) 2006. http://midi-inc.com/pdf/MIS_Technote_101.pdf
- 16.Dione N., Sankar S.A., Lagier J.-C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbe New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu G.C., Xu T.J., Zhu R., Zhang Y., Li S.Q., Wang H.W. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience. 2019;8(1) doi: 10.1093/gigascience/giy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee I., Ouk Kim Y., Park S.-C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]