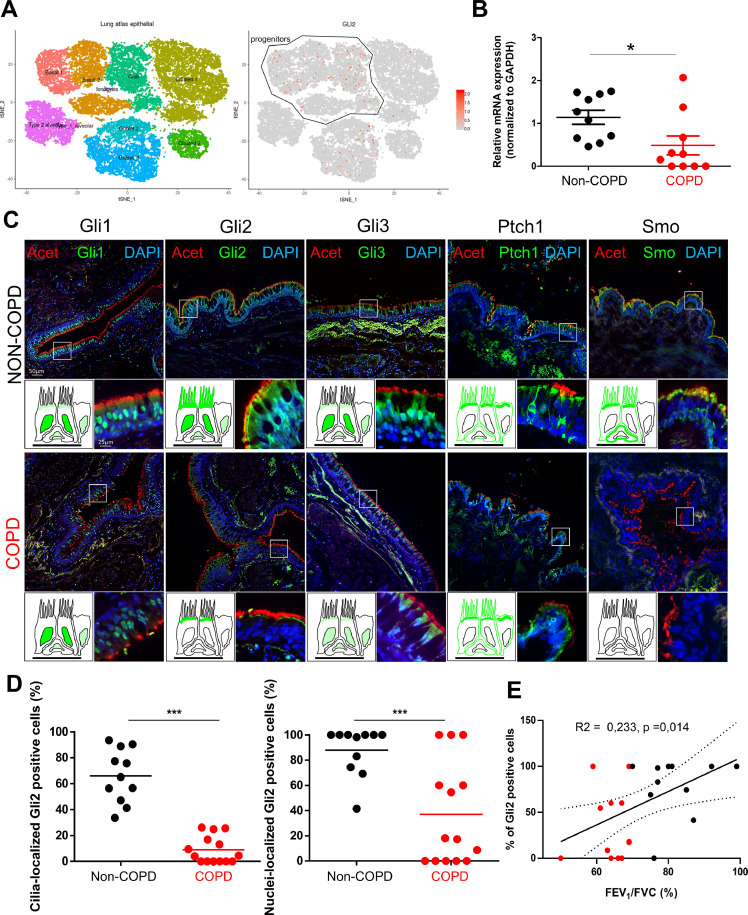
Fig. 5.
Alteration of Gli2 localization in AEC is associated with COPD. A, Data extracted from the Human Lung Atlas. Left, cellular landscape along the airways in human lung analysed by single-cell RNA sequencing from 26154 cells collected from 17 donors to display specific cluster assignment of epithelial cells; right, identification of GLI2-expressing cells by t-distributed Stochastic Neighbour Embedding (tSNE). A progenitor cluster encompassing basal and club-like cells was added. B, Dot plot (mean) presenting the relative mRNAs levels normalized to GAPDH obtained on isolated AEC from bronchial brushings by RT-qPCR (n = 20 non-COPD and COPD patients) for GLI2. Means ±SEM of 2−ΔΔCt; *p < 0.05 non-COPD vs COPD patients. C, Representative micrographs showing the bronchi epithelia of non-COPD and COPD patients stained for the core HH pathway components Gli1, Gli2, Gli3, Ptch1 and Smo (all green); cilia (acetylated tubulin, red) and cell nuclei (DAPI, blue). Magnification corresponding to the selected area is shown. Cartoons depict the localization of each HH pathway element (in green). D, Dot plots (mean) showing the percentages of Gli2 positive cells in cilia (left panel) and nuclei (right panel) in non-COPD (black) and COPD patients (red). *** p < 0.001 non-COPD vs COPD. E, Linear regression of the percentages of nuclear Gli2 positive cells according to the FEV1/FVC (%) for non-COPD (black) and COPD patients (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)