Abstract
The accumulation of lipids is a hallmark of human clear cell renal cell carcinoma (ccRCC). Advanced ccRCC tumors frequently show increased lipid biosynthesis, but the regulation of lipid metabolism in early stage ccRCC tumors has not been studied. Here, we performed combined transcriptomics and metabolomics on a previously characterized transgenic mouse model (TRAnsgenic Cancer of the Kidney, TRACK) of early stage ccRCC. We found that in TRACK kidneys, HIF1α activation increases transcripts of lipid receptors (Cd36, ACVRL1), lipid storage genes (Hilpda and Fabp7), and intracellular levels of essential fatty acids, including linoleic acid and linolenic acid. Feeding the TRACK mice a high-fat diet enhances lipid accumulation in the kidneys. These results show that HIF1α increases the uptake and storage of dietary lipids in this early stage ccRCC model. By then analyzing early stage human ccRCC specimens, we found similar increases in CD36 transcripts and increases in linoleic and linolenic acid relative to normal kidney samples. CD36 mRNA levels decreased, while FASN transcript levels increased with increasing ccRCC tumor stage. These results suggest that an increase in the lipid biosynthesis pathway in advanced ccRCC tumors may compensate for a decreased capacity of these advanced ccRCCs to scavenge extracellular lipids.
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common type of kidney cancer, accounting for over 70% of all primary renal tumors [1]. Clear cell morphology results from the presence of intracellular lipid droplets [2,3]. These lipid droplets are produced in the endoplasmic reticulum (ER), serve as a bioenergetic fuel and for the generation of cell membranes, and may play a role in protecting against oxidative and ER stress [2,3]. The mechanisms involved in lipid droplet accumulation in ccRCC are reported to be increased de novo lipogenesis through reductive glutamine carboxylation, in combination with inhibition of lipid degradation [[4], [5], [6]].
Molecular profiling studies of human primary tumor specimens have revealed the loss of von Hippel Lindau (VHL) as the only consistent clonal event during ccRCC initiation [7,8]. In early stage ccRCC, the expression levels of HIF1α are markedly enhanced by loss or inactivation of VHL tumor suppressor gene [9,10]. Additional genomic events, such as loss of PBRM1, promote the malignant transformation of kidney lesions through activation of HIF1α transcriptional activity [11,12]. To model early stage ccRCC in mice, we previously generated the TRACK (TRAnsgenic Cancer of the Kidney) transgenic mouse model with expression of a mutant, constitutively active HIF1α specifically in the proximal tubules of the kidneys [13]. These mutations in the oxygen-dependent degradation domain of the HIF1α found in TRACK kidneys preclude recognition by VHL, interfere with the proteasomal degradation of this HIF1α, and promote its transcriptional activity in the kidneys. Our subsequent histologic, transcriptomics, and metabolomics analyses showed that this TRACK model shows major similarities to early human ccRCC, including the formation of lipid-filled “clear cells” and a metabolic switch to aerobic glycolysis [[13], [14], [15]]. Here, we analyzed the role of HIF1α in the regulation of lipid uptake and metabolism in early stage ccRCC in TRACK mice, and we compared these results in TRACK mice with human ccRCC patient data. Our results suggest that activation of HIF1α signaling promotes dietary lipid uptake in early stage ccRCC to a greater degree than in late stage ccRCC.
Materials and Methods
Transgenic Mouse Experiments
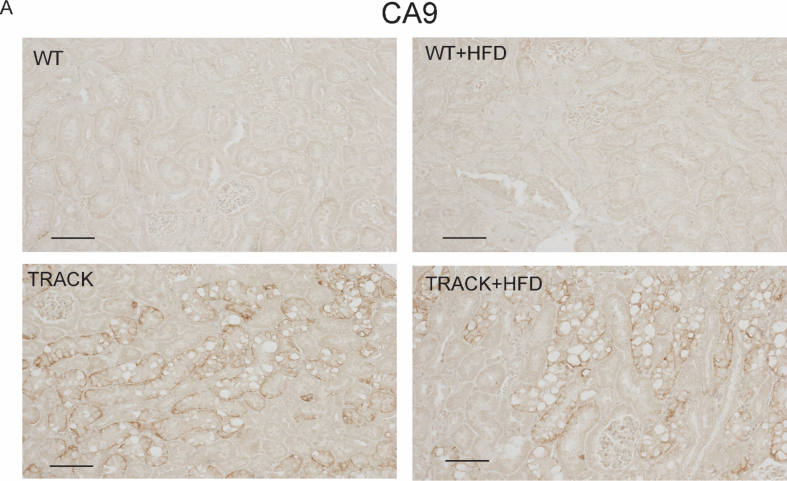
Wild type (WT) C57BL/6 male mice and transgenic lines in the C57BL/6 background carrying constitutively active mutants of HIF1α (P402A, P564A, N803A, γ-HIF1αM3) driven by a truncated γ-glutamyl transpeptidase promoter, as previously characterized, were used for this study [13]. The mice were housed at the Research Animal Resource Center of Weill Cornell Medical College (WCMC). The care and use of these animals was approved by the Institutional Animal Care and Use Committee of WCMC. All mice were fed a regular chow diet after weaning for 1–2 weeks, after which they were randomly assigned to receive a regular chow (#5053, Lab Diet) or high-fat diet (HFD) (#58v8, Test Diet). HFD contains 23.6% fat by weight (versus 5% in the regular diet), is rich in saturated fatty acids (FAs) (9.05% versus 0.78%) and monounsaturated FAs (9.32% versus 0.96%), and has increased amounts of polyunsaturated FAs, including linolenic and linoleic acid. A total of 43 male mice were treated with HFD (γ-HIF1αM3 [TRACK] n = 23, WT n = 20), and 24 mice were used as reference (TRACK n = 16, WT n = 8). We sacrificed mice after 2, 4, 6, 10, 12, and 15 months to study the phenotype. In total, 5 groups of mice, 12–18 months old, were fed a regular diet and used for metabolomics (n = 14 WT, n = 12 TRACK) or transcriptomics (n = 6 WT, n = 3 TRACK). Immunohistochemistry was conducted on the sectioned paraffin- and OCT-embedded kidneys. We used an antibody to CA9 (CA-IX) to show that the TRACK mice were positive for this HIF1α target and ccRCC marker [16] (Suppl. Figure 1). We stained sections with Oil Red O (ORO) (Rowley 1320-06-5). Representative images of each group were selected by a pathologist blinded to the group labels.
