Abstract
Glioblastoma is the most aggressive primary brain tumor. Slowly dividing and therapy-resistant glioblastoma stem cells (GSCs) reside in protective peri-arteriolar niches and are held responsible for glioblastoma recurrence. Recently, we showed similarities between GSC niches and hematopoietic stem cell (HSC) niches in bone marrow. Acute myeloid leukemia (AML) cells hijack HSC niches and are transformed into therapy-resistant leukemic stem cells (LSCs). Current clinical trials are focussed on removal of LSCs out of HSC niches to differentiate and to become sensitized to chemotherapy. In the present study, we elaborated further on these similarities by immunohistochemical analyses of 17 biomarkers in paraffin sections of human glioblastoma and human bone marrow. We found all 17 biomarkers to be expressed both in hypoxic peri-arteriolar HSC niches in bone marrow and hypoxic peri-arteriolar GSC niches in glioblastoma. Our findings implicate that GSC niches are being formed in glioblastoma as a copy of HSC niches in bone marrow. These similarities between HSC niches and GSC niches provide a theoretic basis for the development of novel strategies to force GSCs out of their niches, in a similar manner as in AML, to induce GSC differentiation and proliferation to render them more sensitive to anti-glioblastoma therapies.
Keywords: bone marrow, glioblastoma, glioblastoma stem cells, hematopoietic progenitor cells, hematopoietic stem cells, immunohistochemistry, niches
Introduction
Glioblastoma is the most aggressive primary brain tumor with poor patient survival, which is at least partly due to a small subpopulation of glioblastoma cells, glioblastoma stem cells (GSCs). GSCs are localized in niches where they are maintained as slowly dividing cells, which results in resistance to therapy.1–5 It has been hypothesized that forcing GSCs out of their protective niches has therapeutic potential, because it sensitizes GSCs to irradiation and chemotherapy. Understanding of the functioning of GSC niches, that is, molecular mechanisms that retain GSCs in these protective niches, is required for the development of novel therapeutic strategies targeted against GSCs.
In previous studies, we demonstrated that GSC niches are hypoxic and peri-arteriolar, where GSCs are localized adjacent to the tunica adventitia of a small subset of arterioles in hypoxic areas in glioblastoma tumors.6–9 Hypoxia is one of the prime conditions for stem cell maintenance.10–12 In addition, we showed that similar chemoattractive proteins are expressed in GSC niches and in hematopoietic stem cell (HSC) niches in normal human bone marrow, such as chemoattractants stromal-derived factor-1α (SDF-1α) and osteopontin (OPN) and their receptors C-X-C receptor type 4 (CXCR4) and CD44, respectively, that are involved in retention of stem cells in niches.6–9
In acute myeloid leukemia (AML), the interactions between chemoattractants and their receptors are involved in homing of leukemic cells in HSC niches, where they become quiescent and chemo-resistant leukemic stem cells (LSCs). Ongoing clinical trials are aimed at mobilization of LSCs out of HSC niches to induce LSC differentiation, proliferation, and thus sensitization to chemotherapy.13,14 Similar treatment strategies are a promising approach for anti-glioblastoma therapies. Therefore, this comparative study between HSC niches in human bone marrow and GSC niches in human glioblastoma has been performed to understand the morphological functional features of both niche types.
In bone marrow, mesenchymal stem cells (MSCs) are important for the maintenance of slowly dividing or quiescent HSCs in niches, by producing and secreting multiple chemoattractants, including essential HSC niche factors.15,16 MSCs can differentiate into osteoblasts, adipocytes, chondrocytes, and smooth muscle cells in human bone marrow.15 Bone marrow–derived MSCs infiltrate glioblastoma tumors and are involved in tumor progression. Our previous studies have shown that MSCs have high affinity for glioblastoma tumors, resulting in tumor progression.17,18 In addition, MSCs increase the self-renewal capacity of GSCs as well as invasion and proliferation of GSCs in vitro.19–22 MSCs expressing CD105 have been detected around arterioles in glioblastoma.23 MSCs are known to produce and secrete multiple chemoattractants, such as SDF-1α,20,24,25 which we identified as an important GSC niche factor in our previous studies.6–9,13 Therefore, bone marrow–derived MSCs may be important players in GSC niches in glioblastoma to maintain GSCs in their protective niches.
In the present study, we aimed to determine whether GSC niches in glioblastoma are a functional mimic of normal HSC niches in human bone marrow. In addition, we aimed to localize bone marrow–derived MSCs in both niche types. We performed this study using fluorescence and chromogenic immunohistochemistry on paraffin sections of human glioblastoma samples and human sternum and rib samples. To the best of our knowledge, this is the first human bone marrow study where HSC niches are demonstrated, confirming HSC niche studies in bone marrow that have been conducted in mouse models. Seventeen biomarkers were stained to define HSC niches and GSC niches. Smooth muscle actin (SMA) was stained to detect the smooth muscle cell layer of arteriolar walls.7,9 As HSC biomarkers, we used CD15026–29 and CD13330–32 and as biomarker for hematopoietic progenitor cells, CD244 was stained.26,29,33 CD1337,9,34–36 and SOX237–40 were used to detect GSCs. We stained for the chemoattractive proteins SDF-1α and OPN and their receptors, CXCR4 and CD44, respectively.7,9,12,41,42 In addition, antibodies against CD105, CD73, and stromal factor-1 (STRO-1) were applied to detect MSCs.23,43–45 To confirm hypoxic conditions, we included staining of hypoxia-induced factor (HIF)-1α, HIF-2α, and vascular endothelial growth factor (VEGF).6–8,46–49 VEGF receptor 2 (VEGFR2) and phosphorylated epidermal growth factor receptor (p-EGFR) have been included as these receptors have been found to be highly elevated in GSCs in glioblastoma.50–54
Materials and Methods
Cell Culture
Human NCH421k GSCs55,56 were a generous gift from Prof. Dr. Christel Herold-Mende (Heidelberg University, Heidelberg, Germany) and were cultured in neurobasal medium (Gibco Life Technologies, Carlsbad, CA, USA) containing 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA), 1% l-glutamine (Sigma-Aldrich), 2% B27, 0.08% b-fibroblast growth factor (bFGF; Gibco), 0.01% endothelial growth factor (EGF; Gibco), and 0.01% heparin (Sigma-Aldrich) at 37C in a 5% CO2 incubator.
Human bone marrow-derived MSCs were obtained from Lonza Bioscience (Walkersville, MD USA; Lot number 6F4393). MSCs were cultured in Dulbecco’s medium (DMEM 5921; Sigma-Aldrich) containing 10% fetal bovine serum (Gibco), 100 U penicillin (Thermo Fisher Scientific, Waltham, MA, USA), 1000 U streptomycin (Thermo Fisher Scientific), 2 mM l-glutamine (Thermo Fisher Scientific), sodium-pyruvate (Gibco), and nonessential amino acids (Sigma-Aldrich).
Fluorescence Immunocytochemistry on Human Bone Marrow-derived MSCs and Human GSCs
MSCs were cultured on glass slides in an incubator at 37C. On each glass slide, 500,000 cells were seeded. One million NCH421k GSCs were put in Eppendorf tubes as spheroids and the immunostaining procedure was performed in Eppendorf tubes in order to maintain the 3D structure of the spheroids. Cells were fixed in 4% paraformaldehyde (Merck, Darmstadt, Germany) for 10 min, followed by a washing step using phosphate-buffered saline (PBS; Gibco) containing 1% bovine serum albumin (BSA; Sigma-Aldrich). Cells were permeabilized, and non-specific background staining was reduced by incubating cell preparations in PBS containing 10% normal goat serum (Dako Glostrup, Denmark) and 0.1% Triton-X (Sigma-Aldrich) for 1 hr at room temperature (RT). Cells were incubated overnight at 4C with primary antibodies diluted in PBS containing 1% BSA (Sigma-Aldrich), as indicated in Tables 1 and 2. Cells were washed 3 times using PBS containing 1% BSA.
Table 1.
Details of Primary Antibodies Used for Fluorescence Immunohistochemistry.
| Primary Antibody | Source | Primary Antibody Dilution in PBS + 1% BSA for Fluorescence IHC | Protein Function/Characteristics |
|---|---|---|---|
| Mouse anti-human SMA | Dakoa (1A4) | 1:200 | Expressed by smooth muscle cells of arterioles and venules, myofibroblasts, and pericytes |
| Rabbit anti-human CD150 | Invitrogenb (PA5-21123) | 1:100 | Biomarker to detect HSCs |
| Mouse anti-human CD244 | Invitrogen (PA5-47054) | 1:100 | Biomarker to detect hematopoietic progenitor cells |
| Rabbit anti-human CD133 | Abcamc (ab19898) | 1:100 | Biomarker to detect GSCs in glioblastoma |
| Mouse anti-human SOX2 | Abcam (ab171380) | 1:50 | Transcription factor expressed in GSCs in glioblastoma |
| Rabbit anti-human SDF-1α | Abcam (ab9797) | 1:200 | Chemoattractant for CXCR4-positive (stem) cells |
| Rabbit anti-human CXCR4 | R&D Systemsd
clone 44716 (MAB172) |
1:30 | G-protein-coupled receptor that interacts with ligand SDF-1α |
| Rabbit anti-human OPN | Abcam (ab8448) | 1:100 | Chemoattractant for CD44-positive cells |
| Mouse anti-human CD44 | Biorade (MCA2504) | 1:100 | Receptor that interacts with ligand OPN |
| Rabbit anti-human CD105 | Abcam (ab27422) | Prediluted | Surface biomarker to detect MSCs |
| Mouse anti-human CD73 | Abcam (ab54217) | 1:100 | Surface biomarker to detect MSCs |
| Mouse anti-human STRO-1 | Life Technologiesf (398401) | 1:100 | Surface biomarker to detect MSCs and other stromal cells |
| Mouse anti-human HIF-1α | Abcam (ab8366) | 1:100 | Transcription factor expressed in cells in hypoxic conditions |
| Rabbit anti-human HIF-2α | Abcam (ab109616) | 1:100 | Transcription factor expressed in cells in hypoxic conditions |
| Rabbit anti-human VEGF | Santa Cruzg Biotechnology (sc-152) | 1:50 | Inducer of angiogenesis, upregulates SDF-1α, CXCR4, OPN, and CD44 in glioblastoma and bone marrow |
| Mouse anti-human VEGFR2 | ReliaTechh (101-M34) | 1:100 | Receptor differentially elevated in GSCs in glioblastoma |
| Mouse anti-human phospho-EGFR (Tyr1173) | Merck Milliporei (05-1004) | 1:100 | Receptor differentially elevated in GSCs in glioblastoma |
Abbreviations: PBS, phosphate-buffered saline; BSA, bovine serum albumin; IHC, immunohistochemistry; SMA, smooth muscle actin; HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; SOX2, sex determining region Y-box 2; SDF-1α, stromal–derived factor-1α; CXCR4, C-X-C chemokine receptor type 4; OPN, osteopontin; MSC, mesenchymal stem cell; STRO-1, stromal factor-1; HIF, hypoxia-induced factor; VEGF, vascular endothelial growth factor; VEGFR2, vascular endothelial growth factor receptor 2; EGFR, epidermal growth factor receptor.
Dako, Glostrup, Denmark.
Invitrogen, Waltham, MA, USA.
Abcam, Cambridge, UK.
R&D Systems, Minneapolis, MN, USA.
Biorad, Hercules, CA, USA.
Life Technologies, Carlsbad, CA, USA.
Santa Cruz Biotechnology, Dallas, TX, USA.
ReliaTech, Wolfenbüttel, Germany.
Merck Millipore, Burlington, MA, USA.
Table 2.
Details of Primary Antibodies Used for Chromogenic Immunohistochemistry.
| Primary Antibody | Source | Primary Antibody Dilution in PBS + 1% BSA |
|---|---|---|
| Mouse anti-human SMA | Dakoa (1A4) | 1:2000 |
| Rabbit anti-human CD150 | Invitrogenb (PA5-21123) | 1:200 |
| Mouse anti-human CD133 | Miltenyi Biotechc (130-092-395) | 1:20 |
Abbreviations: PBS, phosphate-buffered saline; BSA, bovine serum albumin; IHC, immunohistochemistry; SMA, smooth muscle actin.
Dako; Glostrup, Denmark.
Invitrogen, Waltham, MA, USA.
Miltenyi Biotech, Bergisch Gladbach, Germany.
