Abstract
Ultrastructural analysis of healthy, diseased, or experimental tissues is essential in diagnostic and investigative pathology. Evaluation of large tissue areas with suborganelle resolution is challenging because biological structures ranging from several millimeters to nanometers in size need to be identified and imaged while maintaining context over multiple scales. Imaging with field emission scanning electron microscopes (FE-SEMs) is uniquely suited for this task. We describe an efficient workflow for the preparation and unobstructed multiscale imaging of tissue sections with backscattered electron scanning electron microscopy (BSE-SEM) for applications in ultrastructural pathology. We demonstrate that a diverse range of tissues, processed by conventional electron microscopy protocols and avoiding the use of mordanting agents, can be imaged on standard glass slides over multiple scales, from the histological to the ultrastructural level, without any visual obstructions. Our workflow takes advantage of the very large scan fields possible with modern FE-SEMs that allow for the acquisition of wide-field overview images which can be explored at the ultrastructural level by digitally zooming into the images. Examples from applications in pulmonary research and neuropathology demonstrate the versatility and efficiency of this method. This BSE-SEM-based multiscale imaging procedure promises to substantially simplify and accelerate ultrastructural tissue analysis in pathology.
Keywords: backscattered electrons, histology, multiscale imaging, pathology, SEM, ultrastructure
Introduction
Ultrastructural imaging of healthy, diseased, or experimental tissues is essential to diagnostic and investigative pathology. Because of the high resolution provided by electron microscopy (EM), tissue structures, cells, organelles, and microorganisms can be visualized and characterized with greater acuity than by light microscopy (LM) alone. For this reason, EM is used as a gold standard diagnostic tool in many fields of pathology, particularly in pulmonary pathology, renal pathology, skeletal and cardiac muscle pathology, and neuropathology, and for the diagnosis of ciliopathies, lysosomal storage disease, tumors, and infectious diseases.1
Identifying regions of interest (ROIs) for investigating tissues with suborganelle resolution is often challenging and time-consuming. Biological structures range in size over multiple scales from several millimeters (organ regions and tissue areas) to few nanometers (suborganelle detail) and need to be identified in an efficient manner, imaged and correlated while maintaining histological context. Many tissues encountered in research pathology are from non-transgenic test animals, or do not contain tagged recombinant proteins or organelles, that could be used as landmarks for the efficient correlation of bright-field or fluorescence microscopy and EM images.2–4
In the field of ultrastructural pathology, these experimental challenges have been addressed by first preparing “survey” sections for LM from samples processed for transmission EM (TEM). These “survey sections” are stained with toluidine blue and analyzed at relatively low overview magnifications by LM for evaluation of processing quality, characterization of the histopathology, and identifying ROIs. After an ROI has been identified, sample blocks are trimmed to generate small block faces for ultrathin sectioning before final ultrastructural analysis in the TEM.5
This procedure has been used for decades with great success. However, it is time-consuming and involves major technical hurdles mainly due to the need for ultrathin sections on small TEM grids that limit the size of the individual tissue sections used for investigation. Furthermore, correlation of ROIs between “survey” LM-sections and ultrathin TEM-sections can be extremely challenging, if not impossible, especially when large and complex tissues are investigated or with homogeneous samples (e.g., cultured cells) that lack distinct “landmarks” for orientation and alignment. Therefore, multiscale imaging, that is the correlated imaging of a sample over multiple magnifications from the millimeter to the nanometer level, has remained challenging using this more traditional approach.
Modern field emission scanning electron microscopes (FE-SEMs) in combination with sensitive backscattered electron detectors (BSDs) and modified tissue processing, mounting, and staining procedures promise to substantially accelerate the workflow for multiscale tissue imaging.6–12 Backscattered electron (BSE) imaging exploits the direct relation between the backscatter coefficient and atomic weight of an element: the higher the atomic number (Z), the more electrons become backscattered and can be detected with a dedicated BSE detector.13
Many popular protocols for processing tissues and cells for backscattered electron scanning electron microscopy (BSE-SEM) imaging, especially for “three-dimensional” volume scanning electron microscopy (SEM) imaging, rely on mordanting agents such as tannic acid or thiocarbohydrazide to increase the deposition of heavy metals to improve membrane contrast and the signal-to-noise ratio and to avoid sample charging.14–16 These protocols have drawbacks when working with tissues other than lipid-rich nervous tissue (e.g., muscle, kidney, lung) and can result in unequal staining and infiltration, overstaining (obscuring fine details by excessive heavy metal deposition), and poor reproducibility due to diverse tissue composition and sample sizes15–18 (and our own observations). Furthermore, there is a lack of reference publications and validation for these protocols for applications in diagnostic and investigative pathology where phenotypes of healthy and diseased tissues need to be compared with published reference “gold standard” EM images in pathology journals, tissue atlases, and ultrastructural pathology text books.1,19
Our goal was to develop a general multiscale imaging method for animal and human tissues which exploits the advantages offered by BSE-SEM and which is specifically targeted for the needs of ultrastructural diagnostic and investigative pathology. Here, we introduce a “universal” processing and imaging workflow that has been tested not only on nervous tissue11 but also on a diverse range of tissues including lung, kidney, muscle, and intestine. This method provides contrast and resolution similar to established TEM applications in ultrastructural pathology, but uses a workflow that dramatically simplifies multiscale imaging. In addition, this workflow uses the large scan field and high resolution (small pixel size) provided by modern FE-SEMs, which allow one to digitally zoom into highly resolved wide-field images to explore ultrastructural detail without losing cellular or histological context.
Materials and Methods
Animals, Tissue Harvesting, and Fixation
All animal studies were performed in accordance with standard regulations and were approved by Genentech’s Institutional Animal Care and Use Committee. Anesthetized animals were perfused with physiological saline, followed by 4% paraformaldehyde in 0.1 M (pH 7.4) phosphate buffer. Organs and tissues were dissected and sectioned into about 1-mm-thick slices. Tissues were fixed by immersion fixation in modified Karnovsky’s fixative (2.5% paraformaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2) for at least 24 hr or longer at 4C.20
Processing of Tissues for BSE-SEM Imaging
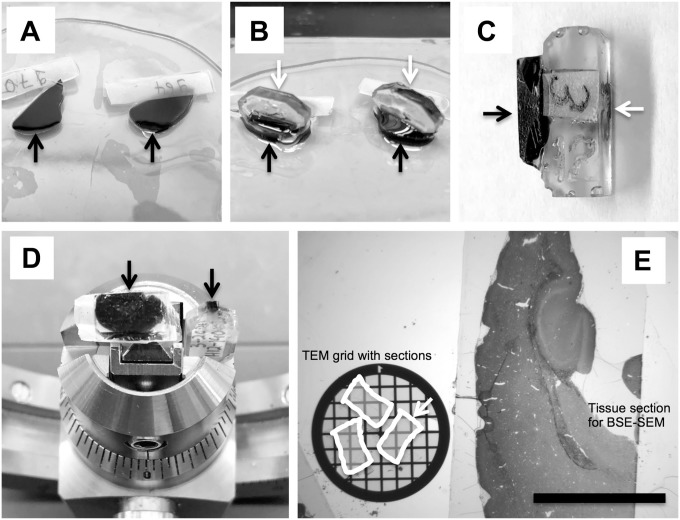
Fixed tissues slices were washed with ultrapure water and postfixed with 1% osmium tetroxide reduced with 1.5% (w/v) potassium ferrocyanide (final concentration) for 2 hr on ice. The samples were then washed in ultrapure water and stained “en block” with 0.5% (w/v) uranyl acetate at 4C overnight. Following staining, samples were dehydrated in a series of ascending ethanol concentrations, rinsed twice with propylene oxide, and finally embedded in epoxy resin Eponate-12 (Ted Pella; Redding, CA). The tissue slices were first flat-embedded between two Aclar plastic sheets with a small weight on top to keep the slices flat and polymerized for at least 24 hr at 65C (Fig. 1A). The top Aclar sheet was then carefully removed and a prepolymerized empty resin block was glued with fresh epoxy resin on top of the tissue slice (Fig. 1B) to serve as a “pin” to be able to clamp the tissue block into the ultramicrotome with the correct orientation (Fig. 1C and D). All reagents and materials for tissue fixation and processing were purchased from Electron Microscopy Sciences (EMS; Hatfield, PA).
