Abstract
BACKGROUND: ESR1 mutations are frequently detected in ER+ MBC, and have been reported to be associated with endocrine therapy resistance. However, there are little researches to validate whether dynamic monitoring of ESR1 mutations could serve as a predictive plasma biomarker of acquired resistance to endocrine therapy. Therefore, in this study, we performed longitudinal circulating tumor DNA (ctDNA) detection to evaluate the clinical implications of monitoring ESR1 mutations. METHODS: We performed longitudinal dynamic mutation analyses of plasma samples from 45 patients with metastatic breast cancer (MBC) and sequencing paired biopsy tissues, using a targeted NGS panel of 425 genes. These patients were treated at the Second Affiliated Hospital of Dalian Medical University between January 2017 and February 2019 with written informed consent. RESULTS: Mutations profiles were highly concordant between plasma and paired tissue samples from 45 MBC patients (r = 0.96, P < 0.0001). ESR1 mutations were enriched in ER+ MBC patients after AI therapy (17.8%, 8/45). The median time from AI endocrine therapies to the initial detection of ESR1 mutation was 39 months (95% CI 21.32–57.57). Some hotspot mutations (Y537S (n = 5), Y537N (n = 1), D538G (n = 2), E380Q (n = 2)) and several rare mutations (L345SfsX7, 24fs, G344delinsGC) were identified in our cohort. In addition, we observed that two patients obtained multiple ESR1 mutations over the course of treatment (Y537N/Y537S/D538G, L345SfsX7/24fs/E380Q). Through dynamically monitoring ESR1 mutations by ctDNA, we demonstrated that the change of allele frequency of ESR1 mutations was an important biomarker, which could predict endocrine resistance of ER+ MBC in our study. We also observed that the combination of everolimus in four cases with acquired ESR1 mutations showed longer PFS than other therapies without everolimus. CONCLUSION: The dynamic monitoring of ESR1 mutations by ctDNA is a promising tool to predict endocrine therapy resistance in ER+ MBC patients.
Introduction
Breast cancer is the most common cancer and the leading cause of cancer death in females globally [1]. It is classified into several molecular subgroups according to hormone receptor and HER2 status. Approximately 70% of breast cancers are estrogen receptor (ER) positive [2], making ER is a key therapeutic target in ER+ breast cancer. Current endocrine therapies primarily target either the ER protein or the ligand binding domain of the receptor [[1], [2], [3]], including (1) selective estrogen receptor modulators (SERMs) that directly inhibit the ER with mixed agonistic/antagonistic activities, such as tamoxifen, (2) selective ER degraders (SERDs), which are more potent antiestrogens and induce ER protein degradation, such as fulvestrant, and (3) aromatase inhibitors (AIs) that block the production of estrogen, such as letrozole or anastrozole [3]. In early and metastatic breast cancer (MBC), endocrine therapies have been proven to improve survival and reduce recurrence [4]. Nevertheless, endocrine resistance is inevitable after prolonged exposure to endocrine therapies [3]. Over the past few years, various mechanisms of endocrine resistance have been discovered, such as loss of ER expression, deregulation of apoptosis and cell cycle signaling, and hyperactive receptor tyrosine kinase (RTK) [5]. Recently, several preclinical studies have demonstrated that mutations in the ER encoding gene, ESR1 are enriched in ER+ MBC patients, while rarely present in primary tumor tissue. ESR1 mutations may account for resistance to aromatase inhibitors [[6], [7], [8], [9]]. However, we are lacking of studies to confirm whether ESR1 mutations could be used to provide a real-time dynamic monitoring of tumor evolution and therapeutic efficacy for ER+ MBC patients who are resistant to endocrine therapy. Therefore, in this study, we performed longitudinal genomic analyses of plasma samples from 45 MBC patients to explore potential clinical utility of tracking ESR1 mutations using circulating tumor DNA (ctDNA).
Materials and Methods
Patients and Samples
A total of 45 MBC patients, treated at the Second Hospital of Dalian Medical University between January 2017 and February 2019, were enrolled in this study. We retrospectively collected the available paraffin-embedded tumor tissues, fresh frozen metastasis tissues, and cerebrospinal fluid (CSF); multiple plasma samples were also drawn for the ctDNA analysis. Basically, routine ctDNA analyses were performed approximately every 3 months during the follow-up period, along with tumor markers and/or imaging diagnosis. The clinicopathological features at the initial diagnosis of the enrolled patients are summarized in Table 1.
Table 1.