Alexa Fluor 488-conjugated goat anti-rabbit antibodies (Life Technologies) and Alexa Fluor 546-conjugated goat anti-mouse antibodies (Thermo Fisher Scientific) were used as secondary antibodies in a dilution of 1:200 in PBS containing 1% BSA for 1 hr at RT. Cells were washed in PBS, followed by nuclear counterstaining using 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) in PBS for 5 min at RT. Cells were washed in PBS for 5 min and coverslipped using Prolong Gold mounting medium (Life Technologies). Control incubations were performed in the absence of primary antibodies.
Bone Marrow Samples
Four sternum and rib samples were obtained through the body donation program from the Department of Medical Biology, Section Clinical Anatomy and Embryology, of the Amsterdam UMC at the location Academic Medical Center in The Netherlands. The bodies from which the samples were taken were donated to science in accordance with Dutch legislation and the regulations of the medical ethical committee of the Amsterdam UMC at the location Academic Medical Center. The samples were decalcified using 11% ethylenediaminetetraacetic acid (EDTA; Thermo Fisher Scientific) in distilled water, pH 7.2, for 21 days and the decalcification buffer was refreshed every 3 days. The sternum and rib samples were formalin-fixed and embedded in paraffin at the Pathology Department of the Amsterdam UMC at the location Academic Medical Center in The Netherlands and 5-µm paraffin sections were provided.
Glioblastoma Samples
Glioblastoma biopsies were obtained from glioblastoma patients who were operated at the Department of Neurosurgery, University Medical Center of Ljubljana, Slovenia. The study was approved by the National Medical Ethics Committee of the Republic of Slovenia (approval no. 0120-190/2018/4). Altogether, 10 patients with glioblastoma (World Health Organization [WHO] glioma grade IV) were included. Tumor diagnoses were established by standard histopathology protocols at the Institute of Pathology of the Medical Faculty, University of Ljubljana. Formalin-fixed, paraffin-embedded tissue was used for immunohistochemical analyses. Clinical data of the glioblastoma patients are shown in Table 3.
Table 3.
Clinical Data of the 10 Glioblastoma Patients.
| Patient Number | Gender | Therapy (Radio- or Chemotherapy With Temozolomide) | Age at the Time of Operation (Years) | Overall Survival (Months) | IDH1 R132H Mutation (Yes/No) | p53 Mutation (Yes/No) |
|---|---|---|---|---|---|---|
| 1 | M | Radiotherapy (60 Gy) + temozolomide, adjuvant temozolomide therapy | 46 | 16 | No | Yes |
| 2 | M | Radiotherapy (60 Gy) + temozolomide, adjuvant temozolomide therapy | 43 | 20 | Yes | Yes |
| 3 | M | Radiotherapy (60 Gy) + temozolomide, without adjuvant temozolomide therapy | 54 | 3 | No | n |
| 4 | M | Radiotherapy (60 Gy) + temozolomide, adjuvant temozolomide therapy | 55 | 26 | No | No |
| 5 | M | Radiotherapy (60 Gy) + temozolomide, adjuvant temozolomide therapy | 66 | 8 | No | n |
| 6 | F | Radiotherapy (60 Gy) + temozolomide, without adjuvant temozolomide therapy | 59 | 16 | No | n |
| 7 | M | Radiotherapy (60 Gy) + temozolomide, adjuvant temozolomide therapy | 62 | 17 | No | No |
| 8 | M | Radiotherapy (30 Gy), adjuvant temozolomide therapy | 71 | 12 | No | No |
| 9 | M | Radiotherapy (60 Gy) + temozolomide, adjuvant temozolomide therapy | 22 | 56 | No | No |
| 10 | n | n | n | n | No | n |
Abbreviations: IDH1, isocitrate dehydrogenase 1; M, male; F, female; n, not known.
Fluorescence Immunohistochemistry Using Human Bone Marrow and Glioblastoma Paraffin Sections
Paraffin sections (5 µm thick) of human bone marrow and glioblastoma were stored at RT until use. Dewaxing was performed by incubation of the sections in xylene (VWR Chemicals, Atlanta, GA, USA) for 5 min and rinsing in 100%, 96% and 70% ethanol (Merck), respectively.
Antigen-retrieval was performed in a microwave using 100 mM citrate buffer containing 0.1% Triton-X, pH 6.0, for 20 min at 98C, followed by cooling down for 20 min and a washing step in PBS.57
Sections were encircled with a PAP pen (Dako) and incubated with PBS containing 10% normal goat serum (Dako) and 0.1% Triton-X for 1 hr at RT to reduce non-specific background staining and for permeabilization of the sections. Sections were subsequently incubated overnight at 4C with primary antibodies diluted in PBS containing 1% BSA, as indicated in Table 1. Sections were washed 3 times using PBS containing 1% BSA.
Alexa Fluor 488-conjugated goat anti-rabbit antibodies and Alexa Fluor 546-conjugated goat anti-mouse antibodies were used as secondary antibodies in a dilution of 1:200 in PBS containing 1% BSA for 1 hr for 1 hr at RT. Sections were then washed in PBS, followed by nuclear counterstaining using DAPI in PBS for 5 min at RT. Next, sections were washed in PBS for 5 min and coverslipped using Prolong Gold mounting medium. Afterwards, sections were sealed with nailpolish and dried overnight. Control incubations were performed in the absence of primary antibodies.
Chromogenic Immunohistochemistry Using Human Bone Marrow and Glioblastoma Paraffin Sections
Paraffin sections (5 µm thick) of human bone marrow and glioblastoma were stored at RT until use. Dewaxing was performed by incubation of the sections in xylene for 10 min and rinsing in 100% ethanol. The sections were treated with 100% methanol (Merck) containing 0.3% H2O2 (Merck) for 10 min to block endogenous peroxidase activity to reduce non-specific background staining, followed by a washing step in distilled water.
Antigen-retrieval was performed in a heating Lab Vision PT Module (Thermo Fisher Scientific) using 100 mM citrate buffer, pH = 6.0, for 20 min at 98C, and afterwards cooling down for 20 min followed by a washing step in distilled water and 3 washing steps of 5 min each using TBS containing 0.1% Triton-X.57
Sections were encircled with a PAP pen and incubated with TBS containing 3% normal goat serum (Dako) and 0.1% Triton-X for 1 hr to further reduce non-specific background staining and for permeabilization of the sections. Sections were incubated overnight at 4C with primary antibodies diluted in normal antibody diluent (Immunologic, Duiven, The Netherlands) as indicated in Table 2. Sections were washed 3 times using TBS.
Sections were incubated with secondary horseradish peroxidase antibodies (goat-anti mouse or goat anti-rabbit) for 1 hr at RT, followed by 3 washing steps of 5 min each using TBS. Next, sections were incubated with either 3,3’-diaminobenzidine (DAB; Dako) or aminoethyl carbazole (AEC; Vector Laboratories, Burlingame, CA, USA) for 10 min, followed by one washing step with tap water to stop the peroxidase activity. Sections were then incubated for 30 s in hematoxylin (Sigma-Aldrich) for nuclear counterstaining and placed in running tap water for 5 min and then in distilled water. Sections were dehydrated by dipping in 70%, 96%, and 100% ethanol and dipping in xylene 3 times, respectively. Finally, sections were covered with the synthetic mountant Pertex (Histolab, Götenburg, Sweden).
Control incubations were performed either in the absence of the primary antibody or in the presence of rabbit serum or mouse serum in the same dilution as the primary antibody to determine the effect of serum on non-specific background staining.
Hematoxylin-eosin (HE) Staining
For HE staining, human bone marrow and glioblastoma paraffin sections were dewaxed in xylene and 100% ethanol. Sections were fixed with freshly-prepared Formol-Macrodex (4% formaldehyde, 7.2 mM CaCl2, 0.12 M Dextran-70, 0.12 M NaCl, and 7.96 mM CaCO3) for 10 min, followed by a washing step in distilled water for 5 min. Nuclei were stained with hematoxylin for 30 s and then, sections were placed in running tap water for 5 min, after which the sections were placed in distilled water. Sections were then stained with eosin (Merck) for 20 s, dipped 5 times in distilled water, 15 times in 70% ethanol, 15 times in 96% ethanol, and 10 times in 100% ethanol. Afterwards, sections were rinsed 3 times for 5 min in xylene. The sections were covered with Pertex. All steps were performed at RT.
Imaging
Fluorescence imaging was performed using a Nikon Eclipse Ti-E inverted microscope (Nikon Instuments, Melville, NY, USA) and the Nikon NIS-Elements AR 4.13.04 software. Fluorescence staining patterns of the glioblastoma sections and bone marrow sections were analyzed by three independent observers (VVVH, BB, and CJFVN).
Fluorescence staining of GSCs in 3D spheroids was imaged using a confocal microscope (Leica DFC 7000 T, Wetzlar, Germany). Staining patterns were analyzed by 4 independent observers (VVVH, BB, MV, and CJFVN).
Chromogenic IHC-stained sections were analyzed using light microscopy and images were taken using an Olympus BX51 microscope (Olympus, Tokyo, Japan), and the Olympus cellSense Standard software. Staining patterns were analyzed by four independent observers (VVVH, ALJ, MK, and CJFVN).
Quantitative Image Analysis of GSC Niches and HSC Niches
In order to obtain quantitative data, image analysis was performed on the images of glioblastoma and bone marrow, using ImageJ software in 5 steps.58,59
Of every GSC niche, the images with CD133-SOX2-DAPI staining were analyzed. The total number of cells was counted using the DAPI staining.
All CD133-SOX2-positive cells (GSCs) were counted and the percentage of GSCs was calculated.
Of every GSC niche, the images with CD105-DAPI staining were analyzed. Again the total number of cells (DAPI) were counted and CD105-positive cells (MSCs) were counted and the percentage of MSCs was calculated.
For HSC niches in bone marrow, image analysis was also performed using steps 1–3. The only difference was that CD133 and CD150 were used as markers for the detection of HSCs.
Statistical analysis (one-way ANOVA test) was performed to determine whether there is a significant difference between the percentage of GSCs in N=13 GSC niches and the percentage of HSCs in N=16 HSC niches. In addition, it was determined whether there is a significant difference between the percentage of MSCs in N=13 GSC niches and percentage of MSCs in N=16 HSC niches. Data were processed in Excel 2013 (Microsoft, Redmond, WA, USA) and GraphPad Prism 6 (La Jolla, CA USA). p values < 0.05 were considered to indicate significant differences.
Results
Structural Characteristics of Human Bone Marrow and Glioblastoma
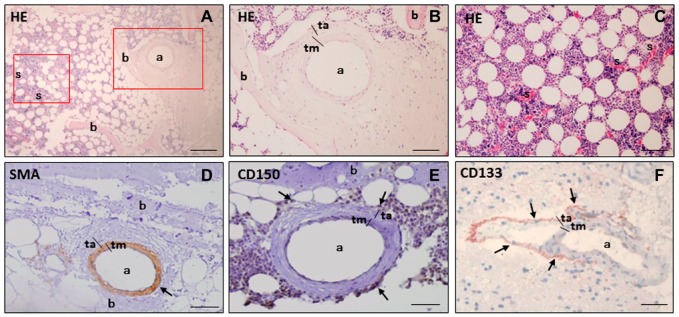
To assess the morphological structures and characteristics of bone marrow, HE staining of bone marrow tissue sections was analyzed. In bone marrow, arterioles were found near bone, whereas sinusoids were found at a distance from bone (Fig. 1A–C). Chromogenic staining using DAB or AEC as chromogens showed that smooth muscle cells in the tunica media of the arteriolar wall expressed SMA (Fig. 1D) and that HSCs expressing CD150 were localized around SMA-positive arterioles (Fig. 1E). In glioblastoma sections, similar patterns were recognized, as CD133-positive GSCs were always found to be localized in niches adjacent to the tunica adventitia of arterioles (Fig. 1F). The only difference between HSC niches and GSC niches was the thick layer of HSCs in a larger area around arterioles in bone marrow (Fig. 1E) and a thin layer of GSCs around arterioles in glioblastoma (Fig. 1F). Therefore, we aimed to determine in more depth the similarities of HSC niches in bone marrow and GSC niches in glioblastoma.
Figure 1.
