Abstract
Objectives
Previous genome-wide association study showed that GLDC/IL33 loci were associated with overall survival in patients with osteosarcoma (OS). We performed a replication study to explore whether variants of GLDC/IL33 are associated with the survival of OS patients and to further verify their functional role in the gene expression.
Methods
A total of 216 patients with OS were enrolled. The overall survival time was calculated from the date of diagnosis till the date of last follow-up or mortality. Two SNPs were genotyped, including rs55933544 and rs74438701. OS specimens were obtained from 72 patients during surgery. The gene expression level of IL33 and GLDC was evaluated by qPCR. Patients were classified into two groups according to the 5-year overall survival (death/survival). The chi-square test was used to analyze difference of genotype frequency. The Student t-test was used to compare the gene expression level between different genotypes. Cumulative survival time was calculated by the Kaplan–Meier method and analyzed by the log-rank test.
Results
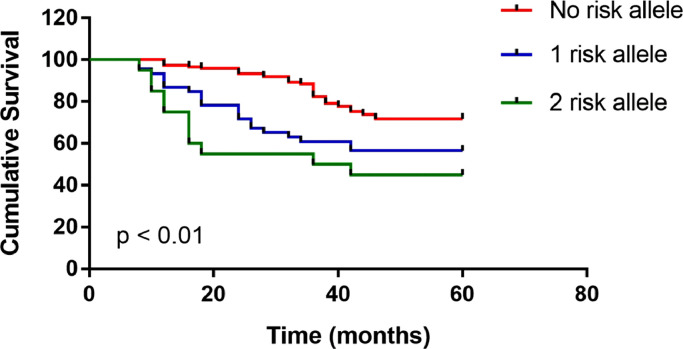
Genotype TT of rs55933544 was significantly associated with the event of death (0.176 vs. 0.061, p < 0.001). Patients with no risk allele T of rs55933544 showed a 5-year overall survival of 81.4% (110/141), which was significantly higher than an overall survival of 55.0% (29/54) for patients with one risk allele and 44.8% (12/21) for patients with two risk alleles (p < 0.01). Genotype TT of rs55933544 were indicative of remarkably lower expression of IL33 than genotype CC (0.00041 ± 0.00025 vs. 0.00065 ± 0.00031, p = 0.04). Patients with low IL33 expression presented remarkably worse survival as compared with the patients with high IL33 expression (p < .01)
Conclusions
Variant rs55933544 was associated with the survival time of OS patients. IL33 may contribute to a poor prognosis of OS. Further investigation into the biological mechanisms by which IL33 influences the overall survival can shed light on the improvement of clinical outcome for OS patients.
Keywords: Osteosarcoma, Variant, IL33, Overall survival, Prognosis
1. Introduction
Osteosarcoma (OS) is the most common malignant bone tumor that primarily originated from primitive malignant bone mesenchymal cells [1]. Typically, OS affects adolescents or adults aged more than 50 years, with an estimated incidence of 0.34/100,000 per year [2]. Before the introduction of the adjuvant chemotherapy and limb salvage surgery, amputation was the main therapeutic choice for OS patients, which produced a poor quality of life and lower overall survival [3,4]. Currently, wide resection combined with adjuvant chemotherapy has been widely used to treat high-grade osteosarcoma, and the overall survival rate has therefore been remarkably raised [5,6]. To be noted, however, approximately 30% patients still cannot survive this malignant disease. A better understanding of the prognostic factors of OS can benefit the patients in terms of advanced therapeutic interventions and prolonged survival.
Several factors have been reported to be associated with the survival of OS patients, such as age at diagnosis, tumor histology, response to chemotherapy, blood alkaline phosphatase levels and rural status [7], [8], [9]. In addition to these factors, the expression level of EHD1 was found negatively correlated with overall survival of OS patients [10]. Increased expression of lncRNA SNHG3 was reported to indicate a poor prognosis of OS patients [11]. Recently, there is increasing interest in whether genetic variants could be predictive of OS outcomes. Several variants associated with prognosis of OS were identified by candidate genetic studies [12], [13], [14], [15]. Liu et al. [14] reported that polymorphism of VEGF may be used as a genetic marker to predict the clinical outcome of OS. Qi et al. [15] reported that the IL6 promoter polymorphism was associated with the risk for metastasis of OS in the Chinese population. These findings should be cautiously interpreted since there was a lack of validation of these candidate gene studies which commonly yielded limited statistical power.
