SUMMARY
Metabolic signaling to chromatin often underlies how adaptive transcriptional responses are controlled. While intermediary metabolites serve as co-factors for histone-modifying enzymes during metabolic flux, how these modifications contribute to transcriptional responses is poorly understood. Here, we utilize the highly synchronized yeast metabolic cycle (YMC) and find that fatty acid β-oxidation genes are periodically expressed coincident with the β-oxidation byproduct histone crotonylation. Specifically, we found that H3K9 crotonylation peaks when H3K9 acetylation declines and energy resources become limited. During this metabolic state, pro-growth gene expression is dampened; however, mutation of the Taf14 YEATS domain, a H3K9 crotonylation reader, results in de-repression of these genes. Conversely, exogenous addition of crotonic acid results in increased histone crotonylation, constitutive repression of pro-growth genes, and disrupted YMC oscillations. Together, our findings expose an unexpected link between metabolic flux and transcription, and demonstrate that histone crotonylation and Taf14 participate in the repression of energy-demanding gene expression.
Graphical abstract
eTOC Blurb
Adaptive responses during environmental nutrient fluctuations are critical for survival. For example, during nutrient limitation, energy-demanding cell growth programs need to be tempered. Gowans et al. report that histone crotonylation, produced by fatty acid β-oxidation, is important for reduction of growth gene expression and promotion of metabolic homeostasis.
INTRODUCTION
To ensure efficient growth and survival, cells must sense diverse and dynamic nutrient environments and rapidly reprogram their metabolic output accordingly. One way this can be achieved is through the synchronization of gene expression with the metabolic environment. Chromatin modifications provide an ideal mechanism to link the metabolic status of the cell to transcriptional output, as these changes are rapid, reversible and are reliant upon metabolic intermediates as cofactors for the modification (Gut and Verdin, 2013). Understanding the relationship between environment, chromatin and gene expression provides insights into general principles of metabolic homeostasis as well as being relevant for a number of disease states where energy metabolism is a major contributing factor, including cancer and diabetes (Gut and Verdin, 2013).
The yeast metabolic cycle (YMC) offers a powerful system for studying the relationships between metabolic flux, chromatin modifications, and transcriptional output (Klevecz et al., 2004; Tu et al., 2005). Similar relationships between energy metabolism and gene expression regulation have been observed during circadian rhythms in mammals (Mellor, 2016; Tu and McKnight, 2006). The YMC occurs in glucose-limited conditions and, therefore, likely simulates low-carbon environments encountered in the wild. A primary characteristic of the YMC is respiration cycles, as yeast rapidly synchronize their metabolic states and oscillate from periods of low oxygen consumption (LOC) to high oxygen consumption (HOC). These phases are accompanied by coordinated changes in gene expression, with over half of all transcripts displaying periodic expression (Kuang et al., 2014), as well as fluctuations in metabolic intermediates (Tu et al., 2007) and histone post-translational modifications (PTMs) (Cai et al., 2011). For example, the HOC phase is characterized by increased histone lysine acetylation and expression of pro-growth genes, including ribosomal subunits and those involved in translation. This temporal regulation coordinates energy production, such as acetyl-CoA, with energy-demanding processes, such as ribosome biogenesis. Disruption of a number of metabolic regulators disturbs YMC profiles and causes a disconnect between metabolism and gene expression. These regulators include chromatin remodelers, transcription factors and TORC1 signaling components (Cai et al., 2011; Gowans et al., 2018; Kuang et al., 2017).
Recent advancements in mass spectrometry have identified a number of novel histone PTMs, greatly expanding the potential complexity of the histone code (Simithy et al., 2017; Strahl and Allis, 2000; Tan et al., 2011; Zhao and Garcia, 2015). Many of these are short-chain lysine acylations with differences in the length of carbon chain or degree of saturation. The precursor metabolites of these acylations are often generated in alternate energy metabolism pathways during low glucose conditions (Sabari et al., 2017; Suganuma and Workman, 2018). For example, crotonyl-CoA, the precursor for histone lysine crotonylation is produced via the fatty acid oxidation pathway (Figure 1A). In yeast, unlike mammals, this occurs in the peroxisome where fatty acids are taken up and processed through a series of reactions to generate acetyl-CoA and a fatty acid of carbon chain length n-2, which can then itself be processed (Hiltunen et al., 2003). During this process, acyl-CoAs are produced as an intermediate and could act as donors for acylation reactions (Simithy et al., 2017). Specifically, the donor crotonyl-CoA, a trans-2-enoyl-CoA, is generated as an intermediate during β-oxidation of butyric or crotonic acid as well as during the processing of longer fatty acid chains.
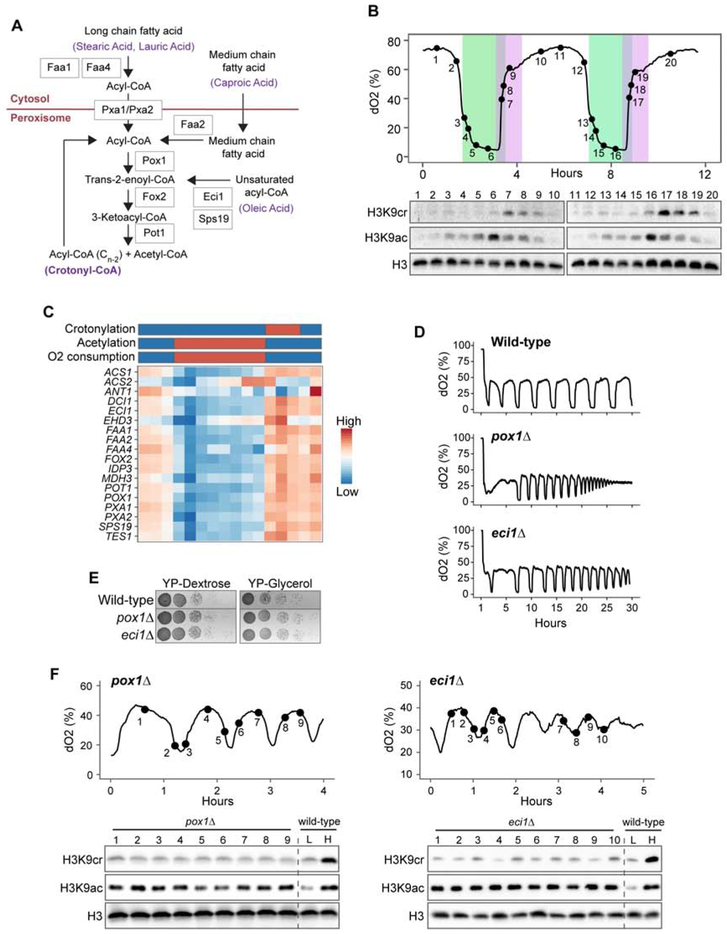
Figure 1. Histone crotonylation is regulated by fatty acid metabolism and is dynamic across the YMC. See also Figure S1.
(A) Fatty acid β-oxidation pathways for generating acyl-CoA are shown (Faergeman et al., 2001; Gurvitz et al., 1998; Hiltunen et al., 2003). Example substrates and products of crotonyl-CoA generation are highlighted in purple. Caproic acid (C6:0) donates 4 carbons to the production of crotonyl-CoA and 2 carbons to acetyl-CoA. Stearic acid (C18:0), lauric acid (C12:0), and oleic acid (C18:1) require multiple rounds of oxidation for production of crotonyl-CoA.
(B) Western blots showing changes in histone H3K9 crotonylation (cr) and acetylation (ac) across two consecutive YMCs. Histone H3 was used as a loading control. Oxygen consumption is inversely correlated with dissolved oxygen (dO2). Peak acetylation and crotonylation are highlighted in green and purple, respectively.
(C) Heatmap showing expression of genes involved in fatty acid oxidation across the YMC. Bars above illustrate changes in crotonylation, acetylation and oxygen consumption. Gene expression data taken from Kuang et al 2014.
(D) Mutants of fatty acid metabolism have disrupted YMCs. Oxygen consumption is inversely correlated with dissolved oxygen (dO2).
(E) Serial dilutions of wild-type (wt) and indicated mutant strains grown on media containing either dextrose (YPD) or glycerol (YPG).
(F) Westerns of pox1Δ (left) and eci1Δ (right) YMC samples using the indicated antibodies (cr is crotonylated; ac is acetylated). Wildtype YMC samples from both high (H) and low (L) crotonylation timepoints are included as a comparison on right of blots.