Histological HE staining and chromogenic immunohistochemical staining of SMA, CD150, and CD133 in paraffin sections of human bone marrow (A-E) and human glioblastoma (F) with DAB (D, E) and AEC (F) as chromogens. HE staining shows an arteriole (a) in the periphery of bone marrow adjacent to bone (b), whereas sinusoids (s) are localized at distance from bone (A-C). SMA is expressed in smooth muscle cells in the tunica media (tm) in the wall of an arteriole (a) adjacent to bone (b) in bone marrow marrow (D). CD150 is expressed as biomarker of HSCs (arrows) around the arteriole (a) near bone (b) in bone marrow (E). The GSC biomarker CD133 is expressed in cells adjacent to the tunica adventitia (ta) of an arteriole (a) in glioblastoma (F). Cell nuclei are counterstained in blue-purple with hematoxylin. Scale bars A, B, and D = 100 µm. Scale bars C, E, and F = 50 µm. Abbreviations: HE, Hematoxylin-eosin; SMA, smooth muscle actin; AEC, aminoethyl carbazole; DAB, 3,3’-diaminobenzidine; a, arteriole; s, sinusoid; HSC, hematopoietic stem cell; tm, tunica media; GSC, glioblastoma stem cell; ta, tunica adventitia.
HSCs and GSCs Are Localized in Peri-arteriolar Hypoxic Niches
Next, we determined whether HSCs and GSCs are exclusively localized around arterioles in bone marrow and glioblastoma, respectively. In bone marrow, HSCs expressing CD133 on the cell surface and CD150 both on the cell surface and in the cytoplasm were found to be localized around SMA-positive arterioles (Fig. 2A–C). In glioblastoma, GSCs expressing CD133 on the cell surface and SOX2 in the nuclei were localized around SMA-positive arterioles (Fig. 2D and E). CD133 and CD150 expression was also detected in endothelial cells and smooth muscle cells in the arteriolar walls in bone marrow and glioblastoma (Fig. 2A–C and E). The cells positive for CD133, CD150, and SOX2 adjacent to the tunica adventitia of arterioles indicate the presence of HSC and GSC niches both in bone marrow and glioblastoma, respectively. In bone marrow, CD150 and CD133 were exclusively expressed around arterioles. In glioblastoma, CD133 was exclusively expressed adjacent to the tunica adventitia of arterioles, whereas SOX2 was also expressed in cells away from peri-arteriolar niches. Sixteen HSC niches were found in serial sections of bone marrow of 4 sternum and rib samples and 13 GSC niches were found in serial sections of 10 glioblastoma samples.
Figure 2.
Fluorescence immunohistochemical staining of hypoxic peri-arteriolar HSCs in human bone marrow (A-C, F, G) and hypoxic peri-arteriolar GSCs in human glioblastoma (D, E, H, I). Yellow rectangle in A is enlarged in B and C. CD133-expressing HSCs are localized around a SMA-positive arteriole (a) adjacent to bone (b) in bone marrow (B, yellow arrows). CD133 is also expressed in endothelial cells and smooth muscle cells of the arteriolar wall (A, B). CD150-positive HSCs are localized around the arteriole adjacent to bone in bone marrow (C, yellow arrows). CD150 expression is detected in endothelial cells and smooth muscle cells in the wall of the arteriole (C). SMA is expressed in smooth muscle cells in the tunica media (tm) of the arteriolar wall in glioblastoma (D). GSCs express CD133 and SOX2 and are localized adjacent to the tunica adventitia (ta) of the arteriole (a) in glioblastoma (E, yellow arrows). HIF-1α is abundantly expressed in the arteriolar wall in bone marrow (F). HIF-2α is highly expressed in HSCs in a HSC niche (F). A subset of HSCs express both HIF-1α and HIF-2α (F, yellow arrows). VEGF is abundantly expressed throughout bone marrow (G). HIF-1α is abundantly expressed in the arteriolar wall in glioblastoma (H). HIF-2α is highly expressed in GSCs (H, yellow arrows). VEGF is abundantly expressed throughout glioblastoma (I). Nuclear counterstaining with DAPI (blue). Scale bar = 100 µm. Abbreviations: HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; SMA, smooth muscle actin; tm, tunica media; SOX2, sex determining region Y-box 2; ta, tunica adventitia; DAPI, 4’,6-diamidino-2-phenylindole; HIF, hypoxia-induced factor; VEGF, vascular endothelial growth factor.
Peri-arteriolar HSC niches were hypoxic as HIF-1α and HIF-2α (Fig. 2F) were expressed in cells around the arterioles. HIF-1α was strongly expressed in the arteriolar wall, whereas HIF-2α was more abundantly expressed in cells around the arteriolar wall. A subset of cells around the arteriolar wall expressed both HIF-1α and HIF-2α (Fig. 2F). VEGF was widely expressed throughout the bone marrow tissue sections (Fig. 2G).
GSC niches were also found to be hypoxic, as HIF-1α, HIF-2α, and VEGF were abundantly expressed in cells around arterioles and in the wall of arterioles as well (Fig. 2H and I). HIF-2α was more abundantly expressed in cells around arterioles than HIF-1α, which was more abundantly expressed in arteriolar walls. A subset of cells around arterioles expressed both HIF-1α and HIF-2α (Fig. 2H). VEGF was widely expressed throughout the glioblastoma tissue sections (Fig. 2I). Collectively, these data demonstrate that HSC niches in bone marrow and GSC niches in glioblastoma are peri-arteriolar and hypoxic.
Chemoattractive Proteins in Peri-arteriolar HSC Niches and Peri-arteriolar GSC Niches
We determined whether the chemoattractants SDF-1α and OPN and their receptors CXCR4 and CD44, respectively, were expressed in HSC niches in bone marrow and GSC niches in glioblastoma. In bone marrow, CD150-positive HSCs also expressed the receptor CXCR4 on their surface (Fig. 3A). CD150 and CXCR4 were co-localized in peri-arteriolar niches (Fig. 3A) where the chemoattractant SDF-1α was abundantly expressed as well (Fig. 3B). CXCR4 was widely expressed on a variety of cell types, including HSCs throughout bone marrow. In glioblastoma, CD133-positive and SOX2-positive GSCs were also expressing CXCR4 and were found in SDF-1α-rich niches (Fig. 3C and D). SDF-1α was expressed intracellularly and extracellularly, also in endothelial cells and smooth muscle cells of arteriolar walls (Fig. 3B and D).
Figure 4.
Fluorescence immunohistochemical staining demonstrates expression of CD44 on CD150-positive HSCs (A, yellow arrows). Chemoattractant OPN and its receptor CD44 are expressed in a peri-arteriolar HSC niche in human bone marrow (B) and in a peri-arteriolar GSC niche in glioblastoma (C). Chemoattractant OPN is expressed intracellularly in HSCs and extracellularly and its receptor CD44 is abundantly expressed on HSCs in a niche in bone marrow (A, yellow arrows). Chemoattractant OPN is expressed intracellularly and extracellularly and its receptor CD44 is abundantly expressed on GSCs in a GSC niche in glioblastoma (C, yellow arrows). OPN expression is also detected in endothelial cells and smooth muscle cells in the wall of an arterioles in both bone marrow and glioblastoma (B, C). CD150-positive HSCs are localized in a HSC niche around an arterioles near bone (D, yellow arrows) and CD244-positive hematopoietic progenitor cells are absent in a HSC niche near bone in bone marrow (D). CD244-positive hematopoietic progenitor cells are detected at distance from bone (E, yellow arrows), where CD150-CD133 positive HSCs are not detected (E, F). DAPI is used for nuclear counterstaining. Scale bar = 100 µm. Abbreviations: HSC, hematopoietic stem cell; OPN, osteopontin; GSC, glioblastoma stem cell; DAPI, 4’,6-diamidino-2-phenylindole.
Figure 3.
Fluorescence immunohistochemical staining demonstrates expression of chemoattractant SDF-1α and its receptor CXCR4 in a peri-arteriolar HSC niche in human bone marrow (A, B) and in a peri-arteriolar GSC niche in glioblastoma (C, D). CD150-CXCR4-positive HSCs are localized around an arteriole in a niche adjacent to bone (A, yellow arrows). CXCR4 is widely expressed throughout the bone marrow tissue section (A). Chemoattractant SDF-1α is expressed intracellularly in HSCs expressing CXCR4 (B, yellow arrows), extracellularly in a HSC niche and in endothelial cells and smooth muscle cells in the wall of the arteriole (B). Chemoattractant SDF-1α and its receptor CXCR4 are abundantly expressed in a GSC niche in glioblastoma. CXCR4 is expressed on CD133-SOX2-positive GSCs (C, yellow arrows). SDF-1α expression is detected intracellularly in GSCs, extracellularly in a GSC niche and in endothelial cells and smooth muscle cells of the arteriolar wall (D). Confocal imaging after immunocytochemical staining demonstrates that GSCs express CD133, SOX2 (E), and CXCR4 (F) in vitro in spheroids. DAPI is used for nuclear counterstaining. Scale bar A-D = 100 µm. Scale bar E, F = 50 µm. Abbreviations: SDF-1α, stromal-derived factor-1α; CXCR4, C-X-C receptor type 4; HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; SOX2, sex determining region Y-box 2; DAPI, 4,’6-diamidino-2-phenylindole.
In vitro, patient-derived GSCs that grow in 3D spheroids, expressed CD133, SOX2, and CXCR4 (Fig. 3E and F), indicating that GSCs in vivo in glioblastoma indeed express CD133, SOX2, and CXCR4.
HSCs are known to express receptor CD44 that enables homing into HSC niches.13,60–62 In bone marrow, CD150-positive HSCs expressed receptor CD44 on their surface (Fig. 4A) as well as its ligand and chemoattractant OPN. OPN was also expressed in endothelial cells and smooth muscle cells of arteriolar walls (Fig. 4B). In glioblastoma, CD44 was found to be expressed by GSCs, but also by differentiated glioblastoma cells. OPN was found to be present intracellularly as well as extracellularly in GSC niches (Fig. 4C).
In conclusion, in both peri-arteriolar HSC niches and peri-arteriolar GSC niches, the chemoattractants SDF-1α and OPN are preferentially expressed in niches, whereas the receptors CXCR4 and CD44 are widely expressed throughout bone marrow and glioblastoma tumors (Supplementary Fig. 1).
HSCs and Hematopoietic Progenitor Cells Are Localized in Separate Regions in Bone Marrow
The exclusive presence of HSCs around arterioles was determined in combination with the localization of hematopoietic progenitor cells in bone marrow. Our fluorescence immunohistochemical data demonstrated that CD150-positive HSCs were localized around arterioles near bone (Fig. 4D), whereas CD244-positive hematopoietic progenitor cells were absent in peri-arteriolar HSC niches near bone (Fig. 4D). The CD244-positive hematopoietic progenitor cells were localized at a larger distance from bone, where CD150-positive HSCs (Fig. 4E) and CD133-positive HSCs (Fig. 4F) were absent. Thus, HSCs are exclusively localized in peri-arteriolar HSC niches and hematopoietic progenitor cells are localized at a distance from bone and arterioles.
MSCs in Peri-arteriolar HSC Niches and Peri-arteriolar GSC Niches
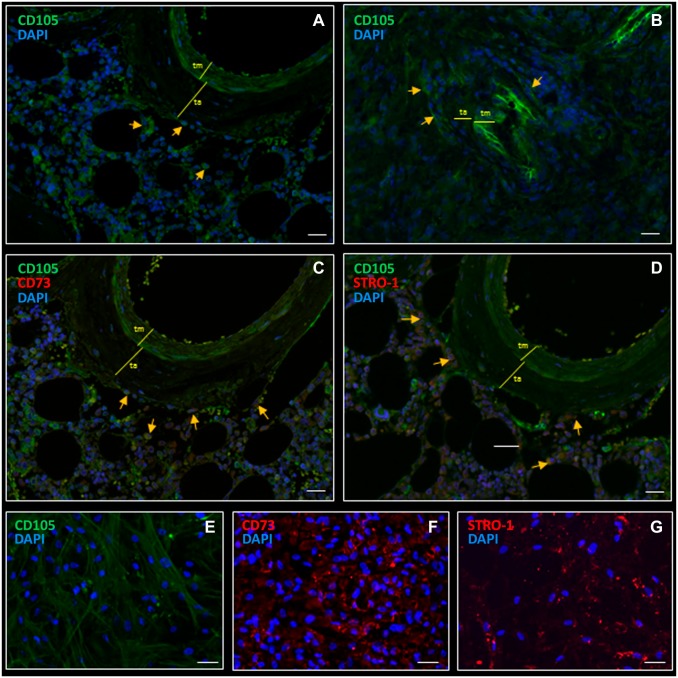
Next, the localization of MSCs was determined in HSC niches in bone marrow and GSC niches in glioblastoma, using the potent MSC marker CD105. In bone marrow, CD105 was exclusively expressed in MSCs in peri-arteriolar HSC niches (Fig. 5A). CD105 was also expressed in endothelial cells and smooth muscle cells in arteriolar walls (Fig. 5A). Similarly, in glioblastoma, CD105-positive MSCs were exclusively detected adjacent to the arteriolar tunica adventitia in peri-arteriolar GSC niches (Fig. 5B). CD105 was also expressed in endothelial cells and smooth muscle cells in the arteriolar wall in glioblastoma.
