Abstract
Background
Abnormal P‐wave morphology (PWM) has been associated with a history of atrial fibrillation (AF) in earlier studies. Although lone AF is believed to have substantial genetic basis, studies on associations between single nucleotide polymorphisms (SNP) linked to lone AF and PWM have not been reported. We aimed to assess whether SNPs previously associated with lone AF (rs2200733, rs13376333, rs3807989, and rs11047543) are also linked to P‐wave abnormalities.
Methods
Four SNPs were studied in 176 unrelated individuals with early‐onset lone AF (age at onset <50 years), median age 38 years (19–63 years), 149 men. Using sinus rhythm ECG, orthogonal PWM was classified as Type 1—positive in leads X and Y and negative in lead Z, Type 2—positive in leads X and Y and biphasic (−/+) in lead Z, Type 3—positive in lead X and biphasic in lead Y (+/−), and the remaining as atypical.
Results
Two SNPs were found to be significantly associated with altered P‐wave morphology distribution: rs3807989 near the gene CAV1/CAV2 and rs11047543 near the gene SOX5. Both SNPs were associated with a higher risk of non‐Type 1 P‐wave morphology (rs3807989: OR = 4.8, 95% CI = 2.3–10.2, p < 0.001; rs11047543: OR = 4.7, 95% CI = 1.1–20.5, p = 0.04). No association was observed for rs2200733 and rs13376333.
Conclusion
In this study, the two variants rs3807989 and rs11047543, previously associated with PR interval and lone AF, were associated with altered P‐wave morphology distribution in patients with early‐onset lone AF. These findings suggest that common genetic variants may modify atrial conduction properties.
Keywords: atrial fibrillation, P‐wave morphology, SNP
1. INTRODUCTION
Atrial fibrillation (AF) is the most common cardiac arrhythmia in the general population. The underlying pathophysiology is not fully understood, but is likely to be multifactorial, including cardiovascular risk factors such as hypertension, valvular heart disease, and ischemic heart disease. However, 10%–20% of the AF population lack the traditional risk factors for AF and are considered having “lone AF” (Fuster et al., 2011). Lone AF with early onset has been suggested to be caused mainly by disturbances in ionic currents with a substantial genetic basis (Mahida, Lubitz, Rienstra, Milan, & Ellinor, 2011).
In recent years, genome‐wide association studies (GWAS) have elucidated the genetic substrate underlying AF (Benjamin et al., 2009; Ellinor et al., 2010, 2012; Gudbjartsson et al., 2007; Lubitz et al., 2014; Sinner et al., 2014). To date, at least six SNPs have been associated with lone AF in association studies (Chu et al., 2013; Olesen et al., 2011, 2012; Wirka et al., 2011; Zang et al., 2013), of which 2 was specifically associated with lone AF in GWAS (Ellinor et al., 2010). Although the role of common genetic variants is receiving increasing attention, the precise electrophysiological mechanisms by which these genetic loci lead to disease still remain unsolved.
Alterations in atrial action potential duration and atrioventricular conduction are believed to influence the AF risk (Olsson, Cotoi, & Varnauskas, 1971). Seven SNPs have been associated with both PR interval and AF risk in GWAS (Holm et al., 2010; Pfeufer et al., 2010), suggesting that several genotypes exert effects on atrial electrophysiology which are translated to surface electrocardiograms (ECGs) and that these changes could be a part of the mechanism leading to AF. If the cause of lone AF is primarily “electrical” and not related to structural heart disease, one may hypothesize that the underlying common genetic variants exert more evident alterations of atrial conductive properties that can be obtained from ECGs in patients with lone AF.
The orthogonal P‐wave morphology has in previous studies been used to explore the atrial conductive properties during sinus rhythm (SR). The P‐wave morphology reflects the duration of the atrial depolarization as well as its three‐dimensional propagation (Figure 1) (Holmqvist et al., 2008; Platonov et al., 2011). Abnormal P‐wave morphologies, most often observed as a pronounced negative terminal phase of the P wave in the right precordial leads or biphasic P waves in the orthogonal lead Z, have been associated with increasing age, advanced cardiac disease, and a history of paroxysmal AF (Havmoller et al., 2007; Holmqvist et al., 2009; Platonov et al., 2000). This abnormal morphology has even been observed in a population of lone AF, suggesting that these changes in atrial conductive properties may be part of the mechanism leading to disease development (Holmqvist et al., 2011).
Figure 1.