As a powerful tool deciphering the genetic architecture of complex disease, genome-wide association study (GWAS) was applied to unveil genes associated with osteosarcomagenesis. Savage et al. [16] performed the first GWAS in OS patients and reported two susceptible loci of OS with genome-wide significance, including rs1906953 in GRM4 at 6p21.3 and rs7591996 in a gene desert at 2p25.2. Mirabello et al. [17] reported that rs7034162 in NFIB was significantly associated with OS metastasis in different populations. Recently, Koster et al. [18] successfully identified the association between common SNPs and the survival in OS through GWAS. For the first time, germline genetic variants in the GLDC/IL33 locus were found associated with overall survival in patients with OS [18]. Imputation across this region showed that two SNPs, including rs55933544 and rs74438701, reached genome-wide significance (p < 5.0 × 10−8) [18]. As a member of the Interleukin family, IL33 functions as a cell endogenous alarm released by damaged barrier cells [19]. It can recognize and interact with specific receptor ST2 which is expressed in immune cells [20]. It was reported that a lack of IL33 signaling could impair the generation of a potent antitumor immune response [21]. To date, whether IL33 is involved in the osteosarcomagenesis remains poorly understood. Moreover, there was lack of study investigating the regulatory role of rs55933544 and rs74438701 on the expression of GLDC/IL33 in OS tissues. We therefore performed a replication study in order to explore whether germline variants of GLDC/IL33 are associated with the survival of OS patients and to further verify their functional role in the gene expression.
2. Methods
2.1. Subjects
A total of 216 patients with OS were enrolled in the current study, who received surgical treatment between January 2008 and May 2018 in our clinic center. All the patients have undergone the same chemotherapy regimen consisted of cisplatin (300 mg/m2), doxorubicin (80 mg/m2) and methotrexate (10 g/m2). A protocol of 2 cycles before surgery and 4 cycles after surgery was prescribed to each patient. The diagnosis was histopathologically verified by two independent pathologists based on the tumor tissues collected by biopsy. The demographic data were obtained from the medical record, including age, gender, surgery type, Enneking stage, histological type, and presence of tumor metastasis. All the patients were followed up for survival every 2 months after the completion of chemotherapy, with a median follow-up period of 38.6 months (range, 8–70 months). The overall survival time was calculated from the date of diagnosis till the date of last follow-up or mortality. Under the approval of the ethics committee of Nanjing Drum Tower Hospital, each patient has given written informed consent to participate in the present study.
2.2. Genotyping of genetic variants
Genomic DNA was extracted from the total blood cells using a commercial kit (Qiagen K.K., Tokyo, Japan) following standard protocol. Genotyping assays were performed on the LightCycler 480 (Roche Applied Science, Mannheim, Germany) with specific TaqMan probes designed for rs55933544 and rs74438701 (Applied Biosystems, Foster City, CA). Real-time PCR was performed with 4 μl of genomic DNA resolved in 16 μl of reaction mixtures which contained 12 μl of FastStart Universal Probe Master (ROX) mix (Roche Diagnostics, Indianapolis, IN, USA) and 4 μl of TaqMan probes. The thermal cycling condition was as follows, denaturation at 95 °C for 10 min, followed by 45 cycles of denaturation at 92 °C for 15 s and annealing/extension at 60 °C for 1 min.
2.3. qPCR
OS specimens were obtained from 72 patients who underwent surgical interventions from June 2012 to May 2016 and stored in liquid nitrogen for expression analysis of target genes. The total RNA was extracted with TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, Tokyo, Japan). The quantitative gene expression level was evaluated on the LightCycler 480 (Roche Applied Science, Mannheim, Germany) with Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) used as the internal control. The primers of the analyzed gene were as follows, forward 5′-AAGAAACATCTCGCCCCGTT-3′, reverse 5′-GCAGTTTCCGTGGCTTGTTT-3′ for GLDC; forward 5′-GTGACGGTGTTGATGGTAAGAT-3′, reverse 5′-AGCTCCACAGAGTGTTCCTTG-3′ for IL33; and 5′-GAGTCAACGGATTTGGTCGT-3′, reverse 5′-TTGATTTTGGAGGGATCTCG-3′ for the endogenous control gene GAPDH. The amplification procedures were composed of an initial denaturation step of 94 °C for 10 min, followed by 40 amplification cycles at 94 °C for 15 s, annealing at 68 °C for 20 s and elongation at 72 °C for 10 s. Melting curve analyses were performed to determine the relative mRNA expression based on the 2−∆∆Ct method.