Figure 5.
Fluorescence immunohistochemical staining demonstrates MSCs in a peri-arteriolar HSC niche in human bone marrow (A) and in a peri-arteriolar niche in GSC niche in glioblastoma (B). CD105-expressing MSCs are localized in a peri-arteriolar HSC niche in bone marrow (A, yellow arrows). CD105-expressing MSCs are localized in a peri-arteriolar GSC niche in glioblastoma (B, indicated by yellow arrows). CD105 expression is also detected in endothelial cells and smooth muscle cells in the wall of the arterioles (A, B). CD105 and CD73 are co-localized in bone marrow (C, yellow arrows). CD105-STRO-1-expressing MSCs are localized in a peri-arteriolar HSC niche in bone marrow (D, yellow arrows). CD105 and STRO-1 are co-localized in bone marrow, but STRO-1 is also expressed in other stormal cells in bone marrow (D). CD105 (E), CD73 (F) and STRO-1 (G) are abundantly expressed in vitro in bone marrow-derived MSCs. DAPI is used for nuclear counterstaining. Scale bar = 100 µm. Abbreviations: MSC, mesenchymal stem cell; HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; STRO-1, stromal factor-1; DAPI, 4’,6-diamidino-2-phenylindole.
MSCs in HSC Niches Bone Marrow
Besides CD105, CD73 and stromal factor STRO-1 have been described as bone marrow–derived MSC biomarkers.23,43–45 CD73 and STRO-1 were not found to be stained in glioblastoma sections (data not shown). Figure 5C shows that CD105 and CD73 are co-localized in MSCs in peri-arteriolar HSC niches. CD105 and STRO-1 also co-localized in a subset of MSCs, but STRO-1 was also expressed on other cell types in bone marrow (Fig. 5D). We confirmed the expression of CD105 (Fig. 5E), CD73 (Fig. 5F), and STRO-1 (Fig. 5G) in vitro, using immunocytochemistry of bone marrow–derived MSCs. All three biomarkers were strongly expressed by bone marrow–derived MSCs. CD105 and CD73 were exclusively expressed in HSC niches, whereas STRO-1 expression was also detected outside niches.
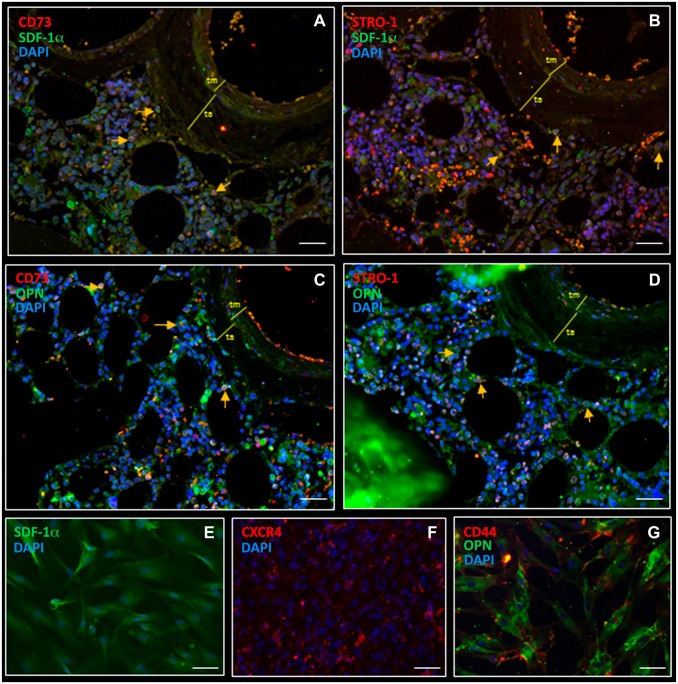
Bone Marrow-derived MSCs Produce Chemoattractants SDF-1α and OPN
MSCs are known to be producers of chemoattractants, such as SDF-1α and OPN.63 Therefore, we determined expression of chemoattractants SDF-1α and OPN and their receptors CXCR4 and CD44 in vitro using immunocytochemistry on bone marrow-derived MSCs (Fig. 6E–G) and found high levels of chemoattractants SDF-1α (Fig. 6E) and OPN (Fig. 6G) and their receptors CXCR4 (Fig. 6F) and CD44 (Fig. 6G), respectively, in MSCs. In bone marrow, MSCs were stained for MSC biomarker CD73 and SDF-1α (Fig. 6A) and MSC biomarker STRO-1 and SDF-1α (Fig. 6B). CD73-STRO-1 positive MSCs contained intracellular SDF-1α and OPN in peri-arteriolar HSC niches (Fig. 6A–D).
Figure 6.
Fluorescence immunohistochemical and immunocytochemical staining shows intracellular expression of chemoattractants SDF-1α and OPN in MSCs in a peri-arteriolar HSC niche in human bone marrow (A–D) and in MSC preparations (E–G). CD73-positive MSCs express intracellular SDF-1α in bone marrow (A, yellow arrows). STRO-1-positive MSCs express intracellular SDF-1α in bone marrow (B, yellow arrows). CD73-positive MSCs express intracellular OPN in bone marrow (C, yellow arrows). STRO-1-positive MSCs express intracellular OPN in bone marrow (D, yellow arrows). Bone marrow-derived MSCs express high levels of SDF-1α intracellularly (E), CXCR4 on the cell surface (F), OPN intracellularly and CD44 on the cell surface in vitro (G). DAPI is used for nuclear counterstaining. Scale bar = 100 µm. Abbreviations: SDF-1α, stromal-derived factor-1α; OPN, osteopontin; MSC, mesenchymal stem cell; HSC, hematopoietic stem cell; STRO-1, stromal factor-1; CXCR4, C-X-C receptor type 4; DAPI, 4’,6-diamidino-2-phenylindole.
P-EGFR and VEGFR Expression in Glioblastoma and Bone Marrow
The membrane receptors EGFR and VEGFR2 have been found to be differentially elevated in glioblastoma.50–54 Therefore, staining of activated EGFR, p-EGFR, and VEGFR2 was analyzed in glioblastoma and in bone marrow (Fig. 7). We found that a subset of CD150-positive HSCs in bone marrow expressed p-EGFR as well as VEGFR2 (Fig. 7A and C), but both receptors were also found on cells outside HSC niches in bone marrow (Fig. 7B and D). In glioblastoma, all CD133-positive GSCs expressed p-EGFR (Fig. 7G), whereas only a fraction of CD133-positive GSCs expressed VEGFR2 (Fig. 7F). Both p-EGFR and VEGFR2 were also found on other cell types in glioblastoma. VEGFR2 expression was found in cells in the arteriolar wall in both GSC niches and HSC niches (Fig. 7C and F). These data show that both p-EGFR and VEGFR2 are not GSC-specific or niche-specific receptors.
Figure 7.
Fluorescence immunohistochemical staining showing p-EGFR and VEGFR2 expression in bone marrow (A–D) and glioblastoma (E–G). A fraction of CD150-positive HSCs in HSC niches express p-EGFR and VEGFR2 (A, C) as well as cells outside HSC niches (B, D). In glioblastoma, a fraction of CD133-positive GSCs in GSC niches express VEGFR2 (F) whereas all CD133-positive GSCs exress p-EGFR (G). Both p-EGFR and VEGFR2 are also expressed in other cell types in glioblastoma. VEGFR2 expression was found in cells in the arteriolar wall in bone marrow and glioblastoma (C, F). DAPI was used for nuclear counterstaining. Scale bar = 100 µm. Abbreviations: p-EGFR, phosphorylated epidermal growth factor receptor; VEGFR2, vascular endothelial growth factor receptor 2; HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; DAPI, 4,’6-diamidino-2-phenylindole.
In Table 4, the localization of all 17 markers in HSC niches and GSC niches is summarized.
Table 4.
Summary of the Localization of 17 Markers in HSC Niches in Bone Marrow and GSC Niches in Glioblastoma.
| Biomarkers | Localization in HSC Niches | Localization in GSC Niches | Exclusively in (Peri)-arteriolar HSC Niches | Exclusively in (Peri)-arteriolar GSC Niches |
|---|---|---|---|---|
| SMA | In SMCs (tm) of arterioles | In SMCs (tm) of arterioles | Yes | No |
| CD150 | On HSCs adjacent to bone around arterioles | Not expressed | Yes | Not expressed |
| CD244 | On hematopoietic progenitor cells | Not expressed | No | Not expressed |
| CD133 | On HSCs adjacent to bone around arterioles | On GSCs adjacent to ta of arterioles | Yes | Yes |
| SOX2 | Not expressed | In GSCs adjacent to ta of arterioles | Not expressed | No |
| SDF-1α | In HSCs and MSCs adjacent to bone around arterioles and
extracellularly in niches In ECs and SMCs of arteriolar walls |
In GSCs and MSCs adjacent to ta of arterioles and
extracellularly in niches In ECs and SMCs of arteriolar walls |
Yes | Yes |
| CXCR4 | In HSCs and MSCs In SMCs and ECs of arteriolar walls |
In GSCs and MSCs In SMCs and ECs of arteriolar walls |
No | No |
| OPN | In HSCs and MSCs In ECs and SMCs of arterioles |
Mainly extracellularly In ECs and SMCs of arterioles |
Yes | Yes |
| CD44 | Intracellularly in HSCs and MSCs | On GSCs | No | No |
| CD105 | In MSCs In ECs and SMCs of arteriolar walls |
In MSCs In ECs and SMCs of arteriolar walls |
Yes | Yes |
| CD73 | In MSCs In SMCs and ECs of arteriolar walls |
Not expressed | Yes | Not expressed |
| STRO-1 | In MSCs In SMCs and ECs of arteriolar walls Other stromal cells |
Not expressed | No | Not expressed |
| HIF-1α | In subsets of HSCs In ECs and SMCs of arterioles |
In subsets of HSCs In ECs and SMCs of arterioles |
No | No |
| HIF-2α | Preferentially in HSCs around arterioles | Preferentially in GSCs around arterioles | Yes | Yes |
| VEGF | Intracellularly in many cell types and
extracellularly In ECs and SMCs of arterioles |
Intracellularly many cell types and
extracellularly In ECs and SMCs of arterioles |
No | No |
| VEGFR2 | On a fraction of HSCs, cells in the arteriolar wall, and other cell types in bone marrow | On a fraction of GSCs, cells in the arteriolar wall, and other cells types in glioblastoma | No | No |
| P-EGFR | On a fraction of HSCs and other cells in bone marrow | On GSCs and other cells types in glioblastoma | No | No |
Abbreviations: HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; SMA, smooth muscle actin; SMC, smooth muscle cell; tm, tunica media; ta, tunica adventitia; SOX2, sex determining region Y-box 2; SDF-1α, stromal–derived factor-1α; EC, endothelial cell; CXCR4, C-X-C receptor type 4; MSC, mesenchymal stem cell; OPN, osteopontin; STRO-1, stromal factor-1; HIF, hypoxia-induced factor; VEGF, vascular endothelial growth factor; VEGFR2, vascular endothelial growth factor receptor 2; p-EGFR, phospho-epidermal growth factor receptor.
Control Incubations
In all tested conditions, control staining in the absence of the primary antibodies was negative in bone marrow sections and glioblastoma sections (Supplementary Fig. 2). In bone marrow sections, erythrocytes were stained aspecifically in fluorescence immunohistochemical experiments (Supplementary Fig. 2).