Histone crotonylation has become intensively studied over the last several years, owing to the fact that this chromatin mark is found on a number of lysine residues across core and linker histones and is associated with regions of active chromatin, including enhancers and transcriptional start sites (Sabari et al., 2015; Tan et al., 2011). Furthermore, histone crotonylation was demonstrated to promote transcription similar to acetylation in an in vitro transcription assay and following crotonic acid addition in cells, suggesting an activating role in gene expression (Li et al., 2016; Sabari et al., 2015). Although the “writers” of histone crotonylation are still being elucidated, a variety of lysine acetyltransferases have been shown to possess histone crotonyl-transferase activity (Sabari et al., 2015; Simithy et al., 2017). Evidence also indicates that non-enzymatic mechanisms for histone acylation can occur (Simithy et al., 2017). In contrast, removal of histone acylation is known to be facilitated by a number of deacetylases in yeast and by Sirt3 in mammalian cells (Andrews et al., 2016; Bao et al., 2014). Thus, the balance between histone acetylation and crotonylation may be determined by the relative intracellular pools of the acyl-CoAs.
Recent reports show that YEATS domain-containing proteins (Yaf9, ENL, AF9, Taf14, Sas5) are prominent readers of histone acylations and are found in a number of chromatin and transcriptional regulators from yeast to humans (Andrews et al., 2016; Li et al., 2016; 2014; Shanle et al., 2015; Zhao et al., 2016). While significant progress has been made in understanding their acyl-binding preferences, our understanding of how histone acylation and its readers contribute to chromatin biology is poorly understood.
In this report, we utilized the YMC to investigate the role of histone crotonylation in metabolic stability and its relationship with histone acetylation. We show that both histone crotonylation and acetylation dynamically fluctuate across the YMC yet have distinct peaks at different points in the metabolic cycle. Genetic and chemical modification of crotonylation patterns alters the periodicity of the YMC and disrupts the expression and timing of metabolic genes. Mechanistically, we found that the ability of Taf14 to read H3K9 crotonylation was required for proper YMC progression and for the periodic repression of growth genes as cells transition to the LOC phase. Together, our data demonstrate a key role for Taf14 and histone crotonylation in linking metabolic state to key gene expression programs.
RESULTS
Histone Crotonylation is Dynamically Regulated Across the YMC and Regulates Metabolic Cycling
While histone acetylation has been intensely studied, our understanding of how other forms of histone acylation contribute to chromatin biology remains obscure. In S. cerevisiae, the peroxisomal fatty acid oxidation pathway produces long and medium acyl-CoA forms, such as crotonyl-CoA, during periods of limited energy availability and low oxygen consumption (Hiltunen et al., 2003) (Figure 1A). In contrast, acetyl-CoA, the cofactor for histone acetylation, is largely generated during high energy availability and utilized during oxygen consumption (Cai et al., 2011). Thus, the molecular pathways and cellular circumstances that result in the deposition of histone acetylation and crotonylation are predicted to be different.
To explore the role of histone crotonylation in the regulation of gene expression and metabolic stability, we employed a well-established continuous culture system wherein yeast cells become synchronized at the metabolic level and cycle through periods of high oxygen and low oxygen consumption (HOC and LOC, respectively) when grown under limited glucose conditions (Klevecz et al., 2004; Tu et al., 2005). Consistent with previous studies (Cai et al., 2011), H3K9 acetylation is dynamic across the YMC and peaks during the HOC phase, when levels of acetyl-CoA are abundant (Figure 1B). Strikingly, we found that H3K9 crotonylation is also dynamic across the YMC but peaks during the HOC to LOC transition when β-oxidation occurs. In addition, the H3K9 crotonylation signal peaks as the H3K9 acetylation signal decreases during the return to the LOC phase, demonstrating that these histone modifications are temporally segregated within the YMC. Notably, and in comparison, only relatively low levels of H3K9 crotonylation were observed in asynchronous yeast cultures grown in nutrient-rich YPD media (Supplementary Figure 1A), demonstrating the utility of the YMC to investigate crotonylation function. Importantly, our H3K9 acetyl- and crotonyl-specific antibodies were validated by peptide dot blot assays and showed no cross reactivity, although some butyrylation reactivity was noted for the anti-crotonyl antibodies (Supplementary Figure 1B). Additionally, the presence of H3K9cr in these YMC samples were supported by mass spectrometric analysis of isolated histones (data not shown).
The transcription of key enzymes involved in fatty acid oxidation and acyl-CoA metabolism correlate well with the distribution of H3K9 crotonylation observed in our western blot data (Figure 1C). In budding yeast, Acs2 is required for the generation of nuclear pools of acetyl-CoA for histone acetylation (Falcón et al., 2010; Takahashi et al., 2006) and as expected, we see an increase in ACS2 transcript levels as histone acetylation levels peak (Figure 1C). An increase in the levels of ACS1 transcript coincide with an increase in histone crotonylation arguing that it may have a role in generating the nuclear pool of crotonyl-CoA for histone crotonylation. ACS1 is the homolog of mammalian ACSS2, which is critical for generation of histone crotonylation in human cells upon addition of exogenous crotonate (Sabari et al., 2015). Deletion of ACS1 has been shown to completely abrogate metabolic cycling arguing for a critical role for histone crotonylation in the YMC (Cai et al., 2011).
The temporal expression of fatty acid oxidation genes, previously reported in high temporal resolution RNA-seq experiments (Kuang et al., 2014), peaks during the HOC to LOC transition (Figure 1C, which are equivalent to time points 6-9 and 16-19 in Figure 1B). Of the fatty acid oxidation genes upregulated during the LOC phase, POX1 and ECI1 are essential to the generation of crotonyl-CoA: POX1 converts acyl-CoAs to trans-2-enoyl-CoA as the first step of the β-oxidation pathway, and ECI1 is part of the auxiliary oxidation pathway involved in processing unsaturated fatty acids (Figure 1A) (Hiltunen et al., 2003). Strikingly, deletion of either POX1 or ECI1 resulted in severe defects to the timing of the YMC, with cycles becoming progressively faster and damped (Figure 1D). Notably, these mutants did not display growth defects on either dextrose- or glycerol-containing medium, indicating that the altered YMC patterns are not a consequence of overall fitness defects but likely due to an inability to correctly regulate and synchronize metabolic states in the chemostat (Figure 1E). Significantly, levels of H3K9 crotonylation were also greatly diminished in these mutants (Figure 1F), suggesting the inability to generate sufficient crotonyl-CoA. Unlike H3K9 crotonylation, the levels of H3K9 acetylation were high throughout the mutant cycles, comparable to a wild-type sample at the peak of acetylation. We also examined the YMC profiles of other mutants in this pathway and found that deletion of FOX2 (converts trans-2-enoyl-CoA to 3-ketoacyl-CoA) or POT1 (converts 3-ketoacyl-CoA to acetyl-CoA and an acyl-CoA containing n-2 carbons) also exhibits disrupted YMC and reduced H3K9 crotonylation (Supplemental Figure 1C and 1D). Together, these data demonstrate an important role for fatty acid oxidation in the YMC, and further, suggests the intriguing possibility that H3K9 crotonylation, which is produced via fatty acid oxidation, may have an important role in regulating the YMC.
Histone Crotonylation and Acetylation are Associated with Highly Expressed Metabolic Genes
The genomic locations of H3K9 crotonylation have not been previously mapped in any organism. Given the significance of Pox1 and Ecl1 for the timely progression of the YMC and for H3K9 crotonylation levels, we next sought to determine where this mark resides across the genome at the peak of H3K9 crotonylation (i.e., during the HOC to LOC transition at time points 3 and 4) (Figure 2A). For comparison and as a control, we also performed H3K9 acetylation ChIP-seq at these same two time points. We found that both H3K9 acetylation and crotonylation are enriched at transcriptional start sites (TSSs) and termination sites (TTSs) of RNA polymerase II (RNAPII)-regulated genes genome-wide (Figure 2B). The location of H3K9 crotonylation at TSSs is consistent with our previous findings that show H3K9 crotonylation is deposited, in part, by Gcn5 and removed by transcription-linked histone deacetylases similar to H3K9 acetylation (Andrews et al., 2016).
