Abstract
The overwhelming list of new bacterial genomes becoming available on a daily basis makes accurate genome annotation an essential step that ultimately determines the relevance of thousands of genomes stored in public databanks. The MicroScope platform (http://www.genoscope.cns.fr/agc/microscope) is an integrative resource that supports systematic and efficient revision of microbial genome annotation, data management and comparative analysis. Starting from the results of our syntactic, functional and relational annotation pipelines, MicroScope provides an integrated environment for the expert annotation and comparative analysis of prokaryotic genomes. It combines tools and graphical interfaces to analyze genomes and to perform the manual curation of gene function in a comparative genomics and metabolic context. In this article, we describe the free-of-charge MicroScope services for the annotation and analysis of microbial (meta)genomes, transcriptomic and re-sequencing data. Then, the functionalities of the platform are presented in a way providing practical guidance and help to the nonspecialists in bioinformatics. Newly integrated analysis tools (i.e. prediction of virulence and resistance genes in bacterial genomes) and original method recently developed (the pan-genome graph representation) are also described. Integrated environments such as MicroScope clearly contribute, through the user community, to help maintaining accurate resources.
Keywords: microbial genome annotation system, gene function curation, comparative genomics, transcriptomics, variant detection, metabolic networks
Introduction
Large-scale genome sequencing and the increasingly massive use of high-throughput approaches produce a vast amount of new information that completely transforms our understanding of thousands of species. However, despite the development of powerful bioinformatics approaches, full interpretation of the content of these genomes remains a difficult task. To address this challenge, several integrated environments that combine and standardize information from a variety of sources and apply uniform (re-)annotation techniques have been developed (i.e. EnsemblGenomes [1], IMG [2], PATRIC [3]). In the context of the French National Sequencing Center (CEA/DRF/Genoscope), we have developed the MicroScope platform, which is a software environment for management, annotation, comparative analysis and visualization of microbial genomes. Published for the first time in 2006 [4], the platform has been under continuous development within the LABGeM group at CEA, and its capacities are now extensive [5–7]. MicroScope serves different used cases in bioinformatics:
It supports the integration of newly sequenced or already available prokaryotic genomes through the offer of a free-of-charge service to the scientific community [genome annotation, RNA sequencing (RNA-seq) and variant analyses].
It performs computational inferences including prediction of metabolic pathways, prediction of resistome and virulome, which can be used for genome analysis.
It provides tools for (comparative) analyses and visualization of prokaryotic genomes.
It supports collaborative expert annotation processes through the use of specific curation tools and graphical interfaces.
The present article provides a comprehensive description of MicroScope from the point of view of the end users. We start with the major objectives for which the platform was designed, and we give an overview of the main categories of MicroScope users and projects. Then we explain how to submit data and interact with the MicroScope team, and how to explore the annotated data, use the various analysis tools and perform expert annotation of gene functions. Technical details on the architecture of the system are given in the last section of this review. Where possible, earlier publications that provide more details are referenced. We conclude by one of the ongoing work that lead to a promising representation of the pan-genome of thousands of prokaryotic genomes.
Who is using MicroScope and for what purposes?
In the era of high-throughput sequencing technologies, a vast majority of genome sequences receive only automatic annotation, mainly based on sequence similarity, that can give spurious results [8]. Indeed, manual expertise of gene functions is a time-consuming and expensive process, but it undoubtedly adds great value to resources. In knowledge bases such as UniProtKB [9], curation efforts remain restricted to large and widespread protein families, and these resources cannot replace expert curations made by specialized biologists in community systems, such as SEED [10], IMG [2] and MicroScope. Our integrated platform supports systematic and efficient revision of microbial genome annotation, data management together with comparative genomics and metabolic analyses [4–7]. The resource provides data from completed and ongoing genome projects together with post-genomic experiments (i.e. transcriptomics; re-sequencing of evolved strains; mutant collections) allowing users to improve the understanding of gene functions.
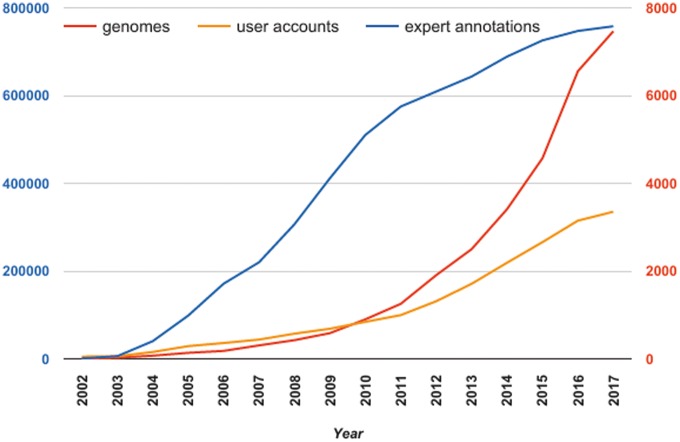
In comparison with other similar systems, MicroScope enables curation in a rich comparative genomic context and is mainly focused on (re-)annotation projects, which are built in close collaboration with microbiologists working on reference species. Indeed, MicroScope was initially dedicated to the annotation and analysis of Acinetobacter baylyi APD1 [11] and to biologists who do not have the required computing infrastructure to perform efficient annotation and analyses of newly sequenced bacterial genomes. Our system rapidly became a ‘service’ free of charge to the scientific community at large. From <400 user accounts in 2006, MicroScope counts >3300 personal accounts at present time (Figure 1). The number of registered users has doubled since 2013, and the platform has even widened its international popularity with 64% of accounts outside France. Many international projects are conducted through the platform involving users from distant geographic areas [7]. Although authentication is not required to navigate in MicroScope, it allows users to annotate genes and save data on their personal session. On average per month, we count 360 active accounts (i.e. the user logged in at least once in the month) and 2200 authentications among ∼1700 monthly unique visitors.
Figure 1.
Evolution of the number of integrated genomes, user accounts and expert annotations stored in MicroScope since 2002. Red scale on the right refers to the number of integrated genomes (red curve) and to the number of user accounts (orange curve). Blue scale on the left refers to the cumulated number of expert annotations.
The platform has been used to perform a complete expert annotation of several reference species such as Escherichia coli [12], Bacillus subtilis 128 [13, 14] and Pseudomonas putida KT2440 [15]. In addition, important pathogens and environmental species have also been extensively curated. The MicroScope system is now also used for variant analysis of re-sequenced bacterial strains (for example, in the context of bacterial evolution experiments) and for the analysis of transcriptomic experiments using RNA-seq sequencing data [6, 7], and finally, the platform is also (and in some cases, exclusively) used for the set of analysis tools pertaining to microbial genomics and metabolism, which have been integrated and made available through the MicroScope Web interface (see next sections). Indeed, the MicroScope platform has been cited 690 times since 13 years.
As shown in Figure 1, although the number of MicroScope users having a personal account has increased significantly since 2011, the number of expert annotations made each year is clearly decreasing, reaching only 21 600 in 2016 (we registered >100 000 expert annotations in 2009). Past year, about one-tenth of the users performed curation of gene function and a third of them made >100 expert annotations. Obviously, with the number of prokaryotic genomes being sequenced today, the time-consuming task of expert annotation is totally unacceptable. This is the reason why our major efforts have been focused on the development of several key functionalities allowing to ease the expert annotation process and to notably improve the final annotation quality of the analyzed genomes, at least, for gene functions of interest.
