Abstract
Simultaneous inhibition of multiple molecular targets is an established strategy to improve the continuance of clinical response to therapy. Here, we screened 49 molecules with dual nanomolar inhibitory activity against BRD4 and PLK1, best classified as dual kinase-bromodomain inhibitors, in pediatric tumor cell lines for their antitumor activity. We identified two candidate dual kinase-bromodomain inhibitors with strong and tumor-specific activity against neuroblastoma, medulloblastoma, and rhabdomyosarcoma tumor cells. Dual PLK1 and BRD4 inhibitor treatment suppressed proliferation and induced apoptosis in pediatric tumor cell lines at low nanomolar concentrations. This was associated with reduced MYCN-driven gene expression as assessed by RNA sequencing. Treatment of patient-derived xenografts with dual inhibitor UMB103 led to significant tumor regression. We demonstrate that concurrent inhibition of two central regulators of MYC protein family of protooncogenes, BRD4, and PLK1, with single small molecules has strong and specific antitumor effects in preclinical pediatric cancer models.
Introduction
Solid tumors are one of the major causes of cancer-related death in children [1]. The MYC family of transcription factors, in particular MYC and MYCN (MYC/N), are one of the most highly investigated oncogenic drivers in high-risk pediatric solid tumors. Amplification of MYC/N often occurs in neuroblastoma, medulloblastoma, and rhabdomyosarcoma, which are among the most prevalent pediatric solid tumors [[2], [3], [4]]. MYC/N amplification is a feature associated with adverse prognosis and is used to clinically stratify patients suffering from neuroblastoma and medulloblastoma [5,6]. In rhabdomyosarcoma, MYC/N is often either genomically amplified or expressed at high levels because of the activity of the PAX3-FOXO1 fusion oncoprotein [7]. Similar as in neuroblastoma and medulloblastoma, MYC/N amplification is associated with adverse prognosis in rhabdomyosarcoma [7]. The means by which MYC/N contributes to features of high-risk tumors are still largely unclear. MYC/N overexpression has pleiotropic effects and can lead to dysregulation of cell proliferation, differentiation, and cell cycle progression leading to increased tumor growth, thus representing an attractive therapeutic target [8].
Given its importance for cancer, the regulation of oncogene expression through transcriptional and posttranscriptional modes is a matter of extensive research([9]). Transcription of oncogenes appears to require regulation by an increased number and size of enhancer elements. Large enhancer regions, called super-enhancers, are abundant nearby genes encoding for lineage-specific transcription factor genes in normal cells and can be found near oncogenes such as MYC/N and PAX3-FOXO1 in cancer cells [7,10]. Regulation of oncogenes by super-enhancers can be achieved through enhancer translocations or other, as of yet unknown mechanisms [11]. Bromodomain-containing protein 4 (BRD4) has emerged as an important factor in super-enhancer recognition through its binding to acetylated lysine K27 of histone 3 [12]. BRD4 is essential for transcriptional regulation of super-enhancer–associated genes [12,13]. When being regulated by large super enhancers, MYC, MYCN, and PAX3-FOXO1 transcription can be potently repressed by pharmacological inhibition of BRD4 [7,14,15]. Although pharmacological BRD4 inhibition shows promising effects against preclinical models of pediatric solid tumors, single agent treatment is not curative in most cases treated so far [[14], [15], [16], [17], [18], [19]].
Simultaneous inhibition of multiple therapeutic targets is an established strategy to improve the durability of clinical responses to targeted therapy. We and others have recently described that combined treatment with small molecule BRD4 and polo-like kinase 1 (PLK1) inhibitors has synergistic antitumor effects in medulloblastoma and acute lymphoid leukaemia [20,21]. PLK1 is overexpressed in pediatric solid tumors and high expression is associated with poor prognosis [[22], [23], [24]]. PLK1 positively regulates the stability of many oncoproteins, including MYC and MYCN and PAX3-FOXO1. PLK1 does so, by increasing ubiquitin ligase SCFFbw7 degradation, thereby preventing SCFFbw7–mediated MYC degradation [25]. Additionally, PLK1-mediated phosphorylation of FOXO1 at Serine 503 directly prevents PAX3-FOXO1 degradation [23]. Consequently, simultaneous inhibition of BRD4 and PLK1 can lead to strong repression of MYC and MYCN mRNA and protein expression resulting in profound antitumor efficacy [21].
In recent years, small molecule inhibitors with dual-targeting activity have been described [[26], [27], [28]]. Intriguingly, some PLK1 kinase inhibitors can simultaneously inhibit BET protein bromodomains [[29], [30], [31], [32], [33], [34]]. Here, we extend previous evidence that combination of BRD4 and PLK1 inhibitors has synergistic antitumor effects in pediatric tumor models and provide evidence for the therapeutic activity of recently developed dual-targeting BRD4 and PLK1 inhibitors in preclinical models of pediatric tumors.
Materials and Methods
Reagents and Chemicals
Materials and reagents were obtained from Carl Roth GmbH & Co.KG unless otherwise specified. MK-8628 and Volasertib (BI 6727) were purchased from Selleckchem (Munich, Germany), GSK461364 was purchased from Cayman chemical (Michigan, US), and dual BRD4/PLK1 inhibitors (UMB analogs 88-161) were produced as previously described [30]. UMB103 corresponds to compound 12 and UMB160 corresponds to compound 23 in our previous report describing the synthesis of these compounds [30]. MK-8628, Volasertib, and UMB88-UMB161 were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mM, and stored at −20 °C for in vitro experiments.
Cell Culture
All cell lines (RH4, RH30, RD, T174, TE381.1, HD-MB03, DAOY, ONS76, UW228, IMR5, GI-ME-N, NBL-S, Kelly, LAN-1, and CHP-212) were obtained at American Type Culture Collection (ATCC, Virginia, US) if not otherwise specified. Rh41, Rh18, Rh36, TE441, and Kym1 were kindly provided by Prof. Simone Fulda. Cell lines were cultured under standard conditions in Roswell Park Memorial Institute medium 1640 (RPMI1640) or Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Calf Serum (FCS, Sigma–Aldrich, Missouri, USA) and 1% Penicillin/Streptomycin (Thermo Fisher Scientific). The identity of all cell lines was verified by STR genotyping (Genetica DNA Laboratories), and cells were periodically checked with Lonza MycoAlert system.
Cell Viability, Proliferation and Cell Cycle
Cell lines were seeded onto 96-well, white, flat bottom plates at a concentration of 500 cells per well and incubated for 24 hours to permit cell surface adherence. For examination of cell viability, cells were treated in triplicates. Final concentration of DMSO was kept below 1%. After 72 hours of treatment, relative ATP abundance was measured using CellTiter Glo® (Promega, Wisconsin, USA) according to the manufacturer's protocol. GloMax® Multi Detection System (Promega) was used to measure the luminescence signal. Measurement of relative BrdU abundance was measured in cells seeded in 96-well, transparent, flat bottom multiwell plates at 500 cells per well. After 24 hours, cells were treated and incubated for 96 hours. Cell fixation and staining was performed according to the manufacturer's instructions (Cell Proliferation ELISA, BrdU, colorimetric, Roche, Basel, Switzerland). For cell cycle analysis, cells were seeded in 100 mm petri dishes at 5 × 105 - 106 cells per dish. After 24 hours, cells were treated and incubated for 72 hours. Dual EdU and propidium iodide labeling were performed according to the manufacturer's protocol (Click-It EdU Alexa Fluor 488 Imaging Kit Thermo Fisher Scientific). Cells were analyzed using the BD LSRFortessa flow cytometer (BD Biosciences, California, USA).
