Chronic infections are frequently caused by polymicrobial biofilms. Importantly, these infections are often difficult to treat effectively in part due to the recalcitrance of biofilms to antimicrobial therapy. Emerging evidence suggests that polymicrobial interactions can lead to dramatic and unexpected changes in the ability of antibiotics to eradicate biofilms and often result in decreased antimicrobial efficacy in vitro.
KEYWORDS: biofilm, polymicrobial, antibiotics, resistance, tolerance, recalcitrance
ABSTRACT
Chronic infections are frequently caused by polymicrobial biofilms. Importantly, these infections are often difficult to treat effectively in part due to the recalcitrance of biofilms to antimicrobial therapy. Emerging evidence suggests that polymicrobial interactions can lead to dramatic and unexpected changes in the ability of antibiotics to eradicate biofilms and often result in decreased antimicrobial efficacy in vitro. In this review, we discuss the influence of polymicrobial interactions on the antibiotic susceptibility of biofilms, and we highlight the studies that first documented the shifted antimicrobial susceptibilities of mixed-species cultures. Recent studies have identified several mechanisms underlying the recalcitrance of polymicrobial biofilm communities, including interspecies exchange of antibiotic resistance genes, β-lactamase-mediated inactivation of antibiotics, changes in gene expression induced by metabolites and quorum sensing signals, inhibition of the electron transport chain, and changes in properties of the cell membrane. In addition to elucidating multiple mechanisms that contribute to the altered drug susceptibility of polymicrobial biofilms, these studies have uncovered novel ways in which polymicrobial interactions can impact microbial physiology. The diversity of findings discussed highlights the importance of continuing to investigate the efficacy of antibiotics against biofilm communities composed of different combinations of microbial species. Together, the data presented here illustrate the importance of studying microbes as part of mixed-species communities rather than in isolation. In light of our greater understanding of how interspecies interactions alter the efficacy of antimicrobial agents, we propose that the methods for measuring the drug susceptibility of polymicrobial infections should be revisited.
BIOFILMS ARE RECALCITRANT TO ANTIMICROBIAL TREATMENT
Biofilms are communities of microbial cells that are attached to a surface and to one another and are surrounded with a self-produced matrix. The biofilm lifestyle is incredibly common in all environments, from natural settings to the human body, both in health and in disease (1, 2). Furthermore, it is estimated that the majority of chronic infections are caused by biofilms (3). Biofilms are 100- to 1,000-fold more tolerant to antibiotics than their planktonic counterparts (2, 4) and promote persistence in the host, which makes successful treatment challenging.
The reason that biofilms are able to withstand such high concentrations of antimicrobial agents is multifactorial and incompletely understood. Antibiotic resistance refers to genetically encoded mechanisms that allow microbes to grow in the presence of a drug, whereas tolerance is the ability to survive transient exposure to an otherwise lethal dose of an antibiotic, by either genetically encoded or phenotypic, nonheritable mechanisms (5–8). Biofilms exhibit both resistance and tolerance to antibiotics, a combination referred to as “recalcitrance” (5–7).
The many genetically encoded resistance mechanisms used by planktonic cells to withstand antimicrobial treatment contribute to the recalcitrance of biofilms. Resistance can arise in various ways, such as by modifying an antibiotic target, enzymatic deactivation, or active efflux of a drug once it has entered the cell. Furthermore, resistance can be acquired by spontaneous mutations or by exchanging antibiotic resistance genes between other cells. These mechanisms are at play within biofilms and are often compounded by other aspects of biofilm physiology to confer even greater levels of recalcitrance. For example, β-lactamases concentrated within the extracellular matrix can degrade the antibiotic before it can reach the cells within the biofilm (4). Additionally, the biofilm lifestyle has been found to facilitate rates of horizontal gene transfer (HGT); bacteria within biofilms exchanged plasmids containing antibiotic resistance genes at a higher rate than their planktonic counterparts (8–10).