Metabolomics Analysis
We harvested the kidneys from six TRACK and five WT mice. Metabolite profiling was performed as previously described [14,17]. Briefly, tissue samples were washed in cold PBS, followed by three cycles of bead beating in 80%−70 °C methanol:water using a tissue lyser cell disrupter. The metabolites in the extraction mixture were separated from proteins by centrifugation. The supernatants were pooled, dried in a speed vac, and stored at −80 °C. The metabolites were solubilized in 0.2 M NaOH and subsequently measured by LC/MS and LC-MS/MS. Untargeted metabolite profiling was performed using aqueous normal phase (ANP) and reverse phase (RP) chromatographic separations, followed by dual spray electrospray ionization and high-resolution accurate mass determination using a time-of-flight (TOF) mass spectrometer (Agilent model 6230). The LC system comprised a Cogent Diamond Hydride™ (ANP) column (MicroSolv Technology Corporation, Eatontown, NJ), a Zorbax SB-AQ (RP) column (Agilent Technologies, Santa Clara, CA), and a Model 1200 Rapid Resolution LC system. An Agilent 6538 UHD Accurate-Mass Q-TOF with the same ANP and RP platform was used to conduct fragmentation analysis for confident molecular identification. Metabolites were normalized to protein as measured with the Bio-Rad DC protein assays. Metabolomics Data Processing was performed using MassHunter Qualitative Analysis software. Statistical analysis was done in Mass Profiler Professional (Agilent Technology, MPP, version B2.02). Aligned molecular features detected in all biological replicates of at least one group were directly applied for statistical analysis across treatment groups by MPP.
Whole Transcriptome RNA Sequencing
We extracted total RNA from one thin, outer slice of the two kidney cortices of each of three TRACK (13 months) and three age-matched, WT mice. RNeasy spin columns (Qiagen) were used to purify RNA. The complete transcriptomes were sequenced using an Illumina HiSeq2000 Sequencer with 51bp single-end reads and 4 samples per lane as previously described [15]. Ingenuity pathway analysis (IPA) was performed with the DESeq2 processed data using the default settings of the software. All genes that displayed a significant change (q < 0.05) were included in the analysis.
Analysis of Publicly Available Human RCC Data
Publicly available transcriptomics data from 66 chromophobe, 533 clear cell, and 290 papillary RCC (pRCC) tumor specimens, along with the results from 129 flanking normal kidney specimens, were used as deposited by The Cancer Genome Atlas (TCGA) on January 28, 2016 with the digital object identifier 10.7908. The preprocessed RNA-Seq by Expectation Maximization (RSEM) normalized data were used. Metabolomics data from 138 patients with ccRCC were used as included in the supplementary data in Hakimi et al. [18]. The levels of individual metabolites from patients with different stage tumors were separated from their levels in corresponding flanking normal kidney tissue, as depicted in the sample output file.
Statistical Analysis
One-way analysis of variance was applied with Tukey's multiple comparison posttest to test statistical significance of the gene expression differences in kidney cancer. All p-values obtained with RNA-Seq were adjusted for the false discovery rate to yield q-values. MPP was used to provide a multivariate statistical platform for comparative metabolite profiling. The two-sided Student's t-test was applied to determine statistical significance of metabolite differences. The association between different gene expression levels and disease stage was assessed by calculation of Spearman's correlation coefficient. A two-sided p < 0.05 or q < 0.05 was considered to indicate significance.