An annotation service to researchers in microbiology
Interface for user data integration
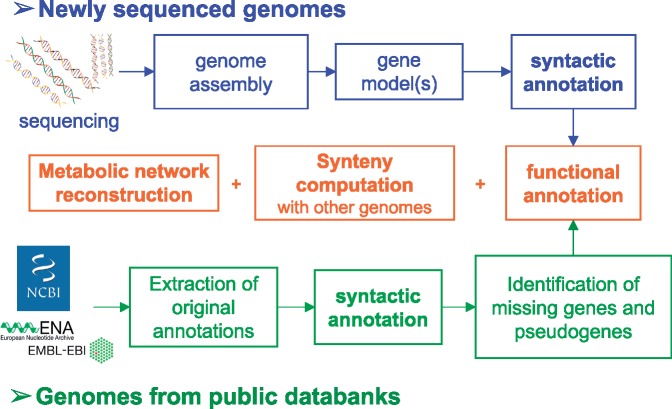
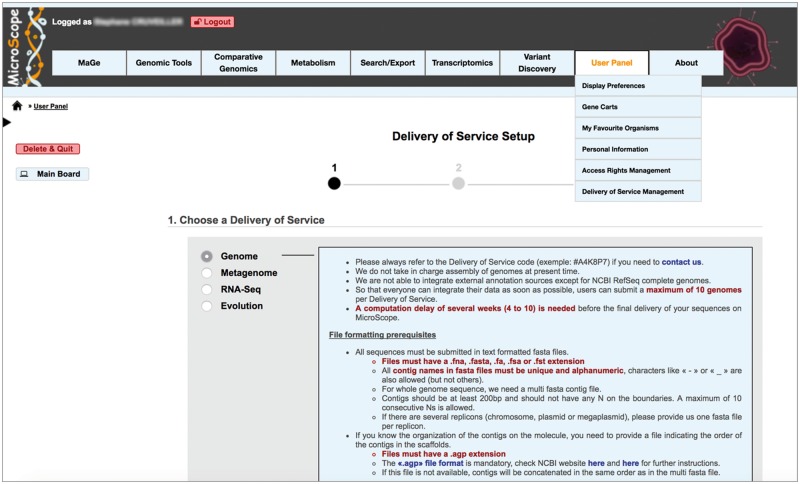
Integration and analysis of genomic data into MicroScope are open and free of charge for the worldwide community of microbiologists. To standardize and make user submission fully automated, we have developed a dedicated Web interface (https://www.genoscope.cns.fr/agc/microscope/about/services.php). The service is mainly used for the annotation of microbial genomes: both newly sequenced genomes (which will remain private till the genome publication and/or their submission to public databanks) and, for comparative analysis purpose, public prokaryotic genomes (Figure 2). Moreover, three other types of services are provided for the integration of (i) genome assemblies (bins) from metagenomic samples (ii) RNA-seq data for quantitative transcriptomics and (iii) DNA sequencing (DNA-seq) data to identify genomic variations in evolved strains (Figure 3). To ease data integration and comparative studies, standardization of contextual data about genome sequences is essential. For metagenomes, we have added a dedicated form that follows the MIMS specifications (minimum information about a metagenome sequence [16]). When submitting assembled metagenomic data in Microscope, the users are invited to select the type of environment (e.g. soil; air; water; human-associated; plant-associated) and to complete the associated fields (e.g. collection date, environment biome, geographic location, etc.). These fields are dynamically loaded and displayed on metagenome type selection. Indeed, the MicroScope database model is flexible enough to store predefined descriptors, like MIMS, or the ones defined by users.
Figure 2.
Annotation pipelines for the analysis of newly sequenced genomes and genomes already annotated in public databanks.
Figure 3.
Submission of genomic data into the MicroScope platform. Four types of services are provided for the integration of (i) newly sequenced or publicly available genomes (Genome), (ii) genome assemblies/bins from metagenomic samples (Metagenome), (iii) RNA-seq data for quantitative transcriptomics (RNA-Seq), (iv) DNA-seq data to identify genomic variations in evolved strains (Evolution). Following the three main steps of the procedure, the user is invited to complete the requested metadata to describe sequencing, genomes and experimental properties, to upload FASTA (genome assemblies) or FASTQ (RNA-seq or DNA-seq reads) files and, finally, to approve the terms of services. Users are then informed by an e-mail about the progress of their integration request.
At present time, an average of eight genomes a day are requested for integration in the platform (this includes bins from metagenomic samples). The resource contains data for >7400 microbial genomes of which ∼3100 are publicly available. In addition, 607 RNA-seq runs and 756 runs corresponding to the re-sequencing of evolved strains have also been requested for integration into MicroScope.
Running the annotation pipelines
About 25 analyses workflows include most of the currently used annotation software, plus some in-house tools and/or annotation strategies (Table 1). The newly sequenced (meta)genomes, generally submitted in several contigs and organized (or not) on the final chromosome(s), are first analyzed by the syntactic annotation pipeline to identify protein genes, transfer RNA (tRNA), ribosomal RNA (rRNA), noncoding RNA (ncRNA) and repeats (Figure 2, Table 1). For a more accurate prediction of small genes and/or atypical gene composition, we have developed a strategy to first construct appropriate gene models that takes into account the codon usage of the studied organism. These models are then used in the core of the AMIGene program [17]. Starting with the set of genomic objects identified during the syntactic annotation process, the next step is to infer biological functions of the predicted genes. Our functional annotation pipeline includes sequence similarity searches tools using generalist (i.e. UniProtKB/Swiss-Prot) or specialized (i.e. Interpro, FIGFAM, etc.) databases (Table 1). Results obtained with high-quality manually curated protein sequence data sets (i.e. Swiss-Prot, E. coli K-12, B. subtilis 168 MicroScope-curated genes) are first considered in the final functional automatic annotation procedure. This procedure also takes into account the results obtained from the computation of synteny groups with complete reference prokaryotic genomes and the one available in MicroScope. Indeed, for assigning function to novel proteins, gene context approaches often complement the classical homology-based gene annotation in prokaryotes. The method we have developed offers the possibility of retaining more than one homologous gene (i.e. not only the bidirectional best hit), to allow for multiple correspondences between genes; that way, paralogy relations and/or gene fusions are easily detected [4].
Table 1.