In addition to the resistance mechanisms outlined above, multiple aspects of a microbe’s physiology contribute to the elevated antibiotic tolerance of cells within a biofilm relative to planktonic cells. These features include reduced growth rate (11–13) due to oxygen-depleted microenvironments (14), nutrient limitation and the subsequent activation of stress responses (15–17), limited diffusion of certain antibiotics, such as via chelation of cationic compounds by extracellular matrix components (18–20), and the frequent emergence of so-called “persister” cells—dormant or slow-growing cells that are genetically identical to the rest of the population but display several phenotypic differences, such as elevated multidrug tolerance and high induction of stress responses (6). In addition, bacterial biofilms upregulate efflux pumps even in the absence of antibiotic exposure, conferring high levels of drug tolerance (21, 22). As another example, the production of periplasmic cyclic glucans in biofilm-grown bacteria can enhance biofilm tolerance to aminoglycoside antibiotics (23). Thus, multifaceted resistance and tolerance mechanisms contribute to the overall recalcitrance of biofilms to antimicrobial therapy.
THE ETIOLOGY OF MOST CHRONIC INFECTIONS IS POLYMICROBIAL
Multispecies biofilm-like communities are responsible for causing persistent infections in a wide range of body sites, including the lung (24–31), oral cavity (32, 33), middle ear (34–36), urinary tract (37–39), and both surgical and chronic wounds (40–45). In several disease settings, polymicrobial infections have been reported to cause worse outcomes than single-species infections (46–54). In particular, polymicrobial bloodstream infections are associated with higher rates of mortality than monospecies infections (46–50). Similarly, polymicrobial lung infections have been shown to lead to worse prognoses. Cystic fibrosis (CF) patients who are coinfected with Pseudomonas aeruginosa and Staphylococcus aureus demonstrate more rapid lung function decline than patients with monospecies infections (55–59). Furthermore, P. aeruginosa and S. aureus have been detected in the same lobe of the lung (60, 61), and they engage in a plethora of interactions in vitro (62–68). Thus, it is possible that these microbes interact during infection.
Interspecies interactions profoundly influence microbial physiology in ways that may alter disease progression and outcome, including the upregulation of virulence factors (69–73), altered biofilm formation (67, 74–76), impaired wound healing (73, 77), and shifted antibiotic susceptibility profiles (63, 67, 74, 75, 77–88). Additionally, interactions between a host and one microbial species may modify immune responses to a coinfecting species (76, 89, 90). Therefore, as others have postulated (91, 92), perhaps the consequences of these interactions on microbial and host physiology contribute to the worse patient outcomes that are frequently observed for coinfections than for monoinfections.
THE DISCONNECT BETWEEN IN VITRO ANTIMICROBIAL SUSCEPTIBILITY AND TREATMENT SUCCESS
An important clinical outcome to consider is whether a given antimicrobial therapy can successfully treat an infection. Various studies have evaluated whether in vitro MIC—still considered the gold standard for drug susceptibility testing—correlated with the success or failure of antimicrobial treatment. Surprisingly, these studies found little or no association between clinical antimicrobial susceptibility testing results (specifically, a pathogen’s in vitro MIC value for a specific drug) and clinical outcomes measured following antibiotic treatment, even in the context of single-species infections (93–98). In other words, patients did no better when they were infected by susceptible organisms (low MIC) than by resistant organisms (high MIC). An even more worrisome observation was made by authors studying the association between Candida fluconazole MICs and treatment outcomes of bloodstream infections. Contrary to expectation, low in vitro MICs actually correlated with treatment failure (99). Approximately one-third of isolates with low MICs (<16 μg/ml) failed to respond to fluconazole therapy, indicating that a drug judged to be effective in vitro was unable to eradicate infections in multiple patients. The reverse was also true, whereby the treatment of four Candida isolates was successful despite having MICs of ≥32 μg/ml (99). A similar correlation was observed for Candida caspofungin MICs and candidiasis outcomes (100). Therefore, these studies illustrate that while the susceptibility methods currently used in the clinical microbiology laboratory are often quite useful, such in vitro tests are not always able to predict a patient’s response to antimicrobial treatment.