Results
Inhibition of Fatty Acid Biosynthesis and β-Oxidation Pathways in the Kidneys of TRACK Mice
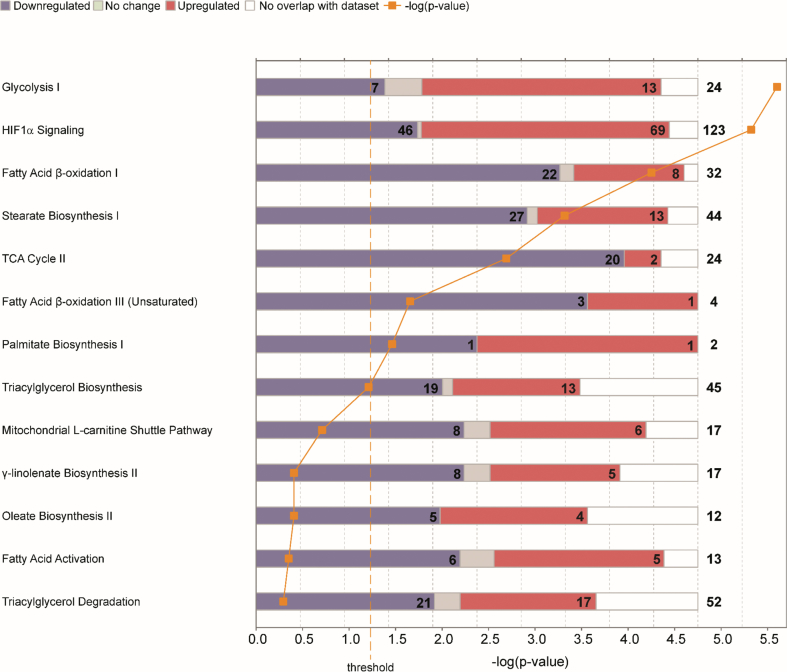
We previously showed that constitutive activation of HIF1α in the kidneys of TRACK mice promotes a metabolic switch to aerobic glycolysis [14]. To investigate further changes in metabolism in this model, we performed IPA. We used differentially expressed genes (q < 0.05) identified in a whole-genome transcriptomics analysis of the kidney cortices from 13-month-old TRACK mice compared with age-matched WT mice (Figure 1). We focused on the canonical pathways with lipid or glucose metabolism annotations. Consistent with our previous analyses, we found the glycolysis I, HIF1α signaling, and TCA cycle II pathways among the most significantly changed pathways in TRACK versus WT kidneys. In addition, the fatty acid β-oxidation I, fatty acid β-oxidation III (unsaturated), stearate biosynthesis I, and palmitate biosynthesis I pathways were perturbed. The fatty acid β-oxidation I and stearate biosynthesis I were among the pathways most significantly altered. The majority of the genes in these two pathways showed lower mRNA levels in TRACK (22/32 genes; 27/44 genes) as compared with WT kidneys, and by IPA, both pathways were predicted to be inhibited.
Figure 1.
Analysis of lipid metabolism pathways in the kidney cortices of transgenic TRACK mice compared with wild-type mice. Ingenuity pathway analysis (IPA) of differentially expressed genes (q < 0.05) in kidney cortices of three TRACK as compared with three WT mice. Stacked bar charts illustrate the number of decreased (blue) and increased (red) transcripts in each individual canonical pathway. The total number of transcripts in each pathway is shown in the far-right column, while the numbers of decreased and increased transcripts are presented in the bars. The right-tailed Fisher's exact test was used to determine statistical significance. The glycolysis I, fatty acid β-oxidation I, stearate biosynthesis I, TCA cycle II, fatty acid β-oxidation III (unsaturated), and palmitate biosynthesis I pathways are significantly (p < 0.05) perturbed in TRACK as compared with WT kidneys. The glycolysis I and HIF1α signaling pathways predominantly contain increased transcripts, indicating activation, while the fatty acid β-oxidation I, stearate biosynthesis I, and TCA cycle II pathways are predicted to be inhibited. These results suggest that constitutive activation of HIF1α in TRACK kidneys increases nonoxidative glucose metabolism, while inhibiting lipid degradation and biosynthesis.
Dietary Lipid Uptake in the Kidneys of TRACK Mice
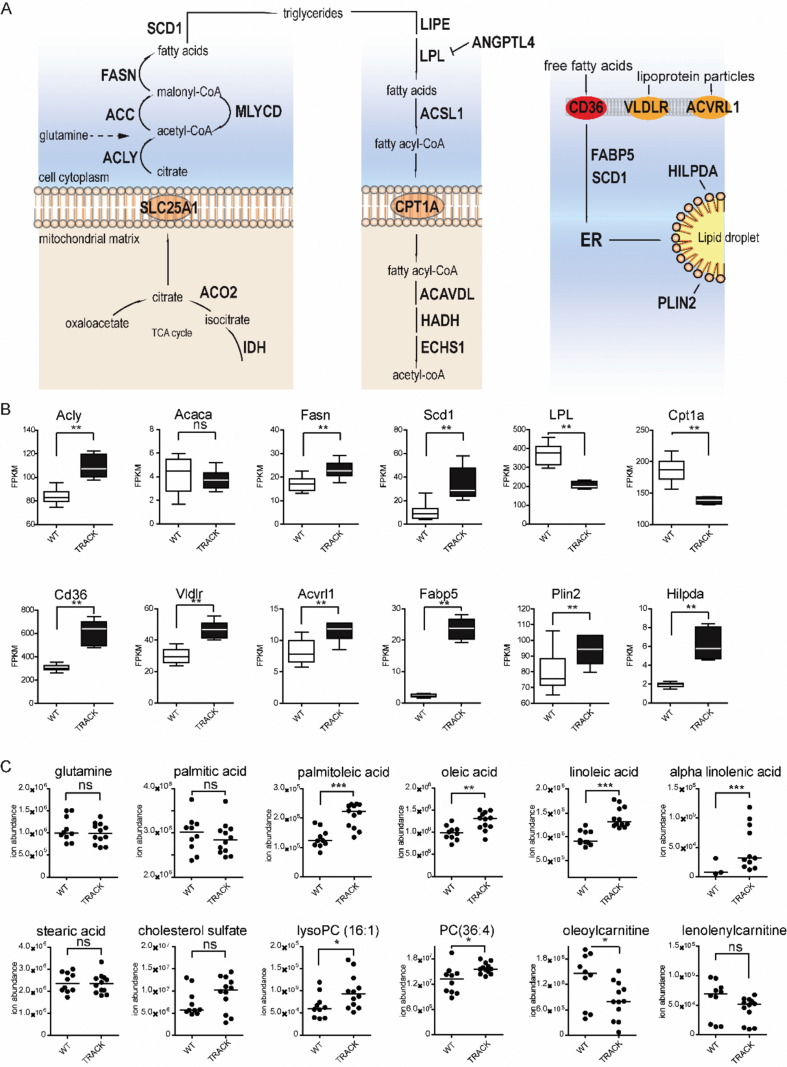
To analyze the regulation of lipid metabolism in the TRACK kidneys, we then measured the levels of critical mRNAs, small molecule intermediates, and products of lipid metabolism. The rate-limiting enzymes in the lipid biosynthesis and β-oxidation pathways have been described (Figure 2A) [19]. Although we detected increased transcript levels of several genes involved in lipid biosynthesis (Acly, Fasn, Scd1, Figure 2B), no significant changes were noted in the expected metabolites (glutamine, palmitic acid, stearic acid, Figure 2C). We detected decreased levels of Lpl and Cpt1a transcripts, as well as decreases in the levels of metabolites in the β-oxidation pathway (oleoylcarnitine; Figure 2B, C) in TRACK compared with WT kidneys.
