Summary
Salmonella is a human and animal pathogen that causes gastro-enteric diseases. The key to Salmonella infection is its entry into intestinal epithelial cells, where the bacterium resides within a Salmonella-containing vacuole (SCV). Salmonella entry also induces the formation of empty macropinosomes, distinct from the SCV, in the vicinity of the entering bacteria. A few minutes after its formation, the SCV increases in size through fusions with the surrounding macropinosomes. Salmonella also induces membrane tubules that emanate from the SCV and lead to SCV shrinkage. Here, we show that these antipodal events are utilized by Salmonella to either establish a vacuolar niche or to be released into the cytosol by SCV rupture. We identify the molecular machinery underlying dynamic SCV growth and shrinkage. In particular, the SNARE proteins SNAP25 and STX4 participate in SCV inflation by fusion with macropinosomes. Thus, host compartment size control emerges as a pathogen strategy for intracellular niche regulation.
Keywords: Salmonella enterica serovar Typhimurium, membrane trafficking, vesicle fusion, membrane rupture, compartmental size control, Salmonella-containing vacuole, macropinosome, SNARE, spacious vacuole-associated tubules (SVAT), magnetic extraction, proteomics
Graphical Abstract
Highlights
-
•
The early SCV simultaneously grows and shrinks through fusion and tubule formation
-
•
SCV shrinkage promotes vacuolar rupture and cytosolic release
-
•
IAMs are enriched in the host SNAREs SNAP25 and STX4, enabling IAM-SCV fusion
-
•
Promoting SNX1-mediated tubule formation, SopB fosters SCV ruptures
When infecting epithelial cells, Salmonella can reside and replicate within vacuolar or cytosolic niches. These co-existing lifestyles provide alternative survival strategies for the bacteria. Stévenin et al. show that the balance between dynamic growth and shrinkage of the Salmonella-containing vacuole determines vacuolar maintenance or rupture and controls the pathogen niche.
Introduction
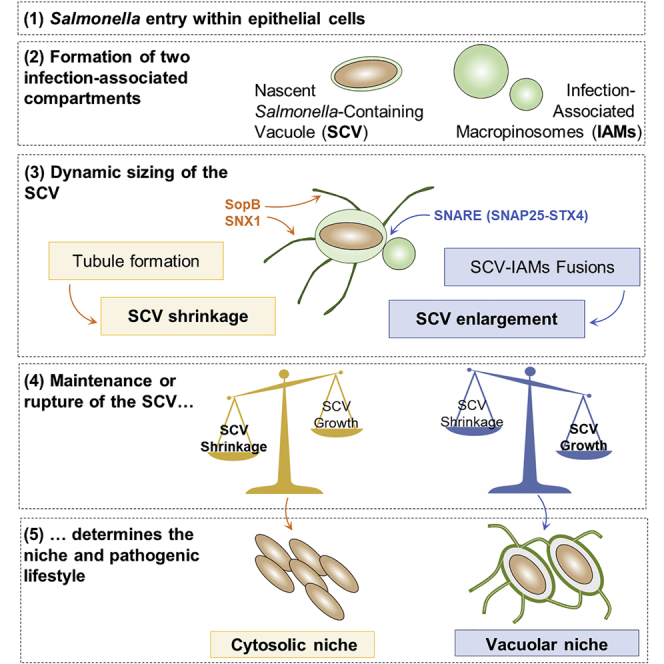
Salmonella enterica serovar Typhimurium (hereafter referred to as Salmonella) is a Gram-negative bacterium that causes salmonellosis, one of the main causes of gastro-enteric disease worldwide (Majowicz et al., 2010). It infects a broad range of animals including humans, mainly through oral ingestion of contaminated food or water (Carter and Collins 1974). At the early infection stage, the intestinal barrier is breached primarily through targeting of barrier-forming epithelial cells (Watson and Holden 2010). Because enterocytes are typically non-phagocytic, Salmonella triggers its own uptake by using a SPI-1-encoded type 3 secretion system (T3SS-1) to inject a cocktail of bacterial effectors into its host cell cytoplasm. These effectors induce an intense remodeling of the actin cytoskeleton and plasma membrane, resulting in the formation of membrane protrusions around the host-cell-bacteria contact site (LaRock et al., 2015). As a consequence, although Salmonella is ingested within the targeted cells, two distinct endomembrane compartments are formed: the tight nascent Salmonella-containing vacuole (SCV) and the surrounding fluid-filled infection-associated macropinosomes (IAMs) (Fredlund et al., 2018, Garcia-del Portillo and Finlay, 1994).
A few minutes after Salmonella entry, the nascent SCV (also called SCC for Salmonella-containing compartment) fuses with surrounding IAMs to form a larger compartment with a distinguishable lumen (Hernandez et al., 2004, Fredlund et al., 2018). Later, the SCV undergoes sequential maturation steps and turns into a vacuolar niche where Salmonella starts replicating within a few hours after entry (LaRock et al., 2015). Alternatively, 10% to 20% of the intracellular bacteria rupture their vacuole within the first 90 min of infection and escapes into the host cytosol (Perrin et al., 2004, Malik-Kale et al., 2012, Knodler et al., 2014). About half of these cytosolic bacteria are targeted by the xenophagy machinery for elimination, and the other half can circumvent this dead end by replicating at very high rates, a state called hyper-replication (Birmingham et al., 2006, Knodler et al., 2010). This bacterial cytosolic lifestyle results in a local burst of infection, host cell pyroptosis, and extrusion of the host cell from the epithelial lumen (Knodler et al., 2010, Sellin et al., 2014). The co-existence of cytosolic and vacuolar salmonellae among the population of infected epithelial cells is a strong advantage for this pathogen to promote its rapid spread in the gut lumen (cytosolic population) while remaining locally hidden from the immune system in infected epithelial cells (vacuolar population). This dual lifestyle also constitutes a challenge for the development of therapeutic treatments. Therefore, it is essential to understand the mechanism that determines the different intracellular niches of Salmonella.
In a recent study, we observed that the fusion between the initially distinct nascent SCVs and IAMs correlates with SCV integrity maintenance and the establishment of vacuolar pathogenic niches (Fredlund et al., 2018). Within the first hour of infection, the early Salmonella vacuole undergoes additional remodeling through the formation of membrane tubules called spacious vacuole-associated tubules (SVATs) (Bujny et al., 2008). These tubules seize a part of the SCV membrane, leading to the shrinkage of the vacuole (Bujny et al., 2008). SCV shrinkage and enlargement have not been studied together, so far. Here, we show that perturbation of SCV size by the inhibition of macropinosome or SVAT formations leads to significant shifts in the percentage of cytosolic bacteria. While linking SCV size control with SCV stability, we investigated the involved host and bacterial molecular players. Considering that SVAT formation depends on the bacterial T3SS-1 effector SopB (Bujny et al., 2008), we performed infections with SopB-depleted Salmonella (ΔsopB). We observed impaired SVAT formations and SCV shrinkage, resulting in a reduced cytosolic Salmonella population. We developed a pipeline to extract a cellular fraction specifically enriched on IAMs by using a highly specific magnetic-based fractionation method. The proteome of these IAM-enriched fractions was analyzed by mass spectrometry and compared with other cellular fractions from infected and non-infected cells. Analyzing the IAM-associated proteome together with the previously published proteome of the SCV (Santos et al., 2015), we identified specific SNAREs required for SCV-IAM fusion and SCV enlargement. SNAP25 and STX4 emerged as central players in this fusion process, promoting the maintenance of the SCV integrity and the vacuolar lifestyle of Salmonella.
Results
Salmonella Regulates the Size of Its Vacuole through Simultaneously Occurring Growth and Shrinkage
To simultaneously investigate the SCV enlargement resulting from IAM fusions and the SCV shrinkage resulting from SVAT formation, we labeled the phospholipid PI(3)P present on the three distinguishable infection-associated structures: the SCVs, IAMs, and SVATs. We transiently expressed the previously described PI(3)P-specific 2xFYVE probe in HeLa cells (Pattni et al., 2001) and infected them with fluorescent Salmonella. This co-labeling procedure provided precise insights on the host membrane trafficking events induced by individual entering bacteria. Time-lapse microscopy at a temporal resolution of 30 s per frame allowed unambiguous observation of the transient SCV growth and shrinkage, events that were so far insufficiently distinguished. During the first 30 min of bacterial challenge, we observed SCV and IAMs fusing homo- and heterotypically, leading to the formation of growing large SCVs, sometimes containing several bacteria (Figure 1A, blue arrows; Video S1A). Besides, we noticed the formation of long and transient PI(3)P-positive tubules emanating from the SCV (i.e., SVATs) concomitantly with the decrease in the SCV volume (Figure 1A, yellow arrows; Video S1A). We observed that SVAT formations and IAM fusions occurred simultaneously rather than sequentially, implicating the remodeling of the SCV through dynamic SCV shrinkage and enlargement.
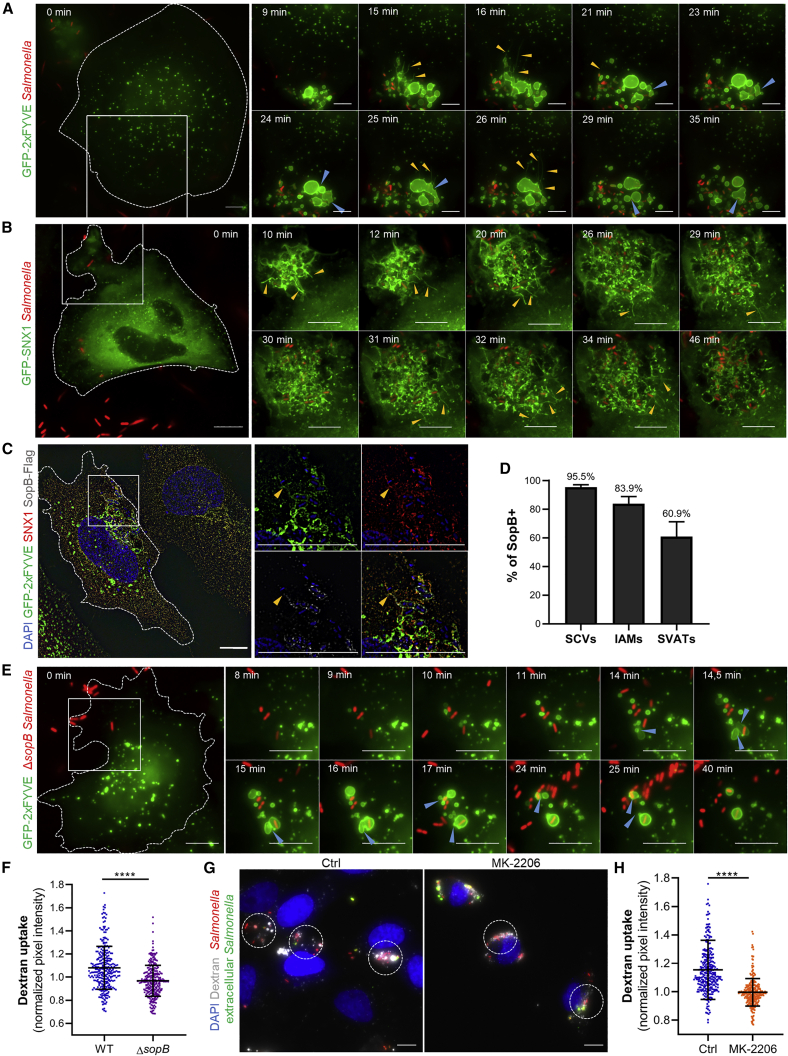
Figure 1.
Concomitant Size Regulation of the Salmonella-Containing Vacuole (SCV)
(A) Time-lapse microscopy of HeLa cells transfected with GFP-2xFYVE (in green) and infected with fluorescent Salmonella (in red). Scale bars: 10 μm. Blue arrowheads designate fusion events; yellow arrowheads designate tubular structures. See also Video S1A.
(B) Time-lapse microscopy of HeLa cells transfected with GFP-SNX1 (in green) and infected with fluorescent Salmonella (in red). Scale bars: 10 μm. Yellow arrowheads designate tubular structures. See also Video S1B and Figure S1A.
(C) Characterization of SopB intracellular localization by super-resolution SIM microscopy. Scale bars: 10 μm. Yellow arrowheads designate tubular structures.
(D) Quantification of the percentage of SopB-positive SCVs, IAMs, and SVATs over more than 150 intracellular salmonellae (3 independent experiments—manual counting based on confocal images). Error bars: ±SEM. Above-bar annotations: mean values.
(E) Time-lapse microscopy of HeLa cells transfected with GFP-2xFYVE (in green) and infected with fluorescent ΔsopB Salmonella (in red). Scale bars: 10 μm. Blue arrowheads designate fusion events. See also Video S1C.
(F) Quantification of dextran uptake over 30 min of incubation with WT or ΔsopB Salmonella.
(G) Fixed microscopy of HeLa cells infected with fluorescent Salmonella (in red) in the presence of fluorescent Dextran (in white) for 30 min. Extracellular bacteria (in green) are labeled by staining the samples without permeabilization. Left panel: untreated cells (Ctrl). Right panel: cells incubated with the allosteric pan-Akt inhibitor MK-2206 for 3 h before infection and during bacterial infection. Dashed circle: infection sites defined as circles with a diameter of 18 μm used for quantification in (F) and (H). Scale bars: 10 μm.
(H) Quantification of dextran uptake over 30 min of incubation with WT Salmonella in untreated cells (Ctrl) or cells treated with MK-2206.
(F and H) Each dot represent the mean fluorescence intensity of the dextran channel normalized by the mean fluorescence intensity of the bacteria channel over an infection site. A total of 100 infection sites were measured per experiment (n = 3 independent experiments). Graphs display all measured infection sites. Error-bars: ±SD. p values were obtained after t test.
A. Related to Figure 1A. Time-lapse of GFP-2xFYVE expressing HeLa cells infected with dsRed Salmonella. Scale bar: 10 μm. B. Related to Figure 1B. Time-lapse of GFP-SNX1 expressing HeLa cells infected with dsRed Salmonella. Scale bar: 10 μm. C. Related to Figure 1E. Time-lapse of GFP-2xFYVE HeLa cells infected with dsRed ΔsopB-Salmonella mutant. Scale bar: 10 μm. D. Related to Figure 2F. Time-lapse of SNX1-KD HeLa cells expressing GFP-2xFYVE and infected with dsRed Salmonella. Scale bar: 10 μm.