Figure 1.
Tissue processing and sectioning for multiscale BSE-SEM imaging. (A) Tissue slices (black arrows) were processed for BSE-SEM and flat-embedded in epoxy resin between two Aclar plastic sheets and then polymerized. (B) The top Aclar sheet was removed and prepolymerized resin blocks (white arrows) were glued on top of the polymerized tissue slices. (C) Side view of a tissue block (black arrow) glued to the empty epon block (white arrow) that served as “pin” for clamping the sample into the ultramicrotome holder. (D) Representative sample block faces (arrows) for BSE-SEM (left, 3 mm × 6 mm) and for TEM (right, 0.5 mm × 1 mm) are shown side by side. (E) A 1-µm-thick section generated from a block face of about 3 mm × 6 mm (area of 18 mm2) of a processed mouse brain was stained with toluidine blue (E, right side) and is shown side by side with a standard TEM grid (E, left side) harboring ultrathin sections of about 1 mm × 0.5 mm (white arrow). Note that many areas in the small TEM sections become obstructed by grid bars. Scale bar: 3 mm. Abbreviations: BSE-SEM, backscattered electron scanning electron microscopy; TEM, transmission electron microscopy.
Preparation of Carbon-coated Histology Glass Slides for BSE-SEM
Clean “superfrost-plus” microscope slides (Thermo Fisher Scientific; Waltham, MA) were sputter-coated with a layer of about 40 to 80 nm carbon from carbon cord, resulting in a surface with shiny brown color, using a sputter coater (EMS150R ES sputter coater; Electron Microscopy Sciences, Hatfield, PA).
Preparation and Staining of Sections on Carbon-coated Glass Slides for BSE-SEM Imaging
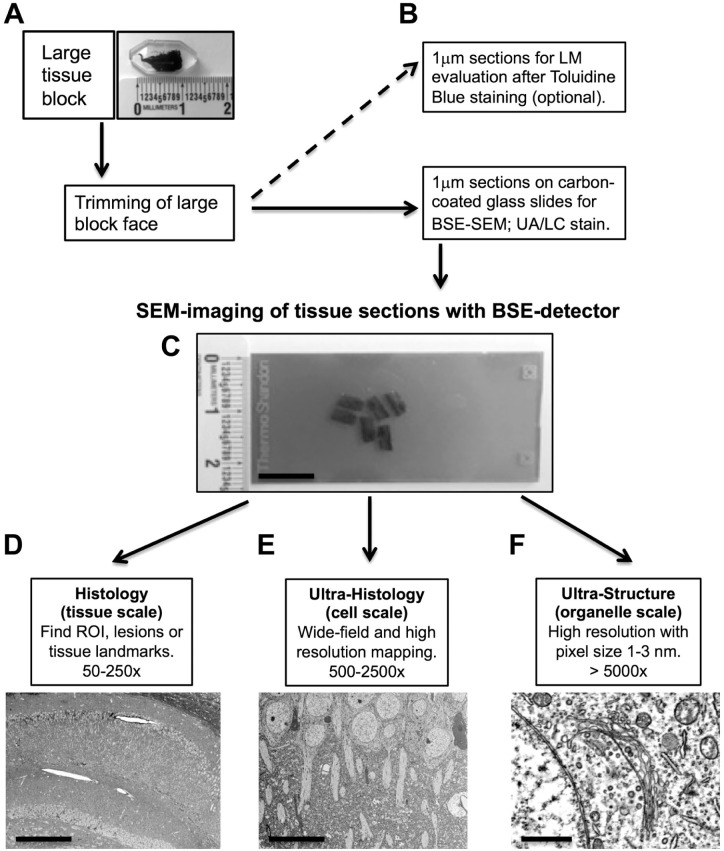
To facilitate unobstructed multiscale imaging of a maximum tissue area, large rectangular block faces with a dimension of up to 4 mm × 8 mm were trimmed (Fig. 1D). Thick sections (1000 nm thickness) were cut with the UMC ultramicrotome (Leica Biosystems; Buffalo Grove, IL) using a DIATOME diamond knife for histology (Electron Microscopy Sciences). Sections were transferred from the knife boat to a drop of ultrapure water on glass slides using a water-wetted wooden stick. The first sections were collected on standard glass slides and stained with toluidine blue to determine whether relevant tissue areas were represented in the sample. When this had been confirmed, sections were collected on carbon-coated glass slides (Fig. 2C). The sections were dried on a heat plate at about 60C. Dried sections firmly adhered to the carbon-coated glass surface, and no pretreatment of the surface of the carbon-coated glass slides was needed. Finally, sections were stained with 4% aqueous uranyl acetate for 15 min and 0.1% Reynold’s lead citrate for 1 min to enhance contrast and prevent charging in the SEM.21 Sections were thoroughly rinsed with ultrapure water and dried on a heat plate before being transferred to the SEM.
Figure 2.
Workflow for multiscale imaging of tissue sections with BSE-SEM. (A) Tissue were processed for BSE-SEM and embedded in epoxy resin. Note the large dimension of the embedded tissue slice. (B) After trimming of a large block face (up to about 4 mm × 8 mm), thick sections (1 µm) were placed on carbon-coated standard glass slides and stained with UA and LC. If required, parallel sections were collected for optional toluidine blue LM analysis. (C) Stained sections on carbon-coated glass slides were imaged in the SEM with a BSE detector. (D) Imaging at low magnifications (tissue scale, 50×–250×) facilitated finding and mapping of ROIs, pathological lesions, and tissue landmarks (e.g., hippocampus in mouse brain). (E) To explore cellular and subcellular structures within their tissue context, an ROI was imaged at moderate magnifications (cell scale, 500×–2500×) (e.g., neurons in hippocampus). (F) For high magnification (organelle scale, >5000×) and resolution of ultrastructure, cells and tissues were imaged with pixel sizes of 1 to 3 nm (e.g., Golgi apparatus in hippocampal neuron). This imaging strategy facilitated the unobstructed observation of tissues over a wide range from the tissue level to the suborganelle level. Scale bars: (C) 10 mm, (D) 300 µm, (E) 20 µm, and (F) 1 µm. Abbreviations: BSE-SEM, backscattered electron scanning electron microscopy; UA, uranyl acetate; LC, lead citrate; LM, light microscopy; SEM, scanning electron microscopy; BSE, backscattered electron; ROI, region of interest.