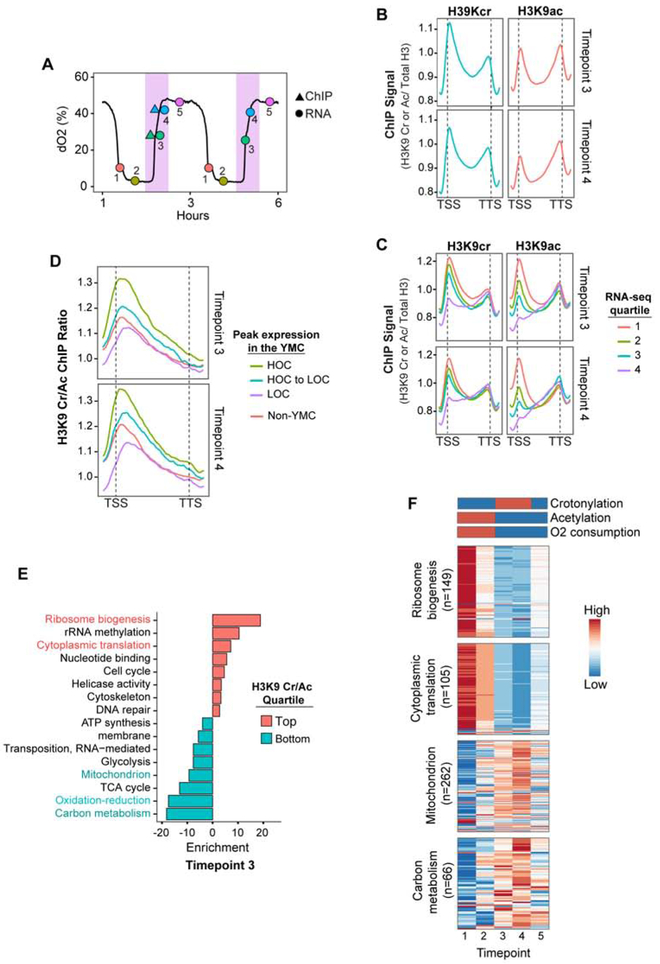
Figure 2. Histone Crotonylation and Acetylation Vary Across the YMC.
(A) H3K9 crotonylation and acetylation ChIP (triangles) and RNA-seq (circles) timepoints are indicated on the YMC trace and the peak of crotonylation (as determined by western blot, Figure 1B) is highlighted in pink.
(B) H3K9 crotonylation (cr) and acetylation (ac) ChIP signals at timepoints 3 and 4 are shown at +/− 200 bp around transcription start site (TSS) or the transcriptional termination site (TTS) of n = 6008 genes. H3K9cr or H3K9ac ChIP signal is normalized by total H3 ChIP.
(C) At timepoints 3 and 4, mean H3K9 crotonylation (cr) and acetylation (ac) ChIP signals are shown for each quartile of gene expression (FPKM, Fragments Per Kilobase of transcript per Million mapped reads) and shown as different colored lines. H3K9cr or H3K9ac ChIP signal is normalized by total H3 ChIP. Transcription start site is TSS, transcriptional termination site is TTS.
(D) Mean H3K9 crotonylation (cr) to acetylation (ac) ChIP ratios at timepoints 3 and 4 on high oxygen consumption (HOC) or low oxygen consumption (LOC) genes in the YMC. Genes that do not have periodic expression in the YMC are labelled as non-YMC. H3K9cr or H3K9ac ChIP signal is normalized by total H3 ChIP. Transcription start site is TSS, transcriptional termination site is TTS.
(E) DAVID functional enrichment analysis of genes in the top and bottom quartiles of H3K9 crotonylation (cr) to acetylation (ac) ChIP ratios at +/− 200 bp around transcription start site at timepoint 3. Enrichment is shown as −log10 p-value.
(F) Heatmaps of genes shown in (E). n= number of genes in each heatmap. YMC oxygen consumption, H3K9 crotonylation and acetylation patterns are illustrated on top of heatmaps from data in Figure 1B.
When analyzed individually, there was a positive association of H3K9 acetylation and crotonylation with YMC gene expression during the HOC to LOC transition (Figure 2C). Specifically, the highest quartiles of gene expression at timepoints 3 and 4 also displayed the highest occupancy of histone acetylation and crotonylation. Furthermore, changes in gene expression correlate with H3K9 modification levels at the TSSs, while modification at the TTSs were predominantly static, suggesting both modifications are linked to the regulation of gene expression.
Further analysis of our ChIP-seq data set showed that H3K9 crotonylation and acetylation occupy many of the same TSSs during the peak of crotonylation (r = 0.88 at timepoint 3), implicating a possible cooperative relationship between these two acylations. To further examine this relationship, we examined the relative ratio of H3K9 crotonylation to H3K9 acetylation at TSSs during time points 3 and 4 (Figure 1B) when both acylations can be detected. This analysis found that genes periodically expressed during the HOC phase (i.e., time points 1 and 2, Figure 2A) were among those with the highest H3K9 crotonylation/acetylation ratios during the transition to LOC (timepoints 3 and 4, Figure 2A and D). These included genes that function in ribosome biogenesis and translation (Figures 2E and 2F), which are pro-growth genes with the highest amplitude of periodic gene expression in the YMC (Tu et al., 2005). These genes have a burst of expression during the HOC that is accompanied with increased H3K9 acetylation and energy production, followed by active repression and H3K9 acetylation loss during the LOC phase when oxygen is no longer consumed (Cai et al., 2011). As such, the high H3K9 crotonylation to acetylation ratios seen on these genes in this study likely represent a combination of decreasing levels H3K9 acetylation concomitate with increasing levels of H3K9 crotonylation.
Collectively, these results demonstrate that H3K9 crotonylation, like H3K9 acetylation, is associated with periodic expression of YMC genes. The high H3K9 crotonylation to acetylation ratios on these genes during the transition from HOC to LOC further implicate a role for these two marks in the precise regulation of energy-demanding, highly expressed HOC genes.
The YEATS Domain of Taf14 is Required for Proper YMC Progression and for the Precise Control of Metabolic Gene Transcription
Given that H3K9 crotonylation is associated with periodic gene expression, we next sought to determine how this mark might function in the YMC. Previous work by our group and others has shown that the YEATS domain of Taf14 is a prominent reader of acylated H3K9 (Andrews et al., 2016; Shanle et al., 2015) with a notable preference for H3K9 crotonylation (Andrews et al., 2016). Consistent with past observations, we confirmed these results quantitatively using isothermal titration calorimetry (ITC), which showed that the YEATS domain of Taf14 has strong preference for H3K9 crotonylation over other acyl-H3K9 forms (Kd of 70 μM versus 171 μM for Kcr versus Kac (Supplementary Figure 2A). These values agree with previously reported ITC values (Li et al., 2016). Interestingly, like bromodomains, the Taf14 YEATS domain binds more tightly to a peptide that is poly-crotonylated or poly-acetylated compared to singly acylated peptides, however the mechanism by which poly-acylation leads to higher affinity binding remains unclear (Supplemental Figure 2A).
To determine if Taf14 reading of H3K9 crotonylation might impact YMC function, we engineered a genomic point mutation (W81A) in the Taf14 YEATS domain (taf14W81A) that abolishes its binding to all forms of acylated histones (Shanle et al., 2015). In vivo Taf14 is a member of multiple chromatin-modifying complexes and the general transcription apparatus (Schulze et al., 2009) and is known to occupy gene promoters marked with H3K9 and H3K14 acetylation (Liu et al., 2005). ChIP-qPCR analysis revealed that the taf14W81A mutant resulted in abrogated binding of Taf14 to the promoters of several cycling genes, indicating an important as well as an undocumented role for H3K9 acylation in the recruitment of Taf14 to genes (Supplemental Figure 2B). ChIP-seq analysis confirmed the enrichment of Taf14 proximal to transcriptional start sites, and to a lesser extent, transcriptional stop sites (Supplemental Figure 2C). Conversely, taf14W81A ChIP did not produce sufficient material for sequencing (data not shown), confirming its reduced ability to localize to chromatin.
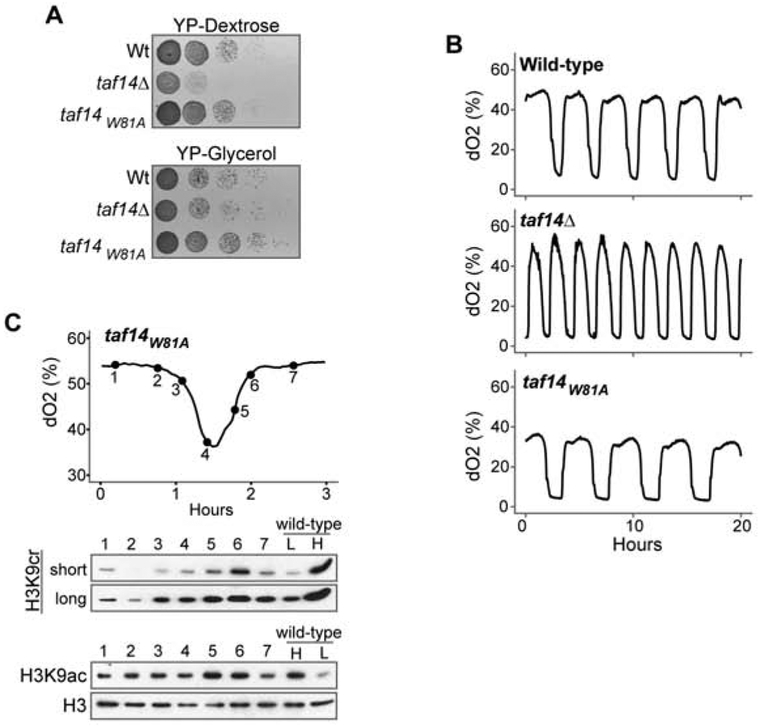
Consistent with our past report (Andrews et al., 2016), complete deletion of TAF14 resulted in a slow growth phenotype on glucose-containing (YPD) medium whereas the YEATS domain pocket mutant that disrupts acyl-H3K9 binding did not (Figure 3A). Interestingly, growth of the taf14Δ strain on non-fermentable glycerol-containing media (YPG) was comparable to the wild-type strain, suggesting that the slow growth phenotype observed in deletions of TAF14 are due to disrupted glycolysis rather than oxidative phosphorylation.