2.4. Immunohistochemistry
For immunohistochemistry (IHC) analysis, paraffin-embedded tissues were cut into 4-µm sections and dried at 70 °C for 4 h. After deparaffinization and hydration, sections were washed in phosphate-buffered saline (PBS, 3 × 3 min). Antigen retrieval was performed in citrate buffer. After blocking endogenous peroxidase, the slides were incubated with primary anti IL33 antibody (1:400, Abcam) overnight at 4 °C, and then incubated with secondary antibody for 30 min at room temperature. The IHC-stained tissue sections were scored by two pathologists blinded to the clinical information. The signal intensity was scored as follows: 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown), and 3 (strong staining, brown). The staining distribution was scored based on the percentage of positive staining cells as follows: score 0, 0%; score 1, 1–30%; score 2, 31–60%; score 3, 61–100%. The final score was equal to the staining distribution score multiply the signal intensity score. A score of < 2 was considered as low expression.
2.5. Statistical analysis
The prognostic factors, including age at diagnosis, gender, presence of metastasis and genetic variants, were evaluated via multivariate Cox proportional hazards regression analyses. Patients were further classified into two groups (death/survival) according to the 5-year overall survival. The chi-square test was used to analyze difference of genotype frequency between the two groups. The odds ratios (OR) and 95% confidence interval (95% CI) were calculated with the risk allele used as reference. In addition, the Student t-test was used to compare the gene expression level between different genotypes. In addition, the inter-group comparison between patients with low IL33 expression and those with high IL33 expression was performed with the Student's t-test. Cumulative survival time was calculated by the Kaplan–Meier method and analyzed by the log-rank test. All the statistical analyses were performed with the SPSS software (SPSS Inc., Chicago, IL). Statistical significance was set at p < 0.05.
3. Results
3.1. Demographic data of the patients
The baseline characteristics of the patients were summarized in Table 1. There were 132 males and 84 females with a mean age of 29.6 ± 18.7 years old. 61 patients (28.2%) had tumor metastasis at the initial visit. Fifty-two (24.1%) patients received limb salvage surgery. At the final follow-up, there were 68 cases of death due to tumor recurrence or metastasis. The 5-year overall survival was 68.5%.
Table 1.
Baseline characteristics of the patients
| Features | Patients (n = 216) |
|---|---|
| Gender | |
| Male | 132 (61.1%) |
| female | 84 (38.2%) |
| Age (years) | |
| >20 | 95 (44.0%) |
| ≤20 | 121 (56.0%) |
| Surgery type | |
| Amputation | 52 (24.1%) |
| Limb-salvage | 164 (75.9%) |
| Enneking stages | |
| I | 18 (8.3%) |
| IIA | 65 (30.0%) |
| IIB | 102 (47.3%) |
| III | 31 (14.4%) |
| Histologic type | |
| Osteoblastic | 123 (56.9%) |
| Chondroblastic | 67 (31.0%) |
| Fibroblastic | 9 (4.2%) |
| Mixed | 17 (7.9%) |
| Tumor metastasis | |
| Presence | 71 (32.9%) |
| Absence | 145 (67.1%) |
3.2. Association of GLDC /IL33 locus with 5-year overall survival
The multivariate Cox analysis revealed that 3 factors, including older age at diagnosis, presence of metastasis, and genotype TT of rs55933544, were independent prognostic factors for the survival time (Table 2). Inter-group comparison of the genotype distribution of rs55933544 and rs74438701 were summarized in Table 3. Genotype TT of rs55933544 was significantly associated with the event of death (0.176 vs. 0.061, p < 0.001). As for rs74438701, no significant association with the 5-year survival was found. As shown in Fig. 1, patients with no risk allele T of rs55933544 showed a 5-year overall survival of 81.4% (110/141), which was significantly higher than an overall survival of 55.0% (29/54) for patients with one risk allele and 44.8% (12/21) for patients with two risk alleles (p < 0.01). Patients with genotype TT of rs55933544 were found to have remarkably lower expression of IL33 than those with genotype CC (0.00041 ± 0.00025 vs. 0.00065 ± 0.00031, p = 0.04). As for GLDC, there was no significant difference regarding gene expression between different genotypes of rs55933544 (0.00062 ± 0.00035 vs. 0.00071 ± 0.00026, p = 0.28).
Table 2.
Multivariate analysis of variables associated with the 5-year overall survival
| Variables | Hazard ratio | 95% confidential interval | P |
|---|---|---|---|
| Gender | 0.56 | ||
| Male | reference | - | |
| Female | 0.87 | 0.72 - 1.11 | |
| Age | 0.03 | ||
| <30 | reference | - | |
| >50 | 1.36 | 1.15 - 1.57 | |
| Metastasis | 0.01 | ||
| Absence | reference | - | |
| Presence | 2.57 | 1.97 - 3.36 | |
| Surgery type | 0.46 | ||
| Amputation | Reference | ||
| Limb-salvage | 1.09 | 0.97 - 1.18 | |
| rs55933544 | <0.001 | ||
| CC | reference | - | |
| TC | 1.67 | 1.32 - 2.05 | |
| TT | 2.86 | 2.12 - 3.67 | |
| rs74438701 | 0.63 | ||
| TT | reference | - | |
| TC | 1.02 | 0.86 - 1.21 | |
| CC | 1.09 | 0.91 - 1.32 |
Table 3.
Comparison of the genotype and allele frequency between the death group and the survival group
| Genotype |
p | Allele |
p | Odds ratio (95% CI a) | ||||
|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | T | C | ||||
| rs55933544 | ||||||||
| Death (n = 68) | 12 (17.6%) | 25 (36.8%) | 31 (45.6%) | <0.001 | 49 (36.0%) | 87 (64.0%) | <0.001 | 2.98 (1.87-4.96) |
| Survival (n = 148) | 9 (6.1%) | 29 (19.6%) | 110 (74.3%) | 47 (15.9%) | 249 (84.1%) | |||
| rs74438701 | ||||||||
| Death (n = 68) | 36 (52.9%) | 24 (35.3%) | 8 (11.8%) | 0.53 | 96 (70.6%) | 40 (29.4%) | 0.56 | 1.17 (0.74-1.82) |
| Survival (n = 148) | 85 (57.5%) | 48 (32.4%) | 15 (10.1%) | 218 (73.6%) | 78 (26.4%) | |||
Fig. 1.
Association of risk allele with 5-year overall survival.
Risk allele T of rs55933544 was significantly associated with a decreased overall survival (p < 0.01). Patients with no risk allele (genotype CC) showed a 5-year overall survival of 81.4% (110/141). By contrast, the overall survival was 55.0% (29/54) in patients with one risk allele (genotype TC) and 44.8% (12/21) in patients with two risk alleles (genotype TT).
3.3. Relationship between gene expression and 5-year overall survival
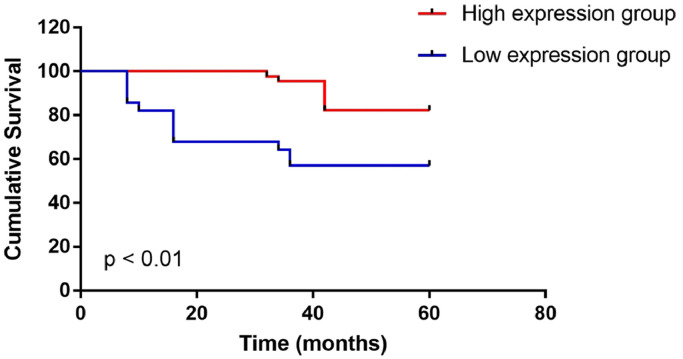
Patients with the event of death were found to have significantly lower expression of IL33 than those without the event (0.00039 ± 0.00023 vs. 0.00068 ± 0.00037, p < 0.001). According to IHC analysis, 44 patients were found to have high IL33 expression and the other 28 patients were found to have low IL33 expression. Inter-group comparison showed that patients with low expression of IL33 had obviously shorter survival time (Table 4). Kaplan–Meier analysis demonstrated that the patients with low IL33 expression presented obviously worse survival as compared with the patients with high IL33 expression (p < 0.01) (Fig. 2).