Multiscale Imaging of Tissue Sections on Glass Slides With BSE-SEM
SEM was performed using a GeminiSEM 300 equipped with a field emission gun (Carl Zeiss AG; Oberkochen, Germany). Carbon-coated glass slides with stained sections were mounted into the correlative SEM holder (Carl Zeiss AG). For operation of the GeminiSEM 300 microscope, the application software SmartSEM (version 6.01) was used (Carl Zeiss AG). Best imaging results were obtained using the “in-chamber” dedicated BSD (BSD1) at 8.5 mm working distance, 30 µm (standard) aperture, 3 to 6 keV acceleration voltage, and with operation of the field emission gun in “high current” mode. For the majority of images, a scan speed of “5,” noise reduction by 4× line averaging, and an image size of at least 4096 × 3072 (4000 × 3000) pixels were chosen. For imaging of ultrastructural detail, pixel sizes between 1 and 3 nm were used. The grayscale of the images was inverted to achieve TEM-like representations.
For multiscale imaging, the following complementary imaging strategies were used. In the default workflow, tissues were imaged by stepwise increasing the magnification starting with a low magnification (50×–250×, wide field of view [FOV]) to find areas of interest (ROIs) at the histology level (Fig. 2D), similar to using “survey” sections in LM. The selected ROIs were then explored at moderate magnification (500×–2500×) to obtain an overview image of the ROI at cellular to subcellular resolution (Fig. 2E). Finally, structures of interest were imaged at ultrastructural resolution by imaging at high magnification (>5000×) with pixel sizes smaller than 3 nm (Fig. 2F).
A different complementary approach for multiscale imaging made use of the very large store resolution (maximum image size in pixels) of the GeminiSEM 300 microscope (Carl Zeiss AG). The maximum store resolution was 32,768 × 24,576 pixels (32,000 × 24,000) per image. This could be used for maximizing the FOVs (wide-field imaging) while keeping the pixel size small to optimize resolution of detail and to digitally “zoom” into an acquired image. For example, using the maximum store resolution of 32,000 × 24,000 pixels per image allowed imaging at a moderate magnification of 1500×, with a large FOV (72 µm × 55 µm) and with ultrastructural resolution (pixel size of 2.3 nm). This allowed for imaging several cells per image and then for digitally “zooming” into each cell for analysis of ultrastructure and suborganelle detail (Fig. 2F).
At the specified magnifications, scanning with the maximum store resolution resulted in the following FOVs and pixel sizes: at 50× magnification—FOV: 2.3 mm × 1.7 mm; 70 nm pixels, at 500× magnification—FOV: 230 µm × 170 µm; 7 nm pixels, at 2500× magnification—FOV: 46 µm × 34 µm; 1.4 nm pixels, and at 5000× magnification—FOV: 23 µm × 17 µm; 0.7 nm)
Photoshop CS4 (Adobe) was used to adjust contrast and brightness of whole images, to crop ROIs, and to reduce the pixel size per area for images that were obtained with a store resolution larger than 4096 × 3072 pixels to produce images at 300 dots per inch (dpi) print resolution for figure preparation.
Results
Large Tissue Sections Can Be Used for Unobstructed Multiscale BSE-SEM Imaging
A major advantage of an SEM is its large sample chamber that accommodates imaging of large samples, such as standard 1″ × 3″ inch histology glass slides, without visual obstructions caused by TEM grids. To fully exploit this space advantage, we developed a procedure to process and flat-embed large tissue slices encompassing areas of more than 50 mm2 (Fig. 1A–E). After polymerization, large rectangular block faces with dimensions of up to 4 mm × 8 mm were trimmed and sectioned (Fig. 1C–E).
Figure 1D shows examples of the dimensions of representative block faces trimmed for multiscale BSE-SEM (left) or TEM (right). The block face for BSE-SEM sections shown here was approximately 3 mm × 6 mm (18 mm2), whereas the block face for TEM was about 1 mm × 0.5 mm (0.5 mm2). The BSE-SEM sample block was then used to prepare 1-µm-thick sections on carbon-coated glass slides. Because the primary electron beam in the SEM generates signals from the surface of the sample,13 thick sections could be used for high-resolution imaging. Preparation of large area sections was much easier to achieve with 1000-nm-thick sections than with ultrathin (50–100 nm) TEM sections. In the example shown in Fig. 1E, the brain tissue area available for unobstructed imaging with BSE-SEM (Fig. 1E, right) was more than 30 times larger than the area of the ultrathin sections on the TEM grid (Fig. 1E, bottom left).
An Efficient Workflow for Multiscale Imaging of Large Tissue Sections With BSE-SEM
Correlating histological features observed in toluidine blue thick sections by LM with ultrastructural features in the TEM is major technical and time-consuming bottleneck of the traditional LM-TEM workflow. These challenges can be circumvented by preparing tissue sections on carbon-coated glass slides and imaging large tissue areas at multiple scales on a single instrument (SEM) from the lowest to the highest magnification and resolution, instead of switching between LM and TEM (Fig. 2).
Imaging a section first at very low magnification (50×–250×) proved helpful to generate organ region and tissue overview maps (Fig. 2D). Further magnification at 500× to 2500× resulted in an FOV wide enough to include several cells with subcellular detail (Fig. 2E). Two different strategies were used to obtain images of cells and tissues with ultrastructural resolution of organelles and suborganelle structures (Fig. 2F) at pixel sizes smaller than 3 nm. In the first strategy, an ROI was imaged with moderate store resolutions (image size: 4000 × 3000 pixels) and by stepwise increasing the magnification setting on the SEM instrument itself. This strategy was useful when a large FOV was not required, when an ROI had already been identified and only needed to be imaged at higher magnification, and to save image acquisition time by reducing the number of scanned pixels per image.
In the second strategy, images were acquired at very large store resolutions (e.g., 32,000 × 24,000 pixels). Increasing the store resolution at a given magnification setting resulted in a smaller pixel size in the corresponding image and improved resolution of ultrastructural details. This strategy was useful to maximize the FOV while still imaging with ultrastructural resolution. The resulting images could then be explored for ultrastructural detail by digitally “zooming” into the images. This helped to identify ROIs or visualize highly magnified and resolved ROIs in the wider histological context.
We found that the smallest useful pixel size for imaging tissues that had been prepared according to our protocols was in the 1 to 2 nm range. At this pixel size, all membrane-bound organelles, including individual cisternae of the Golgi apparatus, vesicles, and mitochondria with cristae, could be unequivocally identified (Fig. 2F), while imaging with smaller pixel sizes did not improve detail and resulted only in a smaller FOV.