SVAT formation depends on the host BAR-domain-containing protein SNX1 that has been localized on the tubules (Bujny et al., 2008). Using GFP-SNX1-transfected cells, we confirmed that SNX1 is recruited at the infection site and the tubules emanating from the shrinking SCV are SNX1 positive (Figures 1B and S1A; Video S1B). SNX1 recruitment and SVAT formation have been reported to depend on the PI(3)P accumulation on the SCV by the diphosphatase activity of the T3SS-1 effector SopB (Bujny et al., 2008, Braun et al., 2010). Using a Salmonella strain expressing FLAG-tagged SopB and super-resolution-structured illumination microscopy (SR-SIM), we localized SopB within the infected epithelial cells during these early time points (i.e., <30 min post-infection [pi]). We observed that SopB localized in a dotted manner at the IAMs and SCV membranes and along the SVATs (Figure 1C). We quantified the percentage of SopB-positive structures and detected SopB on 95% of the SCVs, 83% of the IAMs, and 60% of the SVATs over more than 150 imaged intracellular salmonellae (Figure 1D).
To test the involvement of SopB in SCV size control, we recorded time-lapse infections of HeLa cells by using the Salmonella mutant strain ΔsopB. We found that the absence of SopB significantly decreased but did not abolish the formation of IAMs, nuancing the conclusion of a previous study (Hernandez et al., 2004) (Figure 1E; Video S1C). Using fluorescent dextran to quantify the fluid uptake associated with the formation of IAMs at the infection site, we confirmed a significant decrease of IAM formation upon SopB depletion (Figure 1F). SopB activates the host kinase Akt (also called PKB) among other host pathways (Knodler et al., 2005, Kuijl et al., 2007), which is known to stimulate macropinocytosis in the model organism Dictyostelium (Rupper et al., 2001, Williams et al., 2019). Treating cells with the pan-Akt inhibitor MK-2206, we observed a significant decrease of the dextran uptake at the infection site (Figures 1G and 1H). This result suggests that IAM formation is stimulated by SopB-dependent activation of Akt. Although the number of IAMs decreased, fusion between IAMs and SCV still occurred in the absence of SopB (Figure 1E; Video S1C). We did not observe any PI(3)P-positive tubules emanating from ΔsopB-mutant-containing vacuoles over more than 150 ΔsopB-SCVs followed by time-lapse microscopy, demonstrating that SopB is required for SVAT formation, which agrees with previous studies (Bujny et al., 2008, Braun et al., 2010) (Figure 1E; Video S1C). As a consequence, although ΔsopB-SCVs became larger through IAM fusions, they did not shrink during the time course of the movies (Figure 1E; Video S1C). This corroborates previous reports on the decrease in the SCV size by SopB-dependent SVAT formation and membrane extraction (Bujny et al., 2008). Therefore, SopB plays a central role in SCV size control, whereas its depletion significantly reduces IAM and SVAT formations; the resulting SCVs are enlarged and stable.
Perturbating SCV Size Control Modifies the Rate of Vacuolar Rupture Followed by Cytosolic Releases
Previously, we reported that the absence of SCV enlargement by IAM fusion correlated with an increase in SCV rupture (Fredlund et al., 2018). To establish a causal link between SCV size control and Salmonella lifestyle, we inhibited the formation of IAMs through treatment with millimolar concentrations of amiloride, a Na+/H+ antiport inhibitor, during Salmonella infections. This treatment is specific to macropinocytosis and does not inhibit other entry pathways (West et al., 1989). To confirm the inhibition of IAM formation upon amiloride treatment, we performed infections of HeLa cells with fluorescent salmonellae in the presence of fluorescent dextran, followed by fixation and immunolabeling of the extracellular salmonellae. The cells treated with amiloride did not engulf dextran at their infection site but still contained intracellular bacteria (Figures 2A and 2B). Therefore, amiloride treatment abolished IAM formation but not Salmonella entry, highlighting the distinct nature of IAMs and nascent SCVs. To monitor the intracellular Salmonella lifestyle upon macropinocytosis inhibition, we performed Salmonella infection in the presence or absence of amiloride and incubated the cells in amiloride-free medium supplemented with gentamicin. Using LAMP1 immuno-labeling to distinguish cytosolic (LAMP1-negative) and vacuolar (LAMP1-positive) salmonellae, we observed that amiloride-mediated macropinocytosis inhibition highly increased the number of cells containing cytosolic bacteria at 2 h post-infection (pi) (Figures 2C, S2A, and S2B) and cytosolic hyper-replicating bacteria at 6 h pi (Figure 2D). We quantified this effect by flow cytometry and confirmed a significant increase in the percentage of infected cells containing hyper-replicating salmonellae (Figures 2E and S2C). To exclude potential side effects, we incubated cells with amiloride for 30 min after the infection step. The percentage of hyper-replication was similar in untreated cells and cells treated with amiloride after infection (Figure 2E). We conclude that amiloride treatment specifically affects bacterial replication if it is administrated during Salmonella entry. Thus, in the absence of IAMs, the SCVs have a higher probability to rupture, leading to Salmonella cytosolic hyper-replication.
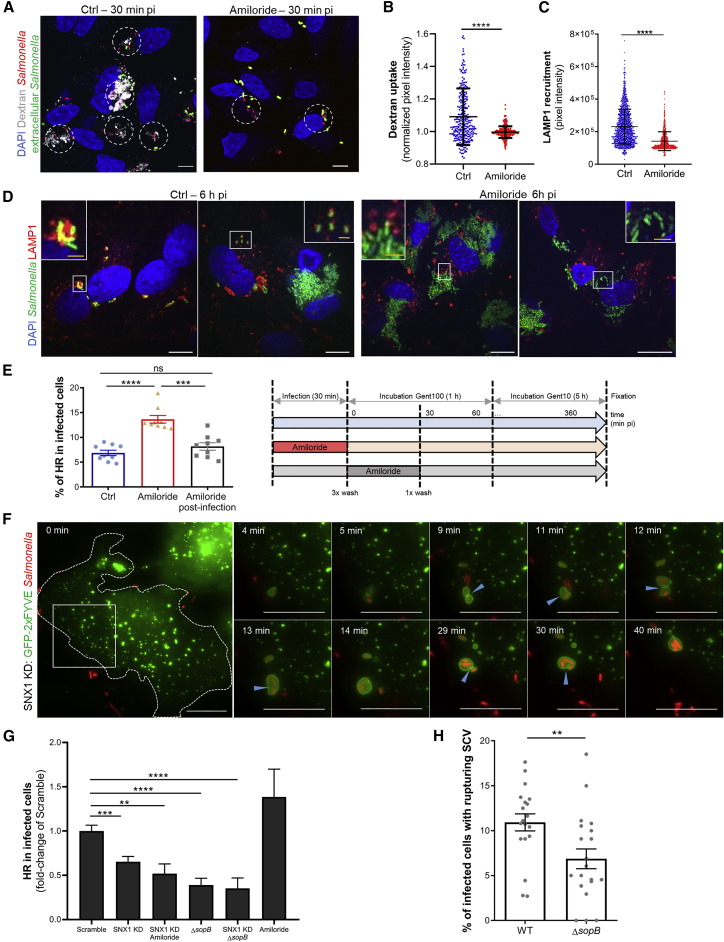
Figure 2.
Perturbating SCV Size Control Modifies the Rate of Vacuolar Rupture Followed by Cytosolic Releases
(A) Fixed microscopy of HeLa cells infected with fluorescent Salmonella (in red) in the presence of fluorescent Dextran (in white) for 30 min in the absence (left panel, Ctrl) or presence (right panel) of amiloride. Extracellular bacteria (in green) are labeled by staining the samples without permeabilization. Dashed circle: infection sites defined as circles with a diameter of 18 μm used for quantification in (B). Scale bars: 10 μm.
(B) Quantification of dextran uptake over 30 min of incubation with WT Salmonella in untreated cells (Ctrl) or cells treated with Amiloride. Each dot represent the mean fluorescence intensity of the dextran channel normalized by the mean fluorescence intensity of the bacteria channel over infection sites defined as circles with a diameter of 18 μm. A total of 100 infection sites were measured per experiment (n = 3 independent experiments). The graph displays all measured infection sites. p values were obtained after t test. Error-bars: ±SD.
(C) Quantification of LAMP1 recruitment around Salmonella at 2 h pi. Individual dots represent mean pixel intensities in the LAMP1 channel at a given bacteria localization. The graph represents measures over 10 fields of view per conditions from 1 out of 3 independent experiments. p values were obtained after t test. Error bars: ±SD. See Figure S2 for details and replicates.
(D) Fixed microscopy of HeLa cells infected with fluorescent Salmonella (in green) at 6 h pi. Infections were performed in the absence (left panels, Ctrl) or presence (right panels) of amiloride. Samples were immunolabeled for LAMP1 (in red). White scale bars: 10 μm; yellow scale bars: 2 μm.
(E) Quantification of the percentage of infected cells containing hyper-replicating (HR) bacteria by flow cytometry. Error-bars: ±SEM. p values were obtained after t test. The individual dots correspond to 3 independent experiments where each condition was analyzed in parallel in triplicate (i.e., 3 samples per conditions) infected in a 12-well plate format before collection for fluorescence-activated cell sorting (FACS) analysis. Timeline of the experimental set-up: blue-arrow, control; red arrow, presence of amiloride during infection; gray arrow, presence of Amiloride from 0 to 30 min post-infection. See Figure S2C for gating details.
(F) Time-lapse microscopy of HeLa cells transfected with siRNA pools against SNX1, transfected with GFP-2xFYVE (in green), and infected with fluorescent Salmonella (in red). Scale-bars: 10 μm. Blue arrowheads designate fusion events. See Figure S3 for knockdown efficiency and Video S1D.
(G) Quantification by high-throughput microscopy of the percentage of infected cells containing hyper-replicative (HR) salmonellae in HeLa cells transfected with a siRNA pool against SNX1 or scramble, infected with WT or ΔsopB Salmonella, in the absence or presence of amiloride. The graph represents data from three independent experiments, normalized against scrambled siRNA. p values were obtained after t test. Error bars: ±SEM.
(H) Quantifications of the percentage of infected cells containing rupturing SCVs performed by time-lapse microscopy of GFP-galectin-3-expressing HeLa cells. Each field was scored for its percentage of infected cells containing at least one rupturing SCV. Data were obtained from three independent experiments. p values were obtained after t test. Error-bars: ±SEM. See Video S4E and Figure S4 for details.
To test the consequence of the SVAT-induced SCV shrinkage on Salmonella infection, we treated the cells with a small interfering RNA (siRNA) pool against SNX1 for 72 h before infection (see Figure S3A for knockdown efficiency). We confirmed by live microscopy that SVATs were not formed in the absence of SNX1 as previously reported (Figure 2F; Video S1D; Bujny et al., 2008, Braun et al., 2010). In addition, we also noted that similarly to ΔsopB-mutant-containing vacuoles, SNX1-depleted SCVs still fused with IAMs to form large SCVs but did not shrink during the time course of the movies (Figure 2F; Video S1D). We performed high-throughput microscopy with more than >1,000 infected cells, and we quantified the percentage of hyper-replication at 6 h pi. Strikingly, hyper-replication significantly decreased under SNX1 knockdown (Figure 2G). Besides, in the absence of SVATs and IAMs (SNX1 KD, amiloride +), the percentage of cells containing hyper-replicating salmonellae remained lower than in the control (scramble) (Figure 2G). This suggests that SVAT-mediated SCV shrinkage directly promotes vacuolar rupture, whereas the fusion with IAMs and SCV enlargement only reduces the probability of rupture. Consequently, SCV enlargement and shrinkage have antagonist effects on the SCV stability, leading to the formation of different intracellular bacterial niches and lifestyles.
We also characterized the role of SopB at the crossroads of the different intracellular Salmonella lifestyles. At 6 h pi, the percentage of infected cells containing hyper-replicating bacteria was lower under ΔsopB Salmonella infection than that of wild-type (WT) Salmonella infection (Figure 2G). This reduction of the hyper-replication percentage was similar to the one observed through the SNX1 depletion. Using GFP-galectin-3 as a reporter of SCV rupture (Paz et al., 2010, Fredlund et al., 2018) (Figure S4; Video S4E), we showed that the percentage of infected cells presenting SCV rupture events significantly decreased for the ΔsopB mutant compared to WT Salmonella infection (Figure 2H). As SopB is required for SNX1 recruitment and SVAT formation, we conclude that SopB acts upstream of SNX1 to trigger the events leading to the vacuolar rupture through SVAT-mediated SCV shrinkage.