Image Analysis of GSC Niches and HSC Niches
Image analysis was performed on all images of 13 GSC niches and 16 HSC niches, using ImageJ software, to determine quantitatively whether GSC niches are similar to HSC niches (Tables 5 and 6). The quantitative image analysis show that there is no significant difference in the percentage of CD133-SOX2-positive GSCs compared to the percentage of CD150-positive HSCs in niches and no significant difference between the percentage of CD105-positive MSCs in GSC niches and the percentage of CD105-positive MSCs in HSC niches (Fig. 8). This confirms that GSC niches and HSC niches are similar.
Table 5.
Image Analysis Data of 13 GSC Niches in Glioblastoma.
| Glioblastoma | NICHE 1 | NICHE 2 | NICHE 3 | NICHE 4 | NICHE 5 | NICHE 6 | NICHE 7 | NICHE 8 | NICHE 9 | NICHE 10 | NICHE 11 | NICHE 12 | NICHE 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total number of cells (DAPI) | 445 | 480 | 390 | 295 | 426 | 394 | 411 | 288 | 402 | 388 | 427 | 456 | 367 |
| Number of CD133/SOX2 GSCs | 180 | 200 | 156 | 140 | 122 | 163 | 153 | 134 | 200 | 176 | 169 | 175 | 145 |
| Number of CD105 MSCs | 102 | 100 | 97 | 86 | 77 | 86 | 106 | 74 | 102 | 88 | 93 | 100 | 73 |
| Percentage GSCs | 40.45 | 41.67 | 40.00 | 47.46 | 28.64 | 41.37 | 37.23 | 46.53 | 49.75 | 45.36 | 39.58 | 38.38 | 39.51 |
| Percentage MSCs | 22.92 | 20.83 | 24.87 | 29.15 | 18.08 | 21.83 | 25.79 | 25.69 | 25.37 | 22.68 | 21.78 | 21.93 | 19.89 |
Abbreviations: GSC, glioblastoma stem cell; DAPI, 4,’6-diamidino-2-phenylindole; SOX2, sex determining region Y-box 2; MSC, mesenchymal stem cell.
Table 6.
Image Analysis Data of 16 HSC Niches in Bone Marrow.
| Bone Marrow | NICHE 1 | NICHE 2 | NICHE 3 | NICHE 4 | NICHE 5 | NICHE 6 | NICHE 7 | NICHE 8 | NICHE 9 | NICHE 10 | NICHE 11 | NICHE 12 | NICHE 13 | NICHE 14 | NICHE 15 | NICHE 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total number of cells (DAPI) | 475 | 523 | 485 | 479 | 501 | 536 | 489 | 497 | 521 | 541 | 473 | 463 | 509 | 467 | 457 | 511 |
| Number of CD150 HSCs | 189 | 203 | 225 | 219 | 187 | 246 | 234 | 197 | 223 | 198 | 201 | 236 | 247 | 243 | 204 | 197 |
| Number of CD105 MSCs | 117 | 121 | 94 | 86 | 99 | 127 | 98 | 112 | 147 | 97 | 88 | 98 | 124 | 97 | 86 | 99 |
| Percentage HSCs | 39.79 | 38.81 | 46.39 | 45.72 | 37.33 | 45.90 | 47.85 | 39.64 | 42.80 | 36.60 | 42.49 | 50.97 | 48.53 | 52.03 | 44.64 | 38.55 |
| Percentage MSCs | 24.63 | 23.14 | 19.38 | 17.95 | 19.76 | 23.69 | 20.04 | 22.54 | 28.21 | 17.93 | 18.60 | 21.17 | 24.36 | 20.77 | 18.82 | 19.37 |
Abbreviations: HSC, hematopoietic stem cell; DAPI, 4,’6-diamidino-2-phenylindole; MSC, mesenchymal stem cell.
Figure 8.

Quantitative image analysis data of HSC niches and GSC niches reveals that both niche types are similar. Quantification of the percentages of HSCs and MSCs in 16 HSC niches in bone marrow were compared with the percentages of GSCs and MSCs in 13 GSC niches in glioblastoma. One-way ANOVA tests revealed that there were no statistical differences between HSC niches and GSC niches. Abbreviations: HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; MSC, mesenchymal stem cell.
Discussion
Our comparison of GSC niches in human glioblastoma and HSC niches in human bone marrow showed that GSC niches in glioblastoma are similar to HSC niches in bone marrow, as both types of niches express biomarkers revealing that they are hypoxic (Fig. 1), peri-arteriolar (Fig. 1) and contain the same functional chemoattractive proteins such as SDF-1α, CXCR4, OPN, and CD44 (Figs. 3 and 4). Figure 9 illustrates our findings and Table 4 summarizes the localization of 17 biomarkers in HSC niches in bone marrow and GSC niches in glioblastoma.
Figure 9.
Overview of hypoxic peri-arteriolar HSC niches in bone marrow (A–C) and hypoxic peri-arteriolar GSC niches in gliobastoma (D–F). Rectangles in A (salmon and green) indicate peri-arteriolar and peri-sinusoidal regions in bone marrow shown in B and C, respectively. Rectangles in D (salmon and green) indicate peri-arteriolar and peri-capillary regions in glioblastoma shown in E and F, respectively. HE staining of human bone marrow (A) shows a peri-arteriolar HSC niche (B) and peri-sinusoidal progenitor cells around sinusoids (A, C) and HE staining of human glioblastoma (D) shows a peri-arteriolar GSC niche (E) and differentiated glioblastoma cells around capillaries (F). HSCs express CD150, CD133, CXCR4, CD44 and HIF-1/2α and GSCs express CD133, SOX2 CXCR4, CD44 and HIF-2α. HSCs and GSCs are localized around SMA-positive arterioles in niches where chemoattractants SDF-1α and OPN are abundantly and exclusively expressed. SDF-1α-CXCR4 and OPN-CD44 facilitate retention of HSCs and GSCs in their niches. MSCs with intracellular SDF-1α and OPN express CD105, CD73 and STRO-1 and are exclusively localized in peri-arteriolar HSC and GSC niches. VEGF is widely expressed in bone marrow and glioblastoma (B, E). At distance from bone, hematopoietic progenitor cells expressing CD244, CXCR4 and CD44 are localized around sinusoids (C). Differentiated glioblastoma cells express CXCR4 and CD44 are localized around capillaries (F). Scale bar A = 200 µm; scale bar D = 100 µm. Abbreviations: HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; HE, Hematoxylin-eosin; CXCR4, C-X-C receptor type 4; HIF, hypoxia-induced factor; SOX2, sex determining region Y-box 2; SMA, smooth muscle actin; SDF-1α, stromal-derived factor-1α; OPN, osteopontin; VEGF, vascular endothelial growth factor; MSC, mesenchymal stem cell; STRO-1, stromal factor-1; OB, osteoblast; ta, tunica adventitia; tm, tunica media.
An essential aspect of HSC niches is hypoxia.7–9,13 Indeed, we demonstrated both in HSC niches and GSC niches expression of HIF-1α and HIF-2α (Fig. 2F and H). In peri-arteriolar HSC niches in human bone marrow, hypoxia, and in particular the expression of HIFs, is crucial for HSC maintenance. HIF-1α is important for the upregulation of a plethora of chemoattractant proteins in HSC niches, such as SDF-1α, CXCR4, OPN, and CD44, stem cell factor, angiopoietin-2, tyrosine kinase TIE2, thrombopoietin, and its receptor MPL.13,64,65 HIF-2α is also important for HSC maintenance, as it interacts with transcription factors HIF-1α and MEIS1, resulting in anaerobic glycolysis that HSCs depend on as a source of energy.11,66–68
In peri-arteriolar GSC niches in glioblastoma, hypoxia induces expression of SOX2, OCT4, and CD133, which are all GSC biomarkers involved in the maintenance of GSC stemness.1,7,8,34–40,69 HIF-1α and VEGF are induced by hypoxia and upregulate SDF-1α, CXCR4, OPN, and CD44,12,41,70–77 which are involved in the maintenance of GSCs in niches.7,12,13,41 In glioblastoma, HIF-2α is directly associated with the GSC phenotype by upregulation of SOX2, CD133, and OCT4 expression,46,78,79 whereas HIF-1α is involved in GSC survival besides upregulation of GSC niche factors.6,12,41 HIF-2α has been shown to be associated with poor prognosis of glioblastoma patients46 and may be a potential therapeutic target for anti-glioblastoma therapies. Currently, the HIF-2α inhibitor PT2385 is tested in a phase II clinical trial in glioblastoma patients (ClinicalTrials.gov NCT03216499). Since GSCs selectively express HIF-2α (Fig. 3), targeting of HIF-2α-positive GSCs may result in a better clinical outcome.
Hypoxic conditions around arterioles can be explained by the fact that arterioles are transport vessels similar to venules, whereas capillaries are exchange vessels. Thus, there is no exchange of oxygen and carbon dioxide between the lumen of arterioles and surrounding tissue and vice versa and thus a hypoxic condition can be created around arterioles and venules in both HSC niches in bone marrow and GSC niches in glioblastoma tumors.6–9
In line with the peri-arteriolar niche as the place to be for stem cells, we exclusively found HSCs around arterioles near bone, whereas the hematopoietic progenitor cells were found at distance from bone and arterioles (Fig. 4D–F). Our data are in line with the study of Kunisaki et al.,80 demonstrating in mice that HSCs are preferentially localized near arterioles in bone marrow.80 HSCs are localized near bone, because bone-lining osteoblasts are crucial for the maintenance of HSC stemness via Notch pathways, transforming growth factor-β (TGF-β) and bone morphogenic protein (BMP) signaling, which results in retention of HSCs in niches. Osteoblasts retain HSCs in niches by the secretion of OPN and SDF-1α, that bind to their receptors CD44 and CXCR4 on HSCs, respectively. OPN and SDF-1α secretion takes place under hypoxic conditions via HIF-1α and VEGF signaling.13,81–83
Sonic hedgehog (Shh), VEGF, and Notch signaling are not only important pathways for HSC maintenance, but also for the maintenance of the arteriolar phenotype, which likely explains the localization of HSCs around arterioles.8,81 A possible explanation for the separate localization of HSCs and hematopoietic progenitor cells was described in our previous report postulating the concept of the continuous compartmentalized HSC niche in human bone marrow.13 According to our concept, HSCs are localized in peri-arteriolar subcompartments of niches near bone around arterioles, which are hypoxic and where levels of reactive oxygen species (ROS) are low, whereas progenitor cells are localized in peri-sinusoidal subcompartments of niches at a distance from bone, where ROS levels are higher.13,84 This phenomenon enables HSCs to proliferate and differentiate into progenitor cells around sinusoids to enter the blood circulation. These findings were established in mouse studies.10,11,13 Whether the peri-arteriolar and peri-sinusoidal subcompartments of HSC niches are indeed two parts of the same continuum in human bone marrow, needs to be elucidated. This hypothesis can be tested using 3D imaging techniques of human bone marrow samples after tissue clearing.85,86
Mobilization of hematopoietic progenitor cells into the circulation requires a reduction of chemoattractants in HSC niches, such as SDF-1α and OPN. This process is mediated by the activation of multiple proteases in bone marrow, among which the bone resorbing protease cathepsin K (CatK), which is able to cleave and inactivate SDF-1α.87 CatK has been shown to be the highest differentially expressed protease in glioblastoma.88,89 CatK can degrade and inactivate SDF-1α by proteolytic cleavages of its N-terminus90 and co-localization of inactive Catk with SDF-1α in peri-arteriolar GSC niches was detected in our previous studies.5,7,90,91 These studies indicate that mobilization of GSCs out of niches in glioblastoma are mediated by similar mechanisms including activation of CatK and possibly other cathepsins as in HSC niches in bone marrow.
MSCs are not well explored as cellular components of GSC niches. Our study has shown that GSCs and MSCs in glioblastoma are localized in peri-arteriolar niches adjacent to the arteriolar tunica adventitia (Fig. 5). The arteriolar tunica adventitia is the place to be for GSCs and MSCs for a number of reasons. First of all, the hypoxic environment around arterioles is required for the maintenance of GSC and MSC stemness. Second, Shh, VEGF, and Notch signaling are important for the arteriolar phenotype.8,92 Exactly these signaling pathways are crucial for GSC77,93 and MSC maintenance,93–100 which may explain the localization of GSCs and MSCs around arterioles. Besides SDF-1α and OPN, MSCs produce a plethora of cytokines when co-cultured with glioblastoma cells, of which CCL2 (or MCP-1) was most relevant in our previous study.18 This was confirmed by Pavon et al.,21 who reported that CCL2- and SDF-1α-mediated tropism of MSCs toward CXCR4-positive and CD133-positive cells in glioblastoma increased tumor growth in vivo.