While alarming, these findings are not entirely surprising, given the enormous difference between controlled laboratory conditions and an infection site within a patient, and others have questioned the clinical predictive value of MIC tests (2, 101). Standard antibiotic susceptibility testing guidelines recommend that sensitivity should be measured when a microbe is grown planktonically, in rich medium, in monoculture. Therefore, test results only actually indicate whether an organism is sensitive to an antimicrobial compound under those precise conditions. These laboratory tests do not consider the conditions that microbes experience within an infection site, including constant assault by the host immune system. In addition, antimicrobial efficacy is also influenced by immunosuppression (102) and drug-drug interactions (103), which may contribute to differences between laboratory results and clinical outcomes. Furthermore, in most chronic infections, microorganisms likely form biofilms and probably interact with a multitude of neighbors (including other microbes and the host) within that infection niche.
Importantly, as we discuss below, in addition to adopting a biofilm lifestyle, interacting with other microbes in these sessile communities can contribute to drug sensitivity profiles that are vastly different from when an organism is grown planktonically in pure culture. Therefore, it is possible that interactions between microbes could influence the success of antimicrobial treatment. Here, we discuss various mechanisms underlying how interspecies interactions alter antimicrobial sensitivity profiles within polymicrobial biofilm communities, which may in part explain why antimicrobial therapies often fail to eradicate chronic infections.
MECHANISMS OF ANTIMICROBIAL RESISTANCE IN POLYMICROBIAL BIOFILMS
Some of the same genetic mechanisms that can cause planktonic cells to become antibiotic resistant also contribute to the ability of biofilm-forming microbes to withstand antibiotic treatment. Here, we review the contributions of HGT and antibiotic-inactivating enzymes to drug resistance within multispecies biofilms.
INTERSPECIES GENETIC EXCHANGE CONFERS ANTIMICROBIAL RESISTANCE
In addition to the occurrence of spontaneous mutations that are genetically inherited by daughter cells, horizontal gene transfer (HGT) is another means whereby microbes can acquire new sources of antibiotic resistance genes. The biofilm lifestyle has been found to promote HGT by increasing the rates of conjugation (8–10, 104, 105) and transformation (106), and to increase the stability of plasmids (107) relative to the planktonic setting. It has been proposed that the ordered structure and high density of cells within a biofilm promote efficient conjugation, although the underlying mechanisms remain unclear (107). However, genetic exchange may not occur uniformly within biofilms. For example, high conjugation frequencies may be confined to subpopulations of cells during initial stages of biofilm development (108, 109). Additionally, conjugative pili were found to promote attachment and biofilm formation by Escherichia coli (110, 111), raising the possibility that maintenance and transfer of conjugative plasmids within biofilms benefit cells growing within these communities.
In addition to conjugation, rates of transformation are also elevated within biofilms. In one study, the transformation frequencies of biofilm-grown Streptococcus mutans were found to be 10- to 600-fold higher than for planktonic populations (112). The induction of competence in many species is influenced by a variety of environmental cues, including cell density, environmental pH, antibiotic exposure, starvation, and the presence or absence of particular carbon sources (113–116). In addition to its role in the induction of competence, quorum sensing plays a role in regulating biofilm formation in many bacterial species (117); therefore, competence and biofilm formation are linked by virtue of being regulated (at least in part) by a common pathway. Furthermore, the presence of extracellular DNA within the biofilm matrix has been proposed to induce competence (118) and may in part contribute to the high rates of transformation observed within biofilms.
There have been multiple reports of HGT of antibiotic resistance genes between species within polymicrobial biofilms. One study found that a plasmid harboring a carbapenemase resistance gene (blaNDM-1) can be transferred from E. coli to either P. aeruginosa or Acinetobacter baumannii via conjugation within dual-species biofilms (119), which was not observed between these organisms in planktonic settings (120). These results highlight the enhanced propensity of genetic exchange within biofilms. Additionally, conjugative transposons have been implicated in transferring antimicrobial resistance genes within a multispecies oral biofilm in vitro (121, 122). For example, Streptococcus spp. acquired a transposon from Veillonella dispar by two distinct mechanisms: either conjugation with V. dispar (121) or transformation of purified V. dispar DNA (122). These data indicate that in addition to conjugation, which requires intimate cell-to-cell contact, uptake of extracellular DNA is another way in which members of a mixed-species community can exchange genetic information while in close proximity.