Clinical Characteristics of the 45 Enrolled MBC Patients
| Variables | No. of Patients (%) (N = 45) |
|---|---|
| Age at Breast Cancer Diagnosis | |
| Median (range) | 48(27–77) |
| Receptor Subtype | |
| ER+/Her2− | 24(53.3%) |
| ER+/Her2+ | 5(11.1%) |
| ER−/Her2+ | 8(17.8%) |
| Triple negative | 8(17.8%) |
| Histology Subtype (Primary tumor) | |
| Invasive ductal | 36(80.0%) |
| Invasive lobular | 2(4.4%) |
| Other | 3(6.7%) |
| Unknown | 4(8.9%) |
| Primary Clinical Stage | |
| I | 3(6.7%) |
| II | 11(24.4%) |
| III | 19(42.2%) |
| IV | 5(11.1%) |
| Unknown | 7(15.6%) |
| Overall Survival | |
| Median (range) month | 79.3(10.8–371.9) |
Median age at primary breast cancer was 48 years (range 22–77 years). The primary breast cancer showed ductal (36, 80%), lobular (2, 4.4%) and other histological subtype (3, 6.7%), except four patients with unavailable tissues. At the clinical stages of primary tumors, 3 patients (6.7%) were categorized as stage I, 11 (24.4%) as stage II, 19 (42.2%) as stage III, and 5 (11.1%) as stage IV, but 7 patients were unknown. The median overall survival was 79.3 months (range, 10.8–371.9) in our cohort. Informed consent was obtained from all patients, and this study was approved by the Ethics Committee of the Second Hospital of Dalian Medical University.
Sample Preparation and Next Generation Sequencing
The paraffin-embedded FFEP white slices or rolled slices without gel and baking, at least 4 four tissue sections with tumor cell proportion of >60% under the microscope, were screened. Five milliliters of peripheral blood was collected by EDTA blood collection tubes and then centrifuged within 1 h of collection at 1800×g for 10 min at 4≥°C or RT to remove blood cells. The supernatant containing the plasma was removed with special care taken as to not disturb the buffy coat. This was then centrifuged at 16,000×g for 10 min to remove any remaining cells. ctDNA was extracted from 2 ml plasma, by digestion in 100 μl proteinase K buffer for 10 min at 37 °C followed by purification with the NucleoSpin Plasma XS kit with modified protocols. The purified ctDNA is quantified by a PicoGreen fluorescence assay using the provided lambda DNA standards (Invitrogen). Then, library construction with the KAPA Hyper DNA Library Prep Kit, containing mixes for end repair, dA addition, and ligation, were performed in 96-well plates (Eppendorf). Dual-indexed sequencing libraries are PCR amplified for 4–7 cycles. Gene panel containing 425 breast cancer related genes was used to monitor the change trend of abundance fraction (Nanjing Geneq Tecnology Inc). The next generation sequencing (NGS), also known as large-scale parallel sequencing, was performed using Illumina Hiseq Technology (Nanjing Geneq Tecnology Inc).
Statistical Analysis
The correlation of primary tumor tissue samples and paired plasma samples of ctDNA was assessed by the Fisher exact test. Overall survival (OS) was performed using the Kaplan Meier method and the long rank test. P value <0.05 was considered statistically significant. All statistical analyses were two-sided and performed by GraphPad Prism 7 (GraphPad Software).
Results
The Genomic Characteristics of MBC Patients in Our Cohort
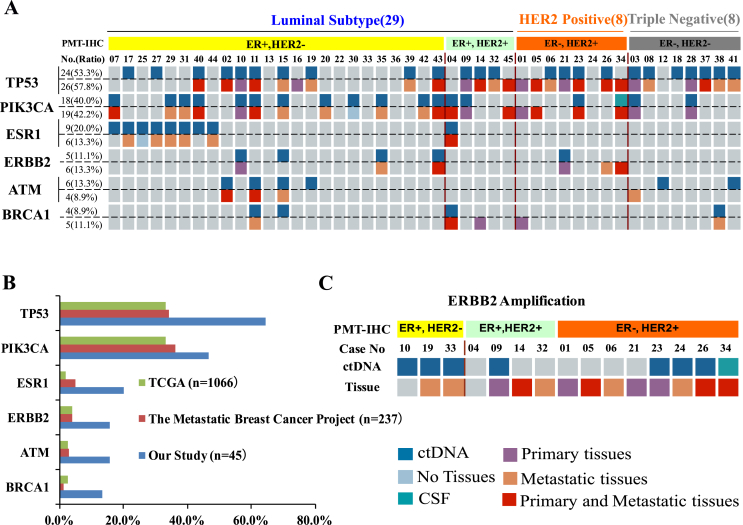
In our study, we retrospectively analyzed mutation profiles of paired plasma and tissue biopsy samples, falling into three distinct molecular subtypes including luminal (n = 29), HER2 positive (n = 8) and triple negative subtypes (n = 8) (Table 1). Six recurrent genes {TP53 (29/45, 64.4%), PIK3CA (21/45, 46.7%), ESR1 (9/45, 20%), ERBB2 (7/45, 15.6%), ATM (7/45, 15.6%), BRCA1 (6/45, 13.3%)} were mutated in more than 13% of patients. The landscape of these highly frequent molecular alterations detected by plasma ctDNA and paired tissue samples is shown for individual patients (Figure 1A). Two of them had no enough tissue because of inadequate tissue or biopsy contraindications.