Software and databases integrated in the MicroScope pipelines
| Topic | Name | Software | Database | Description | Internal | URL |
|---|---|---|---|---|---|---|
| Syntactic annotation | AMIGene | x | CoDing sequences (CDS) prediction | x | http://www.genoscope.cns.fr/agc/tools/amigene | |
| Glimmer | x | https://ccb.jhu.edu/software/glimmer | ||||
| Prodigal | x | http://prodigal.ornl.gov | ||||
| MICheck | x | INSDC genome CDS re-annotation | x | http://www.genoscope.cns.fr/agc/tools/micheck | ||
| tRNAscan-SE | x | tRNA prediction | http://eddylab.org/software/tRNAscan-SE | |||
| RNAmmer | x | rRNA prediction | http://www.cbs.dtu.dk/services/RNAmmer | |||
| Rfam/Infernal | x | x | ncRNA families and prediction | http://rfam.xfam.org, http://eddylab.org/infernal | ||
| RepSeek | x | DNA sequence repeats | http://wwwabi.snv.jussieu.fr/public/RepSeek | |||
| Alien hunter | x | DNA compositional biases to detect HGT regions | http://www.sanger.ac.uk/science/tools/alien-hunter | |||
| SIGI-HMM | x | http://www.brinkman.mbb.sfu.ca/∼mlangill/sigi-hmm | ||||
| GenProtFeat | x | Gene/protein features | x | |||
| Taxonomy | x | NCBI taxonomy database | https://www.ncbi.nlm.nih.gov/taxonomy | |||
| Functional annotation | BLAST+ | x | DNA/protein sequence alignment | https://blast.ncbi.nlm.nih.gov | ||
| Diamond | x | https://github.com/bbuchfink/diamond | ||||
| UniProtKB | x | Protein sequence and function database | http://www.uniprot.org | |||
| InterPro | x | x | Protein signature and family prediction | https://www.ebi.ac.uk/interpro | ||
| COG | x | x | Protein family annotation and prediction | https://www.ncbi.nlm.nih.gov/COG | ||
| FigFam | x | x | http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/FigFam | |||
| MICFAM | x | Protein sequence family classification with SiliX | x | |||
| SiliX | x | Clustering of protein sequences | https://lbbe.univ-lyon1.fr/-SiLiX-.html | |||
| ENZYME | x | Enzymatic activity database | http://enzyme.expasy.org | |||
| PRIAM | x | Enzymatic activity prediction | http://priam.prabi.fr | |||
| dbCAN | x | Carbohydrate-active enzyme prediction | http://csbl.bmb.uga.edu/dbCAN/ | |||
| SignalP | x | Signal peptide cleavage site prediction | http://www.cbs.dtu.dk/services/SignalP | |||
| TMHMM | x | Transmembrane helix prediction | http://www.cbs.dtu.dk/services/TMHMM | |||
| LipoP | x | Lipoprotein prediction | http://www.cbs.dtu.dk/services/LipoP | |||
| PSORTb | x | Subcellular localization prediction | http://www.psort.org | |||
| VFDB | x | Virulence factor database | http://www.mgc.ac.cn/VFs | |||
| VirulenceFinder | x | https://cge.cbs.dtu.dk/services/VirulenceFinder | ||||
| CARD/RGI | x | x | Antibiotic resistance database and prediction | https://card.mcmaster.ca | ||
| AutoFassign | x | Automatic functional annotation of proteins | x | |||
| Relational annotation | Syntonizer | x | Synteny conservation detection | x | http://www.inrialpes.fr/helix/people/viari/cccpart/ | |
| Directon | x | Operon prediction | x | |||
| PhyloProfile | x | Phylogenetic profilef co-evolution score | x | https://dx.doi.org/10.1186/1471-2164-13-69 | ||
| RGP | x | Genomic plasticity region detection | x | |||
| Pathway synteny | x | Synteny involved in metabolic pathways | x | |||
| MIBiG/ antiSMASH | x | x | Biosynthetic Gene Cluster database and prediction | http://www.secondarymetabolites.org/ | ||
| ChEBI | x | Chemical compound database | https://www.ebi.ac.uk/chebi | |||
| Rhea | x | Reaction database | http://www.rhea-db.org | |||
| KEGG | x | Metabolic pathway database | http://www.genome.jp/kegg | |||
| MetaCyc/ Pathway tools | x | x | Metabolic pathway database and prediction | https://metacyc.org, http://brg.ai.sri.com/ptools/ | ||
| Transcriptomics and variant discovery | SSAHA2 | x | Read mapping | http://www.sanger.ac.uk/science/tools/ssaha2-0 | ||
| BWA | x | https://github.com/lh3/bwa | ||||
| SAMtools | x | Mapping analysis | http://www.htslib.org/ | |||
| bedtools | x | http://bedtools.readthedocs.io | ||||
| PALOMA | x | Variant detection | x | |||
| DESeq | x | Differential gene expression analysis | http://bioconductor.org/packages/release/bioc/html/DESeq.html |
Information from the syntactic and functional annotation pipelines can be placed into a biological context to understand how the predicted objects interact in functional modules such as metabolic pathways. Each genome integrated into MicroScope is processed by an in-house workflow based on the MetaCyc reference database [18] and on the Pathway Tools software [19]. This software creates a Pathway Genome DataBase (PGDB) containing the predicted pathways and reactions of an organism. It uses a matching procedure for which we directly use as input the official MetaCyc reaction frame identifiers when available in the genome annotation; this allows to avoid overpredicted or missed enzymatic reactions [20]. The collection of MicroScope PGDBs is made available at the MicroCyc Web site (http://www.genoscope.cns.fr/agc/microcyc) and in the MicroScope database (see ‘Exploration of metabolic data’ section). Moreover, these metabolic networks are synchronized each night with new MicroScope genomes and expert annotations.
When a public prokaryotic genome is integrated into MicroScope, the original annotations are stored in the database, and the syntactic re-annotation process, which uses the MICheck procedure, often allows to identify missing genes or wrongly annotated one [21]. This step is useful to annotate more completely the pseudogenes found in a genome (‘real’ or because of sequencing errors), an important piece of information when comparing closely related species. Data from genomes available in public databanks generally remain with the ‘public’ status too in MicroScope.
A MicroScope staff to support and train a user community
As soon as annotations and comparative analysis results are processed by MicroScope, the user who submitted the genome(s) is alerted by an e-mail; he/she can subsequently use a specific administration tool to grant access to his/her collaborators and to define consultation and modification rights on the sequences (‘User Panel’ menu/‘Access Rights Management’ functionality). Continuing support and assistance to MicroScope users remain an important activity in the context of our services (or collaborative projects). These regular exchanges, together with the satisfaction surveys, are the most efficient way of performing continual evolution of the platform in response to user needs. Indeed, in addition to the user-friendliness of the tools integrated into the platform (see below), the short response time and the quality of feedback to individual queries are highly appreciated aspects of the MicroScope service.
Microbiologists who submitted genomic data to the MicroScope platform are warmly invited to follow a training course organized by our team. Using the data related to their own project, attendees learn how to change or correct the current automatic functional annotations, and how to perform effective searches and analyses with the functionalities available through the Web interface. About twice a year, we provide for new users a four-and-a-half-day training ‘Annotation and analysis of prokaryotic genomes using the MicroScope platform’. Since 2016, we also provide an advanced course for former trainees, so that they can remain up-to-date on recent developments. Since 2008, 450 users from 20 countries have been trained and 13 external sessions have been organized in France and abroad (Tunisia; Denmark; Germany; Switzerland; Spain; the Netherlands; China). More information is available on our Web site: http://www.genoscope.cns.fr/agc/microscope/training.
Data integration, service continuity and data conservation (backups) are currently provided free of charge. MicroScope services follow the quality management system of our laboratory (ISO 9001:2008 and NF X50-900:2013 standards).