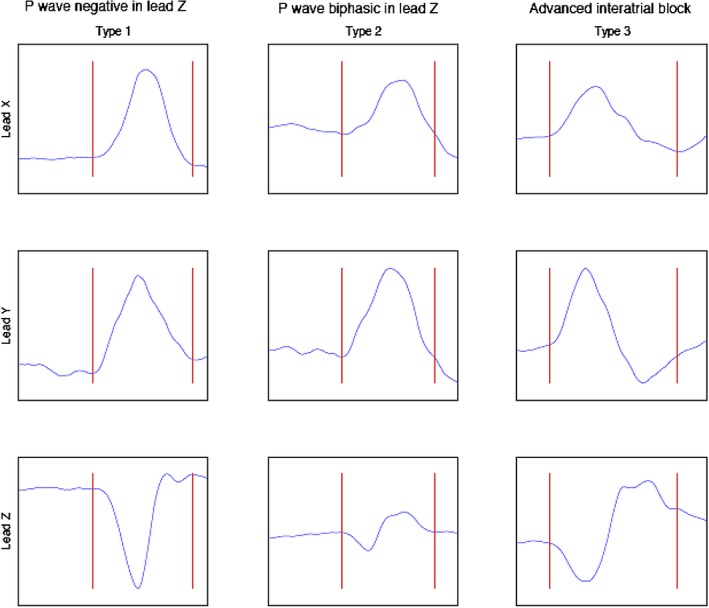
Schematic illustration of the P‐wave morphology classification. Type 1 is characterized by a right‐to‐left (positive signal in Lead X), superior‐to‐inferior (positive signal in Lead Y), and posterior‐to‐anterior (negative signal in Lead Z) activation. Type 2 is characterized by a positive signal in Lead X and Lead Y but a posterior‐to‐anterior‐to‐posterior (biphasic signal in Lead Z). Type 3 is characterized by a positive signal in Lead X, a superior‐to‐inferior‐to‐superior (biphasic signal in Lead Y), and a biphasic signal in Lead Z, reflecting an advanced interatrial block. The remaining P‐wave morphologies are categorized as atypical (Holmqvist et al., 2007)
The association between SNPs linked to lone AF and P‐wave morphology has not been explored before. In the present study, we aimed to assess whether four SNPs previously associated with lone AF (rs2200733 and rs13376333) (Ellinor et al., 2010; Olesen et al., 2012) or lone AF and PR interval (rs3807989 and rs11047543) (Olesen et al., 2012; Pfeufer et al., 2010) were associated with P‐wave abnormalities in patients with early‐onset lone AF.
2. METHODS
2.1. Study population
Unrelated individuals with early‐onset (<50 years old at diagnosis) lone paroxysmal AF (i.e., absence of clinical or echocardiographic findings of other cardiovascular diseases, hypertension, and metabolic or pulmonary disease) were included from two Scandinavian centers (Copenhagen, Denmark; Vestre Viken, Norway). Exclusion criteria were ongoing treatment with class I or class III antiarrhythmic drugs, and missing genotype data for one or more of the SNPs analyzed.
Written informed consent was obtained from all participants. The study was performed in accordance with Helsinki Declarations and was approved by the Scientific Ethics Committee of Copenhagen and Frederiksberg, and by the Regional Ethics Committee in Norway.
2.2. Data acquisition and analysis
Digital 12‐lead ECGs of 10‐second duration were recorded during SR using standard clinical equipment. ECG signals were exported as xml files for processing. The PR‐interval and P‐wave duration were automatically measured using the Glasgow algorithm (Macfarlane et al., 1990), and the P‐wave duration contribution to the overall PR interval (P/PR ratio) was calculated. Details of the method used to obtain orthogonal P‐wave morphology have been published previously (Carlson, 2005; Holmqvist, Platonov, Havmoller, & Carlson, 2007) and will be described briefly below.
Electrocardiograms were mathematically transformed into orthogonal vectorcardiograms, using the pseudo‐inverse of the Dower transform matrix (Carlson et al., 2005). This enables separate analysis of the three orthogonal leads, denoted X, Y, and Z. To reduce low‐frequency artifacts (“baseline wander”), and powerline interference, a 0.5‐Hz high‐pass filter and a 50‐Hz notch filter were applied. QRS complexes were detected automatically. QRS complexes with similar morphology were clustered together using a cross‐correlation coefficient of ρ > 0.9. Only the cluster with the largest number of complexes was used for further analysis in order to exclude artifacts and complexes of different morphologies, for example, ventricular beats. Signal segments of 250 ms preceding the QRS complexes were used to extract the P waves. The segments were shifted in time to achieve maximum correlation and were then sorted into different clusters based on a cross‐correlation coefficient of ρ > 0.9. The cluster with the largest number of P waves was signal‐averaged and used for further analysis. To extract P waves from the signal‐averaged segments, onset and end were defined manually. The morphologies of the P waves were classified as one of three predefined types based on the gross appearance of the three leads as “positive,” “negative,” or “biphasic” (either “positive/negative” or “negative/positive”; Figure 1). P waves that did not match any of the three types were denoted “atypical.”
2.3. SNP genotyping
The SNPs analyzed are presented in Table 1. Detailed description of the genotyping process has been described previously (Olesen et al., 2011). In brief, genomic DNA was extracted from whole blood, using the QIAamp DNA Blood Mini and Maxi Kits (Qiagen, Hilden, Germany). Genotypes were determined using fluorescence‐based real‐time polymerase chain reaction (PCR) (ABI PRISM 7900 Sequence Detection System; Applied Biosystems, Foster City, CA) and TaqMan assay (Applied Biosystems). In order to allow for discrimination between the allele compositions of each sample, an allelic discrimination run was performed.
Table 1.
List of analyzed single nucleotide polymorphisms, nearest gene, chromosome number, risk allele, and risk allele frequency
| SNPs | Nearest gene | Chromosome | Risk allele | RAF |
|---|---|---|---|---|
| rs2200733 | PITX2 | 4 | T | 0.15 |
| rs13376333 | KCNN3 | 1 | T | 0.20 |
| rs3807989 | CAV1/CAV2 | 7 | A | 0.24 |
| rs11047543 | SOX5 | 12 | A | 0.07 |
Abbreviations: RAF: risk allele frequency; SNPs: single nucleotide polymorphisms.
2.4. Statistical analysis
Tests of genetic association, allelic odds ratios (OR), and effect size (beta) were performed using logistic and linear regression in an additive genetic model. All continuous variables are expressed as mean ± one standard deviation unless stated otherwise. Comparison between dichotomized groups was performed using the chi‐square test (categorical variables), and Mann–Whitney test or Student's t test (continuous variables). Comparison of more than two groups was performed using Kruskal–Wallis or ANOVA. Correction for multiple testing was performed by using the Bonferroni method. All statistical analyses were performed using IBM SPSS Statistics, version 21.0.0 (SPSS). All tests were two‐sided, with a p‐value <0.05 considered statistically significant.