Multiscale BSE-SEM Imaging of a Diverse Set of Tissues
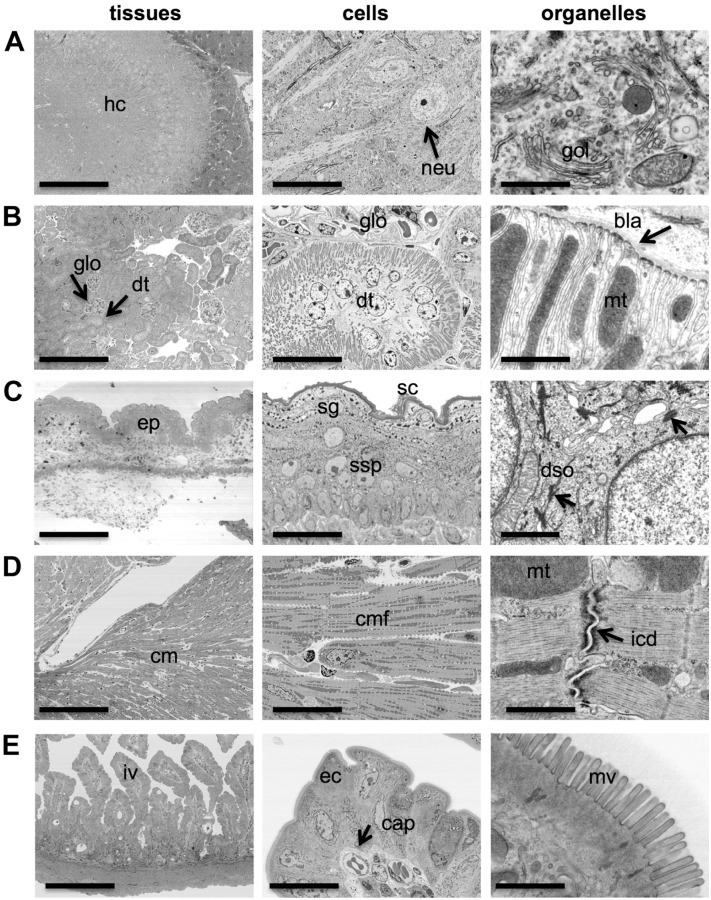
Next, we tested how well several tissues that are frequently encountered in ultrastructural research pathology could be imaged with the BSE-SEM method. We were especially interested to see whether the tissue preservation, staining contrast, and resolution were sufficient for the multiscale BSE-SEM method to be useful in ultrastructural pathology and basic tissue research. This was important to evaluate as the most recent work with BSE-SEM has explored lipid-rich nervous tissue,10,11,13 and much less is known about the processing and imaging quality for alternative tissues with BSE-SEM.12,18 For this purpose, a diverse set of mouse and rat tissues were evaluated, including brain, kidney, skin, cardiac muscle, and intestine (Fig. 3A–E). Other tissues that were also investigated but are not shown here included pancreas, spleen, skeletal muscle, and lung.
Figure 3.
Imaging a diversity of tissues from the tissue level to the organelle level. Tissues that are frequently encountered in research pathology were processed for BSE-SEM and imaged at the tissue level with a magnification of 250× (“tissues,” left panel), at the cellular level with a magnification of 2500× (“cells,” middle panels), and at the organelle level with a magnification of 50,000× (“organelles,” right panels). (A) Mouse hippocampus (hc) with neurons (neu, arrow) and Golgi apparatus (gol). (B) Mouse kidney cortex with marked glomerulus (glo, arrow), distal tubules (dt, arrow), mitochondria (mt), and basal lamina (bla, arrow). (C) Mouse skin epidermis (ep) with marked stratum corneum (sc), stratum granulosum (sg), stratum spinosum (sp), and desmosomes (dso, arrows). (D) Rat cardiac muscle (cm) with marked cardiac muscle fibers (cmf), mitochondria (mt), and intercalated disk (icd, arrow). (E) Mouse small intestine with marked villi (iv), enterocytes (ec), capillaries (cap, arrows), and microvilli (mv). Scale bars: 200 µm (A–E, left panels), 20 µm (A–E, middle panels), and 1 µm (A–E, right panels). Abbreviation: BSE-SEM, backscattered electron scanning electron microscopy.
All tissue samples were first imaged at low magnification (250×) to obtain tissue overviews and determine whether there was sufficient contrast and resolution to identify histological landmarks (Fig. 3A–E, “tissues” panel). The magnification was then increased to 2500× (Fig. 3A–E, “cells” panel) to evaluate ROIs in the tissues at the cellular to subcellular level. Finally, cells and tissue structures were investigated at a high magnification (50,000×) with pixel sizes smaller than 3 nm to resolve ultrastructural detail (Fig. 3A–E, “organelles” panel).
In the hippocampus of the brain, dark layers of myelinated axons and the neurons were recognizable in the tissue overviews (Fig. 3A, “tissue” panel). At 2500× magnification, neuronal soma, nucleus, and larger organelles such as mitochondria were clearly visible (Fig. 3A, “cells” panel). At the highest magnification used (50,000×), we were able to identify the stacked cisternae of the Golgi apparatus, several coated and uncoated vesicles, and mitochondrial cristae (Fig. 3A, “organelles” panel).
In normal kidney tissue, several glomeruli and renal tubules were clearly distinguishable in overview images (Fig. 3B, “tissue” panel). Exploring the morphology of a distal tubule up to a cellular level (2500×) revealed the arrangement of the nuclei and large elongated mitochondria in the epithelial cells of the tubule (Fig. 3B, “cells” panel). At the highest magnification used, the lamina basalis, the membranes of the basal labyrinth, and the mitochondrial cristae were clearly visible (Fig. 3B, “organelles” panel).
When skin tissue was imaged at low (250×) and moderate (2500×) magnifications, the various layers of the epidermis, including the stratum corneum, stratum granulosum, and stratum spinosum, could readily be identified (Fig. 3C, “tissue” and “cells” panel). At high magnification (50,000×), desmosomes were clearly visible (Fig. 3C, “organelles” panel).
Low (250×) and moderate (2500×) magnification of cardiac muscle tissue revealed arrangement of myofibers, including nuclei and intercalated disks (Fig. 3D, “tissue” and “cells” panel). At high magnification, actin–myosin fibrils, intercalated disks, gap junctions, and mitochondria were clearly identifiable (Fig. 3D, “organelles” panel).
Numerous intestinal villi in a cross section of mouse small intestine were imaged simultaneously at 250× magnification (Fig. 3E, “tissue” panel). At 2500× magnification, intestinal epithelial cells (enterocytes) with brush border microvilli were visible, as well as several capillaries (Fig. 3E, “cells” panel). High magnification resolved individual microvilli and membranes of the endoplasmic reticulum, endosomes, coated vesicles, and mitochondria (Fig. 3E, “organelles” panel).
This investigation of several normal mouse and rat tissues demonstrated that multiscale imaging with BSE-SEM was well suited to study a wide range of healthy tissues and cells from the histological overview level up to resolving suborganelle detail with excellent contrast, detail, and no obvious artifacts.
Unobstructed Multiscale Imaging of a Rat Kidney Glomerulus
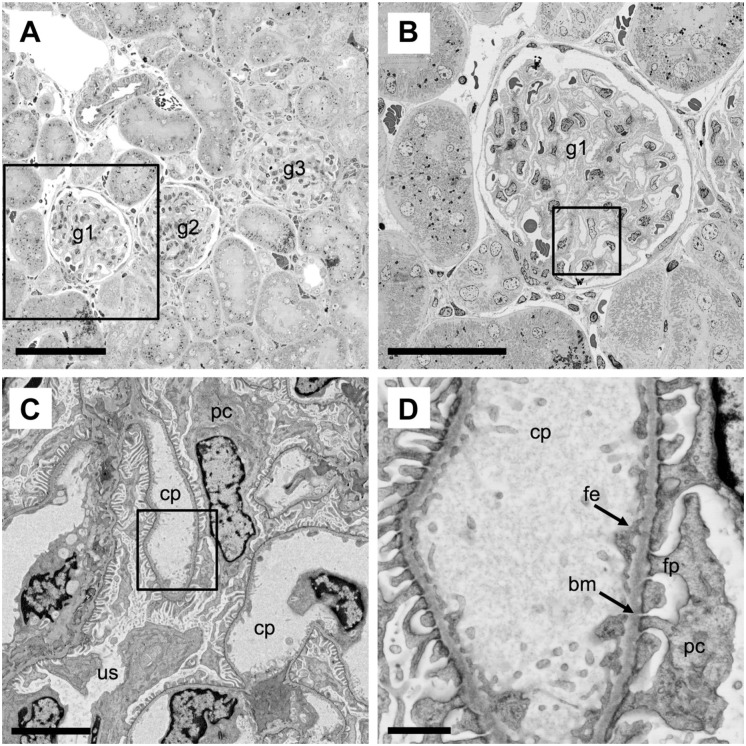
The ultrastructural examination of renal tissue by TEM has contributed significantly to the understanding of renal disease and to disease diagnostics.1 One challenge with traditional TEM is the need to identify multiple glomeruli with representative lesions within a sample. Therefore, we evaluated whether BSE-SEM offered adequate contrast and resolution to successfully characterize ultrastructural hallmarks of rat renal tissue similar to TEM, but with the additional advantage of offering a straightforward multiscale imaging strategy without visual obstructions by TEM grid bars.