Western Immuno Blotting
Whole-cell protein lysate preparation was performed with RIPA buffer (15 mM HEPES, 150 mM NaCl, 10 mM EGTA, 2% Triton X-100) supplemented with protease and phosphatase inhibitors (complete mini EDTA-free and PhosSTOP, Roche) as described before [14,21,35]. Bicinchoninic acid assay (Santa Cruz Biotechnology, Texas, USA) was used for protein quantitation according to the manufacturer's protocol. Laemmli buffer was mixed with cell lysates, denatured for 5 min at 95 °C and loaded in a 10% polyacrylamide gel (Thermo Fisher Scientific). Lysates were separated by electrophoresis at 120 V and transferred to a PVDF membrane. Membranes were blocked in 5% nonfat dry milk in TBS-T and incubated in primary antibody in 5% nonfat dry milk in TBS-T overnight at 4 °C. Antibodies and dilutions are listed in Supplementary Table 1. Membranes were washed three times and incubated with the secondary antibody. Proteins were visualized with ImmunoCruz Western Blotting Luminol Reagent (Santa Cruz Biotechnology) and the Fusion FX7 imaging system was used for detection (Vilber Lourmat).
RNA Sequencing
A total of 105 cells were seeded in 6-well plates and treated in triplicates. Cells were harvested 6 hours and 24 hours after treatment for rhabdomyosarcoma and neuroblastoma cells, respectively. Total RNA was isolated from cell pellets using the RNeasy mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's protocol. Library preparation and sequencing was performed by DKFZ Genomics and Proteomics Core Facility (Deutsches Krebsforschungszentrum, Heidelberg, Germany). Libraries were sequenced on a HiSeq 2000 v4 instruments with 2 × 125 bp paired-end reads (Illumina). Libraries were mapped with STAR (version 2.5.3a) to GRCh38 using the Gencode v27 annotation and the following parameters—sjdbOverhang 300—twopassMode Basic—outReadsUnmapped None—chimSegmentMin 12—chimJunctionOverhangMin 12—alignSJDBoverhangMin 10—alignMatesGapMax 200000—alignIntronMax 200000—limitBAMsortRAM 31532137230. Gene abundance was estimated using featureCounts (version v1.5.1) and the following parameters -F GTF -t exon -g gene_id -C -M –fraction -p -s 0 -O -B -Q 4 allowing for fractional counting of multimapping and multioverlapping reads with both mates aligned and not counting chimeric reads. Standard differential expression analysis for genes with at least 15 reads in at least one sample among the groups considered was conducted using the edgeR Bioconductor package [36].
Treatment of Patient-Derived Xenografts
All experiments were conducted according to the institutional animal protocols and the national laws and regulations. The experiments were conducted as previously described in four replicates [35]. In short, NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac mice (Taconic) were used to perform all patient-derived xenograft experiments. Two neuroblastomas from two independent patients with MYCN-amplified neuroblastoma were serially transplanted in mice at least three times prior to the experiments. Caliper measurement was used to monitor tumor growth. Tumor volume was calculated with the formula length x width 2/2. Mice were sacrificed with cervical dislocation when tumor size exceeded 2000 mm³. Drugs were dissolved in DMSO/Tween/0.9%NaCl and administered intraperitoneally at 20 mg/kg body weight per day.
Immunohistochemistry
Immunopathological analysis was provided by the iPATH.Berlin—Immunopathology for Experimental Models, core unit of the Charité—Universitätsmedizin Berlin (Berlin, Germany). Formalin-fixed tissue samples were embedded in paraffin and 1–2 μm thick sections were cut from paraffin blocks. Sections were dewaxed and histochemically stained with H&E (hematoxylin&eosin) for evaluation of histomorphology or subjected to a heat-induced epitope retrieval step prior to incubation with anti-Ki67 (clone 16A8, BioLegend), anticleaved caspase 3 (clone 5A1E, Cell Signaling). For detection of Ki67 and cleaved caspase3, sections were incubated with secondary antibody (biotinylated rabbit antirat and biotinylated donkey antirabbit, respectively; Dianova). Biotin was detected by streptavidin coupled with alkaline phosphatase (AP) and RED as chromogen (both Dako REAL™ Detection System, Alkaline Phosphatase/RED, Rabbit/Mouse, Agilent Technologies). Nuclei were counterstained with hematoxylin (Merck Millipore) and slides coverslipped with glycerol gelatine (Merck). Whole slide scans were acquired by a Vectra® 3 imaging system (PerkinElmer).
Statistical Analysis
All other data analysis was performed using Excel 2013 and GraphPad Prism 7. Synergism was analyzed with the R package synergyfinder [37] or the free-license software Compusyn [38]. For synergism analysis, only results of > IC10 and < IC90 were used. RNASeq was analyzed as described.
Results
Combination Treatment with BRD4 and PLK1 Inhibitors has Synergistic Antitumor Effects in Pediatric Tumor Cells
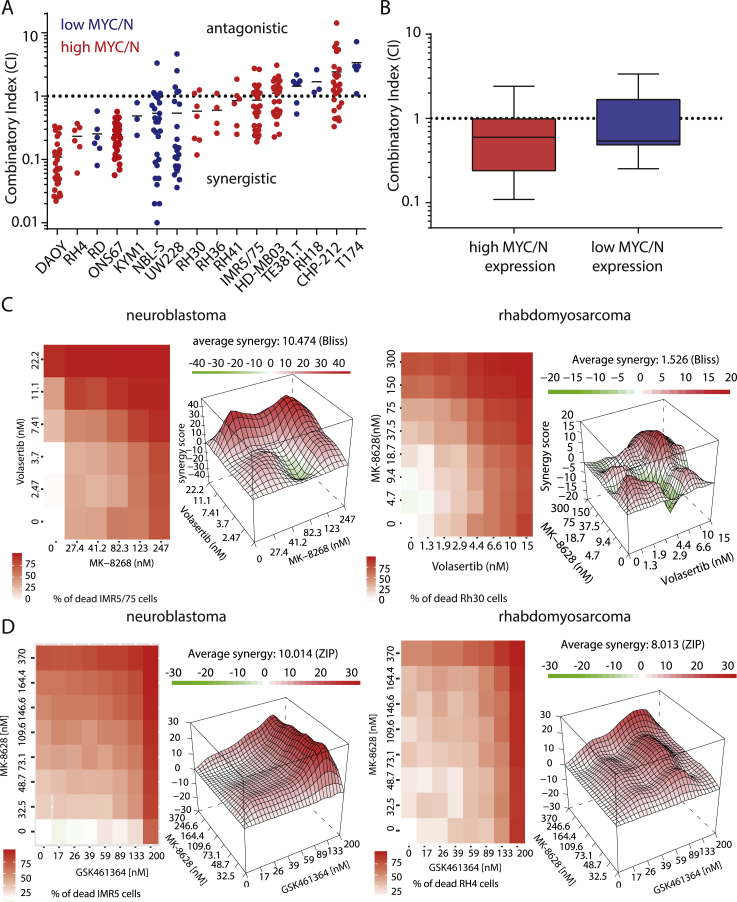
Based on previous reports showing synergistic antitumor effects of combined treatment with BRD4 and PLK1 inhibitors in medulloblastoma and leukaemia [20,21], we hypothesized that combining MK-8628 (BRD4 inhibitor) with Volasertib (PLK1 inhibitor) or GSK461364 (PLK1 inhibitor) could show synergistic effects against MYCN-driven neuroblastoma and rhabdomyosarcoma cells. To test this, we assessed the effect of single agent treatment on neuroblastoma and rhabdomyosarcoma cell line viability. In all cell lines treated with either inhibitor, we observed a significant decrease in the number of viable cells with increasing inhibitor concentrations (Supplementary Figure 1, Table 2). Varying degrees of sensitivity were observed for both inhibitors as evidenced by a wide range of IC50 concentrations (Supplementary Figure 1 A and B). While BRD4 inhibitor, MK-8628, was more effective in cells with high MYC/N expression compared with cells with low MYC/N expression, cells did not respond differentially to PLK1 inhibition with regard to their MYC/N expression levels. To assess the effect of combined Volasertib, GSK461364, and MK-8628 treatment in 12 neuroblastoma and rhabdomyosarcoma cell lines, we calculated combination indices (CI) and compared them with published results in four medulloblastoma cell lines (Figure 1A) [21]. Synergistic effects on cell viability were observed in 11 out of 16 cell lines, as evidenced by CI values below 1 (Figure 1A). No significant difference in synergy was observed between cells with high MYC/N expression compared with cells expressing low levels of MYC/N (Figure 1B). An excess over Bliss analysis confirmed synergistic effects of combined PLK1 and BRD4 inhibitor treatment (Figure 1C and D, Supplementary Figure 2A and B). Thus, combination treatment with BRD4 and PLK1 inhibitors has significant synergistic low nanomolar antitumor activity rhabdomyosarcoma, neuroblastoma and medulloblastoma cells, independent of MYC/N expression levels.