3. RESULTS
3.1. Study population and ECG analysis
Of 244 patients with onset of lone AF before the age of 50, seven patients were excluded due to the treatment with class I or class III antiarrhythmic drugs. In addition, 61 patients were excluded from the study due to missing genotype data for one or more of the SNPs analyzed. Thus, the study population comprised of 176 patients (median age: 38, 85% men). The clinical characteristics of the study population are presented in Table 2.
Table 2.
Clinical characteristics of the lone AF population
| Study population (n = 176) | |
|---|---|
| Clinical characteristics | |
| Age at inclusion (years) | 38; 19–63 |
| Age at AF debut (years) | 33; 16–49 |
| Gender, male (%) | 85 |
| Height (cm) | 183 ± 9 |
| Weight (kg) | 89 ± 17 |
| BMI (kg/m2) | 27 ± 5 |
| Measured parameters | |
| HR, b.p.m. | 64 ± 12 |
| BP, systolic (mmHg) | 128 ± 16 |
| BP, diastolic (mmHg) | 77 ± 11 |
| Medication (%) | |
| Beta‐blocker | 12.0 |
| Calcium blocker | 2.8 |
| Cardiac glycoside | 1.1 |
| Warfarin | 5.1 |
Age presented as median and IQR, and remaining data presented as mean ± standard deviation.
Abbreviations: AF: atrial fibrillation; BMI: body mass index; BP: blood pressure; HR: heart rate; IQR: interquartile range.
Type 1 P‐wave morphology was observed in 29.5%, Type 2 in 46%, Type 3 in 1.7%, and atypical P‐wave morphology in 22.7% of the participants. The mean PR‐interval and P‐wave duration was 160 ± 26 ms and 125 ± 16 ms, respectively.
Compared to the 176 patients included in the study, the 68 patients excluded due to treatment with antiarrhythmic drugs or missing genotype data had a higher heart rate at inclusion (63 ± 12 vs. 59 ± 11 b.p.m, p = 0.012). No differences were observed with regard to age (at inclusion and onset of AF), gender, blood pressure, height, weight, medication (other than antiarrhythmic drugs), or P‐wave distribution.
3.2. Genotype and ECG phenotype correlation
The genotyping and association results are presented in Tables 3 and 4.
Table 3.
Association results for the four single nucleotide polymorphisms analyzed and the distribution of P‐wave morphologies
| SNP; nearby gene | Genotype | PWM Type 1, n (%) | PWM Type 2, n (%) | PWM Type 3, n (%) | Atypical PWM, n (%) | p‐Value |
|---|---|---|---|---|---|---|
| rs2200733; PITX2 | TT | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0.123 |
| TC | 12 (26) | 20 (44) | 2 (4) | 12 (26) | ||
| CC | 37 (29) | 59 (47) | 1 (1) | 30 (24) | ||
| rs13376331; KCNN3 | TT | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0.333 |
| TC | 25 (35) | 31 (43) | 0 (0) | 16 (22) | ||
| CC | 27 (26) | 48 (46) | 3 (3) | 26 (25) | ||
| rs3807989; CAV1/CAV2 | AA | 0 (0) | 5 (39) | 0 (0) | 8 (62) | <0.001*, <0.004** |
| AG | 9 (15) | 34 (56) | 0 (0) | 18 (30) | ||
| GG | 43 (42) | 40 (39) | 3 (3) | 16 (16) | ||
| rs11047543; SOX5 | AA | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0.01*, 0.04** |
| AG | 2 (10) | 9 (43) | 0 (0) | 10 (48) | ||
| GG | 50 (33) | 69 (45) | 3 (2) | 32 (21) |
Abbreviations: PWM, P‐wave morphology; SNP, single nucleotide polymorphism.
p < 0.05.
Bonferroni adjustment, threshold p < 0.0125.
Table 4.
Allelic odds ratios for P‐wave morphology Type 1 vs. Type 2 and P‐wave morphology Type 1 vs. non‐Type 1
| SNP; nearby gene | Allele | P‐wave morphology Type 1 vs. Type 2 | P‐wave morphology Type 1 vs. non‐Type 1 | ||||
|---|---|---|---|---|---|---|---|
| p‐value | OR for PWM Type 2 | 95% CI | p‐value | OR for Non‐Type 1 PWM | 95% CI | ||
| rs2200733; PITX2 | T | 0.302 | 0.7 | 0.35–1.39 | 0.378 | 0.8 | 0.40–1.42 |
| rs13376331; KCNN3 | T | 0.318 | 0.7 | 0.34–1.41 | 0.212 | 0.6 | 0.34–1.27 |
| rs3807989; CAV1/CAV2 | A | <0.001* | 4.4 | 1.98–9.95 | <0.001* | 4.8 | 2.25–10.17 |
| rs11047543; SOX5 | A | 0.104 | 3.5 | 0.77–15.86 | 0.04* | 4.7 | 1.07–20.46 |
Abbreviations: CI, confidence interval; OR, odds ratio; PWM, P‐wave morphology; SNP, single nucleotide polymorphism.
p < 0.05.
Two SNPs were found to be significantly associated with altered P‐wave morphology: rs3807989 located near CAV1/CAV2 (p < 0.001) and rs11047543 located near SOX5 (p = 0.01). When correcting for multiple testing with the Bonferroni method (α = 0.05/4 = 0.0125), the SNP located near CAV1/CAV2 was still significantly associated with altered P‐wave morphology. Both SNPs were associated with a higher risk of non‐Type 1 P‐wave morphology, with ORs of 4.8 (rs3807989) and 4.7 (rs11047543).
rs3807989 was also found to be associated with PR‐interval and P‐wave duration. For each copy of the risk allele (A), the PR interval decreased with 16 ms and the P‐wave duration decreased with 4.3 ms (Tables 5 and 6).