Figure 1.
(A) The landscape of high-frequency genomic alterations from 45 MBC patients detected in plasma and paired tissue samples is presented. The molecular subtyping and the patient ID numbers were at the up panel. For each gene, alteration in both tissue (down) and plasma (up) were presented, with percentage of prevalence shown on the left. (B) The percentage of mutations detected in our patient samples was compared with that reported in the cBioPortal (https://www.cbioportal.org). (C) The status of HER2 expression was measured by immunohistochemistry (IHC). HER2 gene amplification in tissue and ctDNA was detected by NGS.
ESR1 mutations were detected exclusively in the ER+ luminal subtypes, comprising 8 ER+/HER2− and 1 ER+/HER2+ patients (Figure 1A). The overall prevalence of ESR1 mutations was 20%, consistent with Jeselsohn's research in 2013 (39/187, 21%) [6]. In addition, ERBB2 mutations were detected in seven samples, including 4 ER+/HER2− and 3 ER−/HER2+ patients (Figure 1A). Subsequently, we investigated whether the frequency of gene mutations in our cohort is similar with TCGA breast cancer database (Figure 1B). Our six recurrent genes presented significantly higher frequency (Figure 1B). This discrepancy may be partially because of the smaller population in our study.
Evaluation of the Consistency Between Paired Tissue and Plasma Samples
Anti-HER2 therapies have changed the natural biology of HER2 positive breast cancer [10]. Hence, HER2 assessment is very important in the clinic. Next, we compared the expression of HER2 detected by immunohistochemistry (IHC) and HER2 amplification detected by plasma or tissue-based NGS. In our cohort, 13 patients were found to overexpress HER2 by IHC (Figure 1A and C). Among them, five patients were ER+/HER2+, eight were ER−/HER2+, and one was removed from ER+/HER2+ as unavailable plasma tissue (Figure 1C). HER2 amplification was observed in 13(28.9%) tissues, 7(15.6%) plasma, and 1(2.2%) CSF samples. Of these, seven patients were HER2 amp positive for both tissue and liquid biopsy samples (Figure 1C). Furthermore, three samples negative for HER2 by IHC showed HER2 amplification by plasma or tissue-based NGS. Thus, tissue-based sequencing and plasma-based ctDNA might provide extra insights into HER2 status in MBC patients.
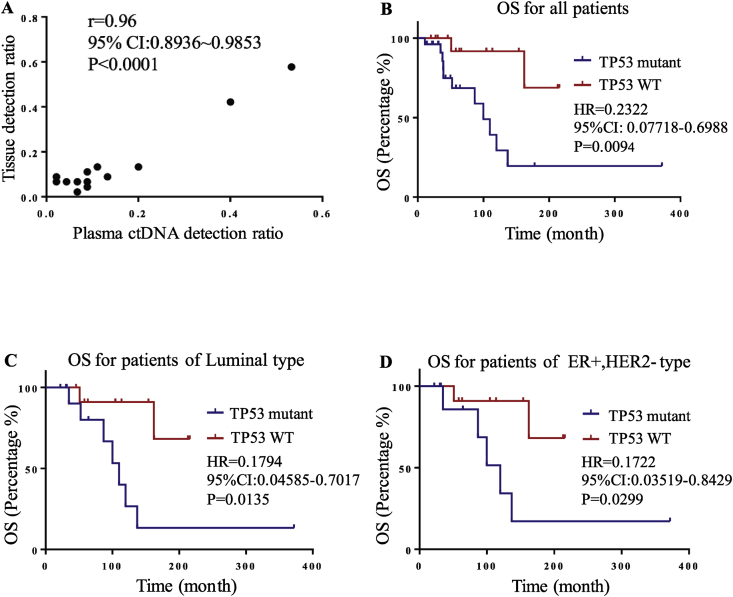
Overall, high concordance was seen in the mutation profiles between tissue and plasma samples (R = 0.96, P < 0.0001) (Figure 2A).
Figure 2.
(A) Correlation analysis between paired tissue and plasma samples. (B∼D) Overall survival analysis according to the state of TP53 mutation.
TP53 Mutations is correlated with poor prognosis
We next assessed whether recurrent gene mutations could predict prognosis in our study. We observed that MBC patients with TP53 mutations had significantly worse OS than the carriers of wild type alleles (P = 0.0094) (Figure 2B). In the luminal subtype or ER+/HER2− subgroup, the presence of TP53 mutations also demonstrated worse OS than wild type patients (P = 0.0135, P = 0.0299) (Figure 2C and D). However, in PIK3CA or ESR1 mutation subgroups, we did not observe any prognostic significance in our cohort, which may be related to the limited sample size in these subgroups.