Figure 1.
Combination of BRD4 and PLK1 inhibition has synergistic antitumoral effects in pediatric tumor cell lines. A Combination indices (CI) of 16 pediatric tumor cell lines treated with varying concentrations of Volasertib and MK-8628 as well as combination of both agents for 72 h. B Comparison of combination indices in cell lines with high and low MYC and/or MYCN expression. C Heatmap of synergy score and excess over Bliss analysis of combination treatment with MK-8628 and Volasertib in MYCN-amplified neuroblastoma cell line IMR5 and rhabdomyosarcoma cell line RH30. D Heatmap of synergy score and excess over Bliss analysis of combination treatment with MK-8628 and GSK461364 in MYCN-amplified neuroblastoma cell line IMR5 and rhabdomyosarcoma cell line RH30.
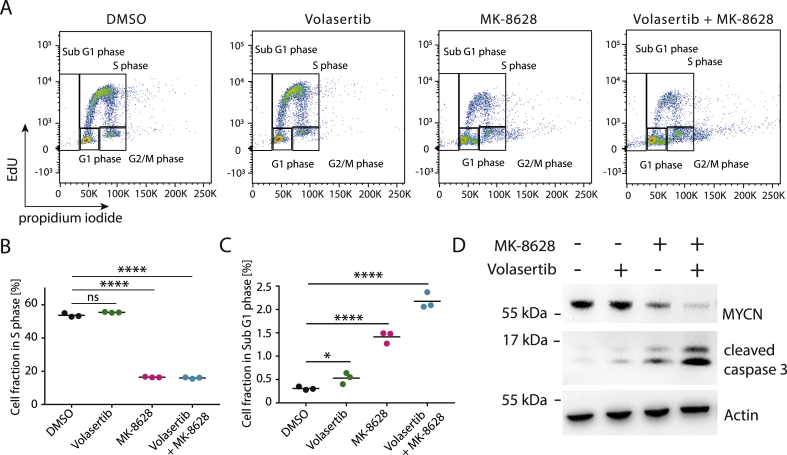
Simultaneous Inhibition of BRD4 and PLK1 Results in Increased Cell Death
Previous reports have suggested that combined BRD4 and PLK1 inhibition leads to greater cellular apoptosis and cell cycle arrest [15,39]. To confirm the effect of combination treatment observed in other tumor entities in our neuroblastoma and rhabdomyosarcoma cell line models, we measured cell cycle distribution of cell lines after treatment with MK-8628 and Volasertib, alone or in combination (Figure 2A). Consistent with previous reports, synergistic effects on cell viability were accompanied by significantly increasing fractions of apoptotic cells (sub G1 phase) and a concomitant decreases in S phase, compared with single agent treatment (Figure 2B and C and Supplementary Figure 3). Combination treatment also led to increased aneuploidy, which may be due to effects of PLK1 inhibition on DNA damage response leading to mitotic failure, in line with previous reports (Figure 2) [40]. The increased cell death observed after combination treatment was accompanied by significantly greater reduction of MYCN protein levels compared with single agent treatment, which was accompanied by increased caspase 3 cleavage, confirming on-target BRD4 and PLK1 inhibition and induction of cellular apoptosis (Figure 2D). In summary, combined PLK1 and BRD4 inhibition disrupts neuroblastoma and rhabdomyosarcoma cell cycling, which is accompanied by repression of BRD4 and PLK1 targets.
Figure 2.
Simultaneous inhibition of BRD4 and PLK1 synergistically represses MYC/N and results in increased cell death. A Fluorescence-activated cell sorting (FACS)-based measurement of IMR5 cell cycle distribution after treatment with DMSO control, MK-8628 (150 nM), Volasertib (5 nM), or combination of both agents (150 nM MK-8628 + 5 nM Volasertib) for 72 h. B Fraction of cells in S-phase as measured in (A). C Fraction of cells in Sub G1 phase as measured in A (A). D Western immunoblotting of MYCN and cleaved caspase 3 after treatment of cells with DMSO control, MK-8628 (500 nM), Volasertib (10 nM), or combination of both (500 nM MK-8628 + 10 nM Volasertib) (Student's t-test: * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001).