Table 5.
Association results for the four single nucleotide polymorphisms analyzed, PR interval, and P‐wave duration
| SNP; nearby gene | Genotype | PR interval (ms) | p‐Value | PWD (ms) | p‐Value |
|---|---|---|---|---|---|
| rs2200733; PITX2 | TT | 145 ± 26 | 0.557 | 120 ± 13 | 0.240 |
| TC | 162 ± 28 | 128 ± 17 | |||
| CC | 161 ± 25 | 124 ± 15 | |||
| rs13376331; KCNN3 | TT | – | 0.3 | – | 0.561 |
| TC | 163 ± 28 | 124 ± 16 | |||
| CC | 159 ± 24 | 126 ± 15 | |||
| rs3807989; CAV1/CAV2 | AA | 149 ± 18 | <0.001* | 126 ± 9 | 0.004* |
| AG | 152 ± 26 | 120 ± 17 | |||
| GG | 167 ± 25 | 128 ± 14 | |||
| rs11047543; SOX5 | AA | 179 | 0.153 | 145 | 0.426 |
| AG | 151 ± 27 | 124 ± 21 | |||
| GG | 162 ± 26 | 125 ± 15 |
Abbreviations: PWD, P‐wave duration; SNP, single nucleotide polymorphism.
p < 0.05.
Table 6.
Association results for the four single nucleotide polymorphisms analyzed, PR interval, and P‐wave duration. Effect size (beta) reported in ms per one copy of the minor allele
| SNP; nearby gene | Allele | PR interval | P‐wave duration | ||||
|---|---|---|---|---|---|---|---|
| Beta | 95% CI | p‐Value | Beta | 95% CI | p‐Value | ||
| rs2200733; PITX2 | T | 0.7 | −8.6 to 7.1 | 0.868 | 2.7 | −2.0 to 7.4 | 0.205 |
| rs13376331; KCNN3 | T | 4.1 | −3.7 to 12.0 | 0.3 | 1.4 | −6.1 to 3.3 | 0.561 |
| rs3807989; CAV1/CAV2 | A | 12.0 | −17.8 to (−6.1) | <0.001* | 4.3 | −7.9 to (−0.7) | 0.02* |
| rs11047543; SOX5 | A | 7.6 | −18.4 to 3.3 | 0.172 | 1.2 | −5.4 to 7.7 | 0.727 |
Abbreviations: CI: confidence interval; PWD: P‐wave duration; SNP: single nucleotide polymorphism.
p < 0.05.
No association was observed in regard to the remaining SNPs analyzed: rs2200733 near PITX2 and rs13376333 near KCNN3.
4. DISCUSSION
4.1. Main findings
We report an association between common genetic variants and P‐wave morphology, in an early‐onset lone AF population. In the present study, two genetic variants, rs3807989 (CAV1/CAV2) and rs11047543 (SOX5), previously associated with PR‐interval duration and AF, as well as lone AF, were found to be associated with an altered P‐wave morphology distribution similar to the one observed in patients with paroxysmal AF (Table 7).
Table 7.
The PR‐interval and P‐wave duration presented for each P‐wave morphology type based on genotype of the SNP located near CAV1/CAV2
| SNP; nearby gene | Genotype | PWM Type 1 | PWM Type 2 | PWM Type 3 | Atypical PWM | ||||
|---|---|---|---|---|---|---|---|---|---|
| PR interval (ms) | PWD (ms) | PR interval (ms) | PWD (ms) | PR interval (ms) | PWD (ms) | PR interval (ms) | PWD (ms) | ||
| rs3807989; CAV1/CAV2 | AA | – | – | 155 ± 20 | 131 ± 6 | – | – | 146 ± 7 | 122 ± 8 |
| AG | 143 ± 22 | 108 ± 18 | 154 ± 27 | 123 ± 13 | – | – | 152 ± 25 | 119 ± 22 | |
| other | 165 ± 22 | 127 ± 14 | 171 ± 27 | 129 ± 12 | 178 ± 26 | 151 ± 18 | 163 ± 25 | 124 ± 18 | |
| p‐value | 0.009* | 0.001* | 0.019* | 0.108 | – | – | 0.118 | 0.259 | |
Abbreviations: PWD, P‐wave duration; PWM, P‐wave morphology; SNP, single nucleotide polymorphism.
*p < 0.05.
4.2. Atrial conductive properties in patients with early‐onset lone AF
4.2.1. P‐wave morphology
The distribution of P‐wave morphologies was as expected in this relatively young population of lone AF without structural heart disease, that is, predominantly Type 2. As an unexpected finding, we observed a rather low prevalence (1.7%) of Type 3 P‐wave morphology, reflecting advanced interatrial block and a high prevalence (23.9%) of atypical P‐wave morphology (i.e., non‐Types 1–3). Notably, 86% (n = 36) of those with atypical P‐wave morphology exhibited the same pattern of positive signals in all the orthogonal leads; the atrial depolarization propagation characterized by right‐to‐left, superior‐to‐inferior, and anterior‐to‐posterior course. Since electro‐anatomical mapping for this pattern remains to be revealed, one can only speculate whether this atypical P‐wave morphology is caused by a different propagation route or an upward shift in the site of the sinus depolarization wave origin similar to what is observed during exercise or heart rate increase (Forfang & Erikssen, 1978; Yokota et al., 1986).