All the data previously described (primarily genomes, analysis results and annotations) should be made appropriately accessible to biologist users, to allow efficient curation of annotations and to develop hypotheses about specific genomes or sets of genes to be experimentally tested. The following sections describe the MicroScope Web interface (http://www.genoscope.cns.fr/agc/microscope), i.e. the components accessible to our users, via secure or anonymous connections. For a complete description of each functionality in terms of input and output data, a complete tutorial is available here: https://microscope.readthedocs.io.
Exploration of the genomic data: simple and advanced queries
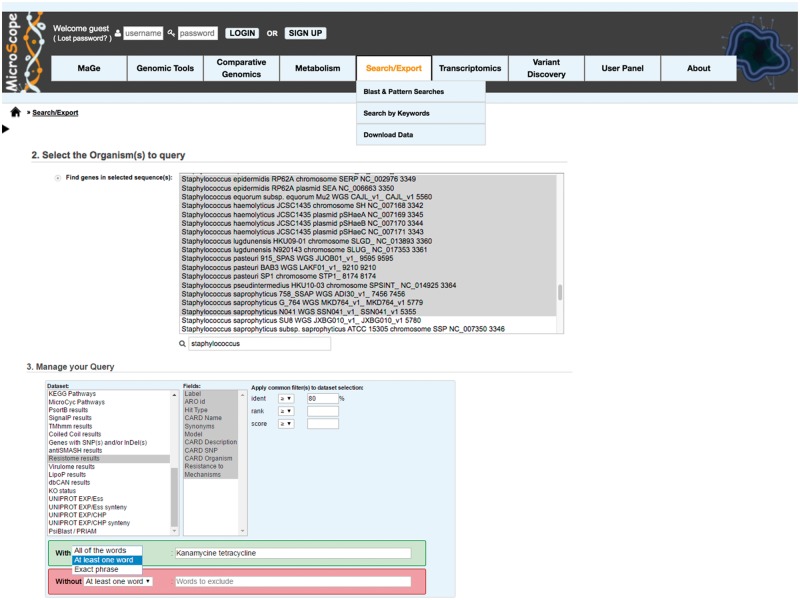
The ‘Search/Export’ menu (Figure 4) allows the user to perform Blast and pattern searches in the MicroScope database, and to download, in standard file formats (Genbank, EMBL, GFF, etc.), sequences, annotation data and the metabolic networks. The ‘Search by keywords’ functionality allows the user to identify genes and functions of interest using a variety of selection filters. The ‘single mode’ is used to query only one chromosome and the ‘multiple mode’ to query several replicons (of one organism) and/or several genomes. A basic keyword search enables the user to quickly retrieve genes having a particular function (i.e. ‘kinase’, ‘transporter’). Each kind of precomputed results (i.e. Blast results on various primary data, InterPro and FigFAM results, etc.) can be queried. Figure 4 shows an example of a query on the similarity searches in the CARD database [22] (‘Resistome’ data set).
Figure 4.
MicroScope interface illustrating the ‘Search by keywords’ functionality. In the ‘multiple’ mode, a set of Staphylococcus species has been selected, and the BLASTP similarity results obtained with well-known resistance genes stored in the CARD database are queried using an amino acid identity threshold of at least 80% and using the keywords ‘kanamycine tetracycline’. The selection of ‘At least one word’ is required to apply an ‘OR’ between the two keywords.
Keyword searches are useful to compare current annotation of the gene functions with the results, in terms of biological function, given by a specific analysis method. Indeed, the result of a query can be refined with a further query. For example, one can search for gene annotated as ‘protein of unknown function’ (first query) and then, search for the one having significant Blast results with proteins annotated with specific functions (second query).
Whatever the query, the result output is a list of candidate genes, the genomic contexts of which can be easily visualized: next to the gene label, a magnify icon can be clicked to come back to the MaGe graphical representation with automatic displacement of the genome browser centered on the gene of interest.
MaGe (Magnifying Genome): a genome browser in the light of synteny results
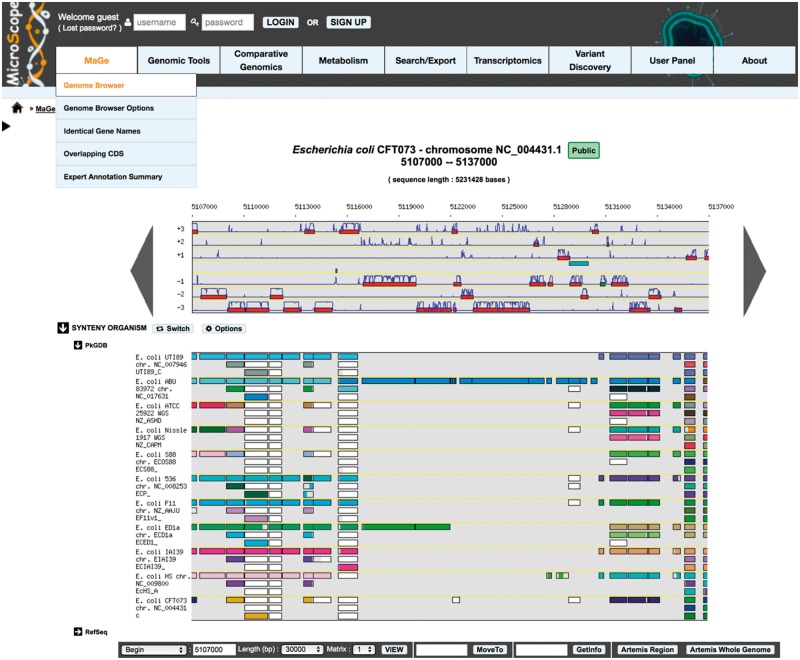
The MaGe graphical interface is one of the functionality that had a strong positive resonance among users: this genome browser offers gene context exploration of the studied genome compared against other microbial genomes. The graphical representation of the synteny groups allows the user to quickly see if part of the genome being annotated shares similarities and locally conserved organization with the selected sequences. As shown in Figure 5, there is a clear synteny break in the visualized part of the E. coli CFT073 strain: the genes located between 5116000 and 5131000 share homologs only with the E. coli pathogenic strain ABU and, partially, with the E. coli commensal strain ED1a. The foreign origin of this region is also obvious if one looks at the coding prediction curves: the gene model used here does not fit well with the codon usage of the genes annotated in this genomic island. The example shown in Figure 5 also indicates possible paralogy relations through multiple correspondences between genes and one case of frameshift (or sequencing error) in E. coli 536 for the idnK gene (D gluconate kinase; see Figure 5). With such graphical representation, the conservation of genomic context is fully integrated in the process of the expert curation of gene function.
Figure 5.