Table 4.
Association of IL33 expression with clinical variables
| Variables | Number |
IL33 expression |
P value | |
|---|---|---|---|---|
| Low expression (n = 28) | High expression (n = 44) | |||
| Gender | 0.38 | |||
| Male | 43 | 19 (67.9%) | 24 (54.5%) | |
| Female | 29 | 9 (32.1%) | 20 (45.5%) | |
| Age (years) | 0.17 | |||
| >50 | 29 | 8 (28.6%) | 21 (47.7%) | |
| <30 | 43 | 20 (71.4%) | 23 (52.3%) | |
| 5-year survival (%) | 0.01 | |||
| Death | 25 | 15 (53.4%) | 10 (22.7%) | |
| Survival | 47 | 13 (46.6%) | 34 (77.3%) | |
Fig. 2.
Association of IL33 expression with 5-year overall survival.
IHC analysis showed that 44 patients had high IL33 expression and the other 28 patients had low IL33 expression. The overall survival was 57.1% (16/28) in patients with low IL33 expression, which was remarkably lower than that in patients with high IL33 expression (81.8%, 36/44, p < 0.01).
4. Discussion
The overall survival of OS has been greatly improved with the advance of neoadjuvant treatment and limb-salvage surgeries. However, many OS patients are confronted with a poor outcome although they have been treated with current mainstay therapeutic protocol. Herein, identification of prognostic factors can help differentiate patients who can benefit from more aggressive therapies before surgery. In the current study, we replicated the results of a GWAS which reported GLDC/IL33 at 9p24.1 was associated with the overall survival of OS patients [18]. Among previously reported signals localized to the 9p24.1 region, rs55933544 was confirmed to be significantly associated with the survival time of OS patients [18]. In line with the finding of Koster et al. [18], we observed that genotype TT of rs55933544 was indicative of an increased incidence of death during the 5-year follow-up. By contrast, we failed to replicate the association of rs74438701 with the prognosis of OS.
To better clarify the biological basis underlying the association between rs55933544 and the clinical outcome, we investigated the tissue expression level of GLDC and IL33. For the first time, genotype TT of rs55933544 was found indicative of lower IL33 expression as compared with genotype CC. As for GLDC, the expression level was comparable between different genotypes. In previous studies, expression quantitative trait locus (eQTL) analyses showed that rs55933544 was associated with expression of IL33 in human skin and brain tissue [22,23]. Based on public dataset, Koster et al. [18] found that allele T of rs55933544 was significantly associated with a decreased expression of IL33 in OS. Taken together, we speculated that IL33 might be the real causative gene contributing to the prognosis of OS.
Low expression of IL33 has been associated with poor survival in many types of cancers [24], [25], [26]. Musolino et al. [25] analyzed the plasma levels of IL33 in multiple myeloma patients and reported that lower expression of IL33 was independently associated with worse patient survival. Consistent with these studies, we confirmed that lower expression of IL33 was associated with poor survival in patients with OS through qPCR and IHC analysis. IL33 is an inhibitor of bone resorption that blocks osteoclastic activity [27]. Reduced IL33 expression could prevent an adequate Th2 response towards tumor cells and compromise the antitumour surveillance [28]. Eissmann et al. [21] reported that IL33 could stimulate IFNγ expression released by tumor infiltrating T cells, while the decrease of IFNγ expression was associated with aggressive colon cancer. Collectively, it appeared that IL33 signaling could be associated with the immune response to tumorigenesis. Interestingly, impairment of antitumor immune response has been previously described for patients with OS [29]. Since the functional role of IL33 in the osteosarcomagenesis remains obscure, more experiments are warranted to unveil the underlying mechanism in the future study.
Previously reported prognostic factors were also verified in the current study. Age at diagnosis was found remarkably associated with the overall survival [30,31]. Comparably, Zheng et al. [31] reported that aged more than 50 years can add to the event of death in OS patients by 2.4-fold. Berner et al. [30] reviewed a nationwide cohort comprising 424 high-grade Norwegian OS patients and reported that metastasis at diagnosis and aged more than 40 years were adverse prognostic factors for overall survival. It was therefore warranted to investigate whether IL33 is associated with the invasion capability of OS cells in future in-vitro experiments.
Two limitations of our study should be mentioned here. As a rare bone tumor, the sample size of OS is relatively small for a genetic association study. More patients need to be recruited through a multi-center study. The second limitation was that we did not perform functional study investigating the regulatory mechanism underlying the outcome of eQTL analyses. To summarize, the association of GLDC/IL33 locus with the survival outcome of OS patients was successfully replicated in our cohorts. Moreover, we demonstrated that the expression level of IL33 could be indicative of a poor survival outcome. Further investigation into the biological mechanisms by which IL33 influences the overall survival can shed light on the improvement of clinical outcome for OS patients.
Conclusions
Variant rs55933544 was associated with the survival time of OS patients. IL33 may contribute to a poor prognosis of OS. Further investigation into the biological mechanisms by which IL33 influences the overall survival can shed light on the improvement of clinical outcome for OS patients.
CRediT authorship contribution statement
Lin Qingxi: Conceptualization, Writing - original draft. Han Jingjing: Data curation. Sun Qi: Writing - review & editing. Wen Li: Formal analysis. Wang Shoufeng: Supervision.
Declaration of Competing Interest
No benefits in any form have been or will be received from a commercial party related directly or indirectly to the subject of this manuscript.
Acknowledgment
This work was supported by the Nanjing Key Program of Medical Science and Technology Development (Grant No. ZKX14021).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jbo.2019.100270.
Appendix. Supplementary materials
References
- 1.Khanna C. Novel targets with potential therapeutic applications in osteosarcoma. Curr. Oncol. Rep. 2008;10:350–358. doi: 10.1007/s11912-008-0054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Mei J., Zhu X.Z., Wang Z.Y. Functional outcomes and quality of life in patients with osteosarcoma treated with amputation versus limb-salvage surgery: a systematic review and meta-analysis. Arch. Orthop. Trauma Surg. 2014;134:1507–1516. doi: 10.1007/s00402-014-2086-5. [DOI] [PubMed] [Google Scholar]
- 4.Tebbi C.K. Psychological effects of amputation in osteosarcoma. Cancer Treat. Res. 1993;62:39–44. doi: 10.1007/978-1-4615-3518-8_7. [DOI] [PubMed] [Google Scholar]
- 5.Robert R.S., Ottaviani G., Huh W.W. Psychosocial and functional outcomes in long-term survivors of osteosarcoma: a comparison of limb-salvage surgery and amputation. Pediatr. Blood Cancer. 2010;54:990–999. doi: 10.1002/pbc.22419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rougraff B.T., Simon M.A., Kneisl J.S. Limb salvage compared with amputation for osteosarcoma of the distal end of the femur. A long-term oncological, functional, and quality-of-life study. J. Bone Joint Surg. Am.Vol. 1994;76:649–656. doi: 10.2106/00004623-199405000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Pakos E.E., Nearchou A.D., Grimer R.J. Prognostic factors and outcomes for osteosarcoma: an international collaboration. Eur. J. Cancer. 2009;45:2367–2375. doi: 10.1016/j.ejca.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Pruksakorn D., Phanphaisarn A., Arpornchayanon O. Survival rate and prognostic factors of conventional osteosarcoma in Northern Thailand: a series from Chiang Mai University Hospital. Cancer Epidemiol. 2015;39:956–963. doi: 10.1016/j.canep.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt A.F., Nielen M., Klungel O.H. Prognostic factors of early metastasis and mortality in dogs with appendicular osteosarcoma after receiving surgery: an individual patient data meta-analysis. Prev. Vet. Med. 2013;112:414–422. doi: 10.1016/j.prevetmed.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Yu H., Qu G., Wang Y. The expression of eps15 homology domain 1 is negatively correlated with disease-free survival and overall survival of osteosarcoma patients. J. Orthop. Surg. Res. 2019;14:103. doi: 10.1186/s13018-019-1137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J., Wu Z., Zhang Y. LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma. Int. J. Immunopathol. Pharmacol. 2019;33 doi: 10.1177/2058738418820743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattinger C.M., Biason P., Iacoboni E. Candidate germline polymorphisms of genes belonging to the pathways of four drugs used in osteosarcoma standard chemotherapy associated with risk, survival and toxicity in non-metastatic high-grade osteosarcoma. Oncotarget. 2016;7:61970–61987. doi: 10.18632/oncotarget.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurucu N., Sahin G., Sari N. Association of vitamin D receptor gene polymorphisms with osteosarcoma risk and prognosis. J. Bone Oncol. 2019;14 doi: 10.1016/j.jbo.2018.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J.Q., Bai X., Duan D.C. Role of five small nucleotide polymorphisms in the VEGF gene on the susceptibility to osteosarcoma and overall survival of patients. Oncol. Lett. 2015;10:1481–1486. doi: 10.3892/ol.2015.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi Y., Zhao C., Li H. Genetic variations in interleukin-6 polymorphism and the association with susceptibility and overall survival of osteosarcoma. Tumour Biol. 2016;37:9807–9811. doi: 10.1007/s13277-016-4876-6. [DOI] [PubMed] [Google Scholar]
- 16.Savage S.A., Mirabello L., Wang Z. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat. Genet. 2013;45:799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirabello L., Koster R., Moriarity B.S. A genome-wide scan identifies variants in NFIB associated with metastasis in patients with osteosarcoma. Cancer Discov. 2015;5:920–931. doi: 10.1158/2159-8290.CD-15-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koster R., Panagiotou O.A., Wheeler W.A. Genome-wide association study identifies the GLDC/IL33 locus associated with survival of osteosarcoma patients. Int. J. Cancer. 2018;142:1594–1601. doi: 10.1002/ijc.31195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J.X., Liu J., Zhang G.J. Interleukin-33 in malignancies: friends or foes? Front. Immunol. 2018;9:3051. doi: 10.3389/fimmu.2018.03051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz J., Owyang A., Oldham E. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Eissmann M.F., Dijkstra C., Wouters M.A. Interleukin 33 signaling restrains sporadic colon cancer in an interferon-gamma-dependent manner. Cancer Immunol. Res. 2018;6:409–421. doi: 10.1158/2326-6066.CIR-17-0218. [DOI] [PubMed] [Google Scholar]
- 22.Consortium G.T. Human genomics. the genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramasamy A., Trabzuni D., Guelfi S. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu W., Wu C., Li X. Serum IL-33 level is a predictor of progression-free survival after chemotherapy. Oncotarget. 2017;8:35116–35123. doi: 10.18632/oncotarget.16627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musolino C., Allegra A., Profita M. Reduced IL-33 plasma levels in multiple myeloma patients are associated with more advanced stage of disease. Br. J. Haematol. 2013;160:709–710. doi: 10.1111/bjh.12146. [DOI] [PubMed] [Google Scholar]
- 26.Rossle M., Cathomas G., Bonapace L. Interleukin-33 expression indicates a favorable prognosis in malignant salivary gland tumors. Int. J. Surg. Pathol. 2016;24:394–400. doi: 10.1177/1066896916633856. [DOI] [PubMed] [Google Scholar]
- 27.Schulze J., Bickert T., Beil F.T. Interleukin-33 is expressed in differentiated osteoblasts and blocks osteoclast formation from bone marrow precursor cells. J. Bone Miner. Res. 2011;26:704–717. doi: 10.1002/jbmr.269. [DOI] [PubMed] [Google Scholar]
- 28.Buzzelli J.N., Chalinor H.V., Pavlic D.I. IL33 is a stomach alarmin that initiates a skewed Th2 response to injury and infection. Cell Mol. Gastroenterol. Hepatol. 2015;1:203–221. doi: 10.1016/j.jcmgh.2014.12.003. e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuohy J.L., Somarelli J.A., Borst L.B. Immune dysregulation and osteosarcoma: staphylococcus aureus downregulates TGF-beta and heightens the inflammatory signature in human and canine macrophages suppressed by osteosarcoma. Vet. Compar. Oncol. 2019 doi: 10.1111/vco.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berner K., Hall K.S., Monge O.R. Prognostic factors and treatment results of high-grade osteosarcoma in norway: a scope beyond the "classical" patient. Sarcoma. 2015;2015 doi: 10.1155/2015/516843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng W., Huang Y., Chen H. Nomogram application to predict overall and cancer-specific survival in osteosarcoma. Cancer Manag. Res. 2018;10:5439–5450. doi: 10.2147/CMAR.S177945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.