Figure 3. Loss of acylated histone binding by Taf14 disrupts the YMC.
(A) Growth tests of wild-type (wt) and indicated mutant strains grown on media containing either dextrose (YPD) or glycerol (YPG).
(B) YMC profiles for wild-type, taf14Δ and taf14W81A point mutant.
(C) Westerns of taf14W81A YMC samples using the indicated antibodies (cr is crotonylated; ac is acetylated). Short and long exposure times are shown for H3K9cr western. Wildtype YMC samples from both high (H) and low (L) crotonylation or acetylation timepoints are included as a comparison on right of blots. Histone H3 is used as a loading control.
Strikingly, a complete loss of Taf14 resulted in a dramatically altered YMC profile, with cycles oscillating rapidly through LOC phases when genes associated with glycolysis peak (Tu et al., 2005) (Figure 3B). Importantly, the Taf14 YEATS domain mutant also altered the timing and amplitude of the YMC, with cycles occurring slightly slower than that found with our wild-type strain. We also confirmed that the taf14W81A strain did not substantially alter H3K9 crotonylation levels in cycling strains (Figure 3C). Thus, the ability of Taf14 to read H3K9 acylation is important for proper regulation of the YMC.
To identify how YMC gene expression would be altered in the absence of Taf14 reading of H3K9 acylation, we performed RNA-seq analysis in wild-type and taf14W81A cells across two consecutive YMC cycles (Figure 4A). Principal component analysis (PCA) of this experiment revealed that both wild-type and taf14W81A samples are generally arranged in a circle, reflecting the cyclical nature of the YMC (Figure 4B). A comparison of both strains at timepoints 1 and 2, which represent the peak of H3K9 acetylation, demonstrates a high degree of similarity between the wild-type and taf14W81A mutant. However, at timepoints 3 and 4, which represent the peak of H3K9 crotonylation during the transition to the LOC quiescent phase (see Figure 1B), the data are further apart, indicative of transcriptional profiles that have more differences between the wild-type and taf14W81A mutant at these times points.
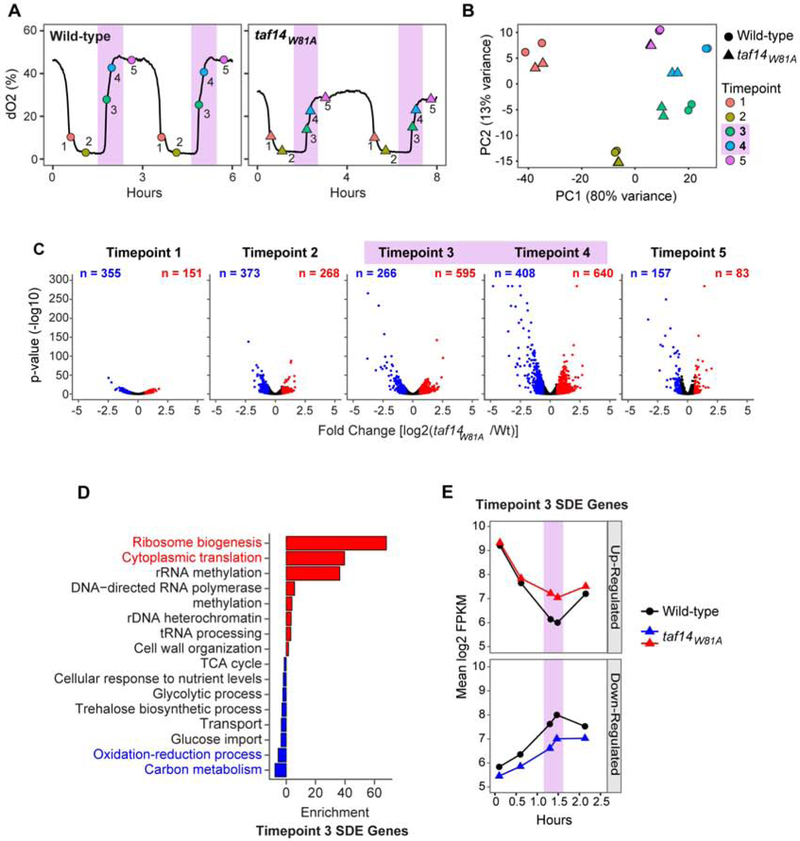
Figure 4. RNA-seq analysis of the taf14W81A Mutant YMC. See also Figure S3.
(A) Samples were collected at the indicated timepoints for RNA-seq analysis. Peak crotonylation, as shown in Figure 1B, is highlighted in pink.
(B) Principal component (PC) analysis of RNA-seq samples. Biological replicates are shown for each timepoint and separated by strain.
(C) Volcano plots displaying the differences in gene expression between wild-type and taf14W81A. log2 fold expression change (x-axis) and significance (-log10 p-value, y-axis) are shown for down-regulated (red) and up-regulated (blue) genes in taf14W81A. Unchanged genes are shown in black.
(D) DAVID functional annotation enrichment analysis of significantly differentially expressed (SDE) genes at timepoint 3. Enrichment is shown as −log10 p-value.
(E) RNA levels (log2 FPKM) of the genes differentially expressed at timepoint 3 across the YMC.
Consistent with the PCA analysis, there were more significantly differentially abundant (SDE) genes, with higher magnitude and significance of difference, during the histone crotonylation peak (timepoints 3 and 4) compared with the histone acetylation peak (timepoints 1 and 2) in the taf14W81A mutant (Figure 4C). More than twice as many up-regulated SDE genes (n=595) compared to down-regulated genes (n=266) were identified in the taf14W81A at the peak of histone crotonylation. Unexpectedly, these up-regulated genes were significantly enriched in pro-growth pathways, such as ribosome biogenesis and translation (Figure 4D). As previously mentioned, pro-growth genes are normally highly expressed during the HOC phase when H3K9 crotonylation levels are low and are repressed in the LOC phase when H3K9 crotonylation levels increase (Figure 2D-F). In the taf14W81A mutant, these genes are still repressed, albeit to a lesser degree during the HOC to LOC transition (Figure 4E and Supplemental Figure 3). Conversely, down-regulated genes in the taf14W81A mutant exhibit broad expression defects throughout the YMC, suggesting these genes may be less related to the direct function of histone crotonylation (Figure 4E).
The data above suggests the possibility that Taf14 association with H3K9 crotonylation directly represses HOC genes; however, loss of Taf14-H3K9 acyl reading might also alter the expression of ‘master’ metabolic transcriptional regulators that influence the expression of these genes. A variety of transcription factors are known to coordinate the expression of numerous metabolic genes in phase with the YMC (Kuang et al., 2017; Rao and Pellegrini, 2011). We, therefore, examined the expression of 41 master transcription factors and the expression of their target genes in the taf14W81A mutant. Of these, Dot6/Tod6-regulated gene expression in the mutant compared to wild-type cells showed a notable increase (Supplemental Figure 3B). In metabolically asynchronous cultures, Dot6/Tod6 are transcriptional repressors of ribosome biogenesis, and are inactivated by TORC1 and PKA-dependent signaling (Huber et al., 2011; Lippman and Broach, 2009). At timepoints when ribosomal gene expression is increased in the taf14W81A mutant, Tod6 expression is elevated while Dot6 is reduced (Supplemental Figure 3C). However, the functional output of these transcription factors in the YMC is not clear, as both TORC1-signaling and Dot6/Tod6 expression are normally elevated during the peak of ribosomal gene expression (Gowans et al., 2018). Thus, it is likely that a combination of repressors and activators are responsible for the tight transcriptional regulation of ribosome biogenesis, the coordination of which is disrupted in the taf14W81A mutant.
Nevertheless, taken together, these findings demonstrate that disrupting the association of Taf14 with H3K9 acylation primarily impacts HOC gene expression patterns during the transition to LOC. They further provide evidence that Taf14-H3K9 crotonylation interaction is important for the proper repression of HOC genes in the LOC phase when oxygen consumption and energy production are depleted (and when H3K9 crotonylation levels are high).
Elevated Histone Crotonylation Alters the YMC and Reduces Pro-Growth Gene Expression
To further investigate the influence of histone crotonylation on YMC gene expression, we manipulated the levels of H3K9 crotonylation by adding a gradual increase of crotonic acid to the YMC medium, which becomes metabolized into crotonyl-CoA and used for histone crotonylation (Li et al., 2016; Sabari et al., 2015). Increasing crotonic acid levels resulted in a dramatic decrease in the periodic length of the YMC (Figure 5A). Conversely, equal molar addition of sodium acetate produced a more gradual decrease in the YMC period. Addition of crotonic acid persistently increased crotonylation on several H3 and H4 lysines, including H3K9, while acetylation levels fluctuated at a reduced amplitude, perhaps as a consequence of altered cycles (Figure 5B and Supplemental Figure 4A). Interestingly, addition of crotonic acid to pox1Δ mutants also increases H3K9 crotonylation (Supplementary Figure 4B and C), suggesting that crotonyl-CoA can be produced by short chain fatty acyl-CoA synthetase conversion of excess crotonic acid.
Figure 5. Increased histone crotonylation alters YMC and reduces ribosomal gene expression. See also Figure S4.
(A) Wild-type YMC profiles with crotonic acid (left) or sodium acetate (right) supplemented media (1 mM). Colored lines indicate concentration in the culture based on dilution rate. A rolling average of the cycle lengths is shown below.
(B) Western analysis of wild-type YMC samples before (left) and after (right) crotonic acid addition probed with the indicated antibodies (cr is crotonylated; ac is acetylated). Histone H3 is used as a loading control.
(C) RT-PCR qPCR analysis of wild-type YMC samples before (left) and after (right) crotonic acid addition using primers targeting the indicated gene. HOC is high oxygen consumption. LOC is low oxygen consumption. Standard deviation is shown of technical replicates.
(D) H3K9 crotonylation (cr) and acetylation (ac) ChIP signals at timepoints 3 and 4 are shown at +/− 200 bp around transcription start site (TSS) or the transcriptional termination site (TTS) of n = 6008 genes. H3K9cr or H3K9ac ChIP signal is normalized by total H3 ChIP.
qPCR analysis of transcript levels demonstrated that ribosomal gene expression was dramatically down-regulated following addition of crotonic acid that was concomitant with increased histone crotonylation (Figure 5C). Conversely, non-cycling genes that were not differentially expressed in the taf14W81A mutant were not significantly changed by the addition of crotonic acid (Figure 5C). Accordingly, these ribosomal genes have higher histone crotonylation to acetylation ratio during the HOC to LOC transition, compared to genes with peak expression in the LOC phase and non-cycling genes (Figure 5D). These results provide further support for the idea that increased crotonylation on genes expressed during the HOC phase reduces expression.
In addition to the influence of Taf14 binding to H3K9 acylations, another way histone crotonylation could reduce expression of HOC genes is through the repulsion of bromodomain-containing transcriptional activators, which have been shown to not tolerate longer acyl lysine forms (Andrews et al., 2016; Li et al., 2016). To examine this possibility, we performed ChIP-qPCR assays for several bromodomain-containing transcriptional regulators including Bdf1/2 and Snf2 in the YMC before and after crotonic acid addition (Supplemental Figure 5). Surprisingly, we observed increased abundance of these transcription factors on several HOC and LOC-regulated genes following the addition of crotonic acid. The reasons for this are currently unknown, however it does demonstrate that histone crotonylation does not simply act by repelling bromodomain-containing factors to facilitate its transcriptional changes. However, we cannot rule out the possibility that H3K9 crotonylation may also contribute to transcription by maintaining these and other bromodomain-containing proteins in a poised state at gene promoters.
Collectively, these data show that disruption of the normal levels of histone crotonylation results in altered metabolic synchronization and the expression of ribosomal genes. Taken together with all of our findings, they suggest an unexpected role for Taf14 association with H3K9 crotonylation in regulating the timely repression of energy-consuming gene expression.
DISCUSSION
Our results demonstrate that the metabolic state of the cell is connected to histone crotonylation and transcriptional regulation. Specifically, we show that histone crotonylation: 1) is dynamically regulated across the YMC; 2) displays a temporally distinct pattern from acetylation; 3) is sensitive to disruption of the fatty acid oxidation pathway; 4) contributes to transcription in a Taf14 YEATS domain-dependent manner; and 5) is linked to the timely control of energy demanding gene expression. These studies also show that short-chain acylations, such as crotonylation, are functionally distinct from acetylation and contributes to the transition from high energy metabolic status (i.e., HOC) to low energy status (i.e., LOC).
Our results show that increased H3K9 crotonylation functions to dampen the expression of pro-growth genes that are normally turned off during the HOC to LOC transition when cellular energy becomes depleted (Figure 6). This is consistent with the idea that metabolic changes lead to the generation of new metabolites that then contribute to the regulation of transcription programs required for cell growth and survival in diverse nutrient conditions (Gut and Verdin, 2013). In the case of H3K9 crotonylation, its distinct role in regulating gene expression is evident by its post-acetylation increase during the HOC to LOC phase transition.
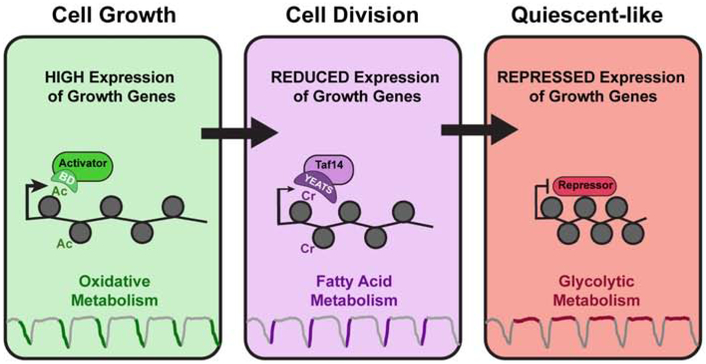
Figure 6. Model for Taf14 and histone crotonylation regulation of metabolic gene expression.
During HOC in the YMC, an abundance of ATP and acetyl-CoA production and consumption occurs along with elevated histone acetylation. Expression of pro-growth genes is facilitated by transcriptional activators containing bromo-domains (BD) that bind acetylated histones. As cells transition from HOC to LOC, fatty acid β-oxidation is activated and histone crotonylation is increased. The YEATS domain of TAF14 binds crotonylated H3K9 to reduce gene expression as energy sources are depleted. During this transition, cell division also occurs. As cells return to the “quiescent-like” LOC phase, reductive metabolism occurs as crotonylation is removed to further repress pro-growth gene expression.
Taf14-mediated repression of genes with H3K9 crotonylation
A major surprise of our studies is the unexpected role of H3K9 crotonylation and Taf14 in gene repression rather than activation, which was our initial expectation based on previous publications (Li et al., 2016; Sabari et al., 2015). While the YEATS domain of Taf14 binds H3K9 crotonylation, this domain is capable of also recognizing H3K9 acetylation, albeit to a lesser degree (Andrews et al., 2016). Taf14 is also a component of several transcriptional complexes, including chromatin remodelers, NuA4 acetyltransferase, and the Pol II pre-initiation (PIC) complex (Schulze et al., 2009). It would be reasonable to think that Taf14 might then function in the YMC through reading both acetylation and crotonylation. However, our findings point to the idea that Taf14 preferentially functions through H3K9 crotonylation, as the greatest number of differentially expressed genes observed in the taf14W81A mutant occurred precisely during the peak of H3K9 crotonylation rather than the peak of H3K9 acetylation.
The idea that Taf14 functions primarily through H3K9 crotonylation is also supported by a recent study that found Taf14-regulated genes can be properly regulated by a form of Taf14 only capable of binding H3K9 crotonylation (Klein et al., 2018). This leads to an important question: how could the binding of crotonylation over acetylation be so profoundly different? Although the answer to this question is currently unknown, we speculate that the unique structural change found in the Taf14 YEATS domain when bound to H3K9 crotonylation (a π-π-π stacking mechanism involving aromatic residues in the YEATS domain (Andrews et al., 2016) may result in either recruitment of Taf14 directly to one or more of its complexes that are already bound on chromatin to block transcriptional initiation or that recognition of H3K9 crotonylation by Taf14 allosterically inhibits some aspect of the PIC or initiation/re-initiation. In support of the idea that Taf14 may be a transient member of repressive transcriptional complexes, a recent mass spectrometry analysis of active RNAPII transcription complexes that bind to yeast promoters found that Taf14 was absent from these RNAPII and TFIID-containing complexes (Joo et al., 2017). Thus, future biochemical and structural studies will be required to determine the distinct mechanisms that regulate Taf14 recruitment and function.
Fatty acid metabolism attenuates energy-consuming transcription in the YMC
The precise regulation of transcriptional processes in the YMC optimizes energy production and consumption with cell growth and division (Wang et al., 2015). During the HOC phase, energy production in the form ATP and acetyl-coenzyme A (CoA) are increased (Cai et al., 2011; Machné and Murray, 2012), concomitant with energy-consuming pro-growth programs. For example, translation is a pro-growth process important for creating components of new cells completing cell division, yet it also necessitates tremendous energy expenditure. Gene expression programs related to translation and ribosome biogenesis often exhibit more than 40-fold increase in expression in the HOC phase compared to the LOC phase. Ribosome biogenesis alone involves nearly 10% of the genome, and has been proposed to account for approximately 60% of total S. cerevisiae transcription (Warner, 1999). Thus, pro-growth transcriptional programs, such as ribosome biogenesis, must be tightly regulated to ensure energy efficiency in the cell.
Our data demonstrate that peroxisomal fatty acid β-oxidation and histone crotonylation increase during the HOC to LOC transition, concomitant with reduction of ATP and acetyl-CoA levels and decreased expression of ribosome biogenesis genes (Cai et al., 2011; Machné and Murray, 2012). Thus, the metabolites of fatty acid β-oxidation, an alternative energy source, can communicate with the transcriptional machinery to signal the depletion of available energy and enact co-coordinated transcriptional responses. Consistent with this idea, as cells prepare to transition from LOC back to HOC, histone crotonylation levels are removed both in wild-type (Figure 1B) and the taf14W81A mutant unable to recognize H3K9 crotonylation (Figure 3C). Although it is currently unknown which deacetylases participate in H3K9 crotonylation removal in the YMC, our previous studies suggest the involvement of Rpd3 given its deletion results in increased H3K9 crotonylation (Andrews et al., 2016).
It should be noted that another cellular pathway for unsaturated crotonyl-CoA production is the conversion of saturated butryl-CoA. However, the dramatic reduction of periodic histone crotonylation in the YMC upon deletion of β-oxidation pathway components suggests that β-oxidation is the primary metabolic pathway that coordinates the transition from HOC to LOC phases (Figures 1F and S1C).
Additionally, our results highlight the benefits of utilizing a synchronized system, as we were unable to detect significant levels of histone crotonylation in asynchronous cultures grown in rich media. Recent work has also characterized other histone modifications derived from short-chain fatty acids, namely histone propionylation (Kebede et al., 2017) and butyrylation (Goudarzi et al., 2016), as enriched at the TSS of highly expressed genes. It would, therefore, be interesting to examine temporal changes in these modifications across the YMC.
Broader implications in fatty acid metabolism
The role of fatty acid metabolism in metabolic stability and transcriptional responses has been somewhat understudied. A currently outstanding question is the nuclear accumulation of intermediary metabolites of fatty acid metabolism, such as crotonyl-CoA. Our preliminary analysis was unable to detect specific peroxisomal proteins, such as Pox1, in the nucleus (data not shown). However, we were able to detect nuclear Acs1 (data not shown), the yeast homologue of mammalian ACSS2 that metabolizes exogenous crotonate (Sabari et al., 2015). Nevertheless, much research is still needed to decipher the connection between histone acylation and diverse metabolic pathways.
Importantly, recent research demonstrates that fatty acid metabolism is critical for human health and may contribute to disease when disrupted (Koh et al., 2016). For example, short-chain fatty acid metabolism occurs in gut microbiota and influences epigenetic profiles in multiple tissues (Donohoe et al., 2011; Krautkramer et al., 2016), including histone crotonylation in the colon that is dynamically regulated by the cell cycle (Fellows et al., 2018). Disruptions in fatty acid metabolism have also been linked to cancer progression (Nath and Chan, 2016). This suggests that fluctuations in histone crotonylation that are linked to metabolic state are likely a normal function of cell physiology and may contribute to disease when disrupted. We predict that the findings we have made in this report will be found to be conserved in other biological contexts.
STAR METHODS
METHOD DETAILS
Yeast strains and metabolic cycling
Saccharomyces cerevisiae strains were constructed in the CEN.PK background using standard genetic techniques (Key Resources Table), and were negative for the petite phenotype caused by mitochondrial dysfunction, as assessed by the ability to grow on glycerol-containing media. The taf14W81A strain was generated scarlessly using the delitto perfetto system and verified by Sanger sequencing (Stuckey and Storici, 2013). Metabolic cycling conditions were performed as described previously (Burnetti et al., 2016; Tu et al., 2005), except starter cultures were grown in glycerol-containing media. Dissolved oxygen percentage shown in YMC plots is relative to starting culture conditions (~100%). Continuous respiration cycles occur following addition of glucose to starved cells. For each experiment, the time (in hours) relative to the start of the experiment is shown.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-H3K9 crotonyl | RevMAb Biosciences | 31-1225-00 |
| Rabbit polyclonal anti-H3K9 acetyl | Millipore | 06-942-S |
| Mouse monoclonal anti-H3K4 crotonyl | PTM Biolabs | PTM527 |
| Mouse monoclonal anti-H3K23 crotonyl | PTM Biolabs | PTM519 |
| Rabbit polyclonal anti-H4K8 crotonyl | PTM Biolabs | PTM522 |
| Rabbit polyclonal anti-H3 C terminus | EpiCypher | 13-0001 |
| Rabbit polyclonal anti-H3K9 crotonyl | PTM Biolabs | PTM516 |
| Rabbit polyclonal anti-Bdf1/2 | Jerome Govin lab | NA |
| Rabbit polyclonal anti-Taf14 | Joseph Reese lab | NA |
| Rabbit polyclonal anti-G6PDH | Sigma Aldrich | A9521 |
| Rabbit polyclonal anti-HA | Bethyl | A190-108A |
| Rabbit polyclonal anti-HA | Abcam | Ab9110 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Crotonic acid | TCI | C0416 |
| Sodium acetate | EMD | SX02651 |
| Critical Commercial Assays | ||
| NEBNext Ultra II DNA Library Prep Kit for Illumina | NEB | E7645 |
| NEB Multiplex Oligos for Illumina (Sets 1 & 2) | NEB | E7355/E7500 |
| MasterPure Yeast RNA Purification Kit | Epicenter | MPY03100 |
| NEB Library Quant Kit for Illumina | NEB | E7630 |
| NEB Ultra Directional RNA Library Prep Kit for Illumina | NEB | E7420 |
| NEB Next Poly (A) mRNA Magnetic Isolation Module | NEB | E7490 |
| Deposited Data | ||
| Raw and analyzed ChIP- and RNA-seq data | This paper | GSE120019 |
| S. cerevisiae genome build (sacCer3) | ENSEMBL | http://www.ensembl.org//useast.ensembl.org/Saccharomyces_cerevisiae/Info/Index?redirectsrc=//www.ensembl.org%2FSaccharomyces_cerevisiae%2FInfo%2FIndex |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae CEN.PK 122-alpha | N/A | |
| S. cerevisiae CEN.PK 113-7D | N/A | |
| CEN.PK 122-alpha taf14::kanMX | This paper | N/A |
| CEN.PK 122-alpha TAF14(W81A) | This paper | N/A |
| CEN.PK 122-alpha HA-SNF2::kanMX | This paper | N/A |
| CEN.PK 122-alpha HA-ACS1::kanMX | This paper | N/A |
| CEN.PK 113-7D eci1::hygMX | This paper | N/A |
| CEN.PK 113-7D pox1::hygMX | This paper | N/A |
| CEN.PK 113-7D fox2::NATMX | This paper | YAT001 |
| CEN.PK 113-7D pot1::NATMX | This paper | YAT003 |
| Oligonucleotides | ||
| See Table S1 | ||
| Software and Algorithms | ||
| Deeptools2 | https://deeptools.readthedocs.io/en/develop/ | N/A |
| DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | N/A |
Crotonic acid (TCI C0416) and sodium acetate (EMD SX02651) were prepared in water and added at the concentrations indicated in figures. Growth assays were performed by plating serial dilutions (1:10) of indicated strains on YP media containing either 2% glucose (YPD) or 3% glycerol (YPG) and grown at 30°C.