MicroScope genome browser and synteny map. The first graphical map contains part of the genome being analyzed (here 30 kb of E. coli CFT073), over which the user can navigate (moving and zooming functionalities). The predicted coding genes are drawn, on the six reading frames, in red rectangles together with the coding prediction curves (computed with the gene model selected by the user; ‘Matrix’ selection menu). Below this genome browser, is represented the synteny map in which each line shows the similarity results between the genome being annotated (E. coli CFT073) and other selected genomes (i.e. 11 pathogenic and commensal E. coli strains; the selection is performed using the ‘Options’ functionality). On this map, a rectangle flags the existence of a gene, somewhere in the compared genome, homolog to the corresponding gene in the genome browser. If, for several co-localized CDSs on the annotated genome, there are several co-localized homologs on the compared genome, the rectangles are all of the same color; otherwise, the rectangle is white. Thus, in this map, a specific color indicates a synteny group. A rectangle is always of the same size as the reference gene in the genome browser; however, it is colored only on part of the gene, which aligns with the compared protein. This allows the user to visualize situations where the alignment is partial. There is one such case in E. coli 536 indicating that the idnK gene in this strain is a pseudogene compared with the idnK gene in CFT073. In contrast with the genome browser, there is no notion of scale on the synteny maps: to see how homologous genes are organized in a synteny group, the user can click on one rectangle in a given synteny group.
Another visualization mode has been added more recently to represent synteny conservation at different taxonomic levels (i.e. phylum, class, order, family or species). In this ‘taxon-synteny’ mode (obtained by clicking the ‘Switch’ button, Figure 5), each line of the synteny map refers to a taxon, and colored boxes represent the percentage of synteny conservation among organisms of the corresponding taxon.
Comparative genomics tools
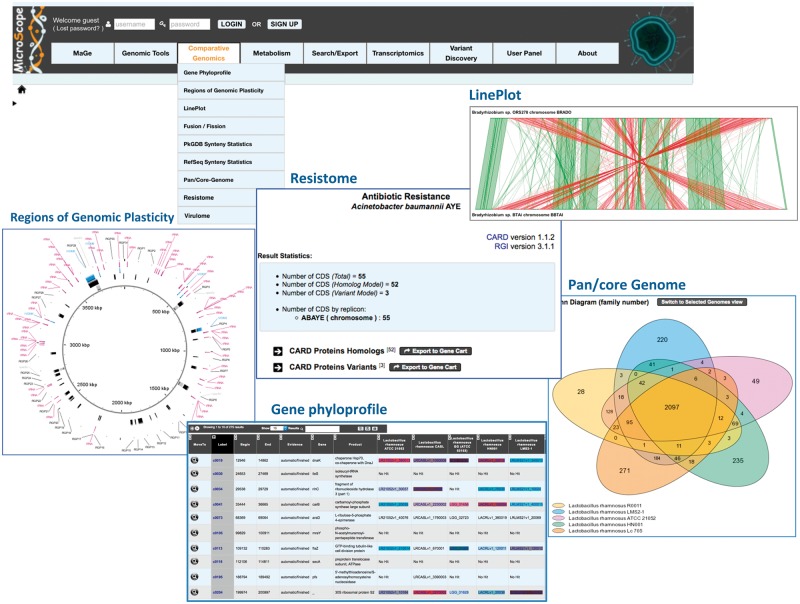
Computations of homologs and synteny groups between microbial genomes are the starting point of several comparative methods available in the ‘Comparative Genomics’ menu (Figure 6).
Figure 6.
Comparative genomics tools of the MicroScope platform. The figure displays some of the tools available to perform in-depth comparative genomics analyses involving the bacterium of interest and one or a set of organisms: ‘Gene Phyloprofile’ (comparison of five Lactobacillus rhamnosus strains), ‘Line Plot’ (shared synteny groups found in the same DNA strand are colored in green, and in red otherwise), ‘Regions of Genomic Plasticity’ (the predicted genomic island is shown in the second layer of the circular representation), ‘Pan-core genome’ and ‘Resistome’. In this last case, the figure shows Acinetobacter baumannii AYE genes having BLASTP hits with proteins from the CARD database.
First, the ‘Fusion/Fission’ functionality provides a list of candidate genes of the selected genome potentially involved in evolutionary events such as gene fusion or fission. Such events involve what is named ‘Rosetta-stone’ proteins, and suggest a high probability of functional interaction between the involved proteins [23]. Second, the ‘Gene phyloprofile’ functionality is used to find unique or common genes in the query genome with respect to other genomes of interest. Homology constraints and inclusions in synteny group criteria may be applied to refine queries. Third, the ‘LinePlot’ functionality draws a global graphical representation of conserved syntenies between two selected genomes, and the ‘Regions of Genomic Plasticity (RGP)’ is used to search for potential horizontal gene transfer (HGT). The method combines (i) the results of algorithms that detect signals in the query sequence indicative of horizontal transfer origin (tRNA hotspots; mobility genes; compositional bias [24]) and (ii) the identification of synteny breaks in the query genome in comparison with closely selected microbial genomes. Results are reported in a tabular form and on a circular representation of the genome (Figure 6). Finally, the ‘Pan/Core Genome’ functionality computes dynamically the pan-genome and its components (core-genome; variable-genome) of a set of selected organisms (up to 200). The method uses the MicroScope gene families (MICFAM) computed with the SiLiX software [25]. The set of common (= core-genome), variable and strain-specific genes of each compared genomes can be exported in a tabular file format or in a ‘Gene Cart’.
Indeed, at any level of the MicroScope Web interface, the gene list that results from the corresponding search/analysis can be selected for inclusion into a ‘Gene Cart’. The user can manage several ‘Gene Carts’ at the same time resulting from different queries. A specific interface has been developed to perform various operations such as the intersection or the difference between two gene carts, to extract sequences or to run multiple alignments via the plugged Jalview software [26] (Functionality ‘Gene Carts’ of the ‘User Panel’ menu).
Two functionalities of the ‘Comparative Genomics’ menu are most specifically related to pathogen analysis (Figure 6): ‘Resistome’, which uses the Comprehensive Antibiotic Resistance Database [22] a manually curated resource containing high-quality reference data on the molecular basis of antimicrobial resistance, and the Resistance Gene Identifier (RGI) tool to predict the resistome of a genome. The ‘Virulome’ functionality gives the results of a Blast similarity searches in three distinct data sets of virulence genes: VFDB, which contains experimentally demonstrated virulence genes [27], VirulenceFinder [28] and a subset of the E. coli main virulence genes.
Exploration of metabolic data
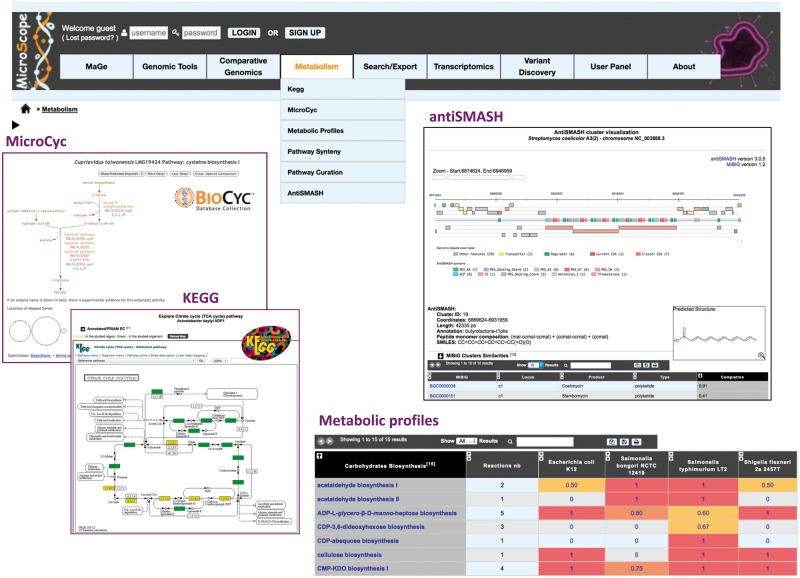
The ‘Metabolism’ menu of MicroScope allows to explore the predicted metabolic pathways using two main resources, KEGG and MetaCyc, and to use analysis tools (Figure 7).