IAMs Are Enriched for the Specific SNARE Proteins VAMP2-3-8 and SNAP23-25
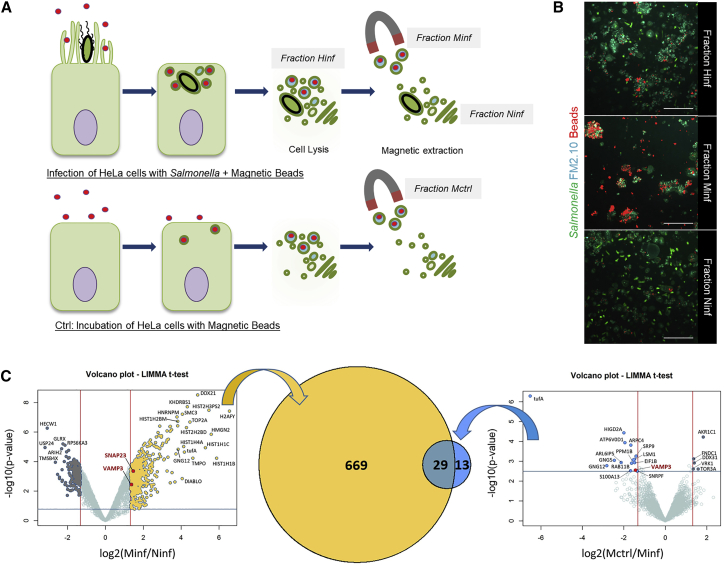
To identify the proteins driving SCV-IAM fusions, we adapted a magnetic-based cell fractionation method (Steinhäuser et al., 2013, Steinhäuser et al., 2014) to isolate IAMs. Briefly, we incubated HeLa cells with Salmonella in the presence of small (100 nm diameter) fluorescent magnetic beads. After Salmonella infection, the beads were non-specifically taken up by the closing ruffles and engulfed inside the IAMs (Figure 3A). The infected cells containing the IAMs—themselves encapsulating the magnetic beads—were subsequently washed, harvested, and homogenized. The obtained homogenate fraction (Hinf) was magnetically purified to separate the magnetic fraction (Minf), strongly enriched in bead-containing IAMs, and the non-magnetic fraction (Ninf), bead and IAM free (Figures 3A and 3B). We used non-infected bead-containing samples as control (Mctrl) by incubating HeLa cells with magnetic beads in the absence of bacteria (Figure 3A). These controls did not present IAMs. The quality of the samples was assessed by microscopy for each experimental condition (Figure 3B). All purified fractions were characterized by mass spectrometry, and we compared the protein enrichment between (1) the magnetic and non-magnetic fractions of infected cells (Minf versus Ninf; Figure 3C, yellow parts; Table 1; see also Table S1) and between (2) the magnetic fractions of infected and non-infected cells (Minf versus Mctrl; Figure 3C, blue parts; Table 1; Table S2). The IAM-containing Minf fraction was enriched in 1,106 proteins compared to Ninf, and we found 698 of them not related to DNA/RNA, nucleus, mitochondria, or ribosome, based on their FASTA headers. Similarly, 97 proteins were enriched in Minf compared to Mctrl, and 42 of them were not related to DNA/RNA, nucleus, mitochondria, or ribosome. When comparing the 698 and 42 proteins enriched in Minf, 29 of them were found in both comparisons (Figure 3C). Most of these common proteins (23 of them) are associated with the Gene Ontology (GO) term “membrane” (GO:0016020). Comparing the IAM proteomic analysis with the previously published proteome of the SCV, we identified a set of SNARE proteins significantly enriched in both the IAM- and SCV-containing cellular fractions while expressed at their endogenous level (Table 1) (Santos et al., 2015). In particular, SNAREs associated with the plasma membrane, early endosome, and recycling endosome, namely, the v-SNAREs VAMP2, VAMP3, and VAMP8 and the t-SNAREs SNAP23 and SNAP25, were found significantly more abundant in the IAM-containing (Minf) fraction (Table 1).
Figure 3.
Magnetic Isolation and Proteomic Analysis of IAMs
(A) Scheme of the IAM extraction (top panel) and the negative control (bottom panel).
(B) Fixed microscopy of the different fractions extracted and labeled with the membrane dye FM2-10 (in cyan). The bacteria are in green, and the magnetic beads are in red. Scale bars: 5 μm.
(C) Volcano plots of the proteomics results showing the respective enrichment in proteins in the Minf fraction compared to the Ninf fraction (yellow) and the Mctrl fraction (blue). Vertical lines correspond to minimal fold-changes of 2.5 between fractions and horizontal lines to a threshold on the p values associated with a false discovery rate (FDR) of 1%. Proteins detected in only one of the two compared fractions are not present on the Volcano plots. For complete protein lists, see Tables S1 and S2. The Venn diagram in the middle represents the numbers of proteins not related to DNA/RNA, nucleus, mitochondria, or ribosome. The yellow part corresponds to the proteins found more abundant in the Minf faction when compared to the Ninf fraction. The blue part corresponds to the proteins found more abundant in the Minf fraction when compared to the Mctrl fraction. 29 of these proteins are common to both comparisons.
Table 1.
Specific SNAREs Are Enriched at the Infection-Associated Compartments
| Proteins More Abundant in the Fraction Minf Than in the Fraction Ninf | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNARE | log2 Fold Change (Minf/Ninf) | p Value | Adjusted p Value | Label-Free Quantification (LFQ) Intensity |
# Peptides | Molecular Weight (kDa) | |||||
| Minf Fraction |
Ninf Fraction |
||||||||||
| xp1 | xp2 | xp3 | xp1 | xp2 | xp3 | ||||||
| VAMP8 | NA | NA | NA | 1.7E+08 | 1.4E+08 | 1.2E+08 | 0 | 0 | 0 | 5 | 11,438 |
| SNAP25 | NA | NA | NA | 3.0E+07 | 2.7E+07 | 2.6E+07 | 0 | 0 | 0 | 2 | 23,336 |
| VAMP3 | 1.35 | 4.4E−03 | 3.0E−04 | 4.5E+08 | 4.2E+08 | 4.6E+08 | 2.9E+08 | 1.8E+08 | 9.9E+07 | 5 | 11,309 |
| SNAP23 | 1.41 | 5.6E−04 | 7.3E−05 | 3.7E+08 | 4.0E+08 | 5.3E+08 | 1.9E+08 | 1.7E+08 | 1.3E+08 | 12 | 23,354 |
| Proteins More Abundant in the Fraction Minf Than in the Fraction Mctrl | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNARE | log2 Fold Change (Mctrl/Minf) | p Value | Adjusted p Value | LFQ Intensity |
# Peptides | Molecular Weight (kDa) | |||||
| Minf Fraction |
Mctrl Fraction |
||||||||||
| xp1 | xp2 | xp3 | xp1bis | xp2bis | xp3bis | ||||||
| VAMP2 | NA | NA | NA | 7.2E+07 | 8.5E+07 | 0 | 0 | 0 | 0 | 5 | 12,251 |
| VAMP3 | −1.42 | 2.9E−03 | 9.9E−03 | 4.5E+08 | 4.2E+08 | 4.6E+08 | 2.5E+08 | 1.7E+08 | 1.1E+08 | 5 | 11,309 |
| Proteins Previously Found Enriched at the SCV by Gradient-Density-Based Segmentation and Proteomic Analysisa | ||||
|---|---|---|---|---|
| SNARE | log2 Fold Change | Fold Change | Adjusted p Value | Standard Error |
| STX4 | 1.07 | 2.09 | 7.8E−4 | 0.129 |
| STX12 | 1.48 | 2.78 | 1.8E−3 | 0.323 |
Quantitative details of the enrichment of selected SNAREs: enrichment within the fraction Minf compared to the fraction Ninf, enrichment within the fraction Minf compared to the fraction Mctrl, and enrichment within the SCV-containing cellular fraction (30 min pi) compared to the cellular fraction of equal density from non-infected cells.
Data extracted from the published proteome of the SCV at 30 min pi. For details, see Santos et al. (2015).
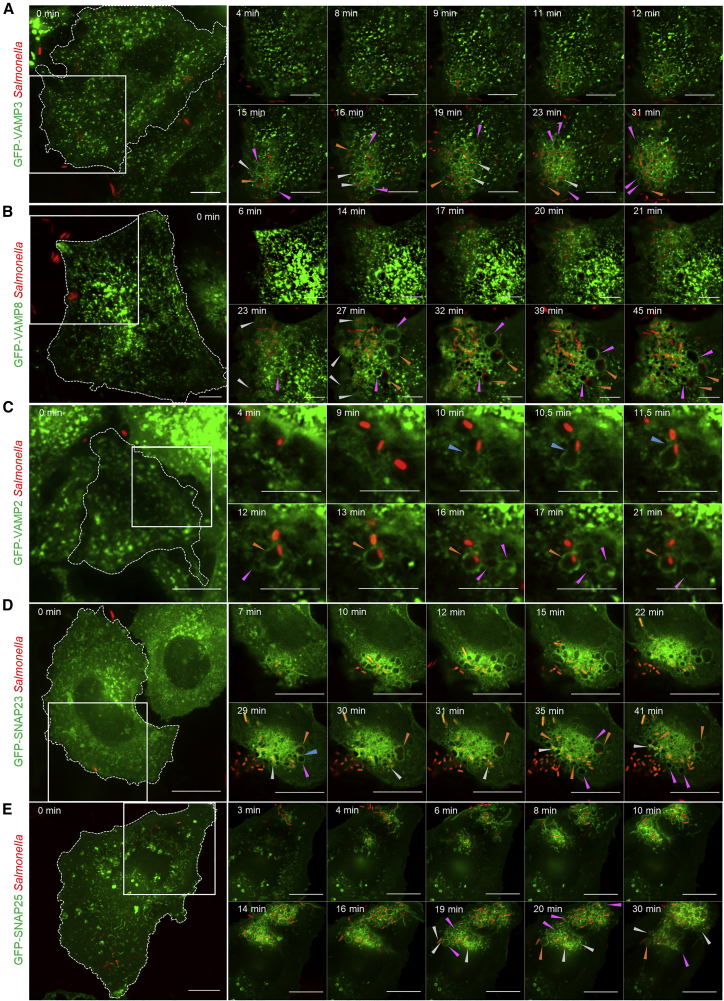
We investigated the recruitment of the identified SNAREs at the infection site by time-lapse microscopy. VAMP3 and VAMP8 are transmembrane proteins localized on vesicles at the cell periphery, such as endosomes (Chen and Scheller, 2001). Upon infection, previous studies have reported that VAMP3 is recruited in the vicinity of the Salmonella infection sites (Coppolino et al., 2001) and VAMP8 at the infection site and the Salmonella-induced macropinosomes (Dai et al., 2007). Here, we characterized the dynamic recruitment of both VAMP3 and VAMP8 at Salmonella infection sites by time-lapse microscopy (Figures 4A, 4B, S1B, and S1C; Videos S2A and S2B). We observed that both VAMP3 and VAMP8 are recruited during ruffle formation and remain locally enriched at the infection site (compared to the rest of the cell) for at least 30 min. They were both found on the IAMs and SCVs. VAMP2 is another transmembrane protein that is localized on vesicles at the cell periphery and, in particular, at recycling endosomes (Chen and Scheller, 2001). Here, we observed that VAMP2 is absent from the ruffles and nascent SCVs and its enrichment at the infection site is limited compared to VAMP3 and VAMP8 (Figures 4C and S1D; Video S2C). Our time-lapse series indicated that VAMP2 is specifically recruited at the IAM surface just (i.e., about 30 s) before the fusion step between the IAMs and the SCV (Figure 4C; Video S2C). Immunolabelling of HeLa cells infected for 30 min before fixation confirmed VAMP2, -3, and -8 localization at the enlarged SCVs (Figure S5). SNAP23 and SNAP25 are soluble proteins mainly present in the cytosol or recruited to the plasma membrane through the interaction with syntaxin (STX) membranous proteins (Vogel et al., 2000). Upon infection, we observed the recruitment of SNAP23 and SNAP25 at the infection site and, in particular, at the IAMs and SCV surface (Figures 4D, 4E, S1E, and S1F; Videos S2D and S2E).
Figure 4.
IAMs Are Enriched for the Specific SNARE Proteins VAMP2-3-8 and SNAP23-25
(A–E) Time-lapse microscopy of HeLa cells transfected with GFP-tagged VAMP3 (A), VAMP8 (B), VAMP2 (C), SNAP23 (D), and SNAP25 (E) (in green) and infected with fluorescent Salmonella (in red). Scale bars: 10 μm. Blue arrowheads designate fusion events, orange arrowheads designate large SCVs, pink arrowheads designate IAMs, and white arrowheads designate tight SCVs. See also Video S2A and Figure S1B for (A); Video S2B and Figure S1C for (B); Video S2C and Figure S1D for (C); Video S2D and Figure S1E for (D); and Video S2E and Figure S1F for (E).
Time-lapse of HeLa cells expressing either GFP-VAMP3 (A., related to Figure 4A), GFP-VAMP8 (B., related to Figure 4B), GFP-VAMP2 (C., related to Figure 4C), GFP-SNAP23 (D., related to Figure 4D), or GFP-SNAP25 (E., related to Figure 4E) and infected with dsRed Salmonella. Scale bar: 10 μm.
Involvement of SNAREs during SCV-IAM Fusion Determines the Intracellular Salmonella Lifestyle
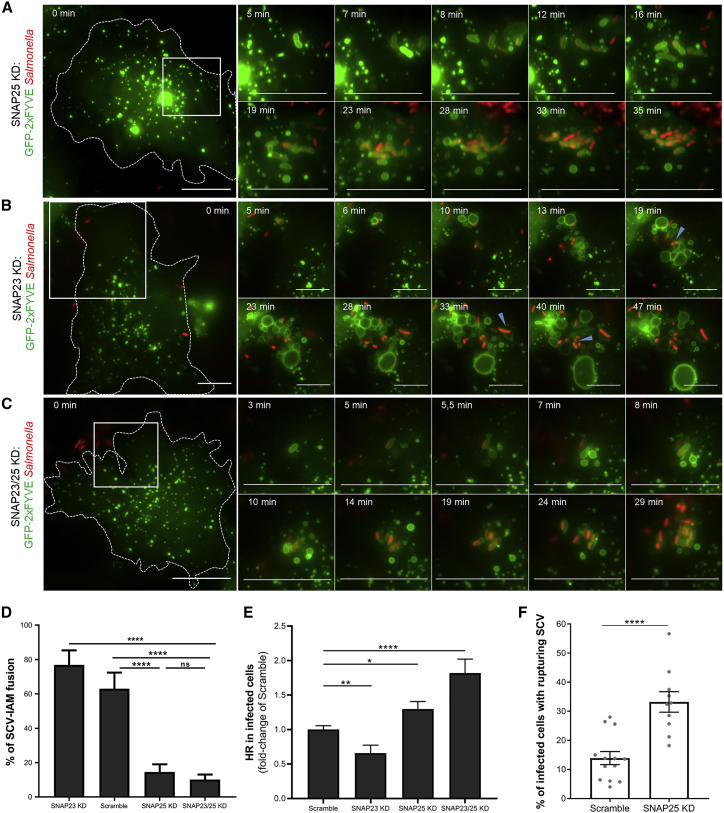
To analyze the involvement of the studied SNAREs on the intracellular lifestyle of Salmonella, we performed siRNA-mediated SNARE knockdowns, and we followed IAM-SCV fusions by time-lapse microscopy of GFP-2xFYVE-transfected cells infected by fluorescent Salmonella (see Figure S3 for siRNA efficiency). SNAP25 knockdown resulted in the most pronounced phenotype, leading to a significant fusion defect and agglomeration of tight PI(3)P-positive SCVs at the infection site (Figures 5A and 5D; Video S3A). On the contrary, the depletion of its homolog SNAP23 did not induce fusion defects and even led to more enlargements of the SCVs (Figures 5B and 5D; Video S3B). Thus, knockdowns of SNAP23 and its homolog SNAP25 induced antagonistic phenotypes. We hypothesized that SNAP25 but not SNAP23 was involved in the SCV enlargement and that the depletion of SNAP23 favored SNAP25 recruitment and activity at the infection site. To investigate this, we performed a double knockdown of SNAP23 and SNAP25. Similar to SNAP25 knockdown, SNAP23/25 double knockdown led to a significant decrease of IAM-SCV fusions, confirming that SNAP25 but not SNAP23 is involved in SCV-IAM fusions (Figures 5C and 5D; Video S3C).