First, low-magnification wide-field imaging at 200× magnification with the BSE-SEM allowed us to quickly find areas in the renal cortex with several glomeruli surrounded by renal tubules (Fig. 4A). Therefore, “survey” imaging by LM of toluidine blue–stained sections from EM sample blocks to find relevant areas was not needed. Next, we selected one glomerulus for further exploration at sequentially higher magnification settings (Fig. 4B–D). An unobstructed view of a complete glomerulus with adjacent proximal and distal renal tubules was obtained at 500× magnification and enabled a global assessment of the glomerular morphology (Fig. 4B). For a detailed examination of the glomerular filtration apparatus, the magnification was further increased to 3000× (Fig. 4C) and subsequently to 13,000× (Fig. 4D). At these magnifications, structures critical for the assessment of normal renal morphology or glomerulopathies became clearly visible and easily resolved. For example, the capillary (cp) and urinary spaces (us), the glomerular lamina basalis (bm), the podocytes (pc) with foot processes (fp), and the fenestrated endothelium (fe) were readily identifiable (Fig. 4C–D). This demonstrated the advantages of multiscale BSE-SEM imaging in renal research and pathology. The method provided large and unobstructed wide-field views over renal tissue that are critical for a global and unbiased morphological assessment, as well as the capacity to resolve ultrastructural details of the glomerular filtration apparatus that are critical for the evaluation of renal pathology.
Figure 4.
Multiscale imaging of a rat kidney glomerulus. (A–D) Normal kidney tissue (rat) was processed for BSE-SEM imaging. Images of the same tissue region were acquired at stepwise higher magnifications: 200× (A), 500× (B), 3000× (C), and 13,000× (D). (A) The unobstructed low magnification and wide field of view shows several glomeruli (g1–g3) surrounded by numerous kidney tubules. The boxed area is shown at higher magnification in panel B. (B) Global unobstructed view of a kidney glomerulus (g1). The boxed area is shown at higher magnification in panel C. (C) Detailed view of a region in the glomerulus (g1) with capillaries (cp), urinary space (us), and podocytes (pc). The boxed area is shown at high magnification in panel D. (D) High magnification and resolution view of a capillary (cp), fenestrated endothelium (fe), podocytes (pc), podocyte foot processes (fp), and lamina basalis (bm). Scale bars: (A) 100 µm, (B) 50 µm, (C) 5 µm, and (D) 1 µm. Abbreviation: BSE-SEM, backscattered electron scanning electron microscopy.
Identification of Amyloid Plaques in a Mouse Model of Alzheimer’s Disease
Next, we explored whether the BSE-SEM workflow was both suitable and provided distinct advantages over the traditional TEM workflow for applications in ultrastructural neuropathology. A major challenge in many ultrastructural pathology studies is finding focal lesions within a tissue section. This problem can also be described as “finding the needle in the haystack.” One test case for the suitability of multiscale BSE-SEM for this type of imaging challenge was to identify and image amyloid plaques in various brain regions of a PS2APP mouse model of Alzheimer’s disease22 and then use the highest reasonable magnification and resolution achievable with our current BSE-SEM instrumentation to define the ultrastructure of an amyloid plaque and its associated cellular or subcellular structures.
One particular goal of this experiment was to determine whether the practical resolution achievable with BSE-SEM was high enough to resolve amyloid fibrils within the core of amyloid plaques as has been shown by TEM.22–24 Resolving fibrils with diameters of 10 to 20 nm in the densely packed amyloid plaques is challenging and requires high-resolution EM imaging.25,26
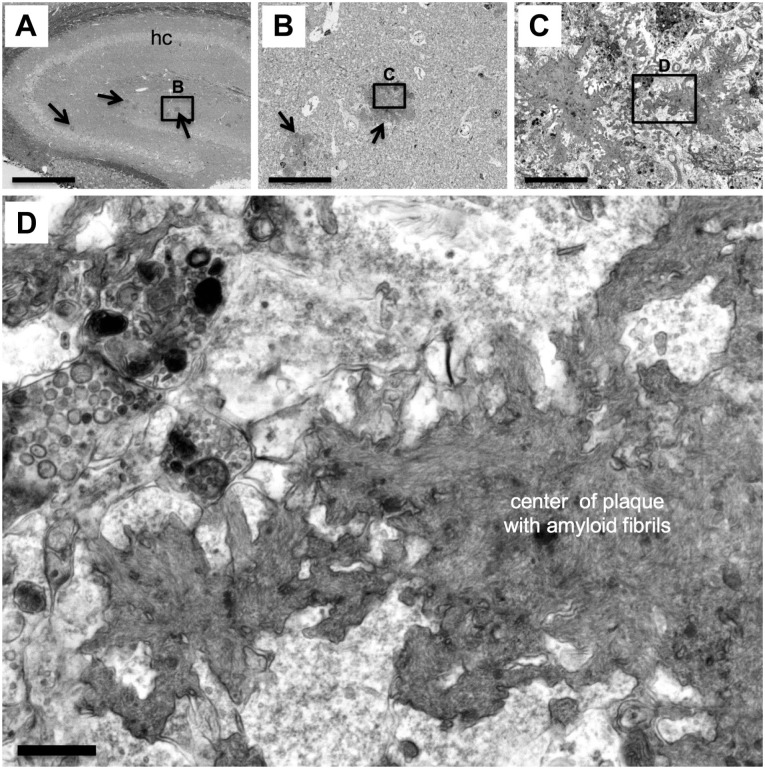
For this purpose, brain slices containing the cerebral cortex and hippocampus of a PS2APP mouse22 were processed for BSE-SEM and imaged at low magnification (100×) to create an overview map of the ROI—in this case, the hippocampal formation (Fig. 5A). In this regional map, various dark patches, putative amyloid plaques, were identified within the hippocampus (Fig. 5A, arrows).
Figure 5.
Identification of amyloid plaques in the PS2APP mouse model of Alzheimer’s disease. (A–D) Slices of brain tissue from the PS2APP mouse model of Alzheimer’s disease were processed for BSE-SEM. (A) The hippocampus (hc) was first investigated at very low magnification (100×) to search and map the position of putative amyloid plaques (arrows). An area in (A) containing putative plaques (black rectangle) was then further investigated. (B) View of the area in the rectangle in (A) at higher magnification (1000×) revealing the dense core and the “lobed” peripheral shape of the putative amyloid plaque. (C) View of the area in the rectangle in (B) at higher magnification (10,000×) showing tree-like dark aggregates in the center of the amyloid plaque. Cytoplasmic processes and structures reminiscent of giant dystrophic neurites (top) filled with dark autolytic vesicles are also visible. (D) At a magnification of 40,000× and a pixel size of 2 nm, individual amyloid fibrils became visible, revealing the “fibrillar texture” of the center of the plaque. Scale bars: (A) 500 µm, (B) 50 µm, (C) 5 µm, and (D) 500 nm. Abbreviation: BSE-SEM, backscattered electron scanning electron microscopy.