Moreover, there is evidence for HGT occurring between pathogens in vivo in the context of a mixed microbial community. It was discovered that the multidrug resistance of an S. aureus isolate within a polymicrobial infection of an indwelling tube was mediated by interspecies interactions (123). The already methicillin-resistant S. aureus isolate likely became resistant to vancomycin in vivo via the acquisition of a vanA-containing plasmid from Enterococcus faecium, one of the constituents of the polymicrobial biofilm. Additionally, the S. aureus isolate harbored a tetracycline resistance gene (tetU) that was not previously detected in S. aureus and was also likely transferred by E. faecium within this mixed-species biofilm during infection.
The studies described above determined that conjugation and transformation can mediate the spread of antimicrobial resistance between species. These findings underscore the potential for interspecies transfer of antibiotic resistance genes within an infection site, which can alter the antimicrobial susceptibility of an entire polymicrobial community. Importantly, a single conjugative event can have wide-ranging consequences for the spread of antimicrobial resistance, both within a single infection and between patients.
β-LACTAMASE-PRODUCING STRAINS PROTECT THEIR NEIGHBORS
Bacteria produce multiple genetically encoded factors that inactivate antibiotics. An important example is the group of proteins called β-lactamases, which are microbially produced enzymes that hydrolyze β-lactam antibiotics, a clinically important class of cell-wall-targeting drugs. These enzymes can be either chromosomally or plasmid encoded and have played a major role in the emergence of multidrug resistance among Gram-negative bacteria (124). β-Lactamases can be either localized to the periplasmic space of the producing cell or released into the extracellular space. Additionally, β-lactamases have been found within the outer membrane vesicles (OMVs) of multiple Gram-negative bacteria (125, 126) as well as the extracellular vesicles of S. aureus (127).
Importantly, inactivation of antibiotics by β-lactamases can protect not only the producing cell but other cells in the vicinity. In addition to protecting sensitive cells of the same species from antibiotics (128, 129), β-lactamase-producing bacteria can alter the antimicrobial sensitivity profiles of entire polymicrobial communities, allowing coinfecting species to survive otherwise lethal doses of such antibiotics (75, 126, 127, 130–132). In particular, the production of β-lactamases was shown to influence the sensitivity of multispecies biofilms involving three predominant etiologic agents of otitis media: Moraxella catarrhalis, Haemophilus influenzae, and Streptococcus pneumoniae (34). β-Lactamases produced by M. catarrhalis conferred protection on both H. influenzae (75) and S. pneumoniae (130, 132) in dual-species biofilms. Furthermore, these interactions were shown to be mediated by M. catarrhalis OMVs (126). Additionally, H. influenzae-produced β-lactamases were shown to influence the sensitivity of S. pneumoniae biofilms to amoxicillin in vitro and in a chinchilla model of otitis media (131). Together, these studies illustrate that three pathogens that are commonly coisolated from inner ear infections and form mixed-species biofilms can greatly influence one another’s susceptibility to β-lactams in a passive, contact-independent manner via antibiotic-degrading enzymes.
Importantly, β-lactamases have been detected in clinical samples from polymicrobial infections, including CF pulmonary infections (133) and otitis media (134, 135). Additionally, one study detected β-lactamase activity in 89% of ear aspirates from patients with infections that failed to respond to amoxicillin (134). These findings indicate the potential for this interspecies mechanism of antimicrobial resistance to occur in vivo, thus potentially contributing to treatment failure.
The above studies demonstrate that enzymatic inactivation of β-lactams can confer antibiotic resistance within mixed-species biofilms. This community-wide mechanism does not require direct interaction between two coinfecting species and has been shown to protect entire biofilms from antibiotic challenge (4). Thus, combined with the threat of plasmid-mediated exchange of resistance genes between species, this type of community-level resistance may have potentially far-reaching impacts on the outcomes of polymicrobial infections.
MECHANISMS UNDERLYING DRUG TOLERANCE OF POLYMICROBIAL BIOFILMS
In addition to the mechanisms leading to antimicrobial resistance typically associated with planktonic organisms (which are discussed above), recent studies have elucidated novel mechanisms explaining how interspecies interactions contribute to the antimicrobial tolerance of polymicrobial biofilm communities. We discuss these mechanisms below in the context of biofilms or biofilm-like infections.
MICROBIALLY SECRETED PRODUCTS ALTER ANTIBIOTIC TOLERANCE
Primary metabolites.