Western Blotting
Cultures were collected from the YMC, fixed in 20% trichloroacetic acid and snap-frozen in liquid nitrogen. Protein was extracted as described in (Kushnirov, 2000). Standard conditions were used for SDS-PAGE and Western blotting. Antibodies used in this study were: anti-H3K9 crotonylation (PTM Biolabs PTM-516, Revmab Biosciences 31-1225-00), anti-H3K4 crotonylation (PTM Biolabs PTM 527), anti-H3K9 acetylation (Millipore 07-352, Millipore 06-942), anti-H3 (Millipore 05-928, EpiCypher 13-0001), anti-HA (Bethyl A190-108A and Abcam Ab9110), anti-G6PDH (Sigma A9521-1VL), and anti-Bdf1/2 (Govin lab).
Dot blotting
Different concentrations of biotinylated histone peptides (0.05-5 μg) were spotted onto a nitrocellulose membrane then probed with the anti-H3K9ac (Millipore, 06–942) at 1:5,000 or H3K9cr (PTM Biolabs, PTM-516; Revmab Biosciences, 31-1225-00) at 1:500 in a 5% nonfat milk solution and detected with an HRP-conjugated anti-rabbit by enhanced chemiluminescence (ECL).
Isothermal titration calorimetry (ITC)
Purified wild-type Taf14 (residues 1-132) and a series of 20-mer histone H3 peptides containing different acyl modifications (H3K9-acetyl, -propionyl, -butyryl, -crotonyl, or H3 peptides acetylated or crotonylated at K9, K14, and K18) were extensively dialyzed into a buffer containing 50mM HEPES pH 7.5, 200mM NaCl, and 1mM tris(2-carboxyethyl)phosphine (TCEP). ITC experimenters were carried out at 10°C using a MicroCal AutoITC-200 microcalorimeter. After an initial delay of 120s, H3 peptide (2 mM) was titrated into Taf14 (70 or 100 μM) over the course of 20 injections using 2 μL per injection. Reference power was set to 8 μcal/s and the stirring speed was 1000 RPM. Control heat of dilution experiments were performed by titrating each H3 peptide (2 mM) into a cell containing buffer only. Each peptide showed negligible heat of mixing (data not shown) therefore each sample titration into Taf14 was not corrected for the minimal background heat. Analysis of data was performed using MicroCal ITC-200 Origin software using a 1:1 binding model. The Kd represents the average of two experiments ± the standard deviation.
RNA-Seq analysis
RNA was prepared from samples (approximately 5 ODs) using the MasterPure™ Yeast RNA Purification Kit (Epicentre, MPY03100). The sequencing libraries were prepared from 1 μg of RNA/sample using the NEB Ultra Directional kit (E7420) with the mRNA isolation module (E7490) and adaptor sets 1/2 (E7355/E7500). Libraries were normalized using the NEB Library Quant Kit for Illumina (E7630). The quality of the pooled library was checked using the Agilent Bioanalyser 2100 HS DNA assay. The library was sequenced on an Illumina HiSeq 2000 Sequencer (50 bp reads). Minimum of 9 million reads per time-point were aligned using Burrows-Wheeler Aligner (bwa) and analyzed using the DESeq2 package (Love et al., 2014). Samples were analyzed in duplicate and replicates were combined for final analysis. PCA plots (log-transformed data) and significantly differentially expressed (SDE) genes (adjusted p-value < 0.05 and log2 fold change > 0.6) were generated using the DESeq2 package (Love et al., 2014). Functional annotation analysis was performed on SDE genes using DAVID with default parameters (Huang et al., 2009a; 2009b). Genes expressed in the wild-type YMC were previously identified as “periodic” or “non-periodic” using a periodicity algorithm on RNA-seq compiled at high resolution across the YMC (Kuang et al., 2014).
Chromatin Immunoprecipitation (ChIP)
Antibodies used for ChIP were: anti-H3K9 crotonylation (PTM Biolabs PTM516), anti-H3K9 acetylation (Millipore 06-942), anti-H3 (Millipore 05-928), anti-HA (Bethyl A190-108A and Abcam Ab9110), anti-Bdf1/2 (Govin lab), and anti-Taf14 (Reese lab). Chemostat samples (15 ml of saturated culture) were collected, fixed in 1% formaldehyde for 15 minutes and quenched in 125 mM glycine for 10 minutes. Cells were washed twice in TBS (300 mM NaCl, 40 mM Tris pH 7.5) and snap frozen. Pellets were resuspended in 1 ml of FA buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% Sodium deoxycholate, Roche complete protease inhibitor cocktail (11697498001), split into two 500 μl aliquots and lysed by beat beating for 10 min at 4°C. Aliquot volumes were adjusted to 1 ml with FA buffer and sonicated (30 seconds on, 30 seconds off) for 25 minutes. Samples were pelleted and the supernatants (whole cell lysate) pooled. Lysates (adjusted to 2 mg/ml protein for histones or 5 mg/ml for TAF14, using FA buffer) were precleared using Dynabeads Protein A (10001D, 50 μl beads per 500 μl lysate, 1 hour at 4°C) and 50 μl was saved as input. Antibody was added (2 μl for histones) and samples rotated overnight at 4°C. Dynabeads were washed three times in FA buffer + 0.5% BSA, with the third wash lasting minimum of two hours to preblock beads. Beads were then added (50 μl per 500 μl sample) and samples incubated for 2 hours at 4°C. Beads were washed as: FA buffer, FA buffer containing 500 mM NaCl, LiCl (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), 1 ml TE pH 8.0. Beads were resuspended in 100 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and DNA was eluted by incubation at 65°C for 15 min. Supernatant was collected and the elution step repeated to yield 200 μl of eluate. Crosslinks were reversed by adding 10 μl of NaCl (5 M) to the eluate and incubating at 65°C overnight. Input samples (50 μl) were treated with 150 μl of elution buffer and 10 μl of NaCl alongside the IP samples. Samples were treated with RNase A (10 μg, 60 min at 37°C) and proteinase K (20 μg, 60 min at 42°C). DNA was purified using the Zymo ChIP DNA Clean & Concentrator (D5205) and eluted in 30 μl of buffer.
ChIP-Seq
Libraries were prepared from 0.5 ng of eluted DNA using the NEBNext Ultra II DNA Library Prep Kit for Illumina (E7645) and adaptor sets 1/2 (E7355/E7500). The quality of the pooled library was checked using the Agilent Bioanalyser 2100 HS DNA assay. The library was sequenced on an Illumina HiSeq 2000 (101 bp reads). Alignments were performed using Burrows-Wheeler Aligner (bwa) with a minimum of 8 million unique reads obtained per sample. Peaks were called using a custom pipeline to generate uniformly processed reads (Beckwith et al., 2018) and the H3K9 acetylation and crotonylation normalized to H3. Fold change signal over H3 was processed using Deeptools2 computeMatrix function (Ramírez et al., 2016). To generate counts around the TSS or TTS using the reference-point setting, +/− 200 bp was used for histone marks. To generate profiles across the gene body, the scale-regions setting was used.
RNA extraction and qPCR
Chemostat samples were collected, pelleted and snap frozen. Pellets were washed in water and resuspended in 400 μl TES buffer (10 mM Tris HCl pH 7.5, 10 mM EDTA, 0.5% SDS). An equal volume of acid phenol added and samples were vortexed for 30 seconds. Samples were then incubated at 65C for 45 minutes with occasional vortexing before being chilled on ice for 5 minutes. Samples were centrifuged (5 min, 4C, 12K RPM) and the upper phase transferred to a fresh tube. Phenol extractions were repeated twice. RNA was precipitated by adding 0.1 volumes of sodium acetate (5M, pH 5.3) and 2.5 volumes of ice-cold ethanol (100%) followed by incubation at −80C for 30 minutes. Samples were centrifuged (5 min, 4C, 12K RPM) and the pellet washed with 70% ice-cold ethanol. Pellets were resuspended in water and the concentration determined. RNA (10 mg) was treated with RQ1 DNase (Promega Corporation Cat# M6101) at 37C for 60 minutes. RNA was purified using RNeasy Mini kit (RNeasy Minikit, Qiagen, Cat# 74104). cDNA was prepared using iScript cDNA synthesis kit (Thermofisher Scientific, 18080051). qPCR was performed using the SYBR green kit (PowerUp SYBR™ Green Mastermix, Biorad, Cat# 170 8880) and the primers in Table S1. Signals were normalized to Actin.
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Brian D. Strahl (brian_strahl@med.unc.edu).
DATA AND CODE AVAILABILITY
RNA-Seq and ChIP-Seq data are available under the NCBI accession GSE120019 available from the Gene Expression Omnibus (GEO) database. Raw data for westerns and dot blots are available at Mendeley: http://dx.doi.org/10.17632/gd9xsh6ywn.1
Supplementary Material
Highlights.