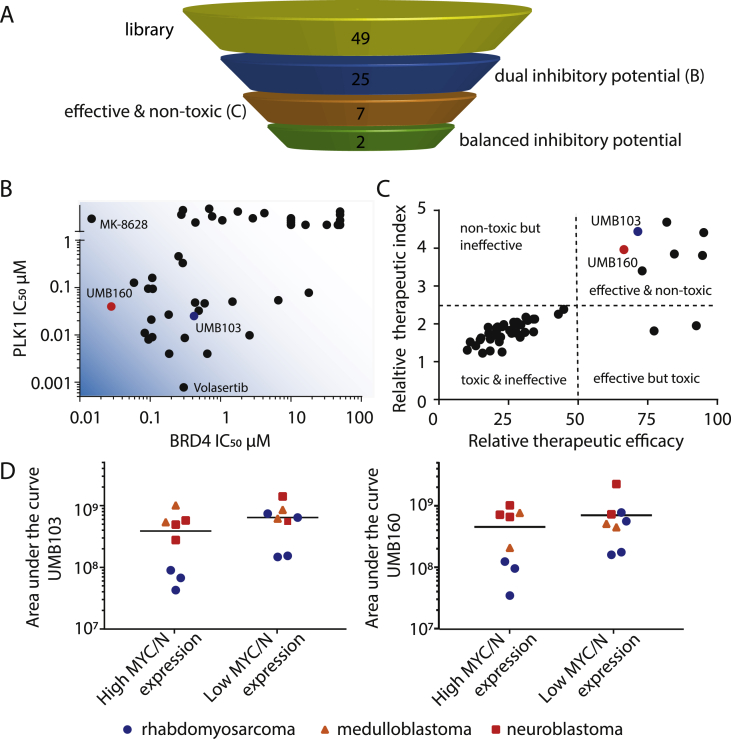
Screening of 49 Small Molecules with Dual Inhibitory Activity Against BRD4 and PLK1 Leads to the Discovery of Molecules with Favourable In Vitro AntiTumor Activity Profiles
Based on the observed synergy of BRD4 and PLK1 inhibitors along with the recent discovery of dual kinase-bromodomain inhibitors [30], we hypothesized that dual BRD4/PLK1 inhibitors might have potent antitumor activity and allow us to simultaneously target two regulators of MYC/N using a single molecule. We performed an in vitro screen with a recently developed small molecule library of dual BRD4/PLK1 inhibitors, called UMB88-162 (Figure 3A and B) [30]. Four tumor cell lines (two rhabdomyosarcoma, one neuroblastoma, one medulloblastoma cell line) as well as one nontransformed human fibroblast cell line were treated with each molecule at fixed drug concentrations (Supplementary Figure 4A–E). Cell viability was assessed after 72 hours of drug treatment and therapeutic efficacy compared. A therapeutic index was calculated by comparing the effect of molecules on tumor cell lines to the effect on nontransformed cells (Figure 3C). Two molecules with high dual BRD4 and PLK1 inhibitory activity, UMB160 and UMB103, showed strong tumor cell-specific effects on cell viability while having a broad therapeutic window as evidenced by low activity against nontransformed cells. All other molecules with high therapeutic window did not have measurable inhibitory potential on BRD4 and were therefore not selected for further testing. Dose response analysis revealed inhibitory concentrations with 50% reduction of cell viability (IC50) values ranging from 2.9 to 87.5 nM and 6.5–178.2 nM for UMB160 and UMB103, respectively (Supplementary Figure 5A to E, Table 3). No difference in sensitivity was observed between cells expressing high levels of MYC/N compared with cells expressing low levels. Notably, rhabdomyosarcoma cell lines were more sensitive to both candidate drugs compared with neuroblastoma and medulloblastoma cells (Figure 3D). Both candidate molecules, UMB160 and UMB103 showed low nanomolar antitumor activity in cell lines, suggesting that dual BRD4/PLK1 inhibitors may have clinically relevant activity.
Figure 3.
A library of dual nanomolar BRD4 and PLK1 inhibitors show varying antitumoral effects against pediatric tumor cell lines. A Schematic of the selection strategy for the screening of 49 dual PLK1/BRD4 inhibitors. B Inhibitory concentrations 50% (IC50) of dual inhibitor library against BRD4 and PLK1 as previously measured in cell free assays [30]. C Therapeutic index compared with relative therapeutic efficacy of dual inhibitor library as measured at 20 nM concentration in pediatric tumor cell lines compared with nontransformed human fibroblasts (candidate molecules are indicated in red and blue). D Dose-response area under the curve (AUC) of 18 different pediatric tumor cell lines treated with dual PLK1/BRD4 inhibitors UMB103 (left) and UMB160 (right).
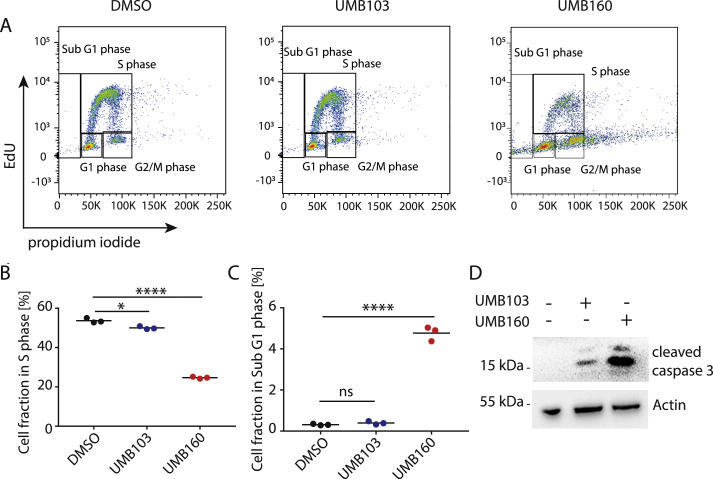
Dual BRD4/PLK1 Inhibitor Treatment Leads to Cell Death and Cell Cycle Arrest in Pediatric Tumor Cell Lines
Based on the results of our screen, we reasoned that UMB160 and UMB103 may affect important BRD4-and PLK1-regulated cellular functions. To test this, we measured changes in cell cycle distribution by performing EdU pulse labeling followed by fluorescence-activated cell sorting (FACS) measurement in cells treated with UMB160 and UMB103 (Figure 4A). Consistent with increased cell death, the fraction of cells in sub G1 phase increased after treatment with UMB160 and UMB103 (Figure 4C). Previous reports have shown that BRD4 inhibition leads to cell cycle arrest in G1 because of BRD4's critical role in cell cycle regulation [15]. Consistently, we observed an increase in the fraction of cells in G1 and a decrease in the fraction of cells S phase after treatment with UMB160 and UMB103 (Figure 4B and Supplementary Figure 6C). Moreover, UMB160 and UMB103 led to an increase in the fraction of cells in G2/M phase (Supplementary Figure 6D and G), in line with the function of PLK1 in G2/M regulation [39]. Consistent with decreased cell viability, both UMB160 and UMB103 treatment led to a significant increase in apoptosis, as evidenced by increased caspase 3 cleavage measured by western immunoblotting (Figure 4D, Supplementary Figure 7A). Taken together, treatment with dual BRD4/PLK1 inhibitors led to reduction in cell viability and cell proliferation and to increased cell death. In line with their ability to inhibit BRD4 and PLK1, cell cycle was disrupted as previously observed for combined treatment with PLK1 and BRD4 inhibitors and in line with known functions of BRD4 and PLK1 in cell cycle regulation [21].
Figure 4.
Treatment with dual BRD4/PLK1 inhibitors UMB103 and UMB160 leads to cell death and cell cycle arrest in pediatric tumor cell lines. A FACS-based measurement of the cell cycle distribution of IMR5 cells after treatment with DMSO, UMB103 (10 nM) or UMB160 (10 nM) for 72 hours. B Fraction of cells in S-phase as measured in (A). C Fraction of cells in Sub G1 phase as measured in (A). D Western immunoblotting of cleaved caspase 3 after incubation of cells with DMSO compared with UMB103 and UMB160 for 24 hours at 20 nM each (Student's t-test: * = ns = not significant, P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001).