One putative plaque was selected and the magnification was increased stepwise by factors of 10 (Fig. 5B and C). At 1000×, the lobe-like periphery of the putative plaque became visible, and neuronal cell nuclei, soma, axons, and dendrites, as well as arterioles and venules, were recognizable (Fig. 5B). Focusing on the center of the putative plaque and increasing the magnification further to 10,000× revealed dark, tree-like structures of aggregates formed by putative amyloid fibrils (Fig. 5C). Adjacent to these aggregates, we found both cytoplasmic processes of putative microglia cells and giant dystrophic axons filled with autophagosome-like vesicles (Fig. 5C), as has been previously reported.22 At the highest magnification used (40,000×), and using a small pixel size of 2 nm for scanning, the amyloid fibrils that constitute the aggregates in the center of the amyloid plaque became visible and appeared as a “fibrillar texture” in the core of the plaque. In addition, synapses with synaptic vesicles could be identified adjacent to the amyloid aggregates (Fig. 5D). This demonstrated that multiscale imaging with BSE-SEM was suitable to investigate neuropathological hallmarks in brain slices from the PS2APP mouse model of Alzheimer’s disease from the millimeter to the nanometer level and could confirm previous results reported from combined histological and TEM investigations of the PS2APP mouse model.22
Investigation of Alveolar Pathology in Monkey Lungs With Multiscale BSE-SEM
Next, we decided to investigate lung samples that had been thoroughly characterized in a published and peer-reviewed TEM study27 to have a baseline reference for image quality, resolution, and comparison of workflows between the traditional TEM method and the novel multiscale BSE-SEM workflow.
For this purpose, we used previously prepared sample blocks of epoxy resin–embedded monkey lung tissue that had been processed for TEM analysis to study the effect of leucine-rich repeat kinase 2 (LRRK2) inhibitors, a proposed therapeutic for Parkinson’s disease, on the lungs of cynomolgus monkeys.27
The traditional TEM workflow previously used for this study had involved the sequential use of thick toluidine blue–stained “survey” sections followed by TEM analysis of parallel ultrathin sections for high-resolution imaging. When compared with vehicle-dosed animals, an increased number of type II pneumocytes with substantially enlarged “multilamellar” surfactant bodies had been observed in the lungs of animals dosed with LRRK2 inhibitors.27
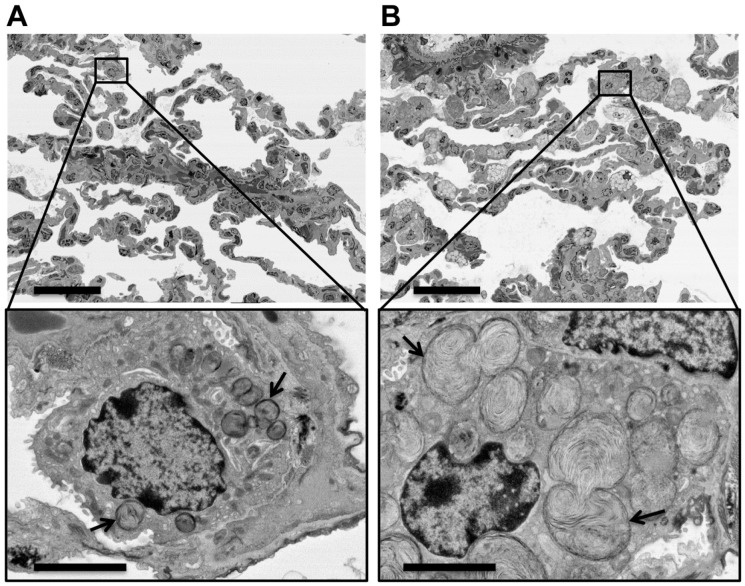
To test the performance of the BSE-SEM method, 1000-nm-thick tissue sections from representative monkeys dosed with either vehicle (Fig. 6A) or the LRRK2 inhibitor GNE-7915 (Fig. 6B) were placed on carbon-coated glass slides and counterstained with uranyl acetate and lead citrate. To acquire unbiased tissue overview images similar to toluidine blue survey sections, we decided on the following strategy. All images were acquired at a fairly low magnification setting of 500× using the largest possible store resolution (maximum number of pixels per image) at 24,000 × 32,000 pixels. These settings produced 230 µm × 172 µm overview images of the lung with a pixel size of 7 nm (Fig. 6A and B). This enabled us to image several alveoli with numerous type I and type II pneumocytes in one wide-field image (Fig. 6A and B, top panels) at a pixel resolution that was high enough to digitally “zoom” into any cell in the acquired images for exploration of the ultrastructural phenotype of the lamellar bodies present in the type II pneumocytes after imaging (Fig. 6A and B, bottom panels). The digitally “zoomed-in” views revealed the distinct phenotype of type II pneumocytes in inhibitor-dosed monkeys, visible as a massive enlargement of multilamellar surfactant bodies (Fig. 6B, bottom panel, arrows), which was not seen in the control animals (Fig. 6A, bottom panel, arrows).
Figure 6.
Effect of LRRK2 kinase inhibition on type II alveolar pneumocytes in monkey lung tissue. Sample blocks of lung tissue from a published study27 that had been processed for TEM analysis were sectioned and stained for reevaluation by BSE-SEM. (A and B, top panels) Sections of tissue blocks from representative monkeys dosed with vehicle (A) or LRRK2 inhibitor GNE-7915 (B) were imaged at an overview magnification of 500× and a pixel size of 7 nm. (A and B, bottom panels) Digitally “zoomed-in” views of the area marked by black boxes in the top panels were created from the original image files by cropping a field of 20 µm × 15 µm. The bottom panels show representative type II alveolar cells found in either the vehicle-treated (A) or inhibitor-treated animals (B), respectively. The digitally “zoomed-in” views revealed the distinct phenotype of type II pneumocytes of inhibitor-dosed monkeys (B) that was characterized by the massive enlargement of multilamellar surfactant bodies (arrows), which were not seen in the control animals (A). The large fields of view in the top panels demonstrated that this phenotype was truly representative for the majority of type II cells in the inhibitor-dosed animals. Note that this multiscale BSE-SEM imaging approach enabled the direct correlation of ultrastructural findings with low-magnification overviews within the same section. Therefore, parallel “survey” sections for light microscopy evaluation were not necessary. Scale bars: 40 µm (A and B, top panels) and 5 µm (A and B, bottom panels). Abbreviations: LRRK2, leucine-rich repeat kinase 2; TEM, transmission electron microscopy; BSE-SEM, backscattered electron scanning electron microscopy.
The wide-field and high-resolution overview images demonstrated directly that the striking phenotype seen at high magnification (after digitally “zooming in”) was truly representative for the majority of type II cells in the inhibitor-dosed animals (Fig. 6A and B, top panels). With the multiscale BSE-SEM approach, a direct correlation of ultrastructural findings with low magnification overviews was achieved simply by capturing a wide-field image at very high pixel resolution and using “digital zoom” to change the view of the acquired image from the tissue level to the ultrastructural level. The BSE-SEM workflow recapitulated the published TEM results and proved to be significantly simpler and faster than the traditional TEM workflow.