4.2.2. PR‐interval and P‐wave duration
In recent years, electrocardiographic PR‐interval prolongation has been identified as a risk factor for AF incidence independent of age, gender, and hypertension (Cheng, Lu, Huang, Zhang, & Gu, 2014). It has also been revealed that not only PR prolongation but also PR‐interval shortening is associated with increased risk of AF (Alonso et al., 2013; Nielsen et al., 2013). The association between PR‐interval variation and risk of lone AF, specifically, has not yet been studied. Only few previous studies report PR‐interval data in lone AF populations and with contradictive results (Holmqvist et al., 2011; Nielsen et al., 2011).
4.3. Genomic predictors of atrial conductive properties
4.3.1. P‐wave morphology and PR interval
In a previous study on P‐wave morphology, comparing early‐onset lone AF patients with age‐ and gender‐matched individuals, the same pattern as seen in the present study, of non‐Type 1 P‐wave morphology in patients with early‐onset lone AF, was revealed (Holmqvist et al., 2011). Furthermore, both SNPs analyzed in the present study have previously been associated with the risk of early‐onset lone AF (Olesen et al., 2012), supporting the hypothesis that these common genetic variants are modifiers of atrial activation, which may lead to early‐onset lone AF..
The SNP located near CAV1/CAV2 (rs3807989) was the only genetic locus found to be associated with PR interval in our relatively small study sample. In previous studies, the A allele of rs3807989 was associated with prolongation of the PR interval and decreased risk of AF (Butler et al., 2012; Holm et al., 2010; Pfeufer et al., 2010). Unexpectedly, the same allele A was associated with a significantly shorter PR interval in our lone AF population. One explanation for the contradictory finding can be differences in comorbidities in the populations studied, that is, the population studied in the AF GWAS by Pfeufer et al. included comorbidity, whereas our study population involves individuals with structurally normal hearts and without comorbidity. Another explanation may be the different genetic architecture of the populations studied. The frequency of the A allele of rs3807989 reported by Pfeufer and colleagues was 0.40 (Pfeufer et al., 2010), compared to 0.24 in our population. Finally, it is possible that the association between rs3807989 and PR interval in our study is spurious due to the rather small sample size.
4.3.2. CAV1/CAV2 and SOX5
The SNP rs3807989 is located near the genes CAV1 and CAV2, known to encode the protein caveolin‐1, required for caveola formation and maintenance within the plasma membrane. Caveolin‐1 is prominently expressed in endothelial cells in addition to atrial myocytes and is the predominant caveolin isoform in the cardiovascular system (Gratton, Bernatchez, & Sessa, 2004; Volonte, McTiernan, Drab, Kasper, & Galbiati, 2008). Caveolae, the cholesterol‐ and sphingolipid‐enriched invaginations of the plasma membrane (Palade, 1953), are increasingly acknowledged due to their involvement in multiple cellular processes and signal transduction, capable of both signal inhibition and enhancement (Roth & Patel, 2011). Furthermore, caveolin‐1 seems to play a role in electrical signal transduction due to its co‐localization with connexin‐43 in the myocyte gap junctions, permitting action potential propagation (Barker, Price, & Gourdie, 2001). Several studies have implicated caveolin‐1 in the pathophysiology of a number of cardiovascular diseases. Overexpression of caveolin‐1 in the endothelial layer was found to accelerate the progression of atherosclerosis (Fernandez‐Hernando, Yu, Davalos, Prendergast, & Sessa, 2010), while caveolin‐1 knockout mice were found to be protected against atherosclerosis (Frank et al., 2004). Caveolin‐1‐deficient mice are noted to develop pulmonary hypertension and dilated cardiomyopathy (Zhao et al., 2002). It is possible that rs3807989, due to its location near CAV1, exerts effects on atrial conductive properties and lone AF, through mechanisms yet to be revealed. One could speculate that the association between rs3807989 and PR interval may be due to the expression of caveolin‐1 in atrial myocytes and downstream effects on electrical signal transduction, leading to possible gain of function/loss of function of different ionic channels. Why the same risk allele is associated with PR prolongation in GWAS and PR‐interval shortening in our population is difficult to say.
The SNP rs11047543 is located near the gene SOX5, known to encode a transcription factor expressed in various tissues, most predominantly the heart, skeletal muscle, and liver (Ikeda et al., 2002). SOX5 plays important roles in regulating processes of embryonic development and cell fate determination (Smits et al., 2001). These developmental roles are achieved by modulating cell proliferation, survival, differentiation, or terminal maturation in different cell lineages (Lefebvre, 2010). The major expression of SOX5 in skeletal muscle and heart tissue suggests potential roles in human myogenesis. In a previous study, mice homozygous for a SOX6 mutation showed abnormal ultrastructure of skeletal and cardiac muscle, developing a widespread myopathy that affected both skeletal and cardiac myocytes (Hagiwara et al., 2000). Since SOX5 and SOX6 are co‐expressed and intimately interacting, it has been hypothesized that SOX5 may exert an effect similar to that of SOX6 on human muscle development (Ikeda et al., 2002). In line with this hypothesis, it has been shown that SOX5‐deficient mice die from heart failure (Smits et al., 2001). rs11047543 may affect the atrial conduction pattern by modifying interatrial muscular connections, due to the potential role of SOX5 in myogenesis.
The precise pathophysiological mechanism by which the two genetic loci represented by rs3807989 and rs11047543 can modify atrial electrical properties still remains to be clarified.
4.4. Clinical application
The role of common genetic variants in lone AF pathophysiology is complex, but crucial for our understanding of interindividual susceptibility to disease. Further research in this important field may help us identify those at risk of AF at an early stage and ultimately prevent the development of the arrhythmia and provide individualized treatment options based on genotypic information and ECG markers aiming at personalized medicine.
5. LIMITATIONS
Compared to previous GWAS, the sample size in this study is small. The risk of spurious associations is increased in a SNP association study with limited sample size, which must be taken into account when interpreting the results from the present study. One can speculate that this is the reason why two of the investigated SNPs did not show any significant association.
We were not able to collect additional information about laboratory values, which limits the interpretation of our results, since abnormal potassium, calcium, and magnesium values may have an impact on atrial conduction properties. Finally, the study participants were all of Scandinavian ancestry, which affects the generalizability across ethnicities.
6. CONCLUSION
The two genetic variants, rs3807989 (CAV1/CAV2) and rs11047543 (SOX5), were associated with abnormal atrial conduction properties in this study. rs3807989 and rs11047543 have previously been associated with PR‐interval prolongation and lone AF, and our findings thus support the hypothesis that lone AF is primarily an “electrical” disease and takes us one step further in elucidating the biological link between common genetic variants and lone AF pathophysiology.
CONFLICT OF INTEREST
Nothing to report.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the financial support from the Swedish Heart‐Lung Foundation, Donations funds at Skåne University Hospital, and governmental funding of clinical research within the Swedish healthcare system, the Norwegian Research Council, the Danish National Research Foundation, the John and Birthe Meyer Foundation, and the Arvid Nilsson Foundation.
Seifert MB, Olesen MS, Christophersen IE, et al. Genetic variants on chromosomes 7p31 and 12p12 are associated with abnormal atrial electrical activation in patients with early‐onset lone atrial fibrillation. Ann Noninvasive Electrocardiol. 2019;24:e12661 10.1111/anec.12661
REFERENCES
- Alonso, A. , Krijthe, B. P. , Aspelund, T. , Stepas, K. A. , Pencina, M. J. , Moser, C. B. , … Benjamin, E. J. (2013). Simple risk model predicts incidence of atrial fibrillation in a racially and geographically diverse population: the CHARGE‐AF consortium. Journal of the American Heart Association, 2(2), e000102 10.1161/JAHA.112.000102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, R. J. , Price, R. L. , & Gourdie, R. G. (2001). Increased co‐localization of connexin43 and ZO‐1 in dissociated adult myocytes. Cell Communication & Adhesion, 8(4–6), 205–208. 10.3109/15419060109080724 [DOI] [PubMed] [Google Scholar]
- Benjamin, E. J. , Rice, K. M. , Arking, D. E. , Pfeufer, A. , van Noord, C. , Smith, A. V. , … Witteman, J. C. (2009). Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nature Genetics, 41(8), 879–881. 10.1038/ng.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A. M. , Yin, X. , Evans, D. S. , Nalls, M. A. , Smith, E. N. , Tanaka, T. , … Avery, C. L. (2012). Novel loci associated with PR interval in a genome‐wide association study of 10 African American cohorts. Circulation: Cardiovascular Genetics, 5(6), 639–646. 10.1161/CIRCGENETICS.112.963991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, J. (2005). Exploration of supraventricular conduction with respect to atrial fibrillation, Methodological aspects on selected techniques. Lund, Sweden: KFS i Lund AB. [Google Scholar]
- Carlson, J. , Havmoller, R. , Herreros, A. , Platonov, P. , Johansson, R. , & Olsson, B. (2005). Can orthogonal lead indicators of propensity to atrial fibrillation be accurately assessed from the 12‐lead ECG? Europace, 7(Suppl 2), 39–48. 10.1016/j.eupc.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Cheng, M. , Lu, X. , Huang, J. , Zhang, S. , & Gu, D. (2014). Electrocardiographic PR prolongation and atrial fibrillation risk: a meta‐analysis of prospective cohort studies. Journal of Cardiovascular Electrophysiology, 26, 36–41. 10.1111/jce.12539 [DOI] [PubMed] [Google Scholar]
- Chu, H. M. , Feng, M. J. , Li, Y. G. , Zhang, Y. X. , Ma, J. F. , He, B. , … Chen, X. M. (2013). Polymorphisms but not mutations of the KCNQ1 gene are associated with lone atrial fibrillation in the Chinese Han population. Scientific World Journal, 2013, 373454 10.1155/2013/373454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinor, P. T. , Lunetta, K. L. , Albert, C. M. , Glazer, N. L. , Ritchie, M. D. , Smith, A. V. , … Kaab, S. (2012). Meta‐analysis identifies six new susceptibility loci for atrial fibrillation. Nature Genetics, 44(6), 670–675. 10.1038/ng.2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinor, P. T. , Lunetta, K. L. , Glazer, N. L. , Pfeufer, A. , Alonso, A. , Chung, M. K. , … Kaab, S. (2010). Common variants in KCNN3 are associated with lone atrial fibrillation. Nature Genetics, 42(3), 240–244. 10.1038/ng.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Hernando, C. , Yu, J. , Davalos, A. , Prendergast, J. , & Sessa, W. C. (2010). Endothelial‐specific overexpression of caveolin‐1 accelerates atherosclerosis in apolipoprotein E‐deficient mice. American Journal of Pathology, 177(2), 998–1003. 10.2353/ajpath.2010.091287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forfang, K. , & Erikssen, J. (1978). Significance of P wave terminal force in presumably healthy middle‐aged men. American Heart Journal, 96(6), 739–743. 10.1016/0002-8703(78)90006-6 [DOI] [PubMed] [Google Scholar]
- Frank, P. G. , Lee, H. , Park, D. S. , Tandon, N. N. , Scherer, P. E. , & Lisanti, M. P. (2004). Genetic ablation of caveolin‐1 confers protection against atherosclerosis. Arteriosclerosis, Thrombosis, and Vascular Biology, 24(1), 98–105. 10.1161/01.ATV.0000101182.89118.E5 [DOI] [PubMed] [Google Scholar]
- Fuster, V. , Ryden, L. E. , Cannom, D. S. , Crijns, H. J. , Curtis, A. B. , Ellenbogen, K. A. , … Wann, L. S. (2011). 2011 ACCF/AHA/HRS focused updates incorporated into the ACC/AHA/ESC 2006 Guidelines for the management of patients with atrial fibrillation: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines developed in partnership with the European Society of Cardiology and in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. Journal of the American College of Cardiology, 57(11), e101–e198. 10.1016/j.jacc.2010.09.013 [DOI] [PubMed] [Google Scholar]
- Gratton, J. P. , Bernatchez, P. , & Sessa, W. C. (2004). Caveolae and caveolins in the cardiovascular system. Circulation Research, 94(11), 1408–1417. 10.1161/01.RES.0000129178.56294.17 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson, D. F. , Arnar, D. O. , Helgadottir, A. , Gretarsdottir, S. , Holm, H. , Sigurdsson, A. , … Stefansson, K. (2007). Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature, 448(7151), 353–357. 10.1038/nature06007 [DOI] [PubMed] [Google Scholar]
- Hagiwara, N. , Klewer, S. E. , Samson, R. A. , Erickson, D. T. , Lyon, M. F. , & Brilliant, M. H. (2000). Sox6 is a candidate gene for p100H myopathy, heart block, and sudden neonatal death. Proceedings of the National Academy of Sciences of the United States of America, 97(8), 4180–4185. 10.1073/pnas.97.8.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havmoller, R. , Carlson, J. , Holmqvist, F. , Herreros, A. , Meurling, C. J. , Olsson, B. , & Platonov, P. (2007). Age‐related changes in P wave morphology in healthy subjects. BMC Cardiovascular Disorders, 7, 22 10.1186/1471-2261-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, H. , Gudbjartsson, D. F. , Arnar, D. O. , Thorleifsson, G. , Thorgeirsson, G. , Stefansdottir, H. , … Stefansson, K. (2010). Several common variants modulate heart rate, PR interval and QRS duration. Nature Genetics, 42(2), 117–122. 10.1038/ng.511 [DOI] [PubMed] [Google Scholar]
- Holmqvist, F. , Husser, D. , Tapanainen, J. M. , Carlson, J. , Jurkko, R. , Xia, Y. , … Platonov, P. G. (2008). Interatrial conduction can be accurately determined using standard 12‐lead electrocardiography: Validation of P‐wave morphology using electroanatomic mapping in man. Heart Rhythm: the Official Journal of the Heart Rhythm Society, 5(3), 413–418. 10.1016/j.hrthm.2007.12.017 [DOI] [PubMed] [Google Scholar]
- Holmqvist, F. , Olesen, M. S. , Tveit, A. , Enger, S. , Tapanainen, J. , Jurkko, R. , … Platonov, P. G. (2011). Abnormal atrial activation in young patients with lone atrial fibrillation. Europace, 13(2), 188–192. 10.1093/europace/euq352 [DOI] [PubMed] [Google Scholar]
- Holmqvist, F. , Platonov, P. G. , Carlson, J. , Zareba, W. , Moss, A. J. , & Investigators, M. I. (2009). Altered interatrial conduction detected in MADIT II patients bound to develop atrial fibrillation. Annals of Noninvasive Electrocardiology, 14(3), 268–275. 10.1111/j.1542-474X.2009.00309.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist, F. , Platonov, P. G. , Havmoller, R. , & Carlson, J. (2007). Signal‐averaged P wave analysis for delineation of interatrial conduction – further validation of the method. BMC Cardiovascular Disorders, 7, 29 10.1186/1471-2261-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, T. , Zhang, J. , Chano, T. , Mabuchi, A. , Fukuda, A. , Kawaguchi, H. , … Ikegawa, S. (2002). Identification and characterization of the human long form of Sox5 (L‐SOX5) gene. Gene, 298(1), 59–68. 10.1016/S0378-1119(02)00927-7 [DOI] [PubMed] [Google Scholar]
- Lefebvre, V. (2010). The SoxD transcription factors–Sox5, Sox6, and Sox13–are key cell fate modulators. International Journal of Biochemistry & Cell Biology, 42(3), 429–432. 10.1016/j.biocel.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz, S. A. , Lunetta, K. L. , Lin, H. , Arking, D. E. , Trompet, S. , Li, G. , … Ellinor, P. T. (2014). Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. Journal of the American College of Cardiology, 63(12), 1200–1210. 10.1016/j.jacc.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane, P. W. , Devine, B. , Latif, S. , McLaughlin, S. , Shoat, D. B. , & Watts, M. P. (1990). Methodology of ECG interpretation in the Glasgow program. Methods of Information in Medicine, 29(4), 354–361. [PubMed] [Google Scholar]
- Mahida, S. , Lubitz, S. A. , Rienstra, M. , Milan, D. J. , & Ellinor, P. T. (2011). Monogenic atrial fibrillation as pathophysiological paradigms. Cardiovascular Research, 89(4), 692–700. 10.1093/cvr/cvq381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J. B. , Olesen, M. S. , Tango, M. , Haunso, S. , Holst, A. G. , & Svendsen, J. H. (2011). Incomplete right bundle branch block: A novel electrocardiographic marker for lone atrial fibrillation. Europace, 13(2), 182–187. 10.1093/europace/euq436 [DOI] [PubMed] [Google Scholar]
- Nielsen, J. B. , Pietersen, A. , Graff, C. , Lind, B. , Struijk, J. J. , Olesen, M. S. , … Holst, A. G. (2013). Risk of atrial fibrillation as a function of the electrocardiographic PR interval: Results from the Copenhagen ECG Study. Heart Rhythm: the Official Journal of the Heart Rhythm Society, 10(9), 1249–1256. 10.1016/j.hrthm.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Olesen, M. S. , Holst, A. G. , Jabbari, J. , Nielsen, J. B. , Christophersen, I. E. , Sajadieh, A. , … Svendsen, J. H. (2012). Genetic loci on chromosomes 4q25, 7p31, and 12p12 are associated with onset of lone atrial fibrillation before the age of 40 years. Canadian Journal of Cardiology, 28(2), 191–195. 10.1016/j.cjca.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Olesen, M. S. , Jabbari, J. , Holst, A. G. , Nielsen, J. B. , Steinbruchel, D. A. , Jespersen, T. , … Svendsen, J. H. (2011). Screening of KCNN3 in patients with early‐onset lone atrial fibrillation. Europace, 13(7), 963–967. 10.1093/europace/eur007 [DOI] [PubMed] [Google Scholar]
- Olsson, S. B. , Cotoi, S. , & Varnauskas, E. (1971). Monophasic action potential and sinus rhythm stability after conversion of atrial fibrillation. Acta Medica Scandinavica, 190(5), 381–387. [DOI] [PubMed] [Google Scholar]
- Palade, G. (1953). Fine structure of blood capillaries. Journal of Applied Physics, 24(144), 1424–1426. [Google Scholar]
- Pfeufer, A. , van Noord, C. , Marciante, K. D. , Arking, D. E. , Larson, M. G. , Smith, A. V. , … Heckbert, S. R. (2010). Genome‐wide association study of PR interval. Nature Genetics, 42(2), 153–159. 10.1038/ng.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platonov, P. G. , Carlson, J. , Ingemansson, M. P. , Roijer, A. , Hansson, A. , Chireikin, L. V. , & Olsson, S. B. (2000). Detection of inter‐atrial conduction defects with unfiltered signal‐averaged P‐wave ECG in patients with lone atrial fibrillation. Europace, 2(1), 32–41. 10.1053/eupc.1999.0072 [DOI] [PubMed] [Google Scholar]
- Platonov, P. G. , Christensen, A. H. , Holmqvist, F. , Carlson, J. , Haunso, S. , & Svendsen, J. H. (2011). Abnormal atrial activation is common in patients with arrhythmogenic right ventricular cardiomyopathy. Journal of Electrocardiology, 44(2), 237–241. 10.1016/j.jelectrocard.2010.08.008 [DOI] [PubMed] [Google Scholar]
- Roth, D. M. , & Patel, H. H. (2011). Role of caveolae in cardiac protection. Pediatric Cardiology, 32(3), 329–333. 10.1007/s00246-010-9881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner, M. F. , Tucker, N. R. , Lunetta, K. L. , Ozaki, K. , Smith, J. G. , Trompet, S. , … Ellinor, P. T. (2014). Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation, 130(15), 1225–1235. 10.1161/CIRCULATIONAHA.114.009892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, P. , Li, P. , Mandel, J. , Zhang, Z. , Deng, J. M. , Behringer, R. R. , … Lefebvre, V. (2001). The transcription factors L‐Sox5 and Sox6 are essential for cartilage formation. Developmental Cell, 1(2), 277–290. 10.1016/S1534-5807(01)00003-X [DOI] [PubMed] [Google Scholar]
- Volonte, D. , McTiernan, C. F. , Drab, M. , Kasper, M. , & Galbiati, F. (2008). Caveolin‐1 and caveolin‐3 form heterooligomeric complexes in atrial cardiac myocytes that are required for doxorubicin‐induced apoptosis. American Journal of Physiology. Heart and Circulatory Physiology, 294(1), H392–H401. 10.1152/ajpheart.01039.2007 [DOI] [PubMed] [Google Scholar]
- Wirka, R. C. , Gore, S. , Van Wagoner, D. R. , Arking, D. E. , Lubitz, S. A. , Lunetta, K. L. , … Smith, J. D. (2011). A common connexin‐40 gene promoter variant affects connexin‐40 expression in human atria and is associated with atrial fibrillation. Circulation. Arrhythmia and Electrophysiology, 4(1), 87–93. 10.1161/CIRCEP.110.959726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, M. , Noda, S. , Koide, M. , Kawai, N. , Yoshida, R. , Mochizuki, K. , … Sotobata, I. (1986). Analysis of the exercise‐induced orthogonal P wave changes in normal subjects and patients with coronary artery disease. Japanese Heart Journal, 27(4), 443–460. 10.1536/ihj.27.443 [DOI] [PubMed] [Google Scholar]
- Zang, X. , Zhang, S. , Xia, Y. , Li, S. , Fu, F. , Li, X. , … Wang, Q. (2013). SNP rs3825214 in TBX5 is associated with lone atrial fibrillation in Chinese Han population. PLoS ONE, 8(5), e64966 10.1371/journal.pone.0064966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. Y. , Liu, Y. , Stan, R. V. , Fan, L. , Gu, Y. , Dalton, N. , … Chien, K. R. (2002). Defects in caveolin‐1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proceedings of the National Academy of Sciences of the United States of America, 99(17), 11375–11380. 10.1073/pnas.172360799 [DOI] [PMC free article] [PubMed] [Google Scholar]