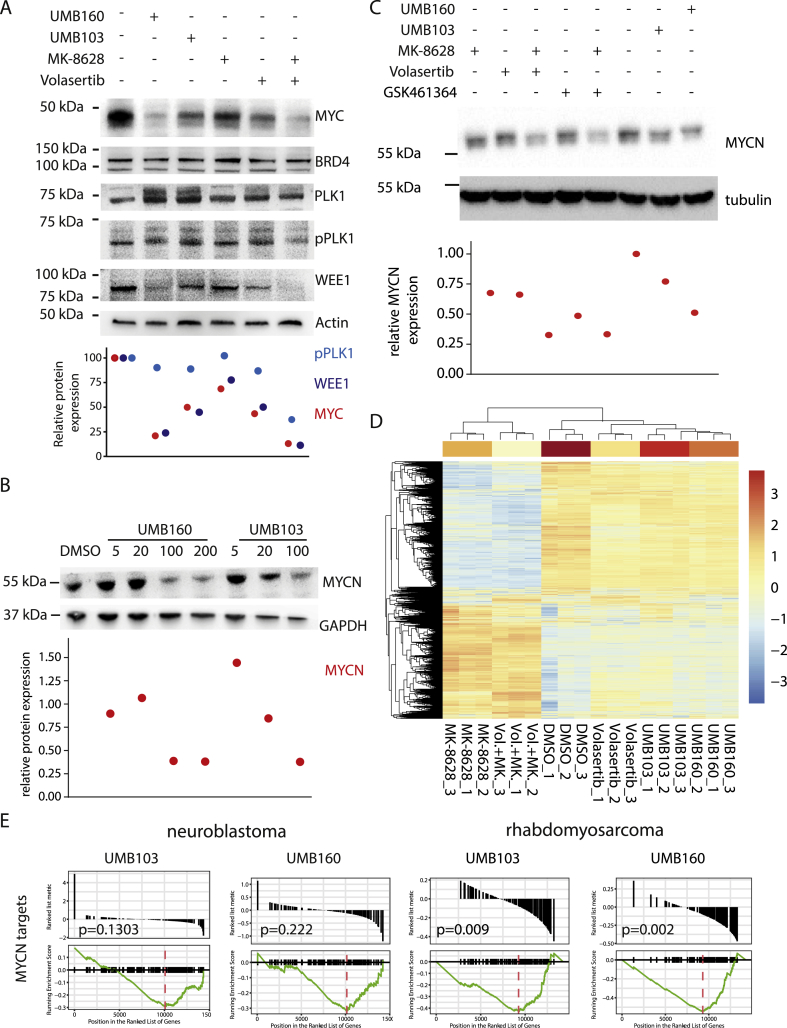
Treatment with Dual BRD4/PLK1 Inhibitors Leads to Repression of BRD4 and PLK1 Activity
To confirm on-target activity of UMB160 and UMB103 we performed western immunoblotting analysis of known PLK1 and BRD4 targets after short-term inhibitor treatment. Both, UMB103 and UMB160 treatment led to a significant repression of MYC in MYC-amplified cells (Figure 5A). In PAX3-FOXO1-driven rhabdomyosarcoma cells, treatment significantly repressed MYCN and FOXO1 expression, phenocopying the effect of BRD4 inhibition (Figure 5B). Furthermore, phospho-PLK1 as well as PLK1 target WEE1 were repressed after treatment with UMB160 but not UMB103, phenocopying the effect of PLK1 inhibition (Figure 5A). BRD4 and PLK1 protein levels did not change after treatment, consistent with the disruption of their activity rather than their stability. To further characterize the molecular activity of the dual inhibitors, we performed RNA sequencing after incubation of neuroblastoma and rhabdomyosarcoma cell lines with dual inhibitors. Consistent with their inhibitory activity against BRD4 and PLK1, UMB160 and UMB103 treatment led to similar changes in gene expression as observed after single PLK1 and BRD4 inhibitor treatment (Supplementary Figure 7). Unsupervised clustering of differentially expressed genes revealed that expression changes induced by UMB160 and UMB103 were more similar to those observed after PLK1 inhibition, suggesting that the impact of PLK1 inhibition on gene expression was stronger compared with effects of BRD4 inhibition (Figure 5C). To confirm the ability of dual BRD4/PLK1 inhibitors to disrupt BRD4-regulated transcriptorial programs we compared our results with previously published gene sets of PAX3-FOXO1 and MYCN target genes, respectively [7,41]. Treatment with dual inhibitors led to significant repression of PAX3-FOXO1 targets in rhabdomyosarcoma (Supplementary Figure 7D) and of MYCN targets in rhabdomyosarcoma cells in line with potent inhibition of BRD4 (Figure 5D, Supplementary Figure 7F). In neuroblastoma cells the effect of dual BRD4/PLK1 inhibitors on MYCN-driven gene expression was less pronounced, consistent with the increased effects of UMB103 and UMB160 on rhabdomyosarcoma cell viability. In conclusion, the disruption of transcriptional programs by dual BRD4/PLK1 inhibitors phenocopies changes induced by single BRD4 and PLK1 inhibitors, suggesting that dual BRD4/PLK1 inhibitors have the potential to disrupt transcriptional programs that are crucial for the survival of pediatric tumors.
Figure 5.
Treatment with dual nanomolar BRD4/PLK1 inhibitors leads to repression of BRD4 and PLK1 functions. A Western immunoblotting of BRD4 and PLK1 targets after incubation of HD-MB03 medulloblastoma cells with DMSO, MK-8628 (500 nM), Volasertib (5 nM), or combination of both, UMB160 (10 nM) or UMB103 (10 nM) for 24 hours. Signal quantification is plotted below. B Western immunoblotting of MYCN after treatment of IMR5 neuroblastoma cells with DMSO, MK-8628 (500 nM), Volasertib (5 nM), or combination of both, GSK461364 (100 nM) or combination of GSK461364 and MK-8628, UMB160 (10 nM) or UMB103 (10 nM) for 24 hours. Signal quantification is plotted below. C Western immunoblotting of MYCN in RH30 rhabdomyosarcoma cells treated for 24 hours with increasing concentrations of UMB160 or UMB103. D Unsupervised clustering of differentially expressed genes as measured by RNA sequencing of MYCN-amplified neuroblastoma cell line IMR5 treated with MK-8628 (500 nM) and/or Volasertib (10 nM) compared with UMB103 (100 nM) or UMB160 (100 nM) for 24 hours. E Gene set enrichment analysis (GSEA) plot for MYCN target genes, as previously defined [41], in neuroblastoma cell line IMR5 and rhabdomyosarcoma cell line RH4 treated with dual PLK1/BRD4 inhibitors.
Dual PLK1/BRD4 Inhibitors Show Significant Anti-Tumor Effects Against High-Risk Neuroblastoma Patient-Derived Xenografts (PDXs)
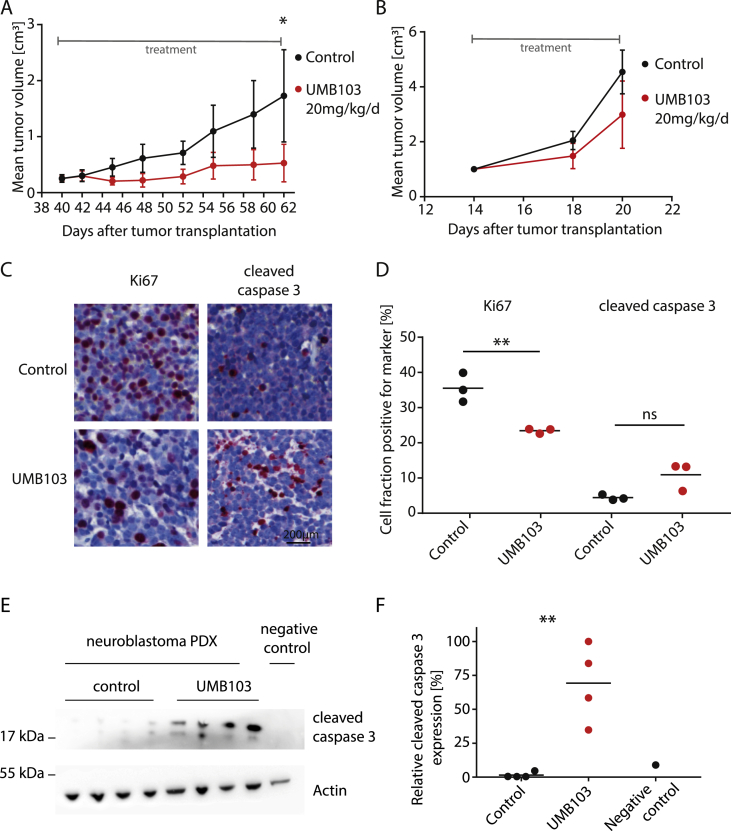
Based on the promising in vitro anti-tumor activity of dual BRD4/PLK1 inhibitors, we hypothesized that these molecules may show clinically relevant therapeutic activity in vivo. We chose UMB103 for further in vivo testing. Based on the assumption of equal drug distribution in mice, the dose applied (20 mg/kg body weight) was expected to lead to a peak plasma concentration equalling 104 times (ca. 20 μM) the estimated IC50 calculated from in vitro experiments. No significant reduction in mouse weight was observed after treatment at these doses. This suggests that concentrations at which inhibitors show strong in vitro anti-tumor effects could be achieved without reaching toxic doses (Figure 3C). To investigate the therapeutic efficacy of UMB103 in vivo we treated two independent MYCN-amplified high-risk neuroblastoma patient-derived xenograft models (PDX) with UMB103. Mice were only treated once the tumor sizes reached 200 mm3 and were exponentially growing, closely mimicking clinical relapse or therapy-refractory situations at which such drugs would be administered. At 20 mg/kg/d, UMB103 treatment led to significant reduction of tumor growth in one of the PDX models (Figure 6A). The fraction of proliferating cells as measured by immunohistological staining for Ki67 significantly decreased in tumors responding to UMB103 treatment whereas apoptotic cells, as measured by cleaved caspase 3 staining increased (Figure 6C). UMB103 treatment was accompanied by increased caspase 3 cleavage, suggesting that treatment showed strong cytostatic and significant cytotoxic effects in vivo (Figure 6E). Together, treatment with UMB103 significantly repressed proliferation and induced apoptosis in vivo at non-toxic doses, which led to reduced tumor growth over time. This suggests that dual PLK1/BRD4 inhibitors have the potential to show clinically relevant therapeutic activity in patients suffering from high-risk pediatric solid tumors.