The Clinical Characteristics of ESR1 Mutations in ER+ MBC
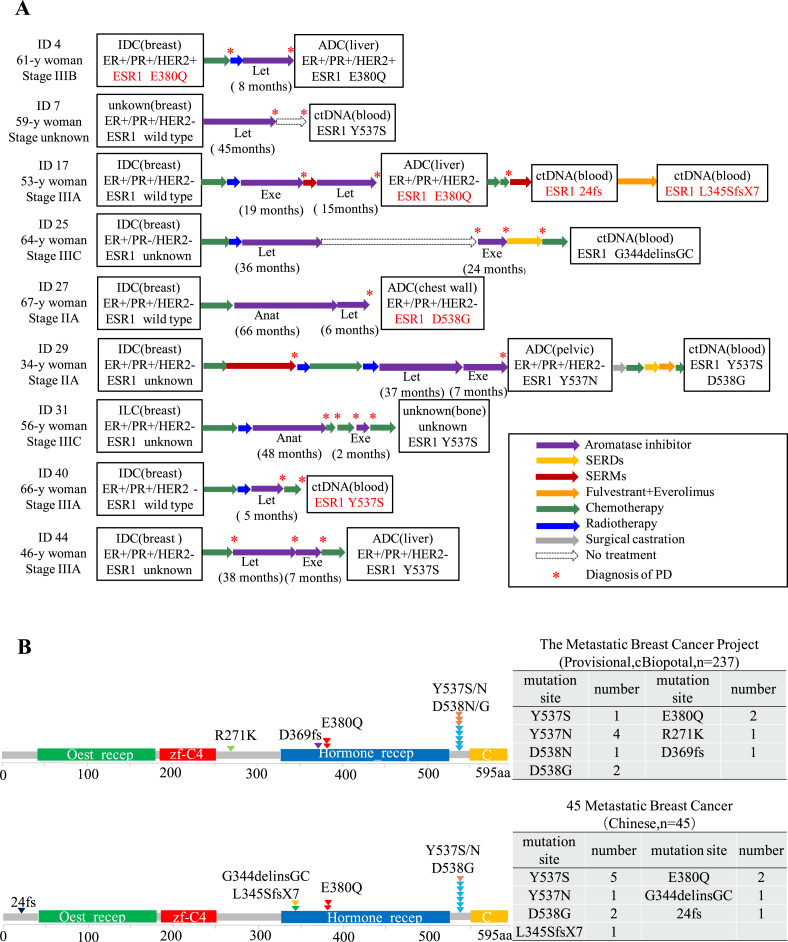
In our study, ESR1 mutations were detected in 20% (9/45) of MBC patients, and all of them were ER+ MBC (Figure 1A). We performed NGS on primary tumor biopsies collected before exposure to any therapies and paired metastatic biopsies in nine patients. To investigate whether ESR1 mutations were acquired after endocrine treatments, we found that 8 patients with ESR1 mutations received aromatase inhibitors before metastasis (except one with mutation in primary cancer) (Figure 3A). Of the 9 patients, ESR1 Y537S (n = 5), Y537N (n = 1), and D538G (n = 2) mutations that located close to the hormone receptor were identified in post-AI samples. E380Q (another LBD mutation) was identified in one baseline and one post-AI tumor samples. We have also uncovered a few rare mutations (L345SfsX7, 24fs, G344delinsGC) in our cohort after SERD or SERM treatment in two patients (Patient 17 and 25) (Figure 3A). Overall, the mutation sites of ESR1 gene in our study were consistent with the TCGA database (Provisional cBioPortal, The Metastatic Breast Cancer Project) (Figure 3B). In addition, concurrent ESR1 mutations were observed in Case29 (Y537N/Y537S/D538G) and Case17 (L345SfsX7/24fs/E380Q) during the course of treatment, which may be associated with the progression of disease and/or drug resistance (Table 2). These results suggested that ESR1 mutations were recurrently enriched in ER+ MBC patients, but were rarely present in primary tumor tissues. The median time from AIs endocrine therapies to the first detection of ESR1 mutations was 39 months (95% CI 21.32–57.57) (Figure 3A). Detailed AI treatment history for all 9 patients is presented in Table 2.
Figure 3.
(A) The clinical treatment courses of the nine patients with ESR1 mutations were summarized. (B) Region of ESR1 mutations detected in our samples (down panel) compared with that reported in the database (The Metastatic Breast Cancer Project, n = 237) (https://www.cbioportal.org) (up panel).
Table 2.