Figure 5.
Involvement of SNAP Proteins during SCV-IAMs Fusion Determining Salmonella Niches
(A–C) Time-lapse microscopy of IAM and SCV formations and interactions in HeLa cells transfected with GFP-2xFYVE (in green) and with a siRNA pool against SNAP25 (A), SNAP23 (B), or both (C). Cells were infected with fluorescent Salmonella (in red). Scale bars: 10 μm. Blue arrowheads designate fusion events. See Video S3A for (A); Video S3B for (B); Video S3C for (C); and Figure S3 for knockdown efficiencies.
(D) Quantification of the percentage of SCVs fusing with IAM upon knockdown of SNAP23, SNAP25, or both. Data were obtained from three independent experiments. p values were obtained after t test. Error bars: ±SEM.
(E) Quantification by high-throughput microscopy of the infected cells containing hyper-replicating salmonellae in HeLa cells transfected with an siRNA pool against SNAP23, SNAP25, or both. The graph represents data from three independent experiments that are normalized with respective values of the scramble. p values were obtained after t test. Error bars: ±SEM.
(F) Quantifications of the percentage of infected cells containing rupturing SCVs in HeLa cells transfected with an siRNA pool against SNAP25 or scrambled siRNA. Rupturing SCVs were quantified by time-lapse microscopy using the rupture reporter GFP-galectin-3. Acquisitions were performed at 20× every 2 min for 1 h pi. Each field was scored for its percentage of infected cells containing at least one rupturing SCV. Data were obtained from three independent experiments. p values were obtained after t test. Error-bars: ±SEM. See Figure S4 for method details and Video S4E.
Time-lapse of HeLa cells expressing GFP-2xFYVE (in green) and infected with dsRed-Salmonella (in red) after 72h of siRNA treatment knocking-down either SNAP25 (A., related to Figure 5A), SNAP23 (B., related to Figure 5B), SNAP23/25 (C., related to Figure 5C), or STX4 (D., related to Figure 6C). Scale bar: 10 μm.
We tested the effects of SNAP23, SNAP25, and SNAP23/25 knockdowns on the intracellular lifestyle of Salmonella. Using high-throughput microscopy, we noted a significant increase in the percentage of hyper-replication in SNAP25 and SNAP23/25 knockdowns (Figure 5E). In contrast, the depletion of SNAP23 led to less hyper-replication (Figure 5E). This suggested that downregulation of the SNAREs involved in SCV-IAM fusion led to more SCV rupture and cytosolic hyper-replicating bacteria. To confirm this, we followed the SCV rupture by time-lapse microscopy using the galectin-3 reporter of membrane damage. Strikingly, SNAP25 knockdown induced a significant increase in the percentage of infected cells presenting SCV rupture events (Figure 5F). Together, our results suggest that SNAP25 belongs to the SNARE complex involved in the SCV enlargement, limiting vacuolar rupture and the Salmonella cytosolic lifestyle.
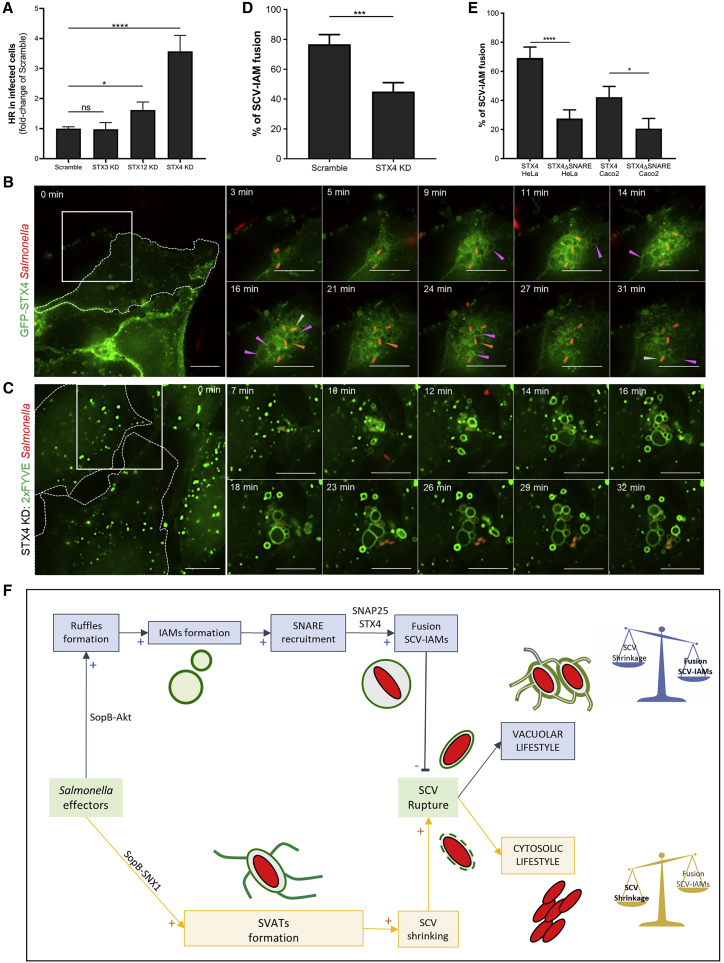
To investigate the other members of this SNARE complex, we examined the previously published SCV proteome obtained after cell fractionation at 30 min pi (Santos et al., 2015). Interestingly, only two SNAREs were significantly enriched at the SCV, namely, the syntaxin proteins STX4 and STX12 (also known as STX13), which can both interact with SNAP25 (Reed et al., 1999, Hirling et al., 2000) (Table 1). STX4 is localized to the basolateral surface of epithelial cells (Low et al., 1996), whereas STX12 shows an endosomal distribution (Prekeris et al., 1998, Koike and Jahn, 2019). STX12 can regulate homotypic early endosome fusion (Brandhorst et al., 2006, McBride et al., 1999), as well as cargo recycling from endosomal tubular extensions to the plasma membrane (Prekeris et al., 1998, Jani et al., 2015). We analyzed the impact of STX4 and STX12 knockdowns on Salmonella intracellular lifestyles compared to the other unrelated syntaxin STX3. High-throughput microscopic quantification revealed a significant increase in the percentage of infected cells containing hyper-replicating bacteria upon both STX12 and STX4 but not STX3 knockdowns (Figure 6A; see Figure S3 for knockdown efficiencies). As STX4 knockdown showed the stronger phenotype, with >3 times more cells containing hyper-replicating Salmonella, we focused on the involvement of STX4 in the SNARE complex mediating IAMs-SCV fusions. Performing time-lapse microscopy of GFP-STX4-expressing HeLa cells infected with fluorescent Salmonella, we observed the recruitment of STX4 at the infection site and, particularly, at the SCVs and IAM surfaces (Figures 6B and S1G; Video S4A). We confirmed GFP-STX4 recruitment at the Salmonella infection site, SCV, and IAMs using the intestinal epithelial Caco-2 cell line (Figures S6A and S6D; Video S4C). Quantifying IAM-SCV interactions by time-lapse microscopy of STX4-depleted HeLa cells, we observed a significant decrease of IAM-SCV fusions compared to cells treated with scrambled siRNA (Figures 6C and 6D; Video S3B). To obtain information on the SNARE motif of STX4 during the SCV-IAM fusion, we established a GFP-tagged STX4 internal truncation mutant depleted for the residues 197–273 (the SNARE motif) and transfected HeLa and Caco-2 cells with either GFP-STX4 or GFP-STX4ΔSNARE (Figures 6B and S6; Videos S4A–S4D). In both cell lines, we observed a significant decrease of SCV-IAM fusions upon transfection with GFP-STX4ΔSNARE compared to GFP-STX4, underlining the importance of the SNARE motif during this process (Figure 6E). Together, our results suggest that SNAP25 and STX4 are involved in SCV enlargement by promoting the SNARE-mediated SCV-IAM fusion determining the intracellular Salmonella niches.
Figure 6.
SCV-IAM Fusion Involves an STX4-Containing SNARE Complex
(A) Quantification of the infected cells containing hyper-replicative salmonellae by high-throughput microscopy of HeLa cells transfected with an siRNA pool against STX3, STX12, or STX4. Graph represents data from three independent experiments, normalized with respective values of scramble. p values were obtained after t test. Error-bars: ±SEM. See also Figure S3 for knockdown efficiencies.
(B) Time-lapse microscopy of HeLa cells transfected with GFP-tagged STX4 (in green) and infected with fluorescent Salmonella (in red). Scale bars: 10 μm. Orange arrowheads designate large SCVs, pink arrowheads designate IAMs, and white arrowheads designate tight SCVs. See also Video S4A and Figure S1G.
(C) Time-lapse microscopy of IAM and SCV formations and interactions using HeLa cells transfected with GFP-2xFYVE (in green), and with an siRNA pool against STX4. Cells were infected with fluorescent Salmonella (in red). Scale bars: 10 μm. See Video S3D.
(D) Quantification of the percentage of SCVs fusing with IAM upon knockdown of STX4. The graph represents data from three independent experiments. p values were obtained after t test. Error-bars: ±SEM.
(E) Quantification of the percentage of SCVs fusing with IAM upon transfection of HeLa and Caco-2 cells with GFP-STX4 or GFP-STX4ΔSNARE. The graph represents data from three independent experiments. p values were obtained after t test. Error-bars: ±SEM.
(F) Model of the SCV size control determining Salmonella intracellular lifestyle mechanism. Salmonella effectors induce ruffles and IAM formation (blue parts). SNAREs are recruited to the IAMs. SNAP25 and STX4 participate in the fusion of the IAMs with the SCV, leading to the enlargement of the bacterial vacuole. Increase in the SCV size prevents its rupture and favors vacuolar lifestyle. The bacterial effector SopB triggers the formation of SVATs, resulting in the shrinking of the vacuole (yellow parts). The decrease in the SCV size promotes its rupture and favors a cytosolic lifestyle.
A-B. Time-lapse of HeLa cells expressing either GFP-STX4 (A., related to Figure 6B) or GFP-STX4ΔSNARE (B., related to Figure S6B) in green and infected with dsRed-Salmonella (in red). C-D. Time-lapse of Caco-2 cells expressing either GFP-STX4 (C., related to Figure S6A) or GFP-STX4ΔSNARE (D., related to Figure S6C) in green and infected with dsRed-Salmonella (in red). Scale bar: 10 μm. E. Time-lapse of HeLa cells transfected with GFP-Galectin3 (in green) and infected with dsRed-Salmonella (in red), related to Figures 2H, 5F and S4. Red circles highlight the rupturing events. Scale bar: 10 μm.
Discussion
Combining mass spectrometry analysis with microscopic acquisitions at high spatiotemporal resolution, we showed that early SCV size control determines Salmonella intracellular lifestyle in epithelial cells. While identifying the main molecular players, we propose a comprehensive model of this mechanism (Figure 6F). Upon injection into the host cytosol, bacterial T3SS-1 effectors induce membrane ruffling and IAM formation (Figure 6F, blue parts). In particular, our study suggests that SopB can promote IAM formation in an Akt-dependent manner. The SNAREs VAMP2, VAMP3, VAMP8, SNAP23, and SNAP25 are recruited to the newly formed IAMs. SNAP25 participates in the fusion of the IAMs with the SCV, most likely by interacting with STX4. This fusion leads to the enlargement of the bacterial vacuole. Concomitantly, SopB also triggers the formation of SVATs, resulting in the shrinkage of the vacuole (Figure 6F, yellow parts). Therefore, at the crossroads between bacterial lifestyles, the SCV undergoes concomitant size increase and decrease, favoring maintenance or rupture, respectively.
Interestingly, SVAT formation is not only involved in the Salmonella cytosolic lifestyle but also required for proper SCV maturation and Salmonella replication within its vacuole (Bujny et al., 2008, Braun et al., 2010). In particular, Bujny and colleagues showed that the SVATs promote the exclusion of mannose-6-phosphate receptor (M6PR) from the SCV, which could set the selective interaction of SCVs with components of the late endosome. Thus, SVAT formation is a critical step for the establishment of the vacuolar lifestyle of Salmonella and the formation of an SCV with its unique identity. Our results suggest that SVAT formation only leads to SCV rupture if its effect on the SCV is not buffered enough by IAM fusions. The SVAT formations promote the rupture event, but the initial membrane damage that culminates in the larger rupturing of the vacuolar membrane remains to be elucidated. One possibility is that the T3SS-1 translocon complex fragilizes the SCV membrane and constitutes the initial damages. Such damages would be amplified by the tubule formation, leading to the complete release of the bacteria into the cytosol. Indeed, although the T3SS has been suggested to form pores in the SCV membrane (Du et al., 2016), damaged SCVs can be repaired in a process involving autophagy-associated proteins (Kreibich et al., 2015). Thus, all damaged SCVs do not result in the bacterial release in the cytosol, and the formation of the tubules may be necessary to trigger this release. In contrast, fusion with the IAMs may provide extra and non-translocon-damaged membranes that would be necessary to compensate for the transient T3SS-induced damage and counterbalance the progression of the membrane damage to full membrane rupture. In a recent study, Lau et al., 2019 have shown that the Salmonella T3SS-1 effector SopF promotes the stability of the nascent SCV. Furthermore, SopF associates with host cell membranes, most likely through phospholipid interactions. Although the authors did not find any interaction between SopF and the previously described host proteins modifying SCV stability (i.e., TBK1, MTMR4, VPS35, and COPII), our study may offer a different avenue to investigate the full mechanism of SopF-mediated SCV stabilization, considering, in particular, the role of SopF in SNARE recruitment at the SCV. So far, the stability of pathogen-containing compartments has not been linked to their size. We envision that the finding of a mechanism of size-related compartment stability will motivate more host-pathogen research studies in this direction. This concept may be extended beyond host-pathogen interactions and find a particular resonance in cell trafficking processes where fusion, tubule formation, and compartment stability are investigated as well.