Figure 6.
Dual nanomolar BRD4/PLK1 inhibitors show significant antitumor effects against patient-derived xenograft models (PDXs) of high-risk MYCN-amplified neuroblastomas. A-B Tumor volumes of two independent neuroblastoma patient-derived models treated with UMB103 at 20 mg/kg/d (N = 4 mice per group, patient A = MYCN-amplified 15 month of age at the time of diagnosis, B = MYCN-amplified, 2 month of age at the time of diagnosis). C Photomicrographs of tumor sections from PDX shown in (A) immunohistochemically stained for Hematoxylin and eosin (H&E, blue) and Ki67 (left, red) and cleaved caspase 3 (right, red). D Quantification of Ki67 and cleaved caspase 3 expression as shown in (C). E Western immunoblotting of cleaved caspase 3 in PDX tumors shown in (A) treated with UMB103 compared with tumors treated with DMSO and compared with untreated IMR5 cells. F Quantification of relative protein expression of cleaved caspase 3 (Student's t-test: ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001).
Discussion
Bromodomain-containing protein 4 (BRD4) inhibitors have shown promising preclinical activity in various tumor models [18,19]. However, as with most small molecule inhibitors, it is likely that single-agent inhibitor treatment will not be curative in most patients. In line with our previous reports in medulloblastoma [21], we here demonstrate that combination of BRD4 and PLK1 inhibitors has synergistic in vitro activity against preclinical models of neuroblastoma and rhabdomyosarcoma, suggesting that the efficacy of this treatment strategy extends to other pediatric solid tumor entities. Through a small molecule in vitro screen for agents with dual BRD4/PLK1 inhibitory activity, we identify two candidate molecules with promising preclinical anti-tumor activity in vitro and in vivo. Consistent with on-target dual BRD4/PLK1 inhibition, treatment with candidate molecules led to concomitant disruption of transcriptional programs controlled by BRD4 and reduced PLK1 kinase activity. We conclude that dual BRD4/PLK1 inhibition represents a promising approach for the treatment of pediatric solid tumors relying on BRD4 and PLK1 activity.
Cancer commonly acquires mutations during the course of treatment that lead to treatment resistance. Targeting more than one process in cancer cells can drastically reduce the probability of treatment resistance [42]. This is supported by the fact that combination of cytotoxic therapies has drastically improved the survival rates in children in the past decades [43]. However, long term side effects of chemotherapy as well as radiation therapy such as organ damage, secondary malignancies, and fatigue constitute considerable sequelae can cause considerable morbidity and mortality after successful first-line treatment [43]. Such adverse side effects occur less commonly when multiple modes of action are synergistically targeted [44]. Additionally, reducing the number of active agents and development of targeted therapies may reduce side effects and lead to reduced usage of cytotoxic conventional chemo- and radiation therapy. Though many highly selective targeted therapies have yet to achieve clinical results, it is widely accepted that synergistic targeted treatment approaches will not only reduce the risk of treatment resistance but have the potential to improve outcome by reducing long term adverse effects.
One example for effective combination of targeted agents are recently proposed combinations of BRD4 inhibitors with other small molecules, which can synergistically increase anti-tumor effects in preclinical models of various tumor entities [[45], [46], [47], [48], [49], [50], [51]]. While these results are promising, a selection of ideal drug combinations for the treatment of pediatric tumors is hampered by the fact that most studies on combination treatments have been performed in tumor models that, on a molecular level, differ significantly from pediatric tumors. Combined inhibition of BRD4 and PLK1 was recently reported to show synergistic antitumor effects in preclinical models of medulloblastoma and acute myeloid leukaemia [20,21]. Consistent with these reports, we observed significant synergistic anti-tumor effects when combining MK-8628 with Volasertib or GSK461364 in preclinical models of three pediatric tumor entities. These effects were accompanied with significant repression of MYC and MYCN expression, reduced PLK1 kinase activity as well as disruption of cell cycling as predicted based on BRD4 and PLK1 functions. Thus, we and others clearly demonstrate on-target synergistic anti-tumor effects of BRD4 and PLK1 inhibitors.
Even though multi target therapies such as combination of BRD4 and PLK1 inhibitors have several excellent rationales, clinical development of combination therapies remains difficult. Testing therapeutic activity and safety is more arduous for combination regimens, as single drugs need to be approved separately prior to clinical testing of combination treatment. Additionally, pharmacokinetics of different molecules are difficult to predict leading to difficulties in planning correct treatment schedules. Notably, complex treatment regimens and multiple different drug administrations lead to lower patient compliance [52]. Thus, novel approaches are clearly needed to enable the rapid and efficient introduction of synergistic therapeutic strategies into clinical settings.
One possibility to circumvent difficulties resulting from combination treatments is the use of molecules that simultaneously and specifically inhibit two or more cancer-relevant targets. Molecules with multiple targets are often more effective against advanced diseases; they have more predictable pharmacokinetic profile compared with combination therapy, especially if adverse effects are molecule-based. Additionally, using a single compound guarantees that synergistic effects occur simultaneously regardless of the pharmacokinetics of the drug [42]. Recent reports suggest that some kinase inhibitors have the ability to bind to the bromodomain of BRD4 and to inhibit kinase activity simultaneously [29]. We recently created a library of small molecules with high affinity to BRD4 as well as PLK1 by chemically modifying BI-2536 analogues [30]. In our in vitro screen presented here, we find that a subset of dual-targeting molecules can suppress the viability, proliferation and cell cycling of pediatric tumor cell lines at low nanomolar concentrations. This is in line with recent reports showing the feasibility of simultaneously targeting bromodomains and Phosphoinositide 3-kinases (PI3K) in pancreatic and neuroblastoma cell lines [34]. As treatment of pediatric tumor cells with dual BRD4/PLK1 inhibitors is accompanied by inhibition of important BRD4 and PLK1 functions in gene expression regulation as well as kinase-mediated cell signalling, we conclude that it is feasible to inhibit both BRD4 and PLK1 with a single molecule in cancer cells. Furthermore, we observe a clear repression of tumor growth in one patient derived high-risk neuroblastoma tumor model treated with the dual BRD4/PLK1 inhibitor UMB103. Importantly, these antitumor effects were not accompanied by significant overall toxicity. This is in line with previous reports of low toxicity in several dual inhibitors [42,52,53]. Even though tumor regression was not complete and only one out of two PDX models responded suggesting yet unknown determinants of treatment response, we conclude that dual BRD4/PLK1 inhibition may have the potential to generate a substantial response in pediatric patients suffering from high-risk solid tumors.