Clinical Characteristics of Nine ER-positive Metastatic Breast Cancer Patients with ESR1 Mutations
| Case No. | Age at Diagnosis | ER/PR/HER-2 | Histologic Type | ESR1-Mutation Types | Prevenient Endocrine Treatment | AI Treatment Overall Time (month) |
|
|---|---|---|---|---|---|---|---|
| 4 | 61 | +/+/+ | Invasive ductal | E380Q | - | 0 | |
| 7 | 59 | +/+/− | Unknown | Y537S | AI | 45 | |
| 17 | 53 | +/+/− | Invasive ductal | E380Q, 24fs, L345SfsX7 | AI, SERMs, SERDs | 34 | |
| 25 | 64 | +/−/- | Invasive ductal | G344 delins GC | AI, SERDs | 60 | |
| 27 | 67 | +/+/− | Invasive ductal | D538G | AI, SERMs, SERDs | 72 | |
| 29 | 34 | +/+/− | Invasive ductal | Y537S, Y537N, D538G | AI, SERMs, SERDs | 44 | |
| 31 | 56 | +/+/− | Invasive lobular | Y537S | AI, SERDs | 50 | |
| 40 | 66 | +/+/− | Invasive ductal | Y537S | AI | 5 | |
| 44 | 46 | +/+/− | Invasive ductal | Y537S | AI | 45 | |
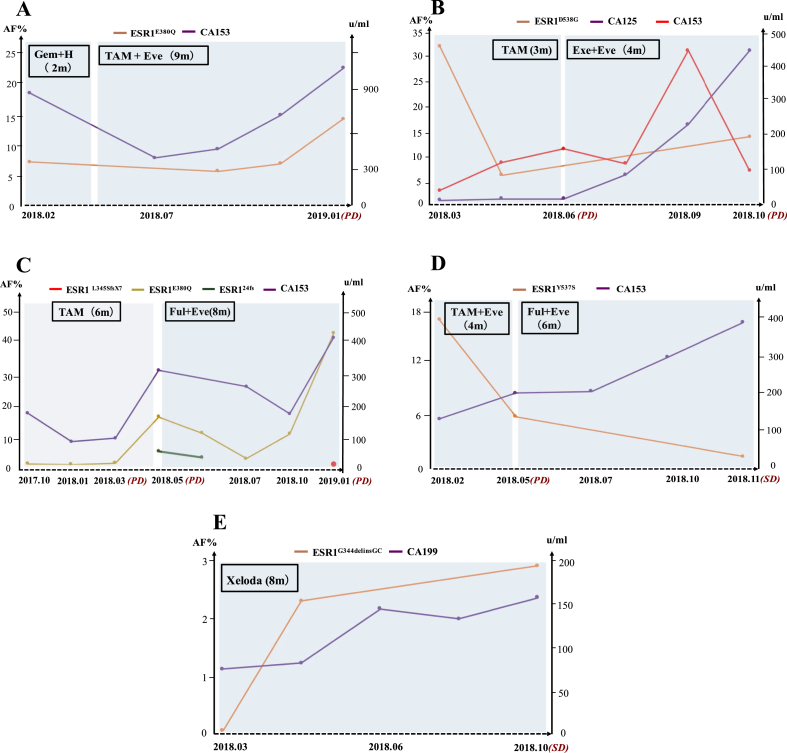
Monitoring ESR1 Mutations
ESR1 mutational status of five ER+ MBC patients was longitudinally monitored by ctDNA during the detailed clinical course. The tumor load was estimated by imaging using RECIST V.1.1. The serum tumor biomarker and ESR1 mutation levels were presented (Figure 4). Overall, dynamically monitoring ESR1 mutations by ctDNA might predict endocrine resistance of ER+ MBC, which was in consistent with dynamic change trend of serum tumor marker CA153/CA199/CA125 (Figure 4). The four ER+ MBC patients treated with the combination of everolimus showed longer PFS than other therapies without everolimus (Figure 4A–D). Strikingly, capecitabine alone also displayed excellent efficacy to against ESR1 mutation (ESR1.G344delinsGC) (Figure 4E). Together, monitoring ESR1 mutations by ctDNA might allow for cessation of ineffective endocrine therapies, and switch to other treatments without the need for tissue biopsy or before the emergence of metastatic disease.
Figure 4.
(A∼E) Clinical timelines for the five representative ER-positive metastatic breast cancer patients harboring ESR1 mutations. The dynamic changes of ESR1 mutation levels were compared with serum tumor marker levels along with treatment.
Discussion
In this study, we performed NGS on plasma and paired tissue samples from MBC patients. Mutation profiles from tissue and plasma samples for these 45 patients were highly concordant. The overall survival seemed to be shorter in MBC patients with TP53 mutations than in TP53 wild type patients, which is in accordance with the analyses in the TP53 mutation-driven subgroup such as luminal subtype or ER+/HER2− subgroup. We also found acquired ESR1 mutations in eight ER+ MBC patients after AI therapy administered, and one primary ESR1 mutation in the baseline tumor tissue. We also demonstrated that dynamically monitoring the change of ESR1 mutations by ctDNA was an important biomarker, which could predict endocrine resistance of ER+ MBC in our study.