MSCs also produce high levels of the cytokine interleukin 6 (IL-6)18 that interacts with co-receptor gp-130 on GSCs, which activates the signal transducer of activation 3 (STAT3) pathway in GSCs, so that their stemness is maintained.23,101 IL-6 and STAT3 activation are found predominantly in the arteriolar tunica adventitia in cardiovascular tissue.101 This may also be the case in glioblastoma, explaining why the arteriolar adventitia is the place to be for MSCs and GSCs.
We show that MSCs are localized in peri-arteriolar HSC niches (Figs. 5A, C, D, and 6A–D) as well as in peri-arteriolar GSC niches (Fig. 5B). Our data are in line with data published by Hossain et al.23 who showed that CD105-positive MSCs expressing high levels of SDF-1α are present around arterioles in glioblastoma tissue sections. Moreover, MSCs were found to produce high levels of the chemoattractant OPN (Fig. 6C, D, G). Therefore, we propose that SDF-1α- and OPN-producing MSCs attract CXCR4-CD44-positive GSCs to GSC niches and protect them from chemotherapy and irradiation, as GSCs are maintained in a quiescent state in their niches.
It has been argued that GSC niches cannot be at distance from endothelial cells. Therefore, it was doubted that peri-arteriolar niches exist.102–105 However, the physiological HSC niches demonstrate that this assumption is not correct, as HSC niches are exclusively present around arterioles. In addition, the direct contact of endothelial cells with GSCs has been supported by the notion that endothelial cells secrete soluble factors that maintain GSC stemness and phenotype.79,102–105 However, secreted soluble factors such as SDF-1α, osteopontin and VEGF are also produced by other cell types, such as smooth muscle cells and MSCs, as we show in the present study.
Our image analysis data revealed that the percentages of GSCs and MSCs in GSC niches in glioblastoma are similar to the percentages of HSCs and MSCs in HSC niches in bone marrow (Fig. 8), which strengthens the notion that the 2 niche types are indeed similar and that treatment strategies for AML patients may also be of benefit for glioblastoma patients. Our finding that HSC niches and GSC niches are similar provides a theoretical basis for the development of novel treatment strategies for glioblastoma, as has been done in AML. In AML, disruption of the SDF-1α-CXCR4 and OPN-CD44 interactions results in the mobilization of CXCR4-CD44-positive LSCs out of HSCs niches and their sensitization to chemotherapy. Phase I/II clinical trials were performed in AML patients who were treated with the CXCR4 inhibitor plerixafor (AMD3100) in combination with chemotherapy. Treatment with plerixafor resulted in a 2-fold increased mobilization of AML cells to the peripheral blood and a better overall response rate to chemotherapy, compared to chemotherapy alone (ClinicalTrials.gov NCT00512252).13,14 On the basis of the similarities between HSC niches in GSC niches, similar treatment approaches may be clinically investigated to improve therapy of glioblastoma patients.
Our data show that all CD133-positive GSCs express p-EGFR (Fig. 7E and G). EGFR activity and amplification has been found to be differentially elevated in GSCs,52,53 which is associated with their resistance to chemotherapy and radiotherapy.54 EGFR is therefore an interesting target for the sensitization of GSCs to therapy besides CXCR4 and CD44. A fraction of CD133-positive GSCs were shown to express VEGFR2 (Fig. 7F), which has been associated with GSC viability, proliferation, and tumor growth via VEGF-VEGFR2-neuropilin-1 signaling.51,106 Furthermore, VEGFR2 plays a role in vascular mimicry by GSCs, thus their ability to form tumor vasculature50,107 as is shown in Fig. 7F. Because only a fraction of CD133-positive GSCs express VEGFR2, it is not likely that anti-VEGFR2 therapy will be successful.
According to the WHO 2016 classification of gliomas,108 infiltrative gliomas of all histological grades can be divided into 3 subgroups: IDH1/2-mutant and 1p/19q-non-co-deleted glioblastoma, IDH1/2-mutant and 1p-19q-codeleted glioblastoma and IDH1/2 wildtype glioblastoma.109,110 Polivka et al.111 describe that IDH1-mutated glioblastoma is associated with lower levels of VEGF expression than IDH1 wild-type but not with angiogenesis. In our study, 1 glioblastoma sample was IDH1 mutated and we did not observe differences in VEGF levels in this tissue sample as compared with the 9 IDH1 wild-type glioblastoma samples. The expression of GSC niche-associated markers in our study was similar in the IDH1-mutated sample compared to the IDH1 wild-type samples. In our future studies, glioblastoma samples will be used for which genetic screening has been performed for IDH1 mutation, EGFR amplification, 1p/19-co-deletion, platelet-derived growth factor receptor (PDGFR) expression, PTEN mutations, CDKN2A mutations, and ATRX mutations for proper classification. Subsequently, expression of GSC niche-associated markers and the metabolic rewiring in niches of IDH1-mutated glioblastoma samples112 will be tested in glioblastoma samples of different subtypes for therapeutic targeting.113 This is the first study demonstrating intact HSC niches in human bone marrow to show that GSC niches in glioblastoma are similar to HSC niches. Both types of niches are hypoxic, peri-arteriolar and contain mesenchymal stem cells and the same functional chemoattractive proteins and their receptors. This similarity implies that therapeutics in clinical trials targeted at LSCs in HSC niches can be tested to target GSCs in their niches in glioblastoma.
Supplemental Material
Supplemental material, DS_10.1369_0022155419878416 for Similarities Between Stem Cell Niches in Glioblastoma and Bone Marrow: Rays of Hope for Novel Treatment Strategies by Vashendriya V.V. Hira, Barbara Breznik, Miloš Vittori, Annique Loncq de Jong, Jernej Mlakar, Roelof-Jan Oostra, Mohammed Khurshed, Remco J. Molenaar, Tamara Lah and Cornelis J.F. Van Noorden in Journal of Histochemistry & Cytochemistry
Acknowledgments
The authors thank Theo Dirksen from the Pathology Department of the Amsterdam UMC at the location Academic Medical Center in The Netherlands, for providing paraffin bone marrow tissue sections. In addition, the authors thank Dr. Tjaša Lukan and Barbara Dušak from the Department of Biotechnology and Systems Biology at the National Institute of Biology (NIB) in Ljubljana, Slovenia, and Dr. Metka Novak from the Department of Genetic Toxicology and Cancer Biology at the NIB, for their assistance with confocal microscopy. We also thank Prof. Dr. Roman Bošnjak (MD) and Andrej Porcˇnik (MD) from the University Medical Center Ljubljana, Slovenia, for providing glioblastoma samples for immunohistochemical analyses. The authors thank Prof. Dr. Eric Reits for his helpful comments on the manuscript.
Footnotes
Competing Interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions: All authors have contributed to this article as follows: conception and design (VVVH, CJFVN), collection and/or assembly of data (VVVH, BB, ALJ, CJFVN), data analysis and interpretation (VVVH, BB, ALJ, MV, MK, RJM, CJFVN) manuscript writing (VVVH, RJM, TL, CJFVN), final approval of manuscript (VVVH, BB, ALJ, MV, MK, JM, RJM, CJFVN, RO, TL), provision of study material (JM, RO), gave intellectual input (RO, RJM, CJFVN), and supervised the entire study (CJFVN).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was financially supported by the Dutch Cancer Society (KWF; UVA 2014-6839 and UVA 2016.1-10460; VVVH, MK, RJM, CJFVN), the European Program of Cross-Border Cooperation for Slovenia-Italy Interreg TRANS-GLIOMA (Program 2017; TL), the IVY Interreg Fellowship (VVVH), the Slovenian Research Agency (Program P10245; TL), and RJM was supported by the Fondation pour la Recherche Nuovo-Soldati 2019.
Contributor Information
Vashendriya V.V. Hira, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia; Department of Medical Biology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands.
Barbara Breznik, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Miloš Vittori, Department of Biology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia.
Annique Loncq de Jong, Department of Medical Biology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands.
Jernej Mlakar, Institute of Pathology, Medical Faculty, University of Ljubljana, Ljubljana, Slovenia.
Roelof-Jan Oostra, Department of Medical Biology, Section Clinical Anatomy and Embryology, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands.
Mohammed Khurshed, Department of Medical Biology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands; Department of Medical Oncology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands.
Remco J. Molenaar, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia Department of Medical Biology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands; Department of Medical Oncology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands.
Tamara Lah, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Cornelis J.F. Van Noorden, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia; Department of Medical Biology, Cancer Center Amsterdam, Amsterdam UMC at the Academic Medical Center, Amsterdam, The Netherlands.
Literature Cited
- 1. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 2. Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71(11):1437–44. doi: 10.1001/jamaneurol. 2014.1701. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. doi: 10.1016/S1470-2045(09)70025. [DOI] [PubMed] [Google Scholar]
- 4. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5. Breznik B, Limback C, Porcnik A, Blejec A, Krajnc MK, Bosnjak R, Kos J, Van Noorden CJF, Lah TT. Localization patterns of cathepsins K and X and their predictive value in glioblastoma. Radiol Oncol. 2018;52(4):433–42. doi: 10.2478/raon-2018-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aderetti DA, Hira VVV, Molenaar RJ, van Noorden CJF. The hypoxic peri-arteriolar glioma stem cell niche, an integrated concept of five types of niches in human glioblastoma. Biochim Biophys Acta Rev Cancer. 2018;1869(2):346–54. doi: 10.1016/j.bbcan.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 7. Hira VV, Ploegmakers KJ, Grevers F, Verbovsek U, Silvestre-Roig C, Aronica E, Tigchelaar W, Turnšek TL, Molenaar RJ, Van Noorden CJ. CD133+ and nestin+ glioma stem-like cells reside around CD31+ arterioles in niches that express SDF-1α, CXCR4, osteopontin and cathepsin K. J Histochem Cytochem. 2015;63(7):481–93. doi: 10.1369/0022155415581689. [DOI] [PubMed] [Google Scholar]
- 8. Hira VVV, Aderetti DA, van Noorden CJF. Glioma stem cell niches in human glioblastoma are periarteriolar. J Histochem Cytochem. 2018;66:349–58. doi: 10.1369/0022155417752676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hira VVV, Wormer JR, Kakar H, Breznik B, van der Swaan B, Hulsbos R, Tigchelaar W, Tonar Z, Khurshed M, Molenaar RJ, Van Noorden CJF. Periarteriolar glioblastoma stem cell niches express bone marrow hematopoietic stem cell niche proteins. J Histochem Cytochem. 2018;66:150–73. doi: 10.1369/0022155417749174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hao Y, Cheng D, Ma Y, Zhou W, Wang Y. The relationship between oxygen concentration, reactive oxygen species and the biological characteristics of human bone marrow hematopoietic stem cells. Transplant Proc. 2011;43(7):2755–61. doi: 10.1016/j.transproceed.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 11. Zhang CC, Sadek HA. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxid Redox Signal. 2014;20(12):1891–901. doi: 10.1089/ars.2012.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zagzag D, Esencay M, Mendez O, Yee H, Smirnova I, Huang Y, Chiriboga L, Lukyanov E, Liu M, Newcomb EW. Hypoxia- and vascular endothelial growth factor-induced stromal cell-derived factor-1alpha/CXCR4 expression in glioblastomas: one plausible explanation of Scherer’s structures. Am J Pathol. 2008;173(2):545–60. doi: 10.2353/ajpath.2008.071197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hira VVV, van Noorden CJF, Carraway HE, Maciejewski JP, Molenaar RJ. Novel therapeutic strategies to target leukemic cells that hijack compartmentalized continuous hematopoietic stem cell niches. Biochim Biophys Acta. 2017;1868(1):183–98. doi: 10.1016/j.bbcan.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 14. Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM, Kulkarni S, Abboud CN, Cashen AF, Stockerl-Goldstein KE, Vij R, Westervelt P, DiPersio JF. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012;119(17):3917–24. doi: 10.1182/blood-2011-10-383406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Charbord P. Bone marrow mesenchymal stem cells: historical overview and concepts. Hum Gene Ther. 2010;21(9):1045–56. doi: 10.1089/hum.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jing D, Fonseca AV, Alakel N, Fierro FA, Muller K, Bornhauser M, Ehninger G, Corbeil D, Ordemann R. Hematopoietic stem cells in co-culture with mesenchymal stromal cells: modeling the niche compartments in vitro. Haematologica. 2010;95(4):542–50. doi: 10.3324/haematol.2009.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oliveira MN, Pillat MM, Motaln H, Ulrich H, Lah TT. Kinin-B1 receptor stimulation promotes invasion and is involved in cell-cell interaction of co-cultured glioblastoma and mesenchymal stem cells. Sci Rep. 2018;8(1):1299. doi: 10.1038/s41598-018-19359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Motaln H, Gruden K, Hren M, Schichor C, Primon M, Rotter A, Lah TT. Human mesenchymal stem cells exploit the immune response mediating chemokines to impact the phenotype of glioblastoma. Cell Transplant. 2012;21(7):1529–45. doi: 10.3727/096368912X640547. [DOI] [PubMed] [Google Scholar]
- 19. Breznik B, Motaln H, Vittori M, Rotter A, Lah Turnsek T. Mesenchymal stem cells differentially affect the invasion of distinct glioblastoma cell lines. Oncotarget. 2017;8(15):25482–99. doi: 10.18632/oncotarget.16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Breznik B, Motaln H, Lah Turnsek T. Proteases and cytokines as mediators of interactions between cancer and stromal cells in tumours. Biol Chem. 2017;398(7):709–19. doi: 10.1515/hsz-2016-0283. [DOI] [PubMed] [Google Scholar]
- 21. Pavon LF, Sibov TT, de Souza AV, da Cruz EF, Malheiros SMF, Cabral FR, de Souza JG, Boufleur P, de Oliveira DM, de Toledo SRC, Marti LC, Malheiros JM, Paiva FF, Tannús A, de Oliveira SM, Chudzinski-Tavassi AM, de Paiva Neto MA, Cavalheiro S. Tropism of mesenchymal stem cell toward CD133(+) stem cell of glioblastoma in vitro and promote tumor proliferation in vivo. Stem Cell Res Ther. 2018;9(1):310. doi: 10.1186/s13287-018-1049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodini CO, Goncalves da, Silva PB, Assoni AF, Carvalho VM, Okamoto OK. Mesenchymal stem cells enhance tumorigenic properties of human glioblastoma through independent cell-cell communication mechanisms. Oncotarget. 2018;9(37):24766–77. doi: 10.18632/oncotarget.25346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hossain A, Gumin J, Gao F, Figueroa J, Shinojima N, Takezaki T, Priebe W, Villarreal D, Kang SG, Joyce C, Sulman E, Wang Q, Marini FC, Andreeff M, Colman H, Lang FF. Mesenchymal stem cells isolated from human gliomas increase proliferation and maintain stemness of glioma stem cells through the IL-6/gp130/STAT3 pathway. Stem Cells. 2015;33(8):2400–15. doi: 10.1002/stem.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brzoska E, Kowalewska M, Markowska-Zagrajek A, Kowalski K, Archacka K, Zimowska M, Grabowska I, Czerwińska AM, Czarnecka-Góra M, Stremińska W, Jańczyk-Ilach K, Ciemerych MA. Sdf-1 (CXCL12) improves skeletal muscle regeneration via the mobilisation of CXCR4 and CD34 expressing cells. Biol Cell. 2012;104(12):722–37. doi: 10.1111/boc.201200022. [DOI] [PubMed] [Google Scholar]
- 25. Yu PF, Huang Y, Xu CL, Lin LY, Han YY, Sun WH, Hu GH, Rabson AB, Wang Y, Shi YF. Downregulation of CXCL12 in mesenchymal stromal cells by TGFbeta promotes breast cancer metastasis. Oncogene. 2017;36(6):840–49. doi: 10.1038/onc.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13(1):102–16. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Birbrair A, Frenette PS. Niche heterogeneity in the bone marrow. Ann N Y Acad Sci. 2016;1370(1):82–96. doi: 10.1111/nyas.13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bennett JA, Singh KP, Welle SL, Boule LA, Lawrence BP, Gasiewicz TA. Conditional deletion of Ahr alters gene expression profiles in hematopoietic stem cells. PLoS ONE. 2018;13(11):e0206407. doi: 10.1371/journal.pone.0206407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 30. Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–12. [PubMed] [Google Scholar]
- 31. Matsuoka Y, Nakamura F, Hatanaka K, Fujioka T, Otani S, Kimura T, Fujimura Y, Asano H, Sonoda Y. The number of CD34(+)CD133(+) hematopoietic stem cells residing in umbilical cord blood (UCB) units is not correlated with the numbers of total nucleated cells and CD34(+) cells: a possible new indicator for quality evaluation of UCB units. Int J Hematol. 2018;108(6):571–9. doi: 10.1007/s12185-018. [DOI] [PubMed] [Google Scholar]
- 32. Kalantari N, Abroun S, Soleimani M, Kaviani S, Azad M, Eskandari F, Habibi H. Effect of the receptor activator of nuclear factor кB and RANK ligand on in vitro differentiation of cord blood CD133(+) hematopoietic stem cells to osteoclasts. Cell J. 2016;18(3):322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iida R, Welner RS, Zhao W, Alberola-lla J, Medina KL, Zhao ZJ, Kincade PW. Stem and progenitor cell subsets are affected by JAK2 signaling and can be monitored by flow cytometry. PLoS ONE. 2014;9(4):e93643. doi: 10.1371/journal.pone.0093643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–8. [PubMed] [Google Scholar]
- 35. Wu B, Sun C, Feng F, Ge M, Xia L. Do relevant markers of cancer stem cells CD133 and Nestin indicate a poor prognosis in glioma patients? a systematic review and meta-analysis. J Exp Clin Cancer Res. 2015;34:44. doi: 10.1186/s13046-015-0163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang M, Song T, Yang L, Chen R, Wu L, Yang Z, Fang J. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 2008;27:85. doi: 10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song WS, Yang YP, Huang CS, Lu KH, Liu WH, Wu WW, Lee YY, Lo WL, Lee SD, Chen YW, Huang PI, Chen MT. Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells. J Chin Med Assoc. 2016;79(10):538–45. doi: 10.1016/j.jcma.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 38. Abdelrahman AE, Ibrahim HM, Elsebai EA, Ismail EI, Elmesallamy W. The clinicopathological significance of CD133 and SOX2 in astrocytic glioma. Cancer Biomark. 2018;23:391–403. doi: 10.3233/CBM-181460. [DOI] [PubMed] [Google Scholar]
- 39. Garros-Regulez L, Garcia I, Carrasco-Garcia E, Lantero A, Aldaz P, Moreno-Cugnon L, Arrizabalaga O, Undabeitia J, Torres-Bayona S, Villanua J, Ruiz I, Egaña L, Sampron N, Matheu A. Targeting SOX2 as a therapeutic strategy in glioblastoma. Front Oncol. 2016;6:222. doi: 10.3389/fonc.2016.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Acanda de la Rocha AM, Lopez-Bertoni H, Guruceaga E, Gonzalez-Huarriz M, Martinez-Velez N, Xipell E, Fueyo J, Gomez-Manzano C, Alonso MM. Analysis of SOX2-regulated transcriptome in glioma stem cells. PLoS ONE. 2016;11(9):e0163155. doi: 10.1371/journal.pone.0163155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zagzag D, Lukyanov Y, Lan L, Ali MA, Esencay M, Mendez O, Yee H, Voura EB, Newcomb EW. Hypoxia-inducible factor 1 and VEGF upregulate CXCR4 in glioblastoma: implications for angiogenesis and glioma cell invasion. Lab Invest. 2006;86(12):1221–32. doi: 10.1038/labinvest.3700482. [DOI] [PubMed] [Google Scholar]
- 42. Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14(3):357–69. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Corselli M, Chen CW, Sun B, Yap S, Rubin JP, Peault B. The tunica adventitia of human arteries and veins as a source of mesenchymal stem cells. Stem Cells Dev. 2012;21(8):1299–308. doi: 10.1089/scd.2011.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Appaix F, Nissou MF, van der Sanden B, Dreyfus M, Berger F, Issartel JP, Wion D. Brain mesenchymal stem cells: the other stem cells of the brain? World J Stem Cells. 2014;6(2):134–43. doi: 10.4252/wjsc.v6.i2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lv FJ, Tuan RS, Cheung KM, Leung VY. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32(6):1408–19. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 46. Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–13. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seidel S, Garvalov BK, Wirta V, von Stechow L, Schänzer A, Meletis K, Wolter M, Sommerlad D, Henze AT, Nistér M, Reifenberger G, Lundeberg J, Frisén J, Acker T. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133(Pt 4):983–95. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 48. Monteiro AR, Hill R, Pilkington GJ, Madureira PA. The role of hypoxia in glioblastoma invasion. Cells. 2017;6(4):45. doi: 10.3390/cells6040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang WJ, Chen WW, Zhang X. Glioblastoma multiforme: Effect of hypoxia and hypoxia inducible factors on therapeutic approaches. Oncol Lett. 2016;12(4):2283–8. doi: 10.3892/ol.2016.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yao X, Ping Y, Liu Y, Chen K, Yoshimura T, Liu M, Gong W, Chen C, Niu Q, Guo D, Zhang X, Wang JM, Bian X. Vascular endothelial growth factor receptor 2 (VEGFR-2) plays a key role in vasculogenic mimicry formation, neovascularization and tumor initiation by glioma stem-like cells. PLoS ONE. 2013;8(3):e57188. doi: 10.1371/journal.pone.0057188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu C, Wu X, Zhu J. VEGF promotes proliferation of human glioblastoma multiforme stem-like cells through VEGF receptor 2. Sci World J. 2013;2013:417413. doi: 10.1155/2013/417413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Howard BM, Gursel DB, Bleau AM, Beyene RT, Holland EC, Boockvar JA. EGFR signaling is differentially activated in patient-derived glioblastoma stem cells. J Exp Ther Oncol. 2010;8(3):247–60. [PubMed] [Google Scholar]
- 53. Liffers K, Lamszus K, Schulte A. EGFR amplification and glioblastoma stem-like cells. Stem Cells Int. 2015;2015:427518. doi: 10.1155/2015/427518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pang LY, Saunders L, Argyle DJ. Epidermal growth factor receptor activity is elevated in glioma cancer stem cells and is required to maintain chemotherapy and radiation resistance. Oncotarget. 2017;8(42):72494–512. doi: 10.18632/oncotarget.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kolosa K, Motaln H, Herold-Mende C, Korsic M, Lah TT. Paracrine effects of mesenchymal stem cells induce senescence and differentiation of glioblastoma stem-like cells. Cell Transplant. 2015;24(4):631–44. doi: 10.3727/096368915X687787. [DOI] [PubMed] [Google Scholar]
- 56. Podergajs N, Motaln H, Rajcevic U, Verbovsek U, Korsic M, Obad N, Espedal H, Vittori M, Herold-Mende C, Miletic H, Bjerkvig R, Turnšek TL. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget. 2016;7(1):593–609. doi: 10.18632/oncotarget.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hira VVV, Loncq de Jong A, Ferro K, Khurshed M, Molenaar RJ, van Noorden CJF. Comparison of different methodologies and cryostat versus paraffin sections for chromogenic immunohistochemistry. Acta Histochem. 2019;121:125–34. doi: 10.1016/j.acthis.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 58. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van Noorden CJF, Chieco P. Image cytometry protocols. J Histochem Cytochem. 2013;61(10):759–60. doi: 10.1369/0022155413500497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cao H, Heazlewood SY, Williams B, Cardozo D, Nigro J, Oteiza A, Nilsson SK. The role of CD44 in fetal and adult hematopoietic stem cell regulation. Haematologica. 2016;101(1):26–37. doi: 10.3324/haematol.2015.135921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zoller M. CD44, hyaluronan, the hematopoietic stem cell, and leukemia-initiating cells. Front Immunol. 2015;6:235. doi: 10.3389/fimmu.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khaldoyanidi S. Directing stem cell homing. Cell Stem Cell. 2008;2(3):198–200. doi: 10.1016/j.stem.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 63. Kowalski K, Kolodziejczyk A, Sikorska M, Placzkiewicz J, Cichosz P, Kowalewska M, Stremińska W, Jańczyk-Ilach K, Koblowska M, Fogtman A, Iwanicka-Nowicka R, Ciemerych MA, Brzoska E. Stem cells migration during skeletal muscle regeneration: the role of SDF-1/CXCR4 and SDF/CXCR7 axis. Cell Adh Migr. 2017;11(4):384–98. doi: 10.1080/19336918.2016.1227911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci U S A. 2007;104(13):5431–6. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, Suda T. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7(3):391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 66. Ochocki JD, Simon MC. Nutrient-sensing pathways and metabolic regulation in stem cells. J Cell Biol. 2013;203(1):23–33. doi: 10.1083/jcb.201303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–56. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vannini N, Girotra M, Naveiras O, Nikitin G, Campos V, Giger S, Roch A, Auwerx J, Lutolf MP. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun. 2016;7:13125. doi: 10.1038/ncomms13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Motegi H, Kamoshima Y, Terasaka S, Kobayashi H, Houkin K. Type 1 collagen as a potential niche component for CD133-positive glioblastoma cells. Neuropathology. 2014;34(4):378–85. doi: 10.1111/neup.12117. [DOI] [PubMed] [Google Scholar]
- 70. Ishikawa T, Nakashiro K, Klosek SK, Goda H, Hara S, Uchida D, Hamakawa H. Hypoxia enhances CXCR4 expression by activating HIF-1 in oral squamous cell carcinoma. Oncol Rep. 2009;21(3):707–12. [PubMed] [Google Scholar]
- 71. Guo M, Cai C, Zhao G, Qiu X, Zhao H, Ma Q, Tian L, Li X, Hu Y, Liao B, Ma B, Fan Q. Hypoxia promotes migration and induces CXCR4 expression via HIF-1alpha activation in human osteosarcoma. PLoS ONE. 2014;9(3):e90518. doi: 10.1371/journal.pone.0090518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schioppa T, Uranchimeg B, Saccani A, Biswas SK, Doni A, Rapisarda A, Bernasconi S, Saccani S, Nebuloni M, Vago L, Mantovani A, Melillo G, Sica A. Regulation of the chemokine receptor CXCR4 by hypoxia. J Exp Med. 2003;198(9):1391–402. doi: 10.1084/jem.20030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Youn SW, Lee SW, Lee J, Jeong HK, Suh JW, Yoon CH, Kang HJ, Kim HZ, Koh GY, Oh BH, Park YB, Kim HS. COMP-Ang1 stimulates HIF-1alpha-mediated SDF-1 overexpression and recovers ischemic injury through BM-derived progenitor cell recruitment. Blood. 2011;117(16):4376–86. doi: 10.1182/blood-2010-07-295964. [DOI] [PubMed] [Google Scholar]
- 74. Jin F, Brockmeier U, Otterbach F, Metzen E. New insight into the SDF-1/CXCR4 axis in a breast carcinoma model: hypoxia-induced endothelial SDF-1 and tumor cell CXCR4 are required for tumor cell intravasation. Mol Cancer Res. 2012;10(8):1021–31. doi: 10.1158/1541-7786.MCR. [DOI] [PubMed] [Google Scholar]
- 75. Liu K, Fang C, Shen Y, Liu Z, Zhang M, Ma B, Pang X. Hypoxia-inducible factor 1a induces phenotype switch of human aortic vascular smooth muscle cell through PI3K/AKT/AEG-1 signaling. Oncotarget. 2017;8(20):33343–52. doi: 10.18632/oncotarget.16448. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76. Erpolat OP, Gocun PU, Akmansu M, Ozgun G, Akyol G. Hypoxia-related molecules HIF-1alpha, CA9, and osteopontin: predictors of survival in patients with high-grade glioma. Strahlenther Onkol. 2013;189(2):147–54. doi: 10.1007/s00066-012-0262-5. [DOI] [PubMed] [Google Scholar]
- 77. Man J, Yu X, Huang H, Zhou W, Xiang C, Huang H, Miele L, Liu Z, Bebek G, Bao S, Yu JS. Hypoxic induction of vasorin regulates notch1 turnover to maintain glioma stem-like cells. Cell Stem Cell. 2018;22(1):104–118. e6. doi: 10.1016/j.stem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20(5):557–70. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Borovski T, De Sousa EMF, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011;71(3):634–9. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 80. Kunisaki Y, Bruns I, Scheiermann C, Ahmed J, Pinho S, Zhang D, Mizoguchi T, Wei Q, Lucas D, Ito K, Mar JC, Bergman A, Frenette PS. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502(7473):637–43. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–6. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 82. Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–34. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yin T, Li L. The stem cell niches in bone. J Clin Invest. 2006;116(5):1195–201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Itkin T, Gur-Cohen S, Spencer JA, Schajnovitz A, Ramasamy SK, Kusumbe AP, Ledergor G, Jung Y, Milo I, Poulos MG, Kalinkovich A, Ludin A, Kollet O, Shakhar G, Butler JM, Rafii S, Adams RH, Scadden DT, Lin CP, Lapidot T. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532(7599):323–28. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Azaripour A, Lagerweij T, Scharfbillig C, Jadczak AE, van der Swaan B, Molenaar M, van der Waal R, Kielbassa K, Tigchelaar W, Picavet DI, Jonker A, Hendrikx EML, Hira VVV, Khurshed M, Van Noorden CJF. Three-dimensional histochemistry and imaging of human gingiva. Sci Rep. 2018;8(1):1647. doi: 10.1038/s41598-018-19685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Azaripour A, Lagerweij T, Scharfbillig C, Jadczak AE, Willershausen B, van Noorden CJF. A survey of clearing techniques for 3D imaging of tissues with special reference to connective tissue. Prog Histochem Cytochem. 2016;51(2):9–23. doi: 10.1016/j.proghi.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 87. Kollet O, Dar A, Shivtiel S, Kalinkovich A, Lapid K, Sztainberg Y, Tesio M, Samstein RM, Goichberg P, Spiegel A, Elson A, Lapidot T. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat Med. 2006;12(6):657–64. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- 88. Verbovsek U, Motaln H, Rotter A, Atai NA, Gruden K, van Noorden CJF, Lah TT. Expression analysis of all protease genes reveals cathepsin K to be overexpressed in glioblastoma. PLoS ONE. 2014;9(10):e111819. doi: 10.1371/journal.pone.0111819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Verbovsek U, van Noorden CJF, Lah TT. Complexity of cancer protease biology: cathepsin K expression and function in cancer progression. Semin Cancer Biol. 2015;35:71–84. doi: 10.1016/j.semcancer.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 90. Hira VV, Verbovsek U, Breznik B, Srdic M, Novinec M, Kakar H, Wormer J, Van der Swaan B, Lenarcˇicˇ B, Juliano L, Mehta S, Van Noorden CJ, Lah TT. Cathepsin K cleavage of SDF-1alpha inhibits its chemotactic activity towards glioblastoma stem-like cells. Biochim Biophys Acta. 2016;1864(3):594–603. doi: 10.1016/j.bbamcr.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 91. Breznik B, Limbaeck Stokin C, Kos J, Khurshed M, Hira VVV, Bošnjak R, Lah TT, Van Noorden CJF. Cysteine cathepsins B, X and K expression in peri-arteriolar glioblastoma stem cell niches. J Mol Histol. 2018;49(5):481–97. doi: 10.1007/s10735-018-9787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Swift MR, Weinstein BM. Arterial-venous specification during development. Circ Res. 2009;104(5):576–88. doi: 10.1161/CIRCRESAHA.108.188805. [DOI] [PubMed] [Google Scholar]
- 93. Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, Nikkhah G, Dimeco F, Piccirillo S, Vescovi AL, Eberhart CG. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Brooks MD, Sengupta R, Snyder SC, Rubin JB. Hitting them where they live: targeting the glioblastoma perivascular stem cell niche. Curr Pathobiol Rep. 2013;1(2):101–10. doi: 10.1007/s40139-013-0012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brooks LJ, Parrinello S. Vascular regulation of glioma stem-like cells: a balancing act. Curr Opin Neurobiol. 2017;47:8–15. doi: 10.1016/j.conb.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 96. Ulasov IV, Nandi S, Dey M, Sonabend AM, Lesniak MS. Inhibition of Sonic hedgehog and Notch pathways enhances sensitivity of CD133(+) glioma stem cells to temozolomide therapy. Mol Med. 2011;17(1–2):103–12. doi: 10.2119/molmed.2010.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog signaling in the maintenance of cancer stem cells. Cancers. 2015;7(3):1554–85. doi: 10.3390/cancers7030851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28(1):17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yu JB, Jiang H, Zhan RY. Aberrant Notch signaling in glioblastoma stem cells contributes to tumor recurrence and invasion. Mol Med Rep. 2016;14(2):1263–8. doi: 10.3892/mmr.2016.5391. [DOI] [PubMed] [Google Scholar]
- 100. Kagiwada H, Yashiki T, Ohshima A, Tadokoro M, Nagaya N, Ohgushi H. Human mesenchymal stem cells as a stable source of VEGF-producing cells. J Tissue Eng Regen Med. 2008;2(4):184–9. doi: 10.1002/term.79. [DOI] [PubMed] [Google Scholar]
- 101. Hou T, Tieu BC, Ray S, Recinos Iii A, Cui R, Tilton RG, Brasier AR. Roles of IL-6-gp130 signaling in vascular inflammation. Curr Cardiol Rev. 2008;4(3):179–92. doi: 10.2174/157340308785160570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schiffer D, Annovazzi L, Casalone C, Corona C, Mellai M. Glioblastoma: microenvironment and niche concept. Cancers. 2018;11(1). doi: 10.3390/cancers11010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schiffer D, Mellai M, Bovio E, Bisogno I, Casalone C, Annovazzi L. Glioblastoma niches: from the concept to the phenotypical reality. Neurol Sci. 2018;39(7):1161–8. doi: 10.1007/s10072-018-3408-0. [DOI] [PubMed] [Google Scholar]
- 104. Sharma A, Shiras A. Cancer stem cell-vascular endothelial cell interactions in glioblastoma. Biochem Biophys Res Commun. 2016;473(3):688–92. doi: 10.1016/j.bbrc.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 105. Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 106. Hamerlik P, Lathia JD, Rasmussen R, Wu Q, Bartkova J, Lee M, Moudry P, Bartek J, Jr, Fischer W, Lukas J, Rich JN, Bartek J. Autocrine VEGF-VEGFR2-neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209(3):507–20. doi: 10.1084/jem.20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wu HB, Yang S, Weng HY, Chen Q, Zhao XL, Fu WJ, Niu Q, Ping YF, Wang JM, Zhang X, Yao XH, Bian XW. Autophagy-induced KDR/VEGFR-2 activation promotes the formation of vasculogenic mimicry by glioma stem cells. Autophagy. 2017;13(9):1528–42. doi: 10.1080/15548627.2017.1336277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 109. Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–50. doi: 10.1111/nan.12432. [DOI] [PubMed] [Google Scholar]
- 110. Pisapia DJ. The updated world health organization glioma classification: cellular and molecular origins of adult infiltrating gliomas. Arch Pathol Lab Med. 2017;141(12):1633–45. doi: 10.5858/arpa.2016-0493-RA. [DOI] [PubMed] [Google Scholar]
- 111. Polívka J, Pešta M, Pitule P, Hes O, Holubec L, Polívka J, Kubíková T, Tonar Z. IDH1 mutation is associated with lower expression of VEGF but not microvessel formation in glioblastoma multiforme. Oncotarget. 2018;9(23):16462–76. doi: 10.18632/oncotarget.24536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Khurshed M, Molenaar RJ, Lenting K, Leenders WP, van Noorden CJF. In silico gene expression analysis reveals glycolysis and acetate anaplerosis in IDH1 wild-type glioma and lactate and glutamate anaplerosis in IDH1-mutated glioma. Oncotarget. 2017;8(30):49165–77. doi: 10.18632/oncotarget.17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Molenaar RJ, Maciejewski JP, Wilmink JW, van Noorden CJF. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene. 2018;37(15):1949–60. doi: 10.1038/s41388-017-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1369_0022155419878416 for Similarities Between Stem Cell Niches in Glioblastoma and Bone Marrow: Rays of Hope for Novel Treatment Strategies by Vashendriya V.V. Hira, Barbara Breznik, Miloš Vittori, Annique Loncq de Jong, Jernej Mlakar, Roelof-Jan Oostra, Mohammed Khurshed, Remco J. Molenaar, Tamara Lah and Cornelis J.F. Van Noorden in Journal of Histochemistry & Cytochemistry