Figure 7.
Tools for the analysis of microbial metabolism. Metabolic data can be explored using the KEGG or MetaCyc metabolic pathway hierarchies. On the left, the figure shows, for one selected MicroScope genome, the mapping of the annotated EC numbers on a KEGG metabolic map (enzymes encoded by genes localized on the current genome browser region are highlighted in yellow, and the ones encoded by genes localized elsewhere are highlighted in green). Predicted PGDBs using the Pathway Tools software are available using the ‘MicroCyc’ functionality. Comparison of metabolic pathways between a set of selected genomes is performed using the ‘Metabolic profiles’ tool: for each metabolic pathway, a completion value is computed, which corresponds to the number of reactions found in the genome × divided by the total number of reactions in the pathway. This value can take into account pseudogenes or not. It ranges between 0 (absence of the pathway) and 1 (complete pathway). The figure also shows an example of antiSMASH, which predicts Biosynthetic Gene Clusters in prokaryotic genomes. For the NRPS/PKS cluster types, the predicted peptide monomer composition and its corresponding SMILES formula are specified. Below the graphical representation of the predicted antiSMASH cluster, a summary of MIBiG cluster similarities, BGC gene composition as well as tailoring cluster similarities is given.
Starting from the set of predicted and/or validated Enzyme Commission numbers (EC numbers), metabolic maps are dynamically drawn via a request to the KEGG Web server (‘KEGG’ functionality). A color-based code enables to see the number of enzymatic activities (i.e. EC number) of the annotated genome found in specific metabolic pathways (Figure 7). The interconnected metabolic pathways represented in KEGG are supplemented by the MicroCyc PGDBs built with the Pathway Tools software using MetaCyc as reference metabolic database (see ‘Running the annotation pipelines’ section). The ‘MicroCyc’ functionality allows the user to browse and query the metabolic network of a target genome using the Pathway Tools Web interface [18].
These two sets of predicted pathways can be used in the ‘Metabolic profiles’ functionality. Starting with a selection of organisms and a subset (or all) of metabolic pathways from the KEGG or MetaCyc classification, the tool computes a pathway completion value for each metabolic pathways (Figure 7). These values can be used by the MeV statistical method (Java Web start application) to cluster genomes according to their metabolic capabilities. Moreover, this table is also a good starting point to find candidate genes for missing gene–reaction associations in specific pathways (see example in [6]). In the same way, the ‘Pathway Synteny’ functionality follows the ‘guilt by association’ strategy [29], as it combines information on synteny groups and metabolic pathways (i.e. it searches for groups of genes, which share conserved synteny and are found on the same metabolic pathway). Using this interface, annotators can quickly check for reaction-hole candidate genes among the conserved miss-annotated genes of a given group.
Finally, the ‘antiSMASH’ functionality relies on the integration of the antiSMASH (antibiotics and Secondary Metabolite Analysis Shell) program, which enables rapid genome-wide identification, annotation and analysis of secondary metabolite Biosynthesis Gene Clusters (BGCs) in microbial genomes [30]. Each predicted cluster and its genomic context are explored in a dedicated visualization window showing also a graphical representation of the gene domain composition (Figure 7). For nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) cluster types, the predicted peptide monomer composition and its corresponding SMILES formula are specified and the corresponding predicted chemical structure is displayed. For each predicted BGC, a summary of similarities with the reference database MIBiG [31], BGC gene composition as well as tailoring cluster similarities is given. This last item relies on a knowledge database provided with antiSMASH about tailoring clusters already described in known BGCs and associated with publications.
Analysis of experimental data
The functionalities available in the ‘Transcriptomics’ and ‘Variant discovery’ menus rely on the results of the pipelines used to analyze data from transcriptomic projects (i.e. RNA-seq experiments) and data from evolution projects (i.e. clones of the same species at different generation times). Exploration of these experimental data has been illustrated in the two last publications of the MicroScope platform [6, 7].
The ‘Transcriptomics’ functionality allows exploring the transcript coverage along genome, expression levels of genomic objects (genes, ncRNAs) and differential expression between samples for distinct experimental conditions. All appropriate pairwise comparisons of experimental conditions can be directly queried from the interface. Differentially expressed genes may be projected on reconstructed metabolic networks to highlight metabolic pathways significantly affected by experimental conditions. The ‘Variant discovery’ functionality offers different tools to explore and analyze the predicted mutations (single nucleotide polymorphisms and small insertions/deletions) in their genomic and functional context. This detection takes into account raw sequencing data and associated read qualities to discriminate between true variations and sequencing errors.
Expert curation of genomic and metabolic data
From the results of the exploration of data and the analysis tools, MicroScope users can review and curate the automatic functional annotation of genes encoded by its genome of interest. This task is performed using the ‘Gene Editor’, which has been illustrated in the 2013 MicroScope publication [6]. Briefly, it is made of three main sections:
The ‘current annotation’ section allows the user to modify, delete and add information. The functional description of gene functions is a free-text field exposed to inconsistencies across genes and genomes. We thus have also integrated enumerated lists of well-defined and nonredundant terms for the product type field (defined in GenProtEC [32]), the functional classifications (MultiFun [33] and TIGRFAMs [34]) and for the class field (inspired from the Pseudomonas Genome database [35]), which helps understanding the origin of the functional annotation (e.g. it comes from the functional description of an homologous gene for which the function has been experimentally demonstrated). The curation of associations between genes coding for enzymatic activities and the biochemical reactions catalyzed by these enzymes is performed using two main enzymatic reactions resources: MetaCyc [18] and Rhea [36]. Finally, to alert users about possible inconsistencies, annotation is checked via an automatic procedure launched when the annotation is saved in the database.
The ‘automatic annotation’ section contains the gene function predicted by our automatic functional annotation procedure (‘MicroScope pipeline annotation’), which involves the transfer of the reliable up-to-date reference annotations to ‘strong’ orthologs, if any [4]. In case of published bacterial genome integrated in MicroScope, the section contains information on the functional annotation in nucleotide sequence databanks and UniProtKB if available.
The ‘method results’ section provides, for each individual annotation tool executed, a summary of the results, visualized in a tabulated form (this includes precomputed lists of homologs and synteny groups). This integrative strategy allows annotators to quickly browse functional evidences, tracking the history of an annotation and checking the gene context conservation with an orthologous gene having an experimentally demonstrated biological function for example. Criteria for entering an expert annotation are based on different level of evidences from direct experimentation to bioinformatics evidences. The confidence status of each gene annotation is available in the class field of the gene editor. The categories are inspired by the ‘protein name confidence’ defined in PseudoCAP (Pseudomonas aeruginosa community annotation project). A set of rules allowing to choose this ‘class’ annotation category according to bioinformatics evidences is proposed in our MicroScope tutorial: https://microscope.readthedocs.io/en/latest/content/mage/info.html (‘How to choose the “Class” annotation category?’ and ‘Annotation Rules’ sections).