ESR1 mutation was first described in a metastatic breast cancer in 1997 [11]. Recently, in several retrospective cohorts, ESR1 mutations have been reported in approximately 11%–55% of ER+ MBC patients treated with AIs, but were found to be extremely rare in primary breast cancers [[6], [7], [8],[12], [13], [14]]. In this study, we also observed the similar incidence rate of ESR1 mutations. In addition, we noticed that the mutation sites of ESR1 gene in our cohort were enriched on Y537 and D538, including missense mutations Y537S, Y537N, D538G, and E380Q. These common ESR1 mutations, located in ligand binding domain (LBD) of ER between amino acids 537–538, account for 74% of all mutations [12]. In the PALOMA-3 trial, the ESR1 mutations in 360 ER-positive MBC patients were reported, including Y537S (23/360, 6%), Y537N (14/360, 4%), Y537C (5/360, 1%), and D538G (51/360, 14%) etc. Furthermore, the similar incidence of ESR1 mutations was also observed in the FERGI, the BOLERO-2 and the SoFEA study [14]. Interestingly, breast cancer patients with the Y537S mutation have a shorter overall survival than patients with the D538G mutation [15]. In this study, we also detected some rare mutations (L345SfsX7, 24fs, G344delinsGC) located outside the LBD's hotspot regions of the ESR1 gene, which has been scarcely described in previous studies or database (Figure 3B).
Previous research showed that acquired ESR1 mutations in ctDNA are rare before adjuvant AI treatments, but are frequently selected by AI therapies for metastatic disease, which provide molecular mechanism of resistance to AI therapy [[16], [17], [18], [19], [20], [21], [22]]. It is worth noting that how long to detect ESR1 mutations after AI exposure would be clinically useful. In this study, we observed that ESR1 mutations became detectable after receiving AIs endocrine therapies for approximately 39 months (95% CI 21.32–57.57) (Figure 3A and Table 2). Together, ESR1 mutations should be assessed before the progress of the disease but not after relapse.
We observed two cases with multiple ESR1 mutations (Case29: Y537N/Y537S/D538G, Case 17: L345SfsX7/24fs/E380Q) (Table 2) by longitudinal analysis of ctDNA. These different types of ESR1 mutations were sequentially acquired during the course of treatment, possibly reflecting tumor heterogeneity or the selection pressure evolution under endocrine treatment. Several longitudinal analyses of ctDNA indicated that ESR1 gene could acquire polyclonal mutations over the course of treatment, possibly reflecting differential response of individual ESR1 mutations to treatments [23,24].
When everolimus was added to tamoxifen, exemestane, or fulvestrant, this resulted in an improved median PFS in ESR1-mutated patients, who previously received other kinds of therapy in our study. Indeed, the previous phase III BOLERO-2 study suggested that ESR1-mutated patients could still benefit from the addition of everolimus [14,23]. In addition, the appearance of ESR1 mutations was proven to have prognostic and predictive significance of poorer outcome on subsequent endocrine treatment [[25], [26], [27]]. Preclinical studies demonstrated the activation of the PI3K pathway after long-term estrogen deprivation of ER+ breast cancer cells, suggesting the effectiveness of combination of mTOR and ER inhibitions [28]. Accordingly, it provides a theoretical basis for mTOR inhibitor in combination with endocrine therapy, which could significantly improve progression-free survival (PFS) of the MBC patients failing previous endocrine therapies [29]. However, the interaction between the presence of ESR1 mutations and activation of the PI3K/AKT/mTOR still needs further research.
It has been shown that monitoring of mutations in ctDNA is a feasible and useful method to predict potential disease progression in MBC patients [30,31]. Some researchers monitored treatment response over time using NGS-based approach, and found that dynamic of ctDNA levels was correlated with changes of CA15-3 or circulating tumor cells in advanced breast cancer patients [[32], [33], [34]]. In this study, our results further demonstrated that dynamically monitoring the change of ESR1 mutations by ctDNA could predict endocrine resistance of ER+ MBC. However, our study was a retrospective, single-institute study with a relatively small patient number, thus a larger sample study is required to confirm our findings.
Ethical Approval
Informed consent was obtained from each patient. This study was reviewed and approved by the Ethics Committee of The Second Hospital of Dalian Medical University.
Authors' Contributions
Conception and design: Xuelu Li, Zuowei Zhao, Man Li.
Acquisition of data: Xuelu Li, Jiawei Lu, Lanxin Zhang, Yaoting Luo.
Analysis and interpretation of data: Xuelu Li, Jiawei Lu, Lanxin Zhang, Yaoting Luo.
Writing, review, and revision of the manuscript: Xuelu Li, Jiawei Lu, Lanxin Zhang, Yaoting Luo, Zuowei Zhao, Man Li.
Study supervision: Zuowei Zhao, Man Li.