Macropinocytosis has been intensively studied as one of the main entry paths for intracellular pathogens (Mercer and Helenius 2009). However, the role of infection-associated but non-pathogen-containing macropinosomes has remained elusive. In the early 1990s, Garcia-del Portillo and Finlay (1994) reported the formation of macropinosomes at the infection site of Salmonella-infected epithelial cells. Yet, their structural difference from the nascent SCV has only been clarified recently (Fredlund et al., 2018). Similarly to Salmonella, other pathogens such as the Gram-negative bacteria Shigella and the Adenovirus 2/5 induce IAM formations while entering in their host cells within distinct pathogen-containing compartments (Weiner et al., 2016, Meier et al., 2002). Shigella-induced IAMs form contacts with the Shigella vacuole and promote its rupture (Weiner et al., 2016). Furthermore, Adenovirus 2/5-induced IAMs are lysed in a virus-dependent manner, and the release of the macropinosome lumen into the cytosol promotes Adenovirus 2/5 escape and subsequent infection (Meier et al., 2002). These examples illustrate the diversity of pathogenic pathways for the exploitation of IAMs. Better knowledge is required to understand this compartment and its contributions to the establishment of pathogenic intracellular niches. We adapted a highly specific magnetic purification method to analyze the IAM proteome upon Salmonella infection. Similar approaches could be performed to study the identity and maturation of IAMs induced by other pathogens.
In this study, we focused on the SNAREs identified in the IAM proteome due to their core function during fusion. Although many intracellular pathogens hijack the host endocytic system for their survival, the SNARE proteins constitute usual suspects targeted by bacteria, either to inhibit potential fusion with the lysosome or to promote fusions allowing nutrient supply and niche maturation. SNARE subversion has been shown for the Gram-negative bacterium Legionella that infects host cells by hijacking endocytic pathways and forming a Legionella-containing vacuole (LCV). This pathogen subverts the function of the SNARE Sec22b and STXs to promote fusion between the LCV and endoplasmic reticulum (ER)-derived vesicles (Arasaki and Roy, 2010, Kagan et al., 2004). Besides, several Legionella effectors functionally mimic SNARE proteins and form complexes with VAMP4 to promote fusion between VAMP4-containing vesicles and the LCV (Shi et al., 2016). The obligate intracellular bacteria Chlamydia also interfere with SNAREs by their effector IncA, presenting SNARE-like motifs. IncA promotes the homotypic fusion of Chlamydia inclusions (Hackstadt et al., 1999, Weber et al., 2016), and it also binds directly to several host SNAREs, inhibiting fusion with other endocytic compartments (Delevoye et al., 2008; Ronzone and Paumet 2013). Similarly, the promotion and inhibition of fusion with other organelles are critical for the establishment of the Salmonella vacuolar niche. In addition to the IAMs, the SCV fuses with many other compartments and remodels the host endosomal system to promote Salmonella replication (Liss et al., 2017). Detoxified lysosomes fuse with the SCV by SifA, SKIP, the small GTPase Arl8b, and the HOPS complex to promote Salmonella replication (McGourty et al., 2012, McEwan et al., 2015, Khatter et al., 2015, Sindhwani et al., 2017). Besides, Santos et al. (2015) reported that the SCV can fuse with VAMP7-positive lysosome-like vesicles between 30 min and 3 h pi to promote SCV maturation and bacterial growth. More recently the Salmonella effector SipA has been reported to promote fusion between the SCV and early endosomes (Singh et al., 2018). Together, these studies highlight the central role of the SNARE proteins as mediators of membrane fusions in the establishment of the Salmonella vacuolar niche.
We went on monitoring the dynamic recruitment of the identified SNAREs on IAMs. Before infection, the SNAREs SNAP23 and SNAP25 are localized mainly in the cytosol and at the plasma membrane of the host cell. Upon bacterial entry, they are both localized at the ruffles and IAMs. We propose that their presence on IAMs is a consequence of their plasma membrane, and therefore ruffle, localization. VAMP3 and VAMP8 are transmembrane proteins localized on vesicles at the cell periphery and are involved in exocytosis (Chen and Scheller, 2001). Upon infection, we observed that VAMP3 and VAMP8 are present at the ruffles and IAMs, which is in agreement with previous studies (Coppolino et al., 2001, Dai et al., 2007). Possibly, the presence of VAMP3 and VAMP8 at the ruffles is due to local exocytosis required to provide the membrane material needed for the ruffle formation (Coppolino et al., 2001, Dai et al., 2007). Therefore, it is likely that VAMP3 and VAMP8 presence at the IAMs is a consequence of their presence at the plasma membrane during ruffle formation, similarly to SNAP23 and SNAP25. VAMP2 is another transmembrane protein that localizes on vesicles at the cell periphery and, in particular, at recycling endosomes (Chen and Scheller, 2001). Here, we found that VAMP2 is not detected at the ruffles, but it is later recruited at the IAMs in a highly transient manner before their fusion with the SCV. However, depletion of VAMP2 did not show a significant impact on the fusion between the SCV and IAMs. Further studies would be needed to understand the mechanism of VAMP2 recruitment and recycling from IAMs, as well as its potential role during the IAMs and SCV interaction. Based on our comparison between the IAM proteome with the one of the SCVs, we propose that SNAP25 interacts with STX4 to mediate the fusion of the IAMs with the SCV. Commonly, two SNAP25 proteins form a four-helix bundle SNARE complex structure together with one VAMP and one STX protein (Chen and Scheller, 2001). Considering that bacterial infections tend to defy canonical pathways, it could be suspected that SNARE complexes of different compositions jointly mediate membrane fusion. This would explain the lack of complete inhibition of SCV-IAM fusion upon individual SNARE depletions. Finally, one of the members of the tetrameric SNARE complex may be a bacterial effector presenting a SNARE motif. Elucidating the structure of the SNARE complex(es) involved in the SCV enlargement will be important to uncover how Salmonella manipulates its intracellular microenvironment at the molecular level.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SNX1 | Abcam | Cat#ab134126 |
| Flag | Sigma | Cat#F3165; RRID: AB_259529 |
| SNAP23 | Abcam | Cat#ab4114; RRID: AB_304287 |
| SNAP25 | Abcam | Cat#ab5666; RRID: AB_305033 |
| STX3 | Abcam | Cat#ab133750 |
| LAMP1 | Abcam | Cat#ab25245; RRID: AB_449893 |
| Rabbit polyclonal anti-S. Typhimurium LPS | Difco laboratories | N/A |
| Rab5 | BD | Cat#610725; RRID: AB_398048 |
| Bacterial and Virus Strains | ||
| SL 1344 WT | ATCC | ATCC SL1344 |
| SL 1344 ΔsopB | Wolf-Dietrich Hardt Lab | Mirold et al., 2001 |
| SL 1344 SopB-Flag | Francisco Garcia-del-Portillo Lab | Ramos-Marquès et al., 2016 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FuGENE® HD Transfection Reagent | Promega | Cat#E2311 |
| Lipofectamine RNAiMAX | Life Technologies | Cat#13778030 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| CellMask DeepRed Plasma Membrane Stain | ThermoFisher Scientific | Cat#C10046 |
| FM2-10 | ThermoFisher Scientific | Cat#T7508 |
| Paraformaldehyde 16% | Electron Microscopy Sciences | Cat#15710 |
| Ampicillin | Sigma-Aldrich | Cat#A9393 |
| Gentamicin | Sigma-Aldrich | Cat#G1397 |
| Kanamycin | Sigma-Aldrich | Cat#K1377 |
| Dulbecco’s Modified Eagle’s Medium, High Glucose, GlutaMax | Thermo Fisher Scientific | Cat#10566016 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat#F7524 |
| DPBS | Thermo Fisher Scientific | Cat#14190144 |
| MEM Non-Essential Amino Acids Solution | Thermo Fisher Scientific | Cat#11140050 |
| HEPES | Thermo Fisher Scientific | Cat#15630080 |
| 0.05% Trypsin-EDTA | Thermo Fisher Scientific | Cat#25300054 |
| ProLong Gold Antifade Mountant | ThermoFisher Scientific | Cat#P10144 |
| TetraSpeck Microspheres, 0.1 μm, fluorescent blue/green/orange/dark red | ThermoFisher Scientific | Cat#T7279 |
| NuPAGE 10% Bis-Tris protein gels | ThermoFisher Scientific | Cat# NP0301BOX |
| 5-(N-ethyl-N-isopropyl)-Amiloride | Sigma-Aldrich | Cat#A3085 |
| Dextran, Alexa Fluor 647, 10,000 MW, Fixable | Life Technologies | Cat#D22914 |
| MK-2206 | ChemieTek | Cat#CT-MK2206 |
| Magnetic bionized nanoferrite particles | Micromod | Cat#94-00-102-S |
| Complete Protease Inhibitor | Roche | Cat#11836170001 |
| Cytochalasin-D | Sigma-Aldrich | Cat#C8273 |
| Micro BCA Protein Assay Kit | ThermoFisher Scientific | Cat#23235 |
| Lys-C | Promega | Cat#VA1170 |
| Sequencing Grade Modified Trypsin | Promega | Cat#V5111 |
| Sep-Pak SPE cartridge | Waters | Cat#WAT054955 |
| Deposited Data | ||
| Proteomic analysis of the IAMs (fraction enrichments) | This study | PRIDE Archive: PXD012825 |
| Human proteome | Uniprot | v20150113 |
| Salmonella Typhimurium proteome | Uniprot | 99287 |
| Experimental Models: Cell Lines | ||
| HeLa cells | Sigma | Cat#11033106 |
| Caco-2 cells | ATCC | C2BBe1 |
| Oligonucleotides | ||
| ON-TARGETplus SMARTpool SNAP23 | Dharmacon | L-017545-00-0005 |
| ON-TARGETplus SMARTpool SNAP25 | Dharmacon | L-011394-01-0005 |
| ON-TARGETplus SMARTpool SNX1 | Dharmacon | L-013503-00-0005 |
| ON-TARGETplus SMARTpool STX3 | Dharmacon | L-015401-00-005 |
| ON-TARGETplus SMARTpool STX4 | Dharmacon | L-016256-00-0005 |
| ON-TARGETplus SMARTpool STX12 | Dharmacon | L-018246-01-0005 |
| ON-TARGETplus Negative Controls | Dharmacon | D-001810-10-05 |
| Recombinant DNA | ||
| GFP-VAMP2 | Martinez-Arca et al., 2003 | Addgene #42308 |
| GFP -VAMP3 | Galli et al., 1998 | Addgene #42310 |
| GFP-VAMP8 | Paumet et al., 2000 | Addgene #42311; |
| GFP-SNAP23 | Kuster et al., 2015 | Addgene #101914 |
| GFP-SNAP25 | Thierry Galli Lab | Verderio et al., 2004 |
| GFP-STX4 | Ludger Johannes Lab | verified by sequencing |
| GFP-STX4ΔSNARE | This study | verified by sequencing |
| GFP-SNX1 | Emmanuel Boucrot Lab | Chan Wah Hak et al., 2018 |
| GFP-galectin-3 | Kreibich et al., 2015, Paz et al., 2010 | N/A |
| pGG2 | Lelouard et al., 2010 | N/A |
| pM965 | Stecher et al., 2004 | N/A |
| Software and Algorithms | ||
| SIMcheck plugin | Ball et al., 2015 | N/A |
| FIJI | Schindelin et al., 2012 | https://fiji.sc |
| CellProfiler |
Carpenter et al., 2006; Kamentsky et al., 2011 |
https://cellprofiler.org |
| Imaris | Oxford Instruments | N/A |
| NIS-Elements Microscope Imaging Software | Nikon | N/A |
| GraphPad Prism | GraphPad Software | N/A |
| FlowJo | FlowJo, LLC | N/A |
| Andromeda | Cox et al., 2011 | https://omictools.com/andromeda-tool |
| MaxQuant software | Cox and Mann, 2008, Tyanova et al., 2016 | https://www.maxquant.org/ |
| R package DAPAR | Wieczorek et al., 2017 | https://cran.r-project.org |
| R package imp4p | Giai Gianetto, 2018 | https://cran.r-project.org |
| R package cp4p | Giai Gianetto et al., 2016 | https://cran.r-project.org |
| PANTHER gene ontology analysis | Mi et al., 2017 | http://www.pantherdb.org/ |
| Other | ||
| Zeiss LSM780 Elyra PS1 microscope equipped with the followings: | Zeiss | N/A |
| 63x/1.4 oil Plan Apo objective | Zeiss | N/A |
| EMCCD Andor Ixon 887 1K camera | Andor | N/A |
| Perkin Elmer UltraView spinning disk confocal microscope equipped with the followings: | Perkin Elmer | N/A |
| 60 × /1.3 oil objective | Nikon | N/A |
| PSU C910-50 camera | Hamamatsu | N/A |
| Inverted widefield microscope equipped with the followings: | Nikon | N/A |
| 20x/0.5NA air objective | Nikon | N/A |
| CoolSnap2 camera | Roeper Scientific | N/A |
| Andor Dragonfly 505 spinning disk confocal microscope equipped with the followings: | Oxford Instruments | N/A |
| 20x/0.75 water Plan Apo objective | Leica | N/A |
| 40x/1.30 oil HCX Plan Apo objective | Leica | N/A |
| 63x/1.40-0.60 oil Plan Apo objective | Leica | N/A |
| Andor Zyla 4.2 PLUS sCMOS Camera | Oxford Instruments | N/A |
| BD FACS CANTO cytometer | BD Biosciences | N/A |
| Deltavision epifluorescence microscope | GE Healthcare | N/A |
| HOKImag free-flow magnetic separation system | Hoock GmbH | N/A |
| Peristaltic pump | Gilson | Minipuls 3 |
| Swinging-bucket centrifuge rotor | Beckman | MLS-50 |
| Q Exactive Plus instrument | Thermo Scientific, Bremen | N/A |
| EASY nLC 1000 chromatography system | Thermo Scientific, Bremen | N/A |
| HPLC column - ReproSil-Pur Basic-C18 | Cluzeau | Cat#REPRO19BC18 |
| 96-well cell culture microplates with clear flat bottom | Greiner Bio One International | Cat#655090 |
| Precision cover glasses thickness No. 1.5H | Marienfeld Superior | Cat#0117580 |
| glass bottom microslide | Ibidi | Cat#80287 |
| 4-well inserts | Ibidi | Cat#80469 |
Lead Contact and Materails Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jost Enninga (jost.enninga@pasteur.fr). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Experimental Model and Subject Details
HeLa and Caco-2 Cells
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) at 37°C with 5% CO2. The Caco-2 C2BBe1 cells were cultured in DMEM supplemented with 10% FBS, 1% HEPES, and 1% MEM non-essential amino acids at 37°C with 10% CO2.