Figure 2.
Analysis of dietary lipid uptake in kidney cortices of transgenic TRACK mice compared with wild-type mice. Cortices from six TRACK and five WT mouse kidneys were compared for RNA expression and small molecule intermediates and products of metabolism (metabolomics). Statistical significance was assessed using a two-sided Student's t-test. A illustrates the specific lipid biosynthesis (left panel), lipid degradation (center panel), and lipid uptake (right panel) products analyzed. B illustrates quantification of mRNA (Fragments Per Kilobase per Million mapped reads [FPKM]) comparing WT and TRACK kidney cortices. Transcripts related to lipid biosynthesis, uptake, and storage are increased, while the mRNA levels of lipid degradation genes are reduced in TRACK compared with WT. C illustrates the relative ion abundance of metabolites. Note a significant (p < 0.05) increase in the levels of (essential) fatty acids (palmitoleic acid, oleic acid, linoleic acid, alpha linolenic acid), as well as a large (p < 0.05) decrease in fatty acid carnitine products (oleoylcarnitine) in TRACK as compared with WT kidneys, showing that HIF1α promotes the uptake of dietary lipids (such as the essential fatty acids linoleic and alpha-linolenic acid) presumably through increased expression of lipid receptors in the kidneys of transgenic mice.
Uptake of extracellular lipids can act as an alternative way to increase the cellular lipid content during hypoxia [20,21]. We thus investigated whether scavenging of extracellular lipids contributes to the lipid accumulation in the kidneys of TRACK mice. We detected increased transcript levels of genes involved in FA transport (Cd36, Fabp5) and cholesterol uptake (Vldlr, ACVRL1) in TRACK compared with WT mice (Figure 2B). We also measured increased levels of unsaturated long-chain FAs (oleic acid, palmitoleic acid) in the kidney cortices of TRACK versus WT mice (Figure 2C). Linoleic acid and linolenic acid are essential, polyunsaturated FAs that cannot be synthesized de novo in humans or mice and therefore, by definition, are obtained from the diet [22]. We detected increased levels of linolenic and linoleic acid in TRACK versus WT kidneys (Figure 2C). We also detected elevated levels of lipid storage genes (Hilpda, Plin2, Figure 2B) and esterified FAs (PC 36:4, lysoPC 16:1, Figure 2C), indicating that lipids are likely stored in lipid droplets. Collectively, these results indicate that constitutively active HIF1α promotes the uptake and storage of dietary lipids in the kidneys of TRACK mice.
A High-fat Diet Enhances Lipid Accumulation in the Kidneys of TRACK Mice
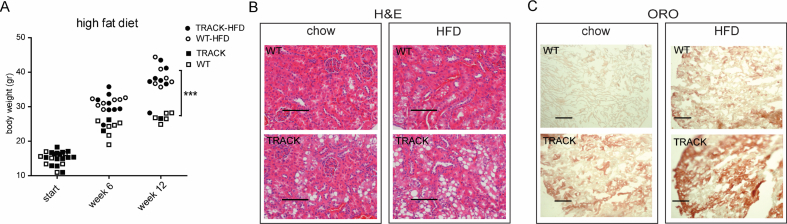
To explore further the role of extracellular lipid uptake, 1-month-old TRACK and WT mice were fed a HFD or a regular chow diet. Weight monitoring showed increases in total body weight in both WT and TRACK mice after 12 weeks on a HFD as compared with a chow diet (38.1 g versus 26.9 g, p < 0.001, Figure 3A). Twenty-three TRACK mice were fed a HFD and sacrificed after 2, 4, 6, 10, 12, and 15 months. Consistent with our previous observations, TRACK mice showed ccRCC precursor lesions, characterized by high carbonic anhydrase (CA-IX) expression, highly disorganized tubular structures, and an abundance of clear cells (Supplementary Figure 1). We did not observe any invasive growth or differences in nuclear morphology in the kidneys of the TRACK mice on a HFD versus a chow diet. The HFD-fed TRACK mice exhibited increased numbers of clear cells in the kidneys, compared with TRACK mice fed a chow diet (Figure 3B). These changes were visible in TRACK kidneys starting after 2 months on a HFD and thereafter, while minimal changes were observed in WT kidneys. Consistent with increases in FA and lipid import resulting from the HFD, the increase in clear cell abundance was accompanied by increased levels of neutral triglycerides (TGs) and lipids, as assessed by ORO staining, in TRACK mice (Figure 3C). These results suggest that the constitutively active HIF1α promotes the accumulation of dietary lipids in the kidneys of TRACK mice. Longer latency periods or alternative dietary formulations may be required to determine the effects of a HFD on tumor development.