In summary, we here present the unique approach of therapeutically targeting two independent cancer-relevant proteins. Based on a screen with a dual kinase-bromodomain small-molecule library, we discovered chemical leads serving as a starting point for therapeutic development. Dual BRD4/PLK1 inhibitors have the potential to generate a measurable response in patients suffering from neuroblastoma. This new insight should provide a translational framework for clinical trial development of dual inhibitors for pediatric patients. Apart from the potential clinical benefit for patients suffering from high-risk solid tumors, introduction of dual-targeting agents may avoid the pharmacological difficulties associated with the administration of two different drugs, thereby expediting clinical development and avoiding accumulation of off-target side effects.
Authors' Contributions
NT investigation, data curation and writing original draft, YH investigation, SL investigation, HOY investigation, HDG investigation and supervision, YB investigation and software, FK investigation, software and supervision, ICMA investigation, AS investigation, VB investigation, CD investigation, AK supervision, JR in vivo studies, PH supervision, AL supervision, GS supervision, AE supervision, JHS supervision, WZ supervision, AGH conceptualization, funding acquisition, writing&editing, supervision. NT and AGH wrote the manuscript with contribution from all authors.
Conflicts of Interest
The authors have no conflict of interest to declare.
Funding
A.G.H. is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—398299703, the Wilhelm Sander Stiftung. A.G.H. and A.K. are participants in the BIH-Charité Clinical Scientist Program funded by the Charité—Universitätsmedizin Berlin and the Berlin Institute of Health. A.G.H. is supported by Berliner Krebsgesellschaft e.V. A.K. is supported by the Else Kröner-Fresenius Stiftung. A.G.H. is supported by TransOnc priority program of the German Cancer Aid within grant “70112951” (ENABLE).
Acknowledgments
The authors are grateful to Alex Kentsis and Nicole Hübener for critical discussions, and Kathy Astrahantseff for editorial advice. We thank the patients and their parents for granting access to the tumor specimen and clinical information that were analyzed in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.09.013.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gatta G., Botta L., Rossi S., Aareleid T., Bielska-Lasota M., Clavel J. Childhood cancer survival in Europe 1999-2007: results of EUROCARE-5--a population-based study. Lancet Oncol. 2014;15(1):35–47. doi: 10.1016/S1470-2045(13)70548-5. [DOI] [PubMed] [Google Scholar]
- 2.Bayani J., Zielenska M., Marrano P., Kwan Ng Y., Taylor M.D., Jay V. Molecular cytogenetic analysis of medulloblastomas and supratentorial primitive neuroectodermal tumors by using conventional banding, comparative genomic hybridization, and spectral karyotyping. J Neurosurg. 2000;93(3):437–448. doi: 10.3171/jns.2000.93.3.0437. [DOI] [PubMed] [Google Scholar]
- 3.Barr F.G., Duan F., Smith L.M., Gustafson D., Pitts M., Hammond S. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children's Oncology Group. Genes Chromosomes Cancer. 2009;48(8):661–672. doi: 10.1002/gcc.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwab M. MYCN in neuronal tumours. Cancer Lett. 2004;204(2):179–187. doi: 10.1016/S0304-3835(03)00454-3. [DOI] [PubMed] [Google Scholar]
- 5.Davidoff A.M. Neuroblastoma. Semin Pediatr Surg. 2012;21(1):2–14. doi: 10.1053/j.sempedsurg.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda Kuzan-Fischer C., Juraschka K., Taylor M.D. Medulloblastoma in the molecular era. J Korean Neurosurg Soc. 2018;61(3):292–301. doi: 10.3340/jkns.2018.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gryder B.E., Yohe M.E., Chou H.C., Zhang X., Marques J., Wachtel M. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 2017;7(8):884–899. doi: 10.1158/2159-8290.CD-16-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustafson W.C., Weiss W.A. Myc proteins as therapeutic targets. Oncogene. 2010;29(9):1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Canc. 2006;94(2):179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S.Y., Chiang C.M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z., He N., Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28(3):967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henssen A., Althoff K., Odersky A., Beckers A., Koche R., Speleman F. Targeting MYCN-driven transcription by BET-bromodomain inhibition. Clin Cancer Res. 2016;22(10):2470–2481. doi: 10.1158/1078-0432.CCR-15-1449. [DOI] [PubMed] [Google Scholar]
- 15.Henssen A., Thor T., Odersky A., Heukamp L., El-Hindy N., Beckers A. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget. 2013;4(11):2080–2095. doi: 10.18632/oncotarget.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noel J.K., Iwata K., Ooike S., Sugahara K., Nakamura H., Daibata M. Vol. 12. 2013. Development of the BET bromodomain inhibitor OTX015. (Proceedings of the AACR-NCI-EORTC International Conference: Molecular Targets and Cancer Therapeutics; 2013 Oct 19-23; Boston, MA). (11 Supplement):Abstract number C244. [Google Scholar]
- 17.Boi M., Gaudio E., Bonetti P., Kwee I., Bernasconi E., Tarantelli C. The BET bromodomain inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res. 2015;21(7):1628–1638. doi: 10.1158/1078-0432.CCR-14-1561. [DOI] [PubMed] [Google Scholar]
- 18.Odore E., Lokiec F., Cvitkovic E., Bekradda M., Herait P., Bourdel F. Phase I population pharmacokinetic assessment of the oral bromodomain inhibitor OTX015 in patients with haematologic malignancies. Clin Pharmacokinet. 2016;55(3):397–405. doi: 10.1007/s40262-015-0327-6. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y., Vakoc C.R. Targeting cancer cells with BET bromodomain inhibitors. Cold Spring Harb Perspect Med. 2017;7(7) doi: 10.1101/cshperspect.a026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tontsch-Grunt U., Rudolph D., Waizenegger I., Baum A., Gerlach D., Engelhardt H. Synergistic activity of BET inhibitor BI 894999 with PLK inhibitor volasertib in AML in vitro and in vivo. Cancer Lett. 2018;421:112–120. doi: 10.1016/j.canlet.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 21.Han Y., Lindner S., Bei Y., Garcia H.D., Timme N., Althoff K. Synergistic activity of BET inhibitor MK-8628 and PLK inhibitor Volasertib in preclinical models of medulloblastoma. Cancer Lett. 2019;445(31):24–33. doi: 10.1016/j.canlet.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Ackermann S., Goeser F., Schulte J.H., Schramm A., Ehemann V., Hero B. Polo-like kinase 1 is a therapeutic target in high-risk neuroblastoma. Clin Cancer Res. 2011;17(4):731–741. doi: 10.1158/1078-0432.CCR-10-1129. [DOI] [PubMed] [Google Scholar]
- 23.Thalhammer V., Lopez-Garcia L.A., Herrero-Martin D., Hecker R., Laubscher D., Gierisch M.E. PLK1 phosphorylates PAX3-FOXO1, the inhibition of which triggers regression of alveolar Rhabdomyosarcoma. Cancer Res. 2015;75(1):98–110. doi: 10.1158/0008-5472.CAN-14-1246. [DOI] [PubMed] [Google Scholar]