Bacterial Culture
All infection experiments were performed using Salmonella enterica serovar Typhimurium SL1344 (WT, ΔsopB, and SopB-Flag), in some instances expressing the pGG2 (dsRed) or pM965 (GFP) plasmids. Bacteria were cultured in Lysogeny Broth medium supplemented with 0.3 M NaCl and with the appropriate antibiotics (ampicillin or kanamycin) at 50 μg/mL.
Method Details
Cell Transfection
HeLa and Caco-2 cells were transfected with plasmid DNA from maxipreps, using the FuGENE® HD Transfection Reagent for 24 or 48 h, according to manufacturer’s instructions. The GFP-tagged VAMP2 (Martinez-Arca et al., 2003), VAMP3 (Galli et al., 1998), VAMP8 (Paumet et al., 2000), SNAP23 (Kuster et al., 2015), and SNAP25 (Verderio et al., 2004) constructs are gifts from Thierry Galli. The GFP-SNX1 construct is a gift from Emmanuel Boucrot (Chan Wah Hak et al., 2018). The pGFP-galectin-3 plasmid was described previously (Kreibich et al., 2015, Paz et al., 2010). The GFP-STX4 construct was a gift from Ludger Johannes. The GFP-STX4ΔSNARE construct was obtained by deletion of the C-terminal SNARE motif (residue 197-273) using the primers 5′-AAGAAAAAGGTCATGATTGCCATC and 5′-CCTTTTTCTTCCGGGTCACCTGTGTGTC and standard procedures. All constructs were checked by sequencing. Caco-2 cells were seeded at confluent density 72 h prior to infection allowing the formation of a continuous epithelial monolayer before infection. 24 h prior siRNA transfection, HeLa cells were seeded in 96- or 12-well plates at a density of 6000 or 45 000 cells per wells, respectively. The cells were transfected with a final siRNA concentration of 10 nM using Lipofectamine RNAiMAX and incubated for 72 h at 37°C, 5% CO2 according to manufacturer’s instruction. Protein knockdown efficiency was controlled by western blot using 10% Bis-Tris protein gels.
Bacterial Infection
Salmonella infections were performed as previously detailed (Voznica et al., 2018). The day before the infection, bacteria were grown overnight in 3 mL Lysogeny Broth medium supplemented with 0.3 M NaCl at 37°C in an orbital shaker. Growth media was supplemented with 50 μg/mL ampicillin or kanamycin. On the day of the infection, bacteria were subcultured at a 1:21 dilution in Lysogeny Broth supplemented with 0.3 M NaCl and incubated at 37°C for 3 h (i.e., until late exponential phase). Bacteria were then washed once and resuspended in EM buffer (120 mM NaCl, 7 mM KCl, 1.8 mM CaCl2, 0.8 mM Mg Cl2, 5 mM glucose, 25 mM HEPES, pH 7.3). Bacterial concentration was determined using OD600, and bacteria were diluted to the desired multiplicity of infection (MOI) in warm EM buffer. When required, EM buffer was supplemented with Dextran 10,000 MW 647 Fixable, 0.5 μM 5-(N-ethyl-N-isopropyl)-amiloride, or 50 μM of the allosteric pan-Akt inhibitor MK-2206. Upon treatment with MK-2206, HeLa cells were also pre-treated 3 h prior infection at the same concentration. Cells were washed once with EM buffer before adding bacteria and incubation for 30 min at 37°C, 5% CO2. Cells were then washed three times with clean EM buffer to eliminate extracellular bacteria. Depending on the time-point of interest, cells were either fixed at that step or incubated with EM buffer supplemented with 10% FBS and gentamicin at 100 μg/mL for 1 h. For longer time courses, media was changed after 1 h for EM buffer supplemented with 10% FBS and gentamicin at 10 μg/mL for the remaining time. Fixation was performed using 4% paraformaldehyde for 15 min at room temperature. Fixed cells were washed 3 times with PBS and stored at 4°C before staining and acquisition.
Flow Cytometry
HeLa cells were challenged with dsRed-expressing salmonellae for 30 min and incubated for 6 h following the infection protocol described above. The cells were trypsinized and fixed with 4% paraformaldehyde for 15 min at room temperature. FACS measurements were performed on a BD FACS CANTO cytometer using the excitation/emission wavelengths of 575/660nm for dsRed fluorescence detection. Doublets were discriminated using SSC/FSC gating. Fluorescence compensation was performed using the FITC and PE channels. The populations of non-infected cells, infected cells, and infected cells containing a high load of bacteria were distinguished using the dsRed fluorescence intensity per cell (see Figure S2C for gating illustration). Data were processed using the FlowJo software.
Fixed Microscopy
After fixation, samples were blocked using PBS supplemented with 20% FBS for 1 h at room temperature. The blocking medium was supplemented with 0.25% saponin when permeabilization was required. The anti-Salmonella antibody was diluted 500 times. The anti-Flag antibody was diluted 2000 times. Other primary antibodies were diluted 200 times. For high-throughput fixed microscopy, HeLa cells were cultured in 96-well cell culture microplates with clear flat bottoms. To enable accurate automatic cell segmentation, cells and nuclei were stained with CellMask DeepRed Plasma Membrane Stain and DAPI, respectively. Microscopic acquisitions were performed using a Nikon inverted widefield microscope equipped with a 20x/0.5NA air objective, an automatic programmable XY-stage, and the Nikon “perfect focus” system. 161 fields were imaged per well using a CoolSnap2 camera. Automatic image segmentation was performed using the free open-source software CellProfiler (Carpenter et al., 2006, Kamentsky et al., 2011).
For high-resolution spinning-disk microscopy and super-resolution structured illumination microscopy (SR-SIM), HeLa cells were cultured on glass support with optimal thickness of 1.5H (170 um ± 5 μm). Depending on the concentration of the drugs, transfection and immunolabeling reagents required, either 12-well plates containing glass coverslips or 2-well glass-bottom microslides containing 4-well inserts were used. After immunolabeling, samples were mounted using ProLong Gold and left at room temperature protected from light for 48 h to allow full polymerization of the mounting medium before sealing with nail polishing. Confocal microscopy was performed on an Andor Dragonfly 505 spinning disk microscope equipped with 20x/0.75 water Plan Apo objective, 40x/1.30 oil HCX Plan Apo objective, 63x/1.40-0.60 oil Plan Apo objective and an Andor Zyla 4.2 PLUS sCMOS Camera for all figures except Figure S5. Confocal microscopy shown in Figure S5 was performed on a Perkin Elmer UltraView spinning disk microscope equipped with a 60 × /1.3 oil objective and a PSU C910-50 camera. SR-SIM was performed on a Zeiss LSM780 Elyra PS1 microscope using 63x/1.4 oil Plan Apo objective with a 1.518 refractive index oil and an EMCCD Andor Ixon 887 1K camera for the detection. 15 images per plane per channel (five phases, three angles) were acquired with a Z-distance of 91 nm to perform SR-SIM images. SIM images were processed with ZEN software and then aligned with ZEN using 100 nm TetraSpeck microspheres embedded in the same conditions as the sample. SIMcheck plugin (Ball et al., 2015) in FIJI (Schindelin et al., 2012) was used to analyze the acquisition and the processing allowing optimizing resolution/signal and noise ratio/artifacts.
Time-Lapse Microscopy
For time‐lapse experiments, fields of view were selected at the microscope in a 37°C chamber. Then, the bacteria were added to the cells before the beginning of the acquisition. The bacteria swam in the culture medium and on the surface of the cells before infection, a process called “near-surface swimming” (Misselwitz et al., 2012). Then, a first bacterium stimulates the initial formation of ruffles that act as a trap for more bacteria to enter the same infection site cooperatively (Fredlund et al., 2018, Lorkowski et al., 2014, Voznica et al., 2017). As a side note, this cooperative entry results in the potential accumulation of a high number of bacteria (> 20) by the end of the movie acquisition. Acquisitions were limited to 1 h due to photobleaching and extracellular bacterial outgrowth. The displayed time-lapse series were cropped to display a few time-frame before each infection. Time‐lapse experiments were performed either on a Nikon inverted widefield microscope equipped with a 20x/0.5NA air objective, and the NIS software (Figures 2H and 5F); or on an inverted Deltavision epifluorescence microscope (GE Healthcare) and a 60x oil objective, with an integrated deconvolution analysis suite. Each dynamic event reported in a time-lapse figure was observed through at least 3 independent infection experiments which included the selection of a minimum of 5 fields of view at a magnification of 60X.
Cell Fractionation and Isolation of the IAMs
For the isolation of the IAMs, approximately 7.5 × 107 HeLa cells were grown in three flasks of 225 cm2. Cells were incubated for 30 min at 37°C with magnetic bionized nanoferrite particles of 100 nm of diameter coated with fluorescent far-red dextran at a concentration of 200 μg/mL, in the presence or absence (control) of Salmonella at MOI 30. Then, the cells were extensively washed with ice-cold homogenization buffer (HB: 250 mM sucrose, 0.5 mM ethylene glycol tetraacetic acid (EGTA), 20 mM HEPES-KOH pH 7.4, supplemented with complete protease inhibitors and 5 μg/mL cytochalasin-D), detached with a cell scraper and then homogenized in HB with a Dounce homogenizer. All steps were performed at 4°C. Between 20 and 30 strokes were performed, until more than 90% free nuclei were visible (controlled under the microscope). Nuclei and intact cells were removed to obtain the homogenate (H-) fraction by performing two sequential centrifugation steps at 200 g for 10 min each at 4°C.
In the cold chamber, a HOKImag free-flow magnetic separation system with a high-gradient 2-Tesla permanent magnetic field and a peristaltic pump were used to perform the magnetic extraction. HB was loaded in the pipe circuit and discarded once to “pre-wet” the pipes and eliminate any electrostatic effect. Then 3 mL of HB, the whole H-fraction, and 1 mL of HB were loaded sequentially in this order with a flow of about 265 μL per min (corresponding to 7.52 cm of pipe per min). Once all the H-fraction passed through the magnetic part of the HOKImag device, the flow was reversed to collect the non-magnetic (N-) fraction, and the sample-containing pipe was washed with 4 mL HB loaded on the other extremity of the pipe circuit (caution was taken to avoid the entry of any bubbles in the flow circuit). Finally, the magnetic (M-) fraction was collected after clamping the part of the pipe circuit localized between the magnet and the content of the pipe and extracted using a plastic syringe.
Small volumes of each fraction were saved for microscopic and fluorimetry analyses (detailed below) while the rest was sonicated with a water sonicator used at maximum power for a 5 min cycle of 13 s sonication followed by 30 s of breaks. This step allowed disrupting the membrane and therefore released the beads from their compartment. Elimination of the beads from the sample was performed using ultracentrifugation with density barrier. Ultracentrifugation tubes were filled with 4 mL of a sucrose solution (PBS supplemented with 818 g/mL of sucrose; density = 45% (m/m)) and 1 mL of every fraction solution was deposited on the top of the sucrose solutions without breaking the gradient. Ultracentrifugation was performed using a swinging-bucket centrifuge rotor at 25 000 g for 2 h at 4°C. As only the magnetic beads were dense enough to cross the density barrier, the first mL of each tube was collected and contained the different fractions deprived of the beads. The protein concentration was measured with a Pierce BCA protein assay using a Micro BCA Protein Assay Kit. 60 μg of protein were precipitate for each fraction using a TCA acetone precipitation. Briefly, cold 40% TCA was added to the sample at equal volume and the mix was vortexed and incubated for 30 min at 4°C before centrifugation for 15 min, at 14 000 g. The supernatant was eliminated and the pellet was washed with −20°C acetone. Samples were vortexed and incubated for 5 min at 4°C before centrifugation for 15 min, at 14 000 g. The supernatant was eliminated and the pellet was air-dried before to be stored at −20°C until mass spectrometry analysis.
The quality of the samples was controlled by fluorimetry, microscopy and Coomassie gel. The concentration of the fluorescent beads was measured using a Tecan fluorescence microplate reader with an excitation filter of 560(+/−10) nm and an emission filter of 665(+/−8) nm, piloted by the software Magellan package. Additionally, the membrane-containing fractions were stained using the dye FM2-10 (excitation/emission wavelengths: 480/610 nm) and imaged on a spinning disk microscope.
Proteomic Analysis
The protein digestion was performed as followed: Protein samples were solubilized in urea 8 M, Tris 100 mM pH 7.5, then disulfide bonds were reduced with 5 mM tris (2-carboxyethyl) phosphine (TCEP) for 20 min at 23°C and alkylated with 20 mM iodoacetamide in the dark for 30 min at room temperature. Subsequently, LysC was added for the first digestion step (protein to Lys-C ratio = 80:1) for 3 h at 30°C. Then the samples were diluted to 1 M urea with 100 mM Tris pH 7.5, and Sequencing Grade Modified Trypsin was added to the sample at a ratio of 50:1 for 16 h at 37°C. A second incubation (1 h at 37°C) with the same amount of trypsin was performed to ensure complete digestion. Proteolysis was stopped by adding 1% formic acid. Resulting peptides were desalted using Sep-Pak SPE cartridge according to manufactures instructions.
The mass spectrometry analysis was performed as followed: Peptides were analyzed on a Q Exactive Plus instrument coupled with an EASY nLC 1000 chromatography system. Samples were loaded on an in-house packed 50 cm nano-HPLC column (75 μm inner diameter) with C18 resin (1.9 μm particles, 100 Å pore size, Reprosil-Pur Basic C18-HD resin) and equilibrated in 98% solvent A (H2O, 0.1% formic acid) and 2% solvent B (acetonitrile, 0.1% formic acid). Peptides were eluted using a 2 to 22% gradient of solvent B during 150 min, then a 22 to 45% gradient of solvent B during 60 min and finally a 45 to 80% gradient of solvent B during 10 min all at 250 nL/min flow rate. The instrument method for the Q Exactive Plus was set up in the data-dependent acquisition mode. After a survey scan in the Orbitrap (resolution 70 000), the 10 most intense precursor ions were selected for higher-energy collisional dissociation (HCD) fragmentation with normalized collision energy set up to 28. Charge state screening was enabled, and precursors with unknown charge state or a charge state of 1 and >7 were excluded. Dynamic exclusion was enabled for 45 s.
The proteomic data processing and analysis were performed as followed: All data were searched using Andromeda (Cox et al., 2011) with the MaxQuant software version 1.5.3.8 (Cox and Mann, 2008, Tyanova et al., 2016) against Uniprot proteome database of Human (v20150113, 89706 entries) and S-typhimurium 99287 (4533 entries). Usual known mass spectrometry contaminants and reversed sequences were also searched. Andromeda searches were performed choosing trypsin as specific enzyme with a maximum number of two missed cleavages. Possible modifications included carbamidomethylation (Cys, fixed), oxidation (Met, variable) and N-terminal acetylation (variable). The mass tolerance in MS was set to 20 parts per million (ppm) for the first search then 6 ppm for the main search and 10 ppm for the MS/MS. Maximum peptide charge was set to 7 and 5 amino acids were required as minimum peptide length. The “match between runs” feature was used between conditions with a maximal retention time window of 1 min. One unique peptide to the protein group was required for the protein identification. A false discovery rate (FDR) cutoff of 1% was applied at the peptide and protein levels. The MaxLFQ, Maxquant’s label-free quantification (LFQ) algorithm was used to calculate protein intensity profiles across samples (Cox et al., 2011). Data were filtered by requiring a minimum peptide ratio count of 2 in MaxLFQ.
Quantification and Statistical Analysis
Following image acquisition, fixed samples and time‐lapse images were analyzed using Fiji, Imaris or CellProfiler. The number of infected cells was assessed using CellProfiler automatic segmentation. LAMP1 recruitment around intracellular salmonellae was measured using Imaris analyses of 3D confocal images. “Large” versus “tight” SCVs were distinguished using the following characteristics: Tight SCVs are rod-shaped following the outline of the bacteria and no lumen can be distinguished between the bacteria and the SCV membrane by standard light microscopy. Large SCVs have semi-spherical to spherical shapes and a lumen can be clearly distinguished between the bacteria and the SCV membrane by standard light microscopy. Statistical analyses of microscopic acquisitions were performed using GraphPad Prism. t tests were used to evaluate the significance of the results, referred like ∗, ∗∗, ∗∗∗, ∗∗∗∗ for p values < 0.05, < 0.01, < 0.001, and < 0.0001, respectively. Statistical details of experiments can be found in the figure legends.
The statistical analysis of the proteomics data was performed as follows: Three biological replicates were acquired per condition. To highlight significantly differentially abundant proteins between two conditions, differential analyses were conducted through the following data analysis pipeline: (1) deleting the reverse and potential contaminant proteins; (2) keeping only proteins with at least two quantified values in one of the two compared fractions to limit misidentifications and ensure a minimum of replicability; (3) log2-transformation of the remaining LFQ intensities of proteins; (4) normalizing the LFQ intensities by median centering within conditions thanks to the normalizeD function of the R package DAPAR (Wieczorek et al., 2017), (5) putting aside proteins without any LFQ value in one of both compared fractions: as they are quantitatively present in a condition and absent in another, they are considered as differentially abundant proteins; (6) imputing the missing values of the remaining proteins using the impute.slsa function of the R package imp4p (Giai Gianetto, 2018); and (7) performing statistical differential analysis on them by requiring a minimum fold-change of 2.5 between conditions and by using a LIMMA t test (Smyth, 2005, Ritchie et al., 2015) combined with an adaptive Benjamini-Hochberg correction of the p values thanks to the adjust.p function of the R package cp4p (Giai Gianetto et al., 2016). The robust method of Pounds and Cheng was used to estimate the proportion of true null hypotheses among the set of statistical tests (Pounds and Cheng 2006). The proteins associated with an adjusted p value inferior to an FDR level of 1% have been considered as significantly differentially abundant proteins. Finally, the proteins of interest are therefore the proteins that emerge from this statistical analysis supplemented by those being quantitatively absent from one condition and present in another. The lists and details of the significantly differentially abundant proteins arising from the analyses between the fraction Minf and Mctrl, and between the fraction Minf and Ninf have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository. To focus on proteins which are not related to DNA/RNA, nucleus, mitochondria or ribosome, we filtered these results by deleting the proteins containing the following keywords in their FASTA headers: “mitochondrial,” “ribosomal,” “DNA,” “RNA,” “nuclear,” “histone.” The filtered results can be found in Tables S1 and S2, and on PRIDE repository. The gene ontology analysis was performed using PANTHER (Protein ANalysis THrough Evolutionary Relationships) tools (Mi et al., 2017) (http://www.pantherdb.org/).
Data and Code Availability
The accession number for the proteomics data reported in this paper is PRIDE: PXD012825. These data are accessible to the readers at https://www.ebi.ac.uk/pride/archive/.
Acknowledgments
V. Stévenin was supported by a PhD fellowship from the University Paris Diderot allocated by the ENS Paris-Saclay, a grant from the Fondation pour la Recherche Médicale (FRM; FDT20170436843), and an extension grant from the BCI Department of Institut Pasteur. V. Sohst and N.R. were supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) Excellence Cluster 306 “Inflammation at Interfaces” (MTP2). J.E. is a member of the LabEx consortia IBEID and MilieuInterieur. J.E. also acknowledges support from the ANR (grant StopBugEntry and AutoHostPath) and the ERC (CoG EndoSubvert). N.R. is very grateful for funding within the DFG priority program (SPP1580) (NR: Re1228 5-1, Re1228 5-2) and also thanks the German Center for Infection (DZIF) for financial support. UTechS PBI is part of the France–BioImaging infrastructure network (FBI) supported by the Agence Nationale pour la Recherche (ANR-10–INSB–04; Investments for the Future), and acknowledges support from ANR/FBI and the Région Ile-de-France (program “Domaine d’Intérêt Majeur-Malinf”) for the use of the Zeiss LSM 780 Elyra PS1 microscope. The authors acknowledge T. Galli, E. Boucrot, L. Johannes, and F. Garcia-del Portillo for providing tools and scientific feedback. The authors are deeply grateful to Sjaak Neefjes and his group for providing equipment, reagents, and scientific feedback to V. Stévenin during the revision period.
Author Contributions
V. Stévenin and J.E. designed the study. V. Stévenin, Y.-Y.C, and Y.L.T. performed the original experiments. V. Stévenin, Y.-Y.C, and C.H.L. performed experiments during revision. V. Stévenin and J.E. wrote the manuscript. J.E. secured funding. M.D., Q.G.G, and M.M. performed, analyzed, and supervised the proteomic analysis. V. Sohst and N.R. provided expertise on magnetic purification. A.S. acquired the SIM images.
Declaration of Interests
The authors declare no competing interests.
Published: December 17, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.11.049.
Supplemental Information
For the full protein list, see PRIDE repository PXD012825.
For the full protein list, see PRIDE repository PXD012825.
References
- Arasaki K., Roy C.R. Legionella pneumophila promotes functional interactions between plasma membrane syntaxins and Sec22b. Traffic. 2010;11:587–600. doi: 10.1111/j.1600-0854.2010.01050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G., Demmerle J., Kaufmann R., Davis I., Dobbie I.M., Schermelleh L. SIMcheck: a Toolbox for Successful Super-resolution Structured Illumination Microscopy. Sci. Rep. 2015;5:15915. doi: 10.1038/srep15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham C.L., Smith A.C., Bakowski M.A., Yoshimori T., Brumell J.H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Brandhorst D., Zwilling D., Rizzoli S.O., Lippert U., Lang T., Jahn R. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. USA. 2006;103:2701–2706. doi: 10.1073/pnas.0511138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V., Wong A., Landekic M., Hong W.J., Grinstein S., Brumell J.H. Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell. Microbiol. 2010;12:1352–1367. doi: 10.1111/j.1462-5822.2010.01476.x. [DOI] [PubMed] [Google Scholar]
- Bujny M.V., Ewels P.A., Humphrey S., Attar N., Jepson M.A., Cullen P.J. Sorting nexin-1 defines an early phase of Salmonella-containing vacuole-remodeling during Salmonella infection. J. Cell Sci. 2008;121:2027–2036. doi: 10.1242/jcs.018432. [DOI] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P.B., Collins F.M. The route of enteric infection in normal mice. J. Exp. Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Wah Hak L., Khan S., Di Meglio I., Law A.L., Lucken-Ardjomande Häsler S., Quintaneiro L.M., Ferreira A.P.A., Krause M., McMahon H.T., Boucrot E. FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat. Cell Biol. 2018;20:1023–1031. doi: 10.1038/s41556-018-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.A., Scheller R.H. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Coppolino M.G., Kong C., Mohtashami M., Schreiber A.D., Brumell J.H., Finlay B.B., Grinstein S., Trimble W.S. Requirement for N-ethylmaleimide-sensitive factor activity at different stages of bacterial invasion and phagocytosis. J. Biol. Chem. 2001;276:4772–4780. doi: 10.1074/jbc.M007792200. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Dai S., Zhang Y., Weimbs T., Yaffe M.B., Zhou D. Bacteria-generated PtdIns(3)P recruits VAMP8 to facilitate phagocytosis. Traffic. 2007;8:1365–1374. doi: 10.1111/j.1600-0854.2007.00613.x. [DOI] [PubMed] [Google Scholar]
- Delevoye C., Nilges M., Dehoux P., Paumet F., Perrinet S., Dautry-Varsat A., Subtil A. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 2008;4:e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Reeves A.Z., Klein J.A., Twedt D.J., Knodler L.A., Lesser C.F. The type III secretion system apparatus determines the intracellular niche of bacterial pathogens. Proc. Natl. Acad. Sci. USA. 2016;113:4794–4799. doi: 10.1073/pnas.1520699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund J., Santos J.C., Stévenin V., Weiner A., Latour-Lambert P., Rechav K., Mallet A., Krijnse-Locker J., Elbaum M., Enninga J. The entry of Salmonella in a distinct tight compartment revealed at high temporal and ultrastructural resolution. Cell. Microbiol. 2018;20:1–13. doi: 10.1111/cmi.12816. [DOI] [PubMed] [Google Scholar]
- Galli T., Zahraoui A., Vaidyanathan V.V., Raposo G., Tian J.M., Karin M., Niemann H., Louvard D. A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol. Biol. Cell. 1998;9:1437–1448. doi: 10.1091/mbc.9.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F., Finlay B.B. Salmonella invasion of nonphagocytic cells induces formation of macropinosomes in the host cell. Infect. Immun. 1994;62:4641–4645. doi: 10.1128/iai.62.10.4641-4645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giai Gianetto Q. 2018. imp4p: Imputation for Proteomics. R package version 0.6. [Google Scholar]
- Giai Gianetto Q., Combes F., Ramus C., Bruley C., Couté Y., Burger T. Calibration plot for proteomics: A graphical tool to visually check the assumptions underlying FDR control in quantitative experiments. Proteomics. 2016;16:29–32. doi: 10.1002/pmic.201500189. [DOI] [PubMed] [Google Scholar]
- Hackstadt T., Scidmore-Carlson M.A., Shaw E.I., Fischer E.R. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell. Microbiol. 1999;1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Hernandez L.D., Hueffer K., Wenk M.R., Galán J.E. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- Hirling H., Steiner P., Chaperon C., Marsault R., Regazzi R., Catsicas S. Syntaxin 13 is a developmentally regulated SNARE involved in neurite outgrowth and endosomal trafficking. Eur. J. Neurosci. 2000;12:1913–1923. doi: 10.1046/j.1460-9568.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- Jani R.A., Purushothaman L.K., Rani S., Bergam P., Setty S.R.G. STX13 regulates cargo delivery from recycling endosomes during melanosome biogenesis. J. Cell Sci. 2015;128:3263–3276. doi: 10.1242/jcs.171165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J.C., Stein M.P., Pypaert M., Roy C.R. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 2004;199:1201–1211. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L., Jones T.R., Fraser A., Bray M.A., Logan D.J., Madden K.L., Ljosa V., Rueden C., Eliceiri K.W., Carpenter A.E. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter D., Raina V.B., Dwivedi D., Sindhwani A., Bahl S., Sharma M. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J. Cell Sci. 2015;128:1746–1761. doi: 10.1242/jcs.162651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Finlay B.B., Steele-Mortimer O. The Salmonella effector protein SopB protects epithelial cells from apoptosis by sustained activation of Akt. J. Biol. Chem. 2005;280:9058–9064. doi: 10.1074/jbc.M412588200. [DOI] [PubMed] [Google Scholar]
- Knodler L.A., Vallance B.A., Celli J., Winfree S., Hansen B., Montero M., Steele-Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. USA. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Nair V., Steele-Mortimer O. Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS One. 2014;9:e84681. doi: 10.1371/journal.pone.0084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S., Jahn R. SNAREs define targeting specificity of trafficking vesicles by combinatorial interaction with tethering factors. Nat. Commun. 2019;10:1608. doi: 10.1038/s41467-019-09617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich S., Emmenlauer M., Fredlund J., Rämö P., Münz C., Dehio C., Enninga J., Hardt W.D. Autophagy Proteins Promote Repair of Endosomal Membranes Damaged by the Salmonella Type Three Secretion System 1. Cell Host Microbe. 2015;18:527–537. doi: 10.1016/j.chom.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Kuijl C., Savage N.D.L., Marsman M., Tuin A.W., Janssen L., Egan D.A., Ketema M., van den Nieuwendijk R., van den Eeden S.J., Geluk A. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450:725–730. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- Kuster A., Nola S., Dingli F., Vacca B., Gauchy C., Beaujouan J.C., Nunez M., Moncion T., Loew D., Formstecher E. The Q-soluble N-Ethylmaleimide-sensitive Factor Attachment Protein Receptor (Q-SNARE) SNAP-47 Regulates Trafficking of Selected Vesicle-associated Membrane Proteins (VAMPs) J. Biol. Chem. 2015;290:28056–28069. doi: 10.1074/jbc.M115.666362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock D.L., Chaudhary A., Miller S.I. Salmonellae interactions with host processes. Nat. Rev. Microbiol. 2015;13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N., Haeberle A.L., O’Keeffe B.J., Latomanski E.A., Celli J., Newton H.J., Knodler L.A. SopF, a phosphoinositide binding effector, promotes the stability of the nascent Salmonella-containing vacuole. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelouard H., Henri S., De Bovis B., Mugnier B., Chollat-Namy A., Malissen B., Méresse S., Gorvel J.P. Pathogenic bacteria and dead cells are internalized by a unique subset of Peyer’s patch dendritic cells that express lysozyme. Gastroenterology. 2010;138:173–184.e1-3. doi: 10.1053/j.gastro.2009.09.051. [DOI] [PubMed] [Google Scholar]
- Liss V., Swart A.L., Kehl A., Hermanns N., Zhang Y., Chikkaballi D., Böhles N., Deiwick J., Hensel M. Salmonella enterica Remodels the Host Cell Endosomal System for Efficient Intravacuolar Nutrition. Cell Host Microbe. 2017;21:390–402. doi: 10.1016/j.chom.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Lorkowski M., Felipe-López A., Danzer C.A., Hansmeier N., Hensel M. Salmonella enterica invasion of polarized epithelial cells is a highly cooperative effort. Infect. Immun. 2014;82:2657–2667. doi: 10.1128/IAI.00023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low S.H., Chapin S.J., Weimbs T., Kömüves L.G., Bennett M.K., Mostov K.E. Differential localization of syntaxin isoforms in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 1996;7:2007–2018. doi: 10.1091/mbc.7.12.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- Malik-Kale P., Winfree S., Steele-Mortimer O. The bimodal lifestyle of intracellular Salmonella in epithelial cells: replication in the cytosol obscures defects in vacuolar replication. PLoS One. 2012;7:e38732. doi: 10.1371/journal.pone.0038732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arca S., Rudge R., Vacca M., Raposo G., Camonis J., Proux-Gillardeaux V., Daviet L., Formstecher E., Hamburger A., Filippini F. A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. USA. 2003;100:9011–9016. doi: 10.1073/pnas.1431910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Rybin V., Murphy C., Giner A., Teasdale R., Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- McEwan D.G., Richter B., Claudi B., Wigge C., Wild P., Farhan H., McGourty K., Coxon F.P., Franz-Wachtel M., Perdu B. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015;17:58–71. doi: 10.1016/j.chom.2014.11.011. [DOI] [PubMed] [Google Scholar]
- McGourty K., Thurston T.L., Matthews S.A., Pinaud L., Mota L.J., Holden D.W. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science. 2012;338:963–967. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O., Boucke K., Hammer S.V., Keller S., Stidwill R.P., Hemmi S., Greber U.F. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Biol. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J., Helenius A. Virus entry by macropinocytosis. Nat. Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirold S., Ehrbar K., Weissmüller A., Prager R., Tschäpe H., Rüssmann H., Hardt W.-D. Salmonella Host Cell Invasion Emerged by Acquisition of a Mosaic of Separate Genetic Elements, Including Salmonella Pathogenicity Island 1 (SPI1), SPI5, and sopE2. Journal of Bacteriology. 2001;183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B., Barrett N., Kreibich S., Vonaesch P., Andritschke D., Rout S., Weidner K., Sormaz M., Songhet P., Horvath P. Near surface swimming of Salmonella Typhimurium explains target-site selection and cooperative invasion. PLoS Pathog. 2012;8:e1002810. doi: 10.1371/journal.ppat.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattni K., Jepson M., Stenmark H., Banting G. A Ptdlns(3)P-specific probe cycles on and off host cell membranes during Salmonella invasion of mammalian cells. Curr. Biol. 2001;11:1636–1642. doi: 10.1016/s0960-9822(01)00486-9. [DOI] [PubMed] [Google Scholar]
- Paumet F., Le Mao J., Martin S., Galli T., David B., Blank U., Roa M. Soluble NSF attachment protein receptors (SNAREs) in RBL-2H3 mast cells: functional role of syntaxin 4 in exocytosis and identification of a vesicle-associated membrane protein 8-containing secretory compartment. J. Immunol. 2000;164:5850–5857. doi: 10.4049/jimmunol.164.11.5850. [DOI] [PubMed] [Google Scholar]
- Paz I., Sachse M., Dupont N., Mounier J., Cederfur C., Enninga J., Leffler H., Poirier F., Prevost M.C., Lafont F., Sansonetti P. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell. Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin A.J., Jiang X., Birmingham C.L., So N.S.Y., Brumell J.H. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr. Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Pounds S., Cheng C. Robust estimation of the false discovery rate. Bioinformatics. 2006;22:1979–1987. doi: 10.1093/bioinformatics/btl328. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Chen Y.A., Scheller R.H. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Marquès E., Zambrano S., Tiérrez A., Bianchi M.E., Agresti A., García-Del Portillo F. Single-cell analyses reveal an attenuated NF-κB response in the Salmonella-infected fibroblast. Virulence. 2016;5594:719–740. doi: 10.1080/21505594.2016.1229727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed G.L., Houng A.K., Fitzgerald M.L. Human platelets contain SNARE proteins and a Sec1p homologue that interacts with syntaxin 4 and is phosphorylated after thrombin activation: implications for platelet secretion. Blood. 1999;93:2617–2626. [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzone E., Paumet F. Two coiled-coil domains of Chlamydia trachomatis IncA affect membrane fusion events during infection. PLoS One. 2013;8:e69769. doi: 10.1371/journal.pone.0069769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupper A., Lee K., Knecht D., Cardelli J. Sequential activities of phosphoinositide 3-kinase, PKB/Aakt, and Rab7 during macropinosome formation in Dictyostelium. Mol. Biol. Cell. 2001;12:2813–2824. doi: 10.1091/mbc.12.9.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.C., Duchateau M., Fredlund J., Weiner A., Mallet A., Schmitt C., Matondo M., Hourdel V., Chamot-Rooke J., Enninga J. The COPII complex and lysosomal VAMP7 determine intracellular Salmonella localization and growth. Cell. Microbiol. 2015;17:1699–1720. doi: 10.1111/cmi.12475. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., Hardt W.D. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16:237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Shi X., Halder P., Yavuz H., Jahn R., Shuman H.A. Direct targeting of membrane fusion by SNARE mimicry: Convergent evolution of Legionella effectors. Proc. Natl. Acad. Sci. USA. 2016;113:8807–8812. doi: 10.1073/pnas.1608755113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhwani A., Arya S.B., Kaur H., Jagga D., Tuli A., Sharma M. Salmonella exploits the host endolysosomal tethering factor HOPS complex to promote its intravacuolar replication. PLoS Pathog. 2017;13:e1006700. doi: 10.1371/journal.ppat.1006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.K., Kapoor A., Lomash R.M., Kumar K., Kamerkar S.C., Pucadyil T.J., Mukhopadhyay A. Salmonella SipA mimics a cognate SNARE for host Syntaxin8 to promote fusion with early endosomes. J. Cell Biol. 2018;217:4199–4214. doi: 10.1083/jcb.201802155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Limma: Linear Models for microarray data. In: Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; 2005. pp. 397–420. [Google Scholar]
- Stecher B., Hapfelmeier S., Müller C., Kremer M., Stallmach T., Hardt W.D. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 2004;72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhäuser C., Heigl U., Tchikov V., Schwarz J., Gutsmann T., Seeger K., Brandenburg J., Fritsch J., Schroeder J., Wiesmüller K.H. Lipid-labeling facilitates a novel magnetic isolation procedure to characterize pathogen-containing phagosomes. Traffic. 2013;14:321–336. doi: 10.1111/tra.12031. [DOI] [PubMed] [Google Scholar]
- Steinhäuser C., Dallenga T., Tchikov V., Schaible U.E., Schütze S., Reiling N. Immunomagnetic isolation of pathogen-containing phagosomes and apoptotic blebs from primary phagocytes. Curr. Protoc. Immunol. 2014;105:1–26. doi: 10.1002/0471142735.im1436s105. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Verderio C., Pozzi D., Pravettoni E., Inverardi F., Schenk U., Coco S., Proux-Gillardeaux V., Galli T., Rossetto O., Frassoni C., Matteoli M. SNAP-25 modulation of calcium dynamics underlies differences in GABAergic and glutamatergic responsiveness to depolarization. Neuron. 2004;41:599–610. doi: 10.1016/s0896-6273(04)00077-7. [DOI] [PubMed] [Google Scholar]
- Vogel K., Cabaniols J.P., Roche P.A. Targeting of SNAP-25 to membranes is mediated by its association with the target SNARE syntaxin. J. Biol. Chem. 2000;275:2959–2965. doi: 10.1074/jbc.275.4.2959. [DOI] [PubMed] [Google Scholar]
- Voznica J., Gardella C., Belotserkovsky I., Dufour A., Enninga J., Stévenin V. Identification of parameters of host cell vulnerability during Salmonella infection by quantitative image amalysis and modeling. Infect. Immun. 2017;86:e00644-17. doi: 10.1128/IAI.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznica J., Enninga J., Stévenin V. High-throughput microscopic analysis of Salmonella invasion of host cells. Bio Protoc. 2018;8:e3017. doi: 10.21769/BioProtoc.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson K.G., Holden D.W. Dynamics of growth and dissemination of Salmonella in vivo. Cell. Microbiol. 2010;12:1389–1397. doi: 10.1111/j.1462-5822.2010.01511.x. [DOI] [PubMed] [Google Scholar]
- Weber M.M., Noriea N.F., Bauler L.D., Lam J.L., Sager J., Wesolowski J., Paumet F., Hackstadt T. A Functional Core of IncA Is Required for Chlamydia trachomatis Inclusion Fusion. J. Bacteriol. 2016;198:1347–1355. doi: 10.1128/JB.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Mellouk N., Lopez-Montero N., Chang Y.Y., Souque C., Schmitt C., Enninga J. Macropinosomes are key players in early Shigella invasion and vacuolar escape in epithelial cells. PLoS Pathog. 2016;12:e1005602. doi: 10.1371/journal.ppat.1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M.A., Bretscher M.S., Watts C. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J. Cell Biol. 1989;109:2731–2739. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek S., Combes F., Lazar C., Giai Gianetto Q., Gatto L., Dorffer A., Hesse A.M., Couté Y., Ferro M., Bruley C., Burger T. DAPAR & ProStaR: software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics. 2017;33:135–136. doi: 10.1093/bioinformatics/btw580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.D., Peak-Chew S.-Y., Paschke P., Kay R.R. Akt and SGK protein kinases are required for efficient feeding by macropinocytosis. J. Cell Sci. 2019;132:jcs224998. doi: 10.1242/jcs.224998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Related to Figure 1A. Time-lapse of GFP-2xFYVE expressing HeLa cells infected with dsRed Salmonella. Scale bar: 10 μm. B. Related to Figure 1B. Time-lapse of GFP-SNX1 expressing HeLa cells infected with dsRed Salmonella. Scale bar: 10 μm. C. Related to Figure 1E. Time-lapse of GFP-2xFYVE HeLa cells infected with dsRed ΔsopB-Salmonella mutant. Scale bar: 10 μm. D. Related to Figure 2F. Time-lapse of SNX1-KD HeLa cells expressing GFP-2xFYVE and infected with dsRed Salmonella. Scale bar: 10 μm.
Time-lapse of HeLa cells expressing either GFP-VAMP3 (A., related to Figure 4A), GFP-VAMP8 (B., related to Figure 4B), GFP-VAMP2 (C., related to Figure 4C), GFP-SNAP23 (D., related to Figure 4D), or GFP-SNAP25 (E., related to Figure 4E) and infected with dsRed Salmonella. Scale bar: 10 μm.
Time-lapse of HeLa cells expressing GFP-2xFYVE (in green) and infected with dsRed-Salmonella (in red) after 72h of siRNA treatment knocking-down either SNAP25 (A., related to Figure 5A), SNAP23 (B., related to Figure 5B), SNAP23/25 (C., related to Figure 5C), or STX4 (D., related to Figure 6C). Scale bar: 10 μm.
A-B. Time-lapse of HeLa cells expressing either GFP-STX4 (A., related to Figure 6B) or GFP-STX4ΔSNARE (B., related to Figure S6B) in green and infected with dsRed-Salmonella (in red). C-D. Time-lapse of Caco-2 cells expressing either GFP-STX4 (C., related to Figure S6A) or GFP-STX4ΔSNARE (D., related to Figure S6C) in green and infected with dsRed-Salmonella (in red). Scale bar: 10 μm. E. Time-lapse of HeLa cells transfected with GFP-Galectin3 (in green) and infected with dsRed-Salmonella (in red), related to Figures 2H, 5F and S4. Red circles highlight the rupturing events. Scale bar: 10 μm.
For the full protein list, see PRIDE repository PXD012825.
For the full protein list, see PRIDE repository PXD012825.
Data Availability Statement
The accession number for the proteomics data reported in this paper is PRIDE: PXD012825. These data are accessible to the readers at https://www.ebi.ac.uk/pride/archive/.