Figure 3.
Impact of a high fat-diet on lipid accumulation in the kidneys of TRACK transgenic mice compared with wild-type mice. Mice (43 total) were fed a high-fat diet (HFD) (TRACK n = 23, wild-type n = 20), and 24 mice were fed with regular chow diet (TRACK n = 16, wild-type n = 8). A illustrates the total body weight of individual mice at baseline, 6 and 12 weeks on the respective diets. Both TRACK and WT mice fed a HFD showed increased body weights compared with chow-fed mice (p < 0.05). B shows representative H&E stained sections of kidney cortex of WT and TRACK mice fed a regular chow (left panels) or HFD (right panels). The scale bar indicates 100 μm. Note the increase in clear cell lipid containing vacuoles in TRACK mice fed a HFD, confirmed by staining mature lipids with Oil Red O (ORO) (C).
Increased Expression of Lipid Uptake and Storage Genes in Human ccRCC
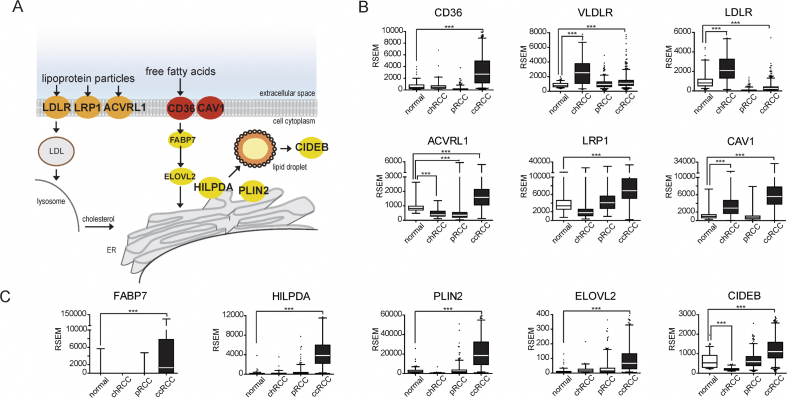
We next compared our TRACK kidney results with data from human ccRCC specimens. The presence of clear cells is a fundamental morphologic feature of human ccRCC that is not often seen in other subtypes of RCC, such as chromophobe RCC (chRCC) and pRCC [23]. Previously, TCGA consortium conducted RNAseq analysis on samples from patients with chRCC (n = 66), pRCC (n = 290), and ccRCC (n = 533) along with 129 normal flanking kidney tissue samples [7,24,25]. To investigate whether dietary lipid uptake contributes to the formation of clear cells in ccRCC, we analyzed these RNAseq datasets. We first focused on the dominant lipid receptors and compared their expression levels in the three types of RCC to normal human kidney tissue (Figure 4A). We found an increase in CD36 (5.1-fold), a modest increase in VLDLR (1.5-fold), and a decrease in LDLR (2.6-fold) transcript levels in ccRCC as compared with normal kidney (Figure 4B). Activin receptor–like type 1 (ACVRL1), LDL receptor–related protein 1 (LRP1), and caveolin 1 (CAV1) are alternative receptors for LDL particles and FAs [26,27]. We found 1.9-fold, 2.0-fold, and 5.1-fold increases, respectively, in ccRCC as compared with the normal kidney tissue (Figure 4B). In contrast, the CD36, ACVRL1, and LRP1 transcripts were either decreased or not significantly changed in chRCC and pRCC compared with the normal kidney tissue. VLDLR, LDLR, and CAV1 also showed significant increases in chRCC (Figure 4B). We conclude that CD36 and VDLR transcripts are increased relative to those in the normal kidneys specifically in human ccRCC, similar to our results in TRACK versus WT kidneys (Figure 2).
Figure 4.
mRNA levels of genes involved in lipid uptake and storage in human kidney cancer. Transcriptomics data obtained by the TCGA from normal human kidney tissue (n = 129), chromophobe (chRCC, n = 66), papillary (pRCC, n = 290), and clear cell renal cell carcinoma (ccRCC, n = 533) specimens were analyzed for lipid uptake and storage genes. A illustrates some of the lipid uptake (orange/red) and storage (yellow) pathways. B illustrates the quantitative mRNA levels (RNA-Seq by Expectation Maximization [RSEM]) of lipid uptake genes in normal tissue and different subtypes of kidney cancer, respectively. Note statistically significant increases in transcripts of lipid uptake genes CD36, VLDLR, ACVRL1, LRP1, and CAV1 in human ccRCC compared with the normal kidney. As shown in C, mRNA levels of lipid storage genes FABP7, HILPDA, PLIN2, ELOVL2, and CIDEB are also increased in ccRCC. Together, these data indicate that increased transcripts of genes involved in both dietary lipid uptake and lipid storage are common in ccRCC compared with both normal kidney and with other histologic subtypes of RCC. Statistical significance of differences in cancer specimens as compared with the normal kidney tissue was assessed using ANOVA (p < 0.05).