Histone lysine crotonylation (Kcr) oscillates in the yeast metabolic cycle (YMC)
Deregulation of crotonyl-CoA metabolism results in YMC defects
Taf14, a histone Kcr reader, is needed for transcription oscillations in the YMC
Kcr reading by Taf14 reduces growth gene expression during nutrient limitation
ACKNOWLEDGMENTS
We thank members of the Morrison and Strahl labs for suggestions and helpful discussions. We also thank Joseph Reese for the Taf14 antibody, Jerome Govin for the Bdf1/2 antibody and Steve Hahn for various Taf antibodies.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andrews FH, Shinsky SA, Shanle EK, Bridgers JB, Gest A, Tsun IK, Krajewski K, Shi X, Strahl BD, and Kutateladze TG (2016). The Taf14 YEATS domain is a reader of histone crotonylation. Nat Chem Biol 12, 396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Wang Y, Li X, Li X-M, Liu Z, Yang T, Wong CF, Zhang J, Hao Q, and Li XD (2014). Identification of “erasers” for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3, e02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith SL, Schwartz EK, Garcia-Nieto PE, King DA, Gowans GJ, Wong KM, Eckley TL, Paraschuk AP, Peltan EL, Lee LR, et al. (2018). The INO80 chromatin remodeler sustains metabolic stability by promoting TOR signaling and regulating histone acetylation. PLoS Genet 14, e1007216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnetti AJ, Aydin M, and Buchler NE (2016). Cell cycle Start is coupled to entry into the yeast metabolic cycle across diverse strains and growth rates. Mol Biol Cell 27, 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, and Tu BP (2011). Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, and Bultman SJ (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metabolism 13, 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman NJ, Black PN, Zhao XD, Knudsen J, and DiRusso CC (2001). The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular Utilization. J Biol Chem 276, 37051–37059. [DOI] [PubMed] [Google Scholar]
- Falcón AA, Chen S, Wood MS, and Aris JP (2010). Acetyl-coenzyme A synthetase 2 is a nuclear protein required for replicative longevity in Saccharomyces cerevisiae. Mol. Cell. Biochem. 333, 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, Balázsi S, Hajnády Z, Liebert A, Kazakevych J, et al. (2018). Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nature Communications 9, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi A, Zhang D, Huang H, Barral S, Kwon OK, Qi S, Tang Z, Buchou T, Vitte A-L, He T, et al. (2016). Dynamic Competing Histone H4 K5K8 Acetylation and Butyrylation Are Hallmarks of Highly Active Gene Promoters. Mol Cell 62, 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowans GJ, Schep AN, Wong KM, King DA, Greenleaf WJ, and Morrison AJ (2018). INO80 Chromatin Remodeling Coordinates Metabolic Homeostasis with Cell Division. Cell Reports 22, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz A, Mursula AM, Firzinger A, Hamilton B, Kilpeläinen SH, Hartig A, Ruis H, Hiltunen JK, and Rottensteiner H (1998). Peroxisomal Delta3-cis-Delta2-trans-enoyl-CoA isomerase encoded by ECI1 is required for growth of the yeast Saccharomyces cerevisiae on unsaturated fatty acids. J Biol Chem 273, 31366–31374. [DOI] [PubMed] [Google Scholar]
- Gut P, and Verdin E (2013). The nexus of chromatin regulation and intermediary metabolism. Nature 502, 489–498. [DOI] [PubMed] [Google Scholar]
- Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, and Gurvitz A (2003). The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiology Reviews 27, 35–64. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huber A, French SL, Tekotte H, Yerlikaya S, Stahl M, Perepelkina MP, Tyers M, Rougemont J, Beyer AL, and Loewith R (2011). Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. Embo J 30, 3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo YJ, Ficarro SB, Soares LM, Chun Y, Marto JA, and Buratowski S (2017). Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes Dev 31, 2162–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebede AF, Nieborak A, Shahidian LZ, Le Gras S, Richter F, Gómez DA, Baltissen MP, Meszaros G, de Fatima Magliarelli H, Taudt A, et al. (2017). Histone propionylation is a mark of active chromatin. Nat. Struct. Mol. Biol 159, nsmb.3490. [DOI] [PubMed] [Google Scholar]
- Klein BJ, Vann KR, Andrews FH, Wang WW, Zhang J, Zhang Y, Beloglazkina AA, Mi W, Li Y, Li H, et al. (2018). Structural insights into the Π-Π-Π stacking mechanism and DNA-binding activity of the YEATS domain. - PubMed - NCBI. Nature Communications 9, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, and Murray DB (2004). A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA 101, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A, Vadder FD, Kovatcheva-Datchary P, and Bäckhed F (2016). From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345. [DOI] [PubMed] [Google Scholar]
- Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, and Denu JM (2016). Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol Cell 64, 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Cai L, Zhang X, Ji H, Tu BP, and Boeke JD (2014). High-temporal-resolution view of transcription and chromatin states across distinct metabolic states in budding yeast. Nat. Struct. Mol. Biol 21, 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Z, Pinglay S, Ji H, and Boeke JD (2017). Msn2/4 regulate expression of glycolytic enzymes and control transition from quiescence to growth. eLife 6, e29938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857–860. [DOI] [PubMed] [Google Scholar]
- Li Y, Sabari BR, Panchenko T, Wen H, Zhao D, Guan H, Wan L, Huang H, Tang Z, Zhao Y, et al. (2016). Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol Cell 62, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SYR, Li W, et al. (2014). AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 159, 558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SI, and Broach JR (2009). Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc Natl Acad Sci USA 106, 19928–19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, and Rando OJ (2005). Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3, e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machné R, and Murray DB (2012). The yin and yang of yeast transcription: elements of a global feedback system between metabolism and chromatin. PLoS ONE 7, e37906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J (2016). The molecular basis of metabolic cycles and their relationship to circadian rhythms. Nat. Struct. Mol. Biol 23, 1035–1044. [DOI] [PubMed] [Google Scholar]
- Nath A, and Chan C (2016). Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci Rep 6, 18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AR, and Pellegrini M (2011). Regulation of the yeast metabolic cycle by transcription factors with periodic activities. BMC Syst Biol 5, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, Dai L, Shimada M, Cross JR, Zhao Y, et al. (2015). Intracellular Crotonyl-CoA Stimulates Transcription Through p300-Catalyzed Histone Crotonylation. Mol Cell 58, 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Zhang D, Allis CD, and Zhao Y (2017). Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Bio 18, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Wang AY, and Kobor MS (2009). YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol 87, 65–75. [DOI] [PubMed] [Google Scholar]
- Shanle EK, Andrews FH, Meriesh H, McDaniel SL, Dronamraju R, DiFiore JV, Jha D, Wozniak GG, Bridgers JB, Kerschner JL, et al. (2015). Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev 29, 1795–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simithy J, Sidoli S, Yuan Z-F, Coradin M, Bhanu NV, Marchione DM, Klein BJ, Bazilevsky GA, McCullough CE, Magin RS, et al. (2017). Characterization of histone acylations links chromatin modifications with metabolism. Nature Communications 8, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, and Allis CD (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Stuckey S, and Storici F (2013). Chapter Eight - Gene Knockouts, in vivo Site-Directed Mutagenesis and Other Modifications Using the Delitto Perfetto System in Saccharomyces cerevisiae In Laboratory Methods in Enzymology: Cell, Lipid and Carbohydrate, Lorsch J, ed. (Academic Press; ), pp. 103–131. [DOI] [PubMed] [Google Scholar]
- Suganuma T, and Workman JL (2018). Chromatin and Metabolism. Annu. Rev. Biochem 87, 27–49. [DOI] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, and Boeke JD (2006). Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell 23, 207–217. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, and McKnight SL (2006). Metabolic cycles as an underlying basis of biological oscillations. 7, 696–701. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, and McKnight SL (2005). Logic of the Yeast Metabolic Cycle: Temporal Compartmentalization of Cellular Processes. Science 310, 1152–1158. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, and McKnight SL (2007). Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA 104, 16886–16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G-Z, Hickey SL, Shi L, Huang H-C, Nakashe P, Koike N, Tu BP, Takahashi JS, and Konopka G (2015). Cycling Transcriptional Networks Optimize Energy Utilization on a Genome Scale. Cell Reports 13, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1999). The economics of ribosome biosynthesis in yeast. Trends in Biochemical Sciences 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Zhao D, Guan H, Zhao S, Mi W, Wen H, Li Y, Zhao Y, Allis CD, Shi X, and Li H (2016). YEATS2 is a selective histone crotonylation reader. Cell Res. 26, 629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, and Garcia BA (2015). Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb Perspect Biol 7, a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq and ChIP-Seq data are available under the NCBI accession GSE120019 available from the Gene Expression Omnibus (GEO) database. Raw data for westerns and dot blots are available at Mendeley: http://dx.doi.org/10.17632/gd9xsh6ywn.1