Discussion
Here, we present an efficient workflow for unobstructed high-resolution and multiscale imaging of large tissue sections on glass slides that enables the straightforward and direct correlation of ultrastructural resolution of ROIs within the histological context and makes full use of the very large scan fields that can be achieved with modern FE-SEMs.
By providing a high-resolution “bird’s-eye view” perspective over vast expanses of the tissue “landscape,” small ROIs, focal lesions, or other “needles in a haystack” could be efficiently detected and investigated at suborganelle resolution.
For this workflow, we developed and integrated various protocols for processing and embedding of large tissue slices (Fig. 1); protocols for preparing, mounting, and staining of large 1-µm-thick sections on carbon-coated glass slides (Figs. 1 and 2); and imaging protocols for the multiscale analysis with BSE-SEM (Figs. 2–6). We have shown that the practical resolution and contrast achieved by BSE imaging of various organelles, cells, and tissues rival that achieved by conventional TEM, but with the additional advantages of unobstructed multiscale views, direct correlation of the ultrastructural scale with the tissue-level scale, and dramatically simpler and faster workflows. The principle of backscatter imaging of tissue sections on solid substrates has been used by several research groups mostly for the analysis of nervous tissue,9,10,11,13,17 or in combination with correlative LM and EM (CLEM) strategies.3,4,12,28 In addition, prior work has shown that BSE-SEM even allows for imaging herpesvirus capsids, demonstrating the resolving power of this approach.7
In contrast, we focused on workflows for the imaging of a wide range of healthy and diseased tissues that are most frequently encountered in ultrastructural research pathology such as lung, kidney, muscle, skin, and intestine and that cannot be analyzed by CLEM strategies that rely on fluorescence or epitope-tagged proteins and organelles. We have shown that the images acquired by detection of BSEs in an SEM of these tissues are essentially indistinguishable from published TEM work in regard to resolution and contrast of cellular and organelle detail.1,19,22,23,27
Here, we have also demonstrated that “historic” sample blocks of resin-embedded lung tissue (as well as heart and kidney tissue, not shown here) that were originally processed for and analyzed by TEM and stored for several years27 could be successfully examined by BSE-SEM (Fig. 6). This opens the avenue for reevaluation of precious historic sample blocks with a much more efficient and multiscale imaging approach. For example, this technique can be used to find rare, focal lesions, cell types, or infectious agents that may have been missed in previous TEM studies.
For all studies reported here, the resolution and contrast have been fully sufficient to address the particular research questions asked. For instance, in the hippocampal formation of the brain from the PS2APP mouse model of Alzheimer’s disease, the densely packed amyloid fibrils were clearly visible in the center of amyloid plaques (Fig. 5). However, for higher resolution structural information about amyloid fibrils in tissues, which goes beyond the scope of diagnostics and identification of amyloid plaques, methods such as EM-tomography of resin-embedded tissues or cryo-electron tomography may be required.25,26,29
In general, with the current contrast and resolution achievable with our BSE-SEM imaging workflow for conventionally processed and stained tissue sections, all membrane-enclosed organelles could unequivocally be identified, including the outer and inner membranes of cell nuclei and mitochondria, the cisternae of the endoplasmic reticulum, the Golgi apparatus, autophagosomes, endosomes, lysosomes, and synaptic vesicles. In addition, many thin filamentous structures and components of the cytoskeleton, such as microtubules, neurofilaments, desmosomes, tight junctions, and the actin–myosin system in muscle tissue, were visible.
Therefore, we anticipate that the vast majority of histopathological and cytopathological phenomena and questions can be addressed by this BSE-SEM workflow. Importantly, our BSE-SEM imaging protocols are fully compatible with a “follow-up” TEM analysis if higher resolution imaging is required. In this case, we recommend preparing large sample blocks and sections for wide-scale BSE-SEM imaging and analysis followed by retrimming the sample blocks for the preparation of ultrathin sections to be further investigated by TEM.
Because our method development focused particularly on the needs in diagnostic and research ultrastructural pathology, we avoided processing protocols that include mordanting reagents such as tannic acid or thiocarbohydrazide,14,15 and instead only used conventional fixation, postfixation, and counterstaining protocols that have been in use for TEM analysis in ultrastructural pathology for decades.1,5,27 This substantially reduces concerns of introducing novel processing, infiltration, or staining artifacts that can interfere with diagnostics and ultrastructural interpretation.
BSE-SEM methods, including our workflow presented here, that image sections on solid substrates can use surface-staining and surface-coating techniques to optimize contrast and resolution and prevent charging, and therefore represent currently the best strategy to combine multiscale imaging with the highest possible resolution suitable for ultrastructural pathology.7,8,10,11,12,28
With our current imaging protocols and equipment, the acquisition of a high-quality image at the largest store resolution (32,000 × 24,000; 800 mega pixels) takes about 1 to 2 hr, depending mainly on the beam dwell time and the averaging method used for noise reduction. However, the hands-on time for a typical imaging setup, including determining the FOV, focusing, and correction of stigmatism, only takes a few minutes, resulting in an overall time savings. Furthermore, one (32,000 × 24,000) image contains the information of 64 (4000 × 3000) images stitched together. Therefore, the actual time investment per image area is very short, and during scanning valuable time is freed for other laboratory or management tasks. The recent development of multibeam SEMs that work with 61 or even 91 electron beams (instead of just one electron beam used in conventional FE-SEMs) promises to further reduce the acquisition time for multiscale and high-resolution images from hours to minutes.30,31
In summary, here we presented a multiscale imaging workflow that makes full use of the advantages offered by BSE-SEM and that is specifically targeted for the variety of tissues encountered in ultrastructural pathology. Using large sections on standard histology glass slides, conventionally processed samples could be imaged without any obstructions, over multiple magnifications from the tissue to the suborganelle scale, in a fully correlated manner. Therefore, this workflow promises to dramatically improve and accelerate the combined histological and ultrastructural analysis of a wide range of resin-embedded tissues for pathology and basic biomedical research.
Acknowledgments
We thank Reina Fuji, Yun-An Chen, Danielle Mandikian, and Cary Austin for critically reviewing the manuscript and helpful comments.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: All authors have contributed to this article as follows: planning (MR, MS), execution (MR, AKK), analysis (MR, MS, JDW), and writing (MR, MS, JDW). All authors have read and approved the final manuscript.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Contributor Information
Mike Reichelt, Department of Pathology, Genentech Inc., South San Francisco, California.
Meredith Sagolla, Department of Pathology, Genentech Inc., South San Francisco, California.