We next investigated markers of lipid storage in the different RCC subtypes. We found a marked and unique increase in the FA transporter FABP7 transcript (113-fold) and the FA elongation enzyme ELOV2 (11-fold) in human ccRCC specimens compared with normal kidneys (Figure 4C). At the same time, the transcripts of genes involved in lipid droplet stability (PLIN2, 7.8-fold), fusion (CIDEB, 1.9-fold), and growth (HILPDA, 21.3-fold) were increased in ccRCC compared with the normal kidney tissue (Figure 4C). The mRNA levels of these genes were either not significantly changed (FABP7, HILPDA, PLIN2, ELOVL2) or decreased (CIDEB) in the other types of RCC. These TCGA data analyses indicate that the elevated transcripts of lipid receptors and lipid storage genes are dominant features of human ccRCC.
HIF1α Promotes Dietary Lipid Uptake in Early Stage Human ccRCC
To investigate whether FA uptake occurs in human ccRCC, we probed metabolomics data obtained from a cohort of 138 patients with different stages of ccRCC [18]. Similar to our observations in TRACK mice, we noted increased amounts of the unsaturated essential FAs, linolenic (1.9 fold, p < 0.0001) and linoleic acid (1.5 fold, p < 0.0001), particularly in stage 1 ccRCC tumors, in comparison with the normal kidney tissue (Figure 5A). In these same stage tumors, we found increased levels of glutamine (1.4 fold, p < 0.0001) and palmitic acid (1.2 fold, p < 0.0001).
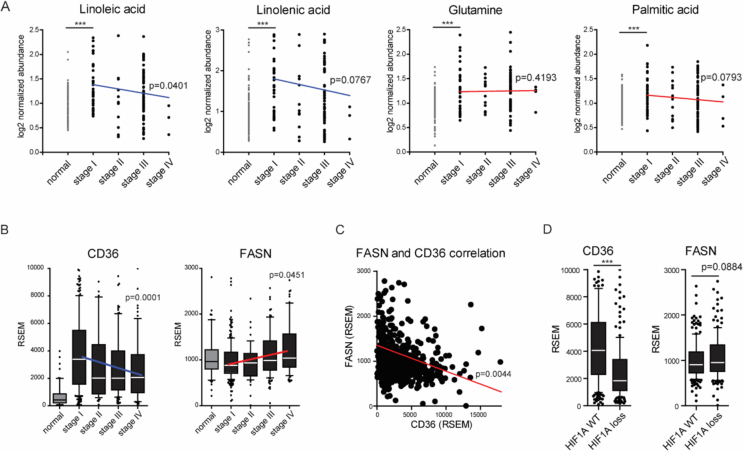
Figure 5.
Lipid metabolism in human ccRCC by tumor stage. The metabolomics (n = 138) (Panel A), transcriptomics (n = 533) (Panel B), and somatic copy number data (n = 418) (Panel D) of stage I, II, III, and IV ccRCC were analyzed. Statistical significance was assessed by the two-sided Student's t-test and Pearson's correlation. A illustrates the levels (log2 normalized abundance) of two essential fatty acids (linoleic and linolenic acid) and intermediates of lipid biosynthesis (glutamine, palmitic acid) as measured by LC-MS/MS. A statistically significant (p < 0.05) increase in essential fatty acids and lipid biosynthesis intermediates was detected in stage I ccRCC tumors as compared with the normal kidney tissue. As stage increased, the level of linoleic acid decreased (p < 0.05), while no significant changes in the glutamine or palmitic acid levels were observed. B illustrates levels of CD36 and FASN mRNA levels (RNA-Seq by Expectation Maximization [RSEM]) by tumor stage. CD36 mRNA levels decreased with increasing tumor stage, while the FASN transcript levels increased with tumor stage. C shows the association between CD36 and FASN mRNA levels within each individual patient with ccRCC. An inverse association was found between the CD36 and FASN expression levels. D shows the CD36 and FASN mRNA levels in patients with or without a HIF1A somatic copy number reduction. We found higher CD36 mRNA levels in patients with wild-type (WT) HIF1A. Collectively, these results suggest that HIF1α increases dietary lipid uptake in patients with early stage ccRCC, while tumors in patients with advanced ccRCC may rely on lipid biosynthesis.
Previous research by the TCGA showed a metabolic shift toward lipid biosynthesis in subgroup of ccRCC patients with a poor prognosis [7]. To gain insight into the relative importance of lipid import during disease progression, we analyzed FA and glutamine levels in the same cohort of 138 patients at different stages of disease. Although the distribution of tumor staging was skewed toward stages 1 and 3, we found a trend toward a decrease in linoleic and linolenic acids with increasing disease stage (Figure 5A, Pearson R −0.1495 and −0.1222, p = 0.0401 and p = 0.0767). In contrast, we detected no clear correlation between glutamine or palmitic acid and tumor stage (R −0.0175, −0.1207, p = 0.4193, p = 0.0793). These results are consistent with the possibility that ccRCC tumors recruit fewer dietary lipid decreases during disease progression. We also compared the CD36 and FASN mRNA levels in patients with different stages of ccRCC. Similar to the decreases in linolenic and linoleic acid, we found a decrease in CD36 mRNA levels with increasing ccRCC tumor stage, while the FASN mRNA levels showed an increasing trend with tumor stage (Figure 5B, Pearson correlation coefficients −0.1594 and 0.0735, p = 0.0001 and p = 0.0451). To determine whether the increase in lipid biosynthesis correlated with a decrease in dietary lipid uptake, we investigated the association of CD36 and FASN within each patient. Indeed, we found an inverse correlation between FASN and CD36 mRNA levels (Figure 5C, Pearson R −0.1594, p = 0.0044). We also compared the CD36 and FASN transcript levels in ccRCC patients with WT HIF1α versus a loss of the HIF1α locus. We detected decreased CD36 mRNA levels, but no change in FASN transcripts in patients that showed loss of at least one allele of the HIF1α gene (Figure 5D). Our results suggest that dietary lipid uptake in early stage ccRCC may be driven by HIF1α signaling in human ccRCC in addition to TRACK mice.
Discussion
The accumulation and storage of lipids are critical for the management of oxidative and ER stress, and lipids promote homeostasis in ccRCC tumors [2,3,28]. Here, we provide evidence from whole genome transcriptomics analyses that HIF1α signaling enhances the accumulation of lipids in early stage ccRCC through the uptake of extracellular lipids. Our results indicate that HIF1α signaling increases the transcript levels of lipid receptors, such as CD36 and ACVRL1, as well as transcripts of genes involved in lipid transport (FABP7) and storage (PLIN2 and HILPDA). We here show that the activation of the lipid biosynthesis pathway may compensate for the decreased ability of more advanced stage ccRCC tumors to scavenge extracellular lipids. These more advanced ccRCC tumors may acquire a relative dependency on the HIF2α or the MTORC1 pathway, which were previously shown to drive the de novo lipogenesis gene network, including FASN [5,29].
Du et al. [30] showed that the rate-limiting enzyme in mitochondrial FA import, carnitine palmitoyltransferase 1a (CPT1a), is directly repressed by HIF1α and HIF2α in human ccRCC cultures and that this repression results in a decrease in FA catabolism. In TRACK kidney cortices, we have shown that CPT1a transcripts are greatly reduced relative to levels in WT kidney cortices (Figure 2B), so a reduction in FA catabolism plus increased lipid uptake resulting from HIF1α activation should result in much greater internal levels of FAs in early stage ccRCC.
Previous lipidomic profiling of human ccRCC specimens showed increased levels of mature TGs, as well as cholesterol esters in cancerous tissues as compared with the normal tissue [31]. Human tumors were also found to be enriched for polyunsaturated, long-chain FAs. These results are in line with preclinical data showing that hypoxic cells preferentially scavenge unsaturated FAs from phospholipids [21,28]. We found increased transcript levels of CD36, SCD1, and ELOVL2 in TRACK kidneys, as well as in early stage human ccRCC tumors. These proteins are known to import, desaturate, and elongate FAs, ultimately yielding the most abundant substrates in the tumors. We also measured increased transcript levels of several cholesterol receptors (VLDLR, ACVRL1, LRP1) in TRACK kidneys and human ccRCC, while we detected one nonsignificantly increased cholesterol ester (cholesterol sulfate) in our metabolomics analysis of the TRACK kidneys. These results suggest that HIF1α activation may not only lead to increased dietary FA uptake but that HIF1α may also enhance the enzymatic processing of FAs and cholesterol uptake. Future research will be needed to determine if (un)saturated FAs, dietary lipids, and cholesterol have distinct roles in ccRCC.
Previous research showed that unsaturated FAs protect against reactive oxygen species in glioblastoma and breast cancer cells [20]. An increase in antioxidant defense mechanisms, with increased glutathione metabolism and somatic alterations of the redox regulators KEAP1 and NRF2, was previously shown in patients with aggressive ccRCC [18]. Although unsaturated FAs may reduce oxidative stress, the reverse has been shown for saturated FAs [32,33]. The results presented here suggest that the composition of dietary lipids may influence the disease course in patients with ccRCC.
Acknowledgments
The authors would like to thank all members of the Gudas lab, K. W. Eng, and Dr. A. J. Dannenberg for stimulating discussions and valuable input. The TRACK kidney RNA-Seq data are deposited in the NCBI database, accession number GSE54390. The metabolomics data are deposited. This research was funded in part by the Weiss family and Turobiner Kidney Cancer Research Fund and was partly supported by U54 CA210184 and Weill Cornell funds.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.10.015.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Supplementary Figure 1.
CA-IX staining in kidneys of high-fat diet fed WT and TRACK mice. CA-IX shows similar positive, membranous staining in the kidneys of the TRACK chow-fed and the TRACK- high-fat diet fed mice, but CA-IX is negative in the wild-type and wild-type-high fat diet mice. The antibody used was sc-25600, 1:50 dilution (Santa Cruz).
References
- 1.Patard J.J., Leray E., Rioux-Leclercq N., Cindolo L., Ficarra V., Zisman A., De La Taille A., Tostain J., Artibani W., Abbou C.C. Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J Clin Oncol. 2005;23:2763–2771. doi: 10.1200/JCO.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu B., Ackerman D., Sanchez D.J., Li B., Ochocki J.D., Grazioli A., Bobrovnikova-Marjon E., Diehl J.A., Keith B., Simon M.C. HIF2alpha-Dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gameiro P.A., Yang J., Metelo A.M., Pérez-Carro R., Baker R., Wang Z., Arreola A., Rathmell W.K., Olumi A., Lopez-Larrubia P. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17:372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., DeBerardinis R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Network C.G.A.R. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turajlic S., Xu H., Litchfield K., Rowan A., Chambers T., Lopez J.I., Nicol D., O'Brien T., Larkin J., Horswell S. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 2018;173:581–594 e12. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 10.Klatte T., Seligson D.B., Riggs S.B., Leppert J.T., Berkman M.K., Kleid M.D., Yu H., Kabbinavar F.F., Pantuck A.J., Belldegrun A.S. Hypoxia-inducible factor 1 alpha in clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:7388–7393. doi: 10.1158/1078-0432.CCR-07-0411. [DOI] [PubMed] [Google Scholar]
- 11.Nargund A.M., Pham C.G., Dong Y., Wang P.I., Osmangeyoglu H.U., Xie Y., Aras O., Han S., Oyama T., Takeda S. The SWI/SNF protein PBRM1 restrains VHL-loss-driven clear cell renal cell carcinoma. Cell Rep. 2017;18:2893–2906. doi: 10.1016/j.celrep.2017.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao W., Li W., Xiao T., Liu X.S., Kaelin W.G. Inactivation of the PBRM1 tumor suppressor gene amplifies the HIF-response in VHL-/- clear cell renal carcinoma. Proc Natl Acad Sci U S A. 2017;114:1027–1032. doi: 10.1073/pnas.1619726114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu L., Wang G., Shevchuk M.M., Nanus D.M., Gudas L.J. Generation of a mouse model of Von Hippel-Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1alpha. Cancer Res. 2011;71:6848–6856. doi: 10.1158/0008-5472.CAN-11-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minton D.R., Fu L., Chen Q., Robinson B.D., Gross S.S., Nanus D.M., Gudas L.J. Analyses of the transcriptome and metabolome demonstrate that HIF1α mediates altered tumor metabolism in clear cell renal cell carcinoma. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu L., Minton D.R., Zhang T., Nanus D.M., Gudas L.J. Genome-wide profiling of TRACK kidneys shows similarity to the human ccRCC transcriptome. Mol Cancer Res. 2015;13(5):870–878. doi: 10.1158/1541-7786.MCR-14-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dornbusch J., Zacharis A., Meinhardt M., Erdmann K., Wolff I., Froehner M., Wirth M.P., Zastrow S., Fuessel S. Analyses of potential predictive markers and survival data for a response to sunitinib in patients with metastatic renal cell carcinoma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q., Park H.C., Goligorsky M.S., Chander P., Fischer S.M., Gross S.S. Untargeted plasma metabolite profiling reveals the broad systemic consequences of xanthine oxidoreductase inactivation in mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A., Aksoy B.A., Liu E.M., Shen R., Lee W. An integrated metabolic Atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Currie E., Schulze A., Zechner R., Walther T.C., Farese R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bensaad K., Favaro E., Lewis C.A., Peck B., Lord S., Collins J.M., Pinnick K.E., Wigfield S., Buffa F.M., Li J.L. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 21.Kamphorst J.J., Cross J.R., Fan J., de Stanchina E., Mathew R., White E.P., Thompson C.B., Rabinowitz J.D. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen A.E., Burr G.O. Essential fatty acids and human nutrition. J Am Med Assoc. 1946;132:855–859. [PubMed] [Google Scholar]
- 23.Srigley J.R., Delahunt B., Eble J.N., Egevad L., Epstein J.I., Grignon D., Hes O., Moch H., Montironi R., Tickoo S.K. The International Society of Urological Pathology (ISUP) vancouver classification of renal neoplasia. Am J Surg Pathol. 2013;37:1469–1489. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 24.Davis C.F., Ricketts C.J., Wang M., Yang L., Cherniack A.D., Shen H., Buhay C., Kang H., Kim S.C., Fahey C.C. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linehan W.M., Spellman P.T., Ricketts C.J., Creighton C.J., Fei S.S., Davis C., Wheeler D.A., Murray B.A., Schmidt L., Vocke C.D. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374:135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowal R.C., Herz J., Goldstein J.L., Esser V., Brown M.S. Low density lipoprotein receptor-related protein mediates uptake of cholesteryl esters derived from apoprotein E-enriched lipoproteins. Proc Natl Acad Sci U S A. 1989;86:5810–5814. doi: 10.1073/pnas.86.15.5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraehling J.R., Chidlow J.H., Rajagopal C., Sugiyama M.G., Fowler J.W., Lee M.Y., Zhang X., Ramirez C.M., Park E.J., Tao B. Genome-wide RNAi screen reveals ALK1 mediates LDL uptake and transcytosis in endothelial cells. Nat Commun. 2016;7:13516. doi: 10.1038/ncomms13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackerman D., Tumanov S., Qiu B., Michalopoulou E., Spata M., Azzam A., Xie H., Simon M.C., Kamphorst J.J. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 2018;24:2596–25605 e5. doi: 10.1016/j.celrep.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duvel K., Yecies J.L., Menon S., Raman P., Lipovsky A.I., Souza A.L., Triantafellow E., Ma Q., Gorski R., Cleaver S. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du W., Zhang L., Brett-Morris A., Aguila B., Kerner J., Hoppel C.L., Puchowicz M., Serra D., Herrero L., Rini B.I. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat Commun. 2017;8:1769. doi: 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito K., Arai E., Maekawa K., Ishikawa M., Fujimoto H., Taguchi R., Matsumoto K., Kanai Y., Saito Y. Lipidomic signatures and associated transcriptomic profiles of clear cell renal cell carcinoma. Sci Rep. 2016;6:28932. doi: 10.1038/srep28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green C.D., Olson L.K. Modulation of palmitate-induced endoplasmic reticulum stress and apoptosis in pancreatic beta-cells by stearoyl-CoA desaturase and Elovl6. Am J Physiol Endocrinol Metab. 2011;300:E640–E649. doi: 10.1152/ajpendo.00544.2010. [DOI] [PubMed] [Google Scholar]
- 33.Listenberger L.L., Han X., Lewis S.E., Cases S., Farese R.V., Jr., Ory D.S., Schaffer J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci U S A. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.