Conflict of Interest
All authors have stated that they have no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NO. 81673762 to Zuowei Zhao, No. 81872156 to Man Li), Provincial Foundation of Liaoning (NO. LR2017012 to Zuowei Zhao, NO. 2019-BS-072 to Xuelu Li), and Innovation Foundation of Dalian (NO. 2018J11CY026 to Zuowei Zhao).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.11.007.
Contributor Information
Zuowei Zhao, Email: dmuzhaozuowei@163.com.
Man Li, Email: dmuliman@163.com.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel RL R.L., Torr L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ariazi E.A., Ariazi J.L., Cordera F., Jordan V.C. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006 February;6(3):181–202. [PubMed] [Google Scholar]
- 3.Williams N., Harris L.N. The renaissance of endocrine therapy in breast cancer. Curr Opin Obstet Gynecol. 2014 Feb;26(1):41–47. doi: 10.1097/GCO.0000000000000039. [DOI] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists' Collaborative Group Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005 May;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 5.Rani A., Stebbing J., Giamas G., Murphy J. Endocrine resistance in hormone receptor positive breast cancer–from mechanism to therapy. Front Endocrinol. 2019 May;10 doi: 10.3389/fendo.2019.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeselsohn R., Buchwalter G., De Angelis C., Brown M., Schiff R. ESR1 mutations—a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol. 2015 Oct;12(10):573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson D.R., Wu Y.M., Vats P., Su F., Lonigro R.J., Cao X., Kalyana-Sundaram S., Wang R., Ning Y., Hodges L. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013 Dec;45(12):1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toy W., Shen Y., Won H., Green B., Sakr R.A., Will M., Li Z., Gala K., Fanning S., King T.A. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013 Dec;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.hou D., Ouyang Q., Liu L., Liu J., Tang Y., Xiao M., Wang Y., He Q., Hu Z.Y. Chemotherapy modulates endocrine therapy-related resistance mutations in metastatic breast cancer. Transl Oncol. 2019 May;12(5):764–774. doi: 10.1016/j.tranon.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slamon D., Eiermann W., Robert N., Pienkowski T., Martin M., Press M., Mackey J., Glaspy J., Chan A., Pawlicki M. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011 Oct;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q.X., Borg A., Wolf D.W., Oesterreich S., Fuqua S.A. An estrogen receptor mutant with strong hormone-independent activity from a metastatic breast cancer. Cancer Res. 1997 Apr;57:1244–1249. [PubMed] [Google Scholar]
- 12.Segal C.V., Dowsett M. Estrogen receptor mutations in breast cancer—new focus on an old target. Clin Cancer Res. 2014 Apr;20(7):1724–1726. doi: 10.1158/1078-0432.CCR-14-0067. [DOI] [PubMed] [Google Scholar]
- 13.Li S., Shen D., Shao J., Crowder R., Liu W., Prat A., He X., Liu S., Hoog J., Lu C. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013 Sep;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeselsohn R., Yelensky R., Buchwalter G., Frampton G., Meric-Bernstam F., Gonzalez-Angulo A.M., Ferrer-Lozano J., Perez-Fidalgo J.A., Cristofanilli M., Gómez H. Emergence of constitutively active estrogen receptor-a mutations in pretreated advanced estrogen receptor- positive breast cancer. Clin Cancer Res. 2014 Apr;20:1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reinert T., Saad E.D., Barrios C.H., Bines J. Clinical implications of ESR1 mutations in hormone receptor-positive advanced breast cancer. Front Oncol. 2017 Mar;7(26) doi: 10.3389/fonc.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiavon G., Hrebien S., Garcia-Murillas I., Cutts R.J., Pearson A., Tarazona N., Fenwick K., Kozarewa I., Lopez-Knowles E., Ribas R. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl. 2015 Nov;7 doi: 10.1126/scitranslmed.aac7551. 313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross D.S., Zehir A., Brogi E., Konno F., Krystel-Whittemore M., Edelweiss M., Berger M.F., Toy W., Chandarlapaty S., Razavi P. Immunohistochemical analysis of estrogen receptor in breast cancer with ESR1 mutations detected by hybrid capture-based next-generation sequencing. Mod Pathol. 