Metabolic interactions between members of a polymicrobial community have been found to influence antibiotic sensitivity of the community’s constituent microbes. For example, P. aeruginosa and anaerobic bacteria, organisms frequently coisolated from CF respiratory infections, were shown to engage in cross-feeding whereby P. aeruginosa consumes metabolites that are produced upon mucin fermentation by anaerobes (136). Interestingly, P. aeruginosa became more susceptible to ampicillin when in the presence of the drug-sensitive anaerobic community than when grown in monoculture (87). These data suggest that the drug sensitivity of an organism can increase when it is participating in particular metabolic interactions. In contrast, the interactions between members of a cross-feeding community composed of E. coli, Salmonella enterica, and Methylobacterium extorquens produced an opposite result, whereby E. coli sensitivity to tetracycline was lower in coculture than in monoculture (87). Together, these data illustrate that polymicrobial interactions can produce unexpected antibiotic sensitivity profiles. These findings also have potentially important implications for patient treatment—when a pathogen is part of a microbial community, the antibiotic dose needed for eradication may be different than what was predicted from monoculture experiments. Examples of such metabolite-driven changes in biofilm tolerance are outlined in the subsequent paragraphs.
Indole, an aromatic compound derived from tryptophan, has been described to shift the antibiotic sensitivity profiles of neighboring organisms both within and between species via transcriptional changes. Indole can impact the antibiotic sensitivity of single-species E. coli populations by inducing the expression of drug transporters (137). Additionally, indole was shown to mediate an interaction between the human commensal E. coli and Salmonella enterica serovar Typhimurium, a pathogen of the gastrointestinal tract. Indole produced by E. coli altered the antibiotic sensitivity of S. Typhimurium during in vitro coculture and in a coinfection model of intestinal infection in part via the induction of the S. Typhimurium OxyR oxidative stress pathway (83). Indole was hypothesized to protect S. Typhimurium from ciprofloxacin by increasing the expression of OxyR-regulated genes that confer protection from oxidative stress, including the catalase gene katG. This interaction illustrates how the production of a metabolite by a commensal can protect a pathogen from antimicrobial treatment by altering its physiology. Since indole is produced in high quantities by the mammalian intestine (138), the host may also influence bacterial drug susceptibility profiles in the same manner. Thus, indole can promote antibiotic tolerance in multiple ways.
The volatile fermentation product 2,3-butanedione has also been implicated in shifting antimicrobial sensitivity profiles by inducing changes in gene expression. 2,3-Butanedione is produced by streptococci as a consequence of acetoin metabolism, and it can be consumed by P. aeruginosa (139). Exposure to 2,3-butanedione was found to protect E. coli from various antibiotics by strongly inducing the expression of hipA (140), which is proposed to promote the generation of persister cells (141). Importantly, this volatile metabolite was detected in high levels in the airways of CF patients by breath gas analysis (139), indicating the possibility that microbially produced volatiles can exert long-range effects on other microbes within the lung.
Finally, the volatile metabolic by-product ammonia was also shown to modulate the antibiotic sensitivity of other microbes at a distance. Exposure to ammonia altered the susceptibility of E. coli to tetracycline (82). Bernier et al. propose that ammonia altered the antibiotic sensitivity of spatially distant bacteria by stimulating the production of polyamines (82)—compounds known to influence bacterial membrane permeability and drug susceptibility profiles (142–144). Additionally, ammonia has the ability to alkalize the surrounding growth medium, which was shown to inactivate ampicillin (145). Thus, it is possible that ammonia and other metabolites can interfere with the activity of antibiotics more generally by chemically modifying the environment. Moreover, the microbially produced gases hydrogen sulfide and nitric oxide were also observed to decrease the antibiotic sensitivity of multiple bacterial species (146, 147), suggesting the common ability of microbially produced volatiles to influence antimicrobial efficacy in vitro (148).