- 24.Triscott J., Lee C., Foster C., Manoranjan B., Pambid M.R., Berns R. Personalizing the treatment of pediatric medulloblastoma: polo-like kinase 1 as a molecular target in high-risk children. Cancer Res. 2013;73(22):6734–6744. doi: 10.1158/0008-5472.CAN-12-4331. [DOI] [PubMed] [Google Scholar]
- 25.Xiao D., Yue M., Su H., Ren P., Jiang J., Li F. Polo-like kinase-1 regulates myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol Cell. 2016;64(3):493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Watts E., Heidenreich D., Tucker E., Raab M., Strebhardt K., Chesler L. Designing dual inhibitors of anaplastic lymphoma kinase (ALK) and bromodomain-4 (BRD4) by tuning kinase selectivity. J Med Chem. 2019;62(5):2618–2637. doi: 10.1021/acs.jmedchem.8b01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonawane V., Mohd Siddique M.U., Jadav S.S., Sinha B.N., Jayaprakash V., Chaudhuri B. Cink4T, a quinazolinone-based dual inhibitor of Cdk4 and tubulin polymerization, identified via ligand-based virtual screening, for efficient anticancer therapy. Eur J Med Chem. 2019;165:115–132. doi: 10.1016/j.ejmech.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Cheng G., Wang Z., Yang J., Bao Y., Xu Q., Zhao L. Design, synthesis and biological evaluation of novel indole derivatives as potential HDAC/BRD4 dual inhibitors and anti-leukemia agents. Bioorg Chem. 2019;84:410–417. doi: 10.1016/j.bioorg.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Ciceri P., Muller S., O'Mahony A., Fedorov O., Filippakopoulos P., Hunt J.P. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol. 2014;10(4):305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S., Yosief H.O., Dai L., Huang H., Dhawan G., Zhang X. Structure-guided design and development of potent and selective dual bromodomain 4 (BRD4)/Polo-like kinase 1 (PLK1) inhibitors. J Med Chem. 2018;61(17):7785–7795. doi: 10.1021/acs.jmedchem.8b00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao F., Li J., Luo Q., Wang R., Kong Y., Carlock C. Plk1 inhibition enhances the efficacy of BET epigenetic reader blockade in castration-resistant prostate cancer. Mol Cancer Ther. 2018;17(7):1554–1565. doi: 10.1158/1535-7163.MCT-17-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H., Mao J.H., Wang Y., Gu D.H., Pan X.D., Shan Y. Dual inhibition of BRD4 and PI3K-AKT by SF2523 suppresses human renal cell carcinoma cell growth. Oncotarget. 2017;8(58):98471–98481. doi: 10.18632/oncotarget.21432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ember S.W., Lambert Q.T., Berndt N., Gunawan S., Ayaz M., Tauro M. Potent dual BET bromodomain-kinase inhibitors as value-added multitargeted chemical probes and cancer therapeutics. Mol Cancer Ther. 2017;16(6):1054–1067. doi: 10.1158/1535-7163.MCT-16-0568-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrews F.H., Singh A.R., Joshi S., Smith C.A., Morales G.A., Garlich J.R. Dual-activity PI3K-BRD4 inhibitor for the orthogonal inhibition of MYC to block tumor growth and metastasis. Proc Natl Acad Sci U S A. 2017;114(7):E1072–E1080. doi: 10.1073/pnas.1613091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henssen A.G., Reed C., Jiang E., Garcia H.D., von Stebut J., MacArthur I.C. Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Sci Transl Med. 2017;9(414) doi: 10.1126/scitranslmed.aam9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson M.D., Smyth G.K. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 37.deCarvalho A.C., Kim H., Poisson L.M., Winn M.E., Mueller C., Cherba D. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet. 2018;50(5):708–717. doi: 10.1038/s41588-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Erikson R.L. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci U S A. 2003;100(10):5789–5794. doi: 10.1073/pnas.1031523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marder B.A., Morgan W.F. Delayed chromosomal instability induced by DNA damage. Mol Cell Biol. 1993;13(11):6667–6677. doi: 10.1128/mcb.13.11.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westermann F., Muth D., Benner A., Bauer T., Henrich K.O., Oberthuer A. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9(10):R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy A.S., Zhang S. Polypharmacology: drug discovery for the future. Expert Rev Clin Pharmacol. 2013;6(1):41–47. doi: 10.1586/ecp.12.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kizilocak H., Okcu F. Late effects of therapy in childhood acute lymphoblastic leukemia survivors. Turk J Haematol. 2019;36(1):1–11. doi: 10.4274/tjh.galenos.2018.2018.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calhoun D.A. Use of single-pill combination therapy in the evolving paradigm of hypertension management. Expert Opin Pharmacother. 2009;10(12):1869–1874. doi: 10.1517/14656560902980210. [DOI] [PubMed] [Google Scholar]
- 45.Mazur P.K., Herner A., Mello S.S., Wirth M., Hausmann S., Sanchez-Rivera F.J. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21(10):1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paoluzzi L., Hanniford D., Sokolova E., Osman I., Darvishian F., Wang J. BET and BRAF inhibitors act synergistically against BRAF-mutant melanoma. Cancer Med. 2016;5(6):1183–1193. doi: 10.1002/cam4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vazquez R., Riveiro M.E., Astorgues-Xerri L., Odore E., Rezai K., Erba E. The bromodomain inhibitor OTX015 (MK-8628) exerts anti-tumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget. 2017;8(5):7598–7613. doi: 10.18632/oncotarget.13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L., Zhang Y., Shan W., Hu Z., Yuan J., Pi J. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci Transl Med. 2017;9(400) doi: 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lam L.T., Lin X., Faivre E.J., Yang Z., Huang X., Wilcox D.M. Vulnerability of small-cell lung cancer to apoptosis induced by the combination of BET bromodomain proteins and BCL2 inhibitors. Mol Cancer Ther. 2017;16(8):1511–1520. doi: 10.1158/1535-7163.MCT-16-0459. [DOI] [PubMed] [Google Scholar]
- 50.Enssle J.C., Boedicker C., Wanior M., Vogler M., Knapp S., Fulda S. Co-targeting of BET proteins and HDACs as a novel approach to trigger apoptosis in rhabdomyosarcoma cells. Cancer Lett. 2018;428:160–172. doi: 10.1016/j.canlet.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 51.Karakashev S., Zhu H., Yokoyama Y., Zhao B., Fatkhutdinov N., Kossenkov A.V. BET bromodomain inhibition synergizes with PARP inhibitor in epithelial ovarian cancer. Cell Rep. 2017;21(12):3398–3405. doi: 10.1016/j.celrep.2017.11.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters J.U. Polypharmacology - foe or friend? J Med Chem. 2013;56(22):8955–8971. doi: 10.1021/jm400856t. [DOI] [PubMed] [Google Scholar]
- 53.Carlino L., Rastelli G. Dual kinase-bromodomain inhibitors in anticancer drug discovery: a structural and pharmacological perspective. J Med Chem. 2016;59(20):9305–9320. doi: 10.1021/acs.jmedchem.6b00438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.