Anand K. Katakam, Department of Pathology, Genentech Inc., South San Francisco, California
Joshua D. Webster, Department of Pathology, Genentech Inc., South San Francisco, California
Literature Cited
- 1. Stirling JW, Curry AS, Eyden B. Diagnostic electron microscopy: a practical guide to interpretation and technique. Chichester: John Wiley; 2013. [Google Scholar]
- 2. Müller-Reichert T, Verkade P. Introduction to correlative light and electron microscopy. Methods Cell Biol. 2012;111:xvii–xix. doi: 10.1016/B978-0-12-416026-2.03001-6. [DOI] [PubMed] [Google Scholar]
- 3. de Boer P, Hoogenboom JP, Giepmans BN. Correlated light and electron microscopy: ultrastructure lights up! Nat Methods. 2015;12:503–13. [DOI] [PubMed] [Google Scholar]
- 4. Santarella-Mellwig R, Haselmann U, Schieber NL, Walther P, Schwab Y, Antony C, Bartenschlager R, Romero-Brey I. Correlative light electron microscopy (CLEM) for tracking and imaging viral protein associated structures in cryo-immobilized cells. J Vis Exp. 2018. September 7;139:e58154. doi: 10.3791/58154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mackay B. Introduction to diagnostic electron microscopy. New York: Appleton-Century-Crofts; 1981. [Google Scholar]
- 6. Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2(11):e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reichelt M, Joubert L, Perrino J, Koh AL, Phanwar I, Arvin AM. 3D reconstruction of VZV infected cell nuclei and PML nuclear cages by serial section array scanning electron microscopy and electron tomography. PLoS Pathog. 2012;8(6):e1002740. doi: 10.1371/journal.ppat.1002740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horstmann H, Aydin D, Kuner T. Serial section scanning electron microscopy (S3EM) on silicon wafers for ultra-structural volume imaging of cells and tissues. PLoS ONE. 2012;7(4):e35172. doi: 10.1371/journal.pone.0035172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, Lichtman JW. Saturated reconstruction of a volume of neocortex. Cell. 2015. July 30;162(3):648–61. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 10. Koga D, Kusumi S, Shodo R, Dan Y, Ushiki T. High-resolution imaging by scanning electron microscopy of semithin sections in correlation with light microscopy. Microscopy (Oxf). 2015. December;64(6):387–94. doi: 10.1093/jmicro/dfv042. [DOI] [PubMed] [Google Scholar]
- 11. Rodríguez JR, Turégano-López M, DeFelipe J, Merchán-Pérez A. Neuroanatomy from mesoscopic to nanoscopic scales: an improved method for the observation of semithin sections by high-resolution scanning electron microscopy. Front Neuroanat. 2018. February 27;12:14. doi: 10.3389/fnana.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wacker IU, Veith L, Spomer W, Hofmann A, Thaler M, Hillmer S, Gengenbach U, Schröder RR. Multimodal hierarchical imaging of serial sections for finding specific cellular targets within large volumes. J Vis Exp. 2018. March 20;133:e57059.doi: 10.3791/57059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goldstein JI. Scanning electron microscopy and x-ray microanalysis. New York: Springer; 2009. [Google Scholar]
- 14. Giddings TH. Freeze-substitution protocols for improved visualization of membranes in high-pressure frozen samples. J Microsc. 2003;212:53–61. [DOI] [PubMed] [Google Scholar]
- 15. Tapia JC, Kasthuri N, Hayworth KJ, Schalek R, Lichtman JW, Smith SJ, Buchanan J. High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nat Protoc. 2012. January 12;7(2):193–206. doi: 10.1038/nprot.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mikula S, Binding J, Denk W. Staining and embedding the whole mouse brain for electron microscopy. Nat Methods. 2012. December;9(12):1198–201. doi: 10.1038/nmeth.2213. [DOI] [PubMed] [Google Scholar]
- 17. Mikula S, Denk W. High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nat Methods. 2015. June;12(6):541–6. doi: 10.1038/nmeth.3361. [DOI] [PubMed] [Google Scholar]
- 18. Shami GJ, Cheng D, Huynh M, Vreuls C, Wisse E, Braet F. 3-D EM exploration of the hepatic microarchitecture—lessons learned from large-volume in situ serial sectioning. Sci Rep. 2016. November 11;6:36744. doi: 10.1038/srep36744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheville NF, Ames IA. Ultrastructural pathology: the comparative cellular basis of disease. Ames, IA: Wiley-Blackwell; 2009. [Google Scholar]
- 20. Karnovsky MJ. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron-microscopy. J Cell Biol. 1965;27:137–8A. [Google Scholar]
- 21. Reynolds E. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963. April;17(1):208–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richards JG, Higgins GA, Ouagazzal AM, Ozmen L, Kew JN, Bohrmann B, Malherbe P, Brockhaus M, Loetscher H, Czech C, Huber G, Bluethmann H, Jacobsen H, Kemp JA. PS2APP transgenic mice, coexpressing hPS2mut and hAPPswe, show age-related cognitive deficits associated with discrete brain amyloid deposition and inflammation. J Neurosci. 2003. October 1;23(26):8989–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamaguchi H, Nakazato Y, Hirai S, Shoji M, Harigaya Y. Electron micrograph of diffuse plaques. Initial stage of senile plaque formation in the Alzheimer brain. Am J Pathol. 1989. October;135(4):593–7. [PMC free article] [PubMed] [Google Scholar]
- 24. Yamaguchi H, Nakazato Y, Shoji M, Takatama M, Hirai S. Ultrastructure of diffuse plaques in senile dementia of the Alzheimer type: comparison with primitive plaques. Acta Neuropathol. 1991;82(1):13–20. [DOI] [PubMed] [Google Scholar]
- 25. Han S, Kollmer M, Markx D, Claus S, Walther P, Fandrich M. Amyloid plaque structure and cell surface interactions of β-amyloid fibrils revealed by electron tomography. Sci Rep. 2017. February 27;7:e43577. doi: 10.1038/srep43577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kollmer M, Meinhardt K, Haupt C, Liberta F, Wulff M, Linder J, Handl L, Heinrich L, Loos C, Schmidt M, Syrovets T, Simmet T, Westermark P, Westermark GT, Horn U, Schmidt V, Walther P, Fändrich M. Electron tomography reveals the fibril structure and lipid interactions in amyloid deposits. Proc Natl Acad Sci U S A. 2016. May 17;113(20):5604–9. doi: 10.1073/pnas.1523496113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuji RN, Flagella M, Baca M, Baptista MA, Brodbeck J, Chan BK, Fiske BK, Honigberg L, Jubb AM, Katavolos P, Lee DW, Lewin-Koh SC, Lin T, Liu X, Liu S, Lyssikatos JP, O’Mahony J, Reichelt M, Roose-Girma M, Sheng Z, Sherer T, Smith A, Solon M, Sweeney ZK, Tarrant J, Urkowitz A, Warming S, Yaylaoglu M, Zhang S, Zhu H, Estrada AA, Watts RJ. Effect of selective LRRK2 kinase inhibition on nonhuman primate lung. Sci Transl Med. 2015;7(273):273ra15. doi: 10.1126/scitranslmed.aaa3634. [DOI] [PubMed] [Google Scholar]
- 28. Smith SJ. Q&A: array tomography. BMC Biol. 2018. September;16(1):98. doi: 10.1186/s12915-018-0560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Milanesi L, Sheynis T, Xue WF, Orlova EV, Hellewell AL, Jelinek R, Hewitt EW, Radford SE, Saibil HR. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc Natl Acad Sci U S A. 2012. December 11;109(50):20455–60. doi: 10.1073/pnas.1206325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keller AL, Zeidler D, Kemen T. High throughput data acquisition with a multi-beam SEM. Proc SPIE 9236. 2014. September;9236:92360B. doi: 10.1117/12.2069119. [DOI] [Google Scholar]
- 31. Pereira AF, Hageman DJ, Riedesel C, Knothe U, Zeidler D, Tate ML. Creating high-resolution multiscale maps of human tissue using multi-beam SEM. PLoS Comput Biol. 2016. November 21;12(11):e1005217. doi: 10.1371/journal.pcbi.1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]