Together, these findings suggest that by-products of microbial metabolism can have diverse effects on the drug susceptibility of other organisms, both within and between species. However, it remains unclear in many instances how exposure to metabolites leads to altered antibiotic sensitivity profiles. Is it by directly influencing microbial physiology or the growth environment, or both? Further research is needed to elucidate the mechanisms underlying these observations. Additionally, multiple compounds, including ammonia, nitric oxide, and indole, were also reported to influence biofilm formation (149–152), which can further exacerbate the recalcitrance of microbes. Finally, the detection of volatile metabolites in vivo suggests that they have the potential to influence bacteria at a distance from the producing cell, which may lead to widespread changes in antibiotic sensitivity within an infection site.
Quorum sensing signals and other secondary metabolites.
Quorum sensing molecules have been shown to mediate interactions between microbes residing within multispecies biofilms. It is thought that microbes can distinguish between signals from different species, since different concentrations and combinations of autoinducers lead to distinct behaviors in responding species (153–155). Interspecies communication using quorum sensing signals was shown to impact drug sensitivity within mixed-species biofilms. Ryan et al. (74) discovered that Stenotrophomonas maltophilia and Pseudomonas aeruginosa, two organisms that coinfect CF patients, can communicate using quorum sensing molecules. The S. maltophilia-derived diffusible signal factor (DSF) was shown to be recognized by a P. aeruginosa sensor kinase and to cause the induction of P. aeruginosa PmrAB, a two-component system that regulates resistance to cationic peptides (74). As a result, P. aeruginosa biofilms became less sensitive to polymyxins. In addition, P. aeruginosa exposed to DSF adopts a striking filamentous phenotype (74).
A different interspecies interaction involving quorum sensing molecules was found to influence biofilm formation as well as to confer protection from antibiotic challenge (75). The authors observed that the thickness of M. catarrhalis biofilms, as well as the number of viable cells within the biofilm, increased in response to the quorum sensing signal AI-2 produced by H. influenzae (75). Whether and how M. catarrhalis senses and processes the AI-2 signal remain to be elucidated. Furthermore, the H. influenzae-mediated enhancement in M. catarrhalis biofilm biomass benefited both organisms, resulting in increased tolerance of both species to multiple antibiotics (75), perhaps by inhibiting drug diffusion through the biofilm. In addition, H. influenzae promoted enhanced persistence of M. catarrhalis in a chinchilla model of infection in an AI-2-dependent manner (75), suggesting that quorum sensing signals are mediating interactions between these bacterial pathogens in vivo. Together, the above studies suggest that interspecies communication via quorum sensing can impact antibiotic efficacy within multispecies biofilms.
Furthermore, quorum sensing signals and related small molecules can shift the antimicrobial susceptibility of neighboring microbes via mechanisms other than classical quorum sensing-mediated transcriptional changes. It has been recognized that quorum sensing signals can impact multiple aspects of microbial physiology (156), and emerging evidence suggests that these physiological changes can, in turn, lead to unexpected changes in drug tolerance. One study showed that farnesol, a quorum sensing molecule produced by Candida albicans, protected S. aureus from vancomycin within a polymicrobial biofilm (157). The authors propose a mechanism whereby exposure to farnesol induces the production of reactive oxygen species in S. aureus, which activates a general stress response and causes the upregulation of efflux pumps, thereby enhancing S. aureus antibiotic resistance (157). These data suggest that a quorum sensing molecule can shift drug sensitivity within polymicrobial biofilms by stimulating a cascade of physiological changes.
A different secondary metabolite has been described to mediate polymicrobial interactions and alter antibiotic susceptibility. The P. aeruginosa-secreted small molecule 2-heptyl-4-hydroxyquinolone N-oxide (HQNO) has been shown to strongly influence S. aureus physiology in vitro. HQNO is part of the Pseudomonas quinolone signal (PQS) quorum sensing system pathway and acts as a potent inhibitor of the S. aureus electron transport chain (ETC) (158). Inhibition of the S. aureus ETC causes a reduction in the electrochemical gradient, which is required for the cellular uptake of multiple classes of protein synthesis inhibitors (159). Consequently, exposure to HQNO decreases the susceptibility of S. aureus to the aminoglycosides dihydrostreptomycin and tobramycin (78, 80) and to multiple tetracycline and macrolide antibiotics (85).