Following the integration of novel functionalities into MicroScope, the ‘Gene Editor’ is constantly evolving. First, new interfaces allowing to ease the curation of resistance and virulence genes are under development, especially using defined ontologies such as ARO, the Antibiotic Resistance Ontology [22]. Second, to fully exploit the results of the different tools dedicated to genomic region analysis (e.g. antiSMASH or RGPfinder), we are currently working on the development of a specific editor to annotate gene clusters such as operons, BGCs, genomic islands, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) regions, secretion systems and phages.
Expert annotations are continuously gathered in the MicroScope database. Indeed, ∼35 000 annotations are made in a year (Figure 1), and >370 000 genes have been curated so far. A third of these annotations correspond to the description of precise molecular functions supported by direct or indirect (i.e. from homology relationships) experimental evidences. Biologists generally focused their annotations on proteins/functions of interest; however, it is interesting to note that about 50 genomes integrated in MicroScope are near completely curated (≥80% of the genes were expertly annotated), and 124 additional genomes got >300 curated genes.
MicroScope annotations are submitted to INSDC databanks when the genomes get published and can be easily downloaded via the Web interface (‘Search/Export->Download Data’ functionality). Moreover, we provide a RESTful API to access programmatically public genome data, and semantic Web approaches are currently used to work on the interoperability of MicroScope curated data with other European resources such as UniProtKB [9], HAMAP [37], EnsemblBacteria [1] and Rhea [36]. These developments are performed in the context of the ELIXIR bioinformatics infrastructure (https://www.elixir-europe.org).
Software and database architecture
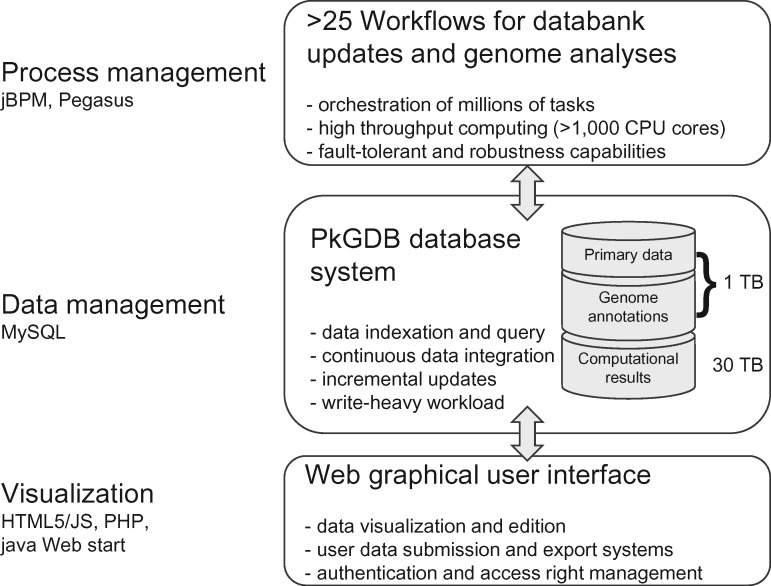
The technical architecture of the MicroScope platform is shown on Figure 8. Its three components have been described and updated in the previous publications of MicroScope [5, 6]. In summary:
Figure 8.
Technical architecture of the MicroScope platform. The MicroScope platform is made of three components: (i) a ‘Process management’ system to organize workflow execution, (ii) a ‘Data management’ system, called PkGDB, to store information from databanks, genomes and computational results and (iii) a ‘Visualization’ system for textual and graphical representation of PkGDB data.
Process management system
The annotation pipelines are organized in a robust automated workflow management system using the jBPM framework (java Business Process Management; http://jbpm.org), which allows us to handle simultaneously millions of tasks for the analysis of several new microbial genomes. These tasks are parallelized on hundreds of CPU cores using Pegasus MPI cluster module (https://pegasus.isi.edu). The pipelines for the structural, functional and relational annotation orchestrate >50 external/internal bioinformatics software (see section ‘Running the MicroScope pipelines’). A large part of these analyses are updated at regular intervals to take into account primary databases growth and new expert annotations.
Data management system
The results of these analysis tools, together with the primary data used as inputs, are stored in a relational database named PkGDB and based on the open-source MySQL relational database management system and the InnoDB (for continuous data integration and incremental updates) and MyISAM (for large bulk inserts) table engines. The PkGDB architecture supports integration of automatic and human-curated functional annotations and records a history of all the modifications. Finally, for metabolic comparative analyses purposes (see the ‘Metabolic profiles’ functionality in the ‘Exploration of metabolic data’ section), relational tables have been designed in PkGDB to store information of the MicroCyc PGDBs, together with the KEGG metabolic pathways and modules. The size of PkGDB is today 1 TB for databanks and genome data, and 30 TB for the computational results (Figure 8). Only one instance of the database gathers all genome analyses, which eases collaborative annotation process.
The Web visualization component
The MicroScope Web interface (http://www.genoscope.cns.fr/agc/microscope) is developed using the Apache/PHP server-based language and consists of numerous dynamic Web pages containing textual and graphical representations for accessing and querying data. Several useful graphical applications, such as Artemis [38], MeV [39] and IGV [40], are also available in the MicroScope interface through plugged Java applications. As shown in this article, the tools are organized in a menu bar to facilitate the exploration and the curation process. At any level of the interface, a ‘Help’ functionality is available, and a complete tutorial can be found in the ‘About’ menu.
Conclusion
In this article, we have described the MicroScope platform from the point of view of the end user, i.e. following one of the main objectives of our prokaryotic genome annotation and comparative system: to allow biologists to submit their genomic data in a simple way and, then, to perform analysis and make relevant assessments of the predicted gene functions using (i) the functionalities for querying and browsing the computed data, (ii) the synteny results and metabolic network predictions, the combination of which can be helpful in formulating hypotheses on the biological function of nonannotated genes and (iii) a gene annotation editor giving access to the results of each method applied, together with links to several useful public resources.
Among the ongoing developments described in the last update of the platform [7], we have currently made great progresses in the consensus representation of thousands bacterial genomes to provide a better analysis workflow of prokaryotic species. The idea is to structure the pan-genome of an organism into the set of ‘persistent’ genes (relaxed core definition, that is to say genes found in the great majority of the genomes), the ‘shell’, which gathers moderately conserved genes and the ‘cloud’ corresponding to rare and unique genes [41]. To organize pangenomic information, we are using a graph data model, where the nodes represent the protein families, and the edges represent the genome co-localization of the two protein families (weighted by the number of the genomes sharing this co-localization). A statistical method is then used to divide the pan-genome into the three main classes (persistent, shell and cloud). The next step is the integration of this representation in MicroScope to facilitate comparative analysis and data visualization of thousands of strains. We will also add functionalities allowing users to select, at any level of this pan-genome graph, a subpart of this graph and, using one genome as reference, to come back to the MaGe genome browser. We are starting to work on an instance of MicroScope based on this novel pan-genome representation that will contain most of the reference species found in the human gut microbiota.
Key Points
MicroScope is open to microbiologists interested in extended analyses of species of interest.
MicroScope is an integrated environment allowing to perform comparative genomic and metabolic analyses.
Tools and graphical interfaces for the curation of gene function are part of the specificities of the MicroScope platform.
MicroScope provides a collaborative environment to share and improve knowledge on genomes.
Acknowledgements
The authors would like to thank all MicroScope users for their feedback, which helped greatly in optimizing and improving many functionalities of the system. The authors also thank the entire IT system team of Genoscope for its essential contribution to the efficiency of the platform.
Biographies
Claudine Médigue, PhD, is a research director at CNRS. She is the head of the Laboratoire d’Analyse Bioinformatiques en Génomique et Métabolisme located at Genoscope. She has worked on the annotation and comparative analysis of prokaryotic genomes >25 years.
Alexandra Calteau is a senior researcher at CEA. She contributes to different bioanalysis projects and the development of functionalities in the MicroScope platform, mainly in the Comparative Genomics field. She is responsible for the MicroScope professional training organization, and for the quality management of the LABGeM.
Stéphane Cruveiller is a senior researcher at CEA. He is managing the MicroScope services and developments for the analysis of variants discovery, transcriptomics and metagenomics data. He has specific research activities in microbial evolution.
Mathieu Gachet is a master student at CEA. He is working on the improvement of metagenomic data integration in MicroScope.
Guillaume Gautreau is a PhD student at CEA. He works on the development of pan-genome graphs in MicroScope and their application in metagenomics.
Adrien Josso is an engineer in bioinformatics at CEA. He works on MicroScope software development for workflow management and metabolic data integration.
Aurélie Lajus is an engineer in bioinformatics at CEA. She works mainly on (meta)genome project management and software integration in the MicroScope platform.
Jordan Langlois is an engineer in bioinformatics at CEA. He works on software integration and Web developments in the MicroScope platform.
Hugo Pereira is an engineer in bioinformatics at CEA. He works on MicroScope software development for workflow management of NGS projects.
Rémi Planel is an engineer in bioinformatics at CEA. He works on MicroScope software development for pan-genome computation and Web visualization.
David Roche is an engineer in bioinformatics at CEA. He works on NGS project management, software integration and Web developments in the MicroScope platform. He is also involved in the training of MicroScope users.
Johan Rollin is an engineer in bioinformatics at CEA. He works on software integration and Web developments in the MicroScope platform.
Zoe Rouy is an engineer in bioinformatics at CEA. She works mainly on (meta-)genome project management, software integration and Web developments in the MicroScope platform.
David Vallenet is a senior researcher at CEA. He is managing all the technological developments of the MicroScope platform and has specific research activities in the development of methods for enzyme function prediction and metabolic network analysis.
Funding
French Government ‘Investissements d’Avenir programmes’, namely, FRANCE GENOMIQUE (grant number ANR-10-INBS-09-08); INSTITUT FRANCAIS DE BOINFORMATIQUE (grant number ANR-11-INBS-0013).
References
- 1. Kersey PJ, Allen JE, Armean I, et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res 2016;44:D574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen I-MA, Markowitz VM, Palaniappan K, et al. Supporting community annotation and user collaboration in the integrated microbial genomes (IMG) system. BMC Genomics 2016;17:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wattam AR, Davis JJ, Assaf R, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 2017;45:D535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vallenet D, Labarre L, Rouy Z, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 2006;34:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vallenet D, Engelen S, Mornico D, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database 2009;2009:bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vallenet D, Belda E, Calteau A, et al. MicroScope–an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res 2013;41:D636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vallenet D, Calteau A, Cruveiller S, et al. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res 2017;45:D517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilson CA, Kreychman J, Gerstein M.. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J Mol Biol 2000;297:233–49. [DOI] [PubMed] [Google Scholar]
- 9. The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Overbeek R, Begley T, Butler RM, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 2005;33:5691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barbe V, Vallenet D, Fonknechten N, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res 2004;32:5766–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Touchon M, Hoede C, Tenaillon O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 2009;5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barbe V, Cruveiller S, Kunst F, et al. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 2009;155:1758–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belda E, Sekowska A, Le Fèvre F, et al. An updated metabolic view of the Bacillus subtilis 168 genome. Microbiology 2013;159:757–70. [DOI] [PubMed] [Google Scholar]
- 15. Belda E, van Heck RG, José Lopez-Sanchez M, et al. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 2016;18:3403–24. [DOI] [PubMed] [Google Scholar]
- 16. Field D, Garrity G, Gray T, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008;26:541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bocs S, Cruveiller S, Vallenet D, et al. AMIGene: annotation of MIcrobial genes. Nucleic Acids Res 2003;31:3723–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 2016;44:D471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karp PD, Latendresse M, Paley SM, et al. Pathway Tools Version 19.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform 2015;17:877–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vieira G, Sabarly V, Bourguignon PY, et al. Core and panmetabolism in Escherichia coli. J Bacteriol 2011;193:1461–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cruveiller S, Le Saux J, Vallenet D, et al. MICheck: a web tool for fast checking of syntactic annotations of bacterial genomes. Nucleic Acids Res 2005;33:W471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jia B, Raphenya AR, Alcock B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017;45:D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suhre K. Inference of gene function based on gene fusion events: the Rosetta-Stone method. Methods Mol Biol 2007;396:31–41. [DOI] [PubMed] [Google Scholar]
- 24. Vernikos GS, Parkhill J.. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 2006;22:2196–203. [DOI] [PubMed] [Google Scholar]
- 25. Miele V, Penel S, Duret L.. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 2011;12:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waterhouse AM, Procter JB, Martin DM, et al. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009;25:1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen L, Zheng D, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res 2016;44:D694–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 2014;52:1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aravind L. Guilt by association: contextual information in genome analysis. Genome Res 2000;10:1074–7. [DOI] [PubMed] [Google Scholar]
- 30. Blin K, Wolf T, Chevrette MG, et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 2017;45:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medema MH, Kottmann R, Yilmaz P, et al. Minimum information about a Biosynthetic Gene cluster. Nat Chem Biol 2015;11:625–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Serres MH, Goswami S, Riley M.. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res 2004;32:D300–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Serres MH, Riley M.. MultiFun, a multifunctional classification scheme for Escherichia coli K-12 gene products. Microb Comp Genomics 2000;5:205–22. [DOI] [PubMed] [Google Scholar]
- 34. Haft DH, Selengut JD, Richter RA, et al. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 2013;41:D387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winsor GL, Griffiths EJ, Lo R, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 2016;44:D646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgat A, Lombardot T, Axelsen KB, et al. Updates in Rhea—an expert curated resource of biochemical reactions. Nucleic Acids Res 2017;45:4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pedruzzi I, Rivoire C, Auchincloss AH, et al. HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic Acids Res 2015;43:D1064–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carver T, Harris SR, Berriman M, et al. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012;28:464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003;34:374–8. [DOI] [PubMed] [Google Scholar]
- 40. Thorvaldsdóttir H, Robinson JT, Mesirov JP.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lobkovsky AE, Wolf YI, Koonin EV.. Gene frequency distributions reject a neutral model of genome evolution. Genome Biol Evol 2013;5:233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]