2019 Jan;32(1):81. doi: 10.1038/s41379-018-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allouchery V., Beaussire L., Perdrix A., Sefrioui D., Augusto L., Guillemet C., Sarafan-Vasseur N., Di Fiore F., Clatot F. Circulating ESR1 mutations at the end of aromatase inhibitor adjuvant treatment and after relapse in breast cancer patients. Breast Canc Res. 2018 May;20(1):40. doi: 10.1186/s13058-018-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahreini A., Li Z., Wang P., Levine K.M., Tasdemir N., Cao L., Weir H.M., Puhalla S.L., Davidson N.E., Stern A.M. Mutation site and context dependent effects of ESR1 mutation in genome-edited breast cancer cell models. Breast Canc Res. 2017 May;19(1):60. doi: 10.1186/s13058-017-0851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beije N., Sieuwerts A.M., Kraan J., Van N.M., Onsten W., Vitale S.R., van der Vlugt-Daane M., Dirix L.Y., Brouwer A., Hamberg P. Estrogen receptor mutations and splice variants determined in liquid biopsies from metastatic breast cancer patients. Mol Oncol. 2018 Aug;12(1):48–57. doi: 10.1002/1878-0261.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busonero C., Leone S., Bartoloni S., Acconcia F. Strategies to degrade estrogen receptor α in primary and ESR1 mutant-expressing metastatic breast cancer. Mol Cell Endocrinol. 2019 Jan;15(480):107–121. doi: 10.1016/j.mce.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Lei J.T., Gou X., Seker S., Ellis M.J. ESR1 alterations and metastasis in estrogen receptor positive breast cancer. J Cancer Metastasis Treat. 2019 May;38 doi: 10.20517/2394-4722.2019.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandarlapaty S., Chen D., He W., Sung P., Samoila A., You D., Bhatt T., Patel P., Voi M., Gnant M. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer. JAMA Oncol. 2016 Oct;2(10):1310–1315. doi: 10.1001/jamaoncol.2016.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeshita T., Yamamoto Y., Yamamoto-Ibusuki M., Tomiguchi M., Sueta A., Murakami K., Omoto Y., Iwase H. Comparison of ESR1 mutations in tumor tissue and matched plasma samples from metastatic breast cancer patients. Transl Oncol. 2017 Oct;10(5):766–771. doi: 10.1016/j.tranon.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Knowles E., Pearson A., Schuster G., Gellert P., Ribas R., Yeo B., Cutts R., Buus R., Garcia-Murillas I., Haynes B. Molecular characterisation of aromatase inhibitor-resistant advanced breast cancer: the phenotypic effect of ESR1 mutations. Br J Cancer. 2019 Jan;120(2):247–255. doi: 10.1038/s41416-018-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K., Hong R., Xu F., Xia W., Kaping L., Qin G., Zheng Q., Lu Q., Shi Y.X., Yuan Z.Y. Clinical value of circulating ESR1 mutations for patients with metastatic breast cancer: a meta-analysis. Cancer Manag Res. 2018 Aug;10:2573–2580. doi: 10.2147/CMAR.S173193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du Y., Li N., Jiao X., Li K., Yan S. The predictive ability of plasma ESR1 mutations for the efficacy of endocrine therapy in hormone-receptor-positive advanced breast cancer. Onco Targets Ther. 2018 Sep;11:6023–6029. doi: 10.2147/OTT.S171465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez C.G., Ma C.X., Crowder R.J., Guintoli T., Phommaly C., Gao F., Lin L., Ellis M.J. Preclinical modeling of combined phosphatidylinositol-3-kinase inhibition with endocrine therapy for estrogen receptor-positive breast cancer. Breast Cancer Res. 2011 Mar;13(2):R21. doi: 10.1186/bcr2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baselga J., Campone M., Piccart M., Burris H.A., Rugo H.S., Sahmoud T., Noguchi S., Gnant M., Pritchard K.I., Lebrun F. Everolimus in postmenopausal hormone-receptor–positive advanced breast cancer. N Engl J Med. 2011 Feb;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeshita T., Yamamoto Y., Yamamoto-Ibusuki M., Inao T., Sueta A., Fujiwara S., Omoto Y., Iwase H. Clinical significance of monitoring ESR1 mutations in circulating cell-free DNA in estrogen receptor positive breast cancer patients. Oncotarget. 2016 May;7(22):32504–32518. doi: 10.18632/oncotarget.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttery D.S., Page K., Hills A., Woodley L., Marchese S.D., Rghebi B., Hastings R.K., Luo J., Pringle J.H., Stebbing J. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor–positive metastatic breast cancer. Clin Chem. 2015 Jul;61(7):974–982. doi: 10.1373/clinchem.2015.238717. [DOI] [PubMed] [Google Scholar]
- 32.Dawson S.J., Rosenfeld N., Caldas C. Circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013 Jul;369(1):93–94. doi: 10.1056/NEJMc1306040. [DOI] [PubMed] [Google Scholar]
- 33.Yanagawa T., Kagara N., Miyake T., Tanei T., Naoi Y., Shimoda M., Shimazu K., Kim S.J., Noguchi S. Detection of ESR1 mutations in plasma and tumors from metastatic breast cancer patients using next-generation sequencing. Breast Cancer Res Treat. 2017 Jun;163(2):231–240. doi: 10.1007/s10549-017-4190-z. [DOI] [PubMed] [Google Scholar]
- 34.Dawson S.J., Tsui D.W., Murtaza M., Biggs H., Rueda O.M., Chin S.F., Dunning M.J., Gale D., Forshew T., Mahler-Araujo B. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013 Mar;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.