Furthermore, HQNO has been shown to protect S. aureus biofilms from cell wall-targeting antibiotics, including the front-line drug vancomycin and multiple β-lactams (85). HQNO-mediated inhibition of the ETC forces S. aureus to grow by fermentation (64), leading to a decrease in ATP levels (86) and reduction in growth (85). Since many drug classes are effective against only actively growing cells, it is likely that slow growth renders S. aureus less susceptible to cell wall-targeting antibiotics. Additionally, prolonged exposure to HQNO has been shown to select for small-colony variants (SCVs) of S. aureus (80). SCVs have a nonfunctional ETC, which renders them highly tolerant to several classes of antibiotics and promote persistence within the host (160). Infection sites are often oxygen limited (161, 162), which can also induce a metabolic shift to fermentation and slow growth (163) and may further contribute to antibiotic tolerance within chronic infections. Together, these data illustrate that inhibition of electron transport can have wide-ranging effects on drug efficacy and can explain how the same microbe grown under different conditions might display differential susceptibility to the same antimicrobial agent.
Moreover, quorum sensing signals and related small molecules can impact antimicrobial efficacy by altering properties of bacterial membranes. For example, HQNO was shown to increase S. aureus cell membrane fluidity, rendering S. aureus biofilms more susceptible to several membrane-targeting antibiotics and antiseptics, including chloroxylenol (88). Additionally, the C. albicans quorum sensing signal farnesol can disrupt the integrity of the S. aureus cell membrane (164, 165), which, in turn, was hypothesized to contribute to farnesol’s ability to shift S. aureus drug sensitivity profiles (164). It has been proposed that the hydrophobic character of both of these molecules may allow for their accumulation within the S. aureus membrane (88, 164). These findings raise the possibility that other hydrophobic molecules with well-known roles in interbacterial signaling also have the ability to alter membrane properties and influence antibiotic sensitivity.
Two additional P. aeruginosa quorum sensing-regulated products, LasA and rhamnolipids, have been found to shift the susceptibility of S. aureus planktonic populations to various antibiotics. The endopeptidase LasA was observed to potentiate the efficacy of vancomycin, while rhamnolipids were proposed to increase the efficacy of tobramycin by facilitating uptake of the antibiotic (86, 166). In the future, it will be important to investigate whether LasA and rhamnolipids are able to modulate the drug susceptibility of biofilm cells in addition to that of planktonic cells.
Together, the above studies illustrate that quorum sensing signals and downstream products regulated by these pathways can elicit a suite of physiological changes that greatly alter the antimicrobial susceptibility profiles of polymicrobial biofilm communities. It is important to note that a single molecule can act via more than one mechanism to alter antibiotic sensitivity, as in the case of HQNO or farnesol. Finally, it is possible that other quorum sensing molecules modulate drug efficacy via mechanisms other than their canonical roles in signaling.
BIOFILM MATRIX COMPONENTS WITHIN POLYMICROBIAL BIOFILMS MODULATE DRUG SENSITIVITY
Multiple studies have described cross-domain interactions that modify the structure of microbial biofilms and lead to altered sensitivity to antimicrobial agents. C. albicans is well known to form robust, polymicrobial biofilms with several bacterial species, including staphylococci (81, 84, 167–169). In particular, S. aureus cells can use C. albicans hyphae as a scaffold for attachment and subsequent microcolony formation (81). As a consequence, residence within this mixed-species biofilm protected S. aureus from miconazole (84) and from elevated doses of vancomycin (up to 1,600 μg/ml) (81). In contrast, the drug susceptibility of C. albicans to multiple antifungal drugs was unaltered (81, 84). Extracellular DNA and β-1,3-glucan components of the fungal matrix were shown to contribute to the observed protection of S. aureus within the mixed-species biofilm (81, 84, 170), but the underlying mechanisms are still unclear. Similarly, protection of E. coli from ofloxacin within a dual-species biofilm with C. albicans is also dependent on the presence of the polysaccharide β-1,3-glucan (171).
As another example, formation of a mixed-species biofilm can also shield both microbes from antimicrobial treatment. A matrix-nonproducing strain of Staphylococcus epidermidis is normally highly sensitive to vancomycin; however, when grown in coculture with C. albicans, S. epidermidis was protected from the drug, while C. albicans was protected from fluconazole (172). Thus, while it is well appreciated that biofilm matrix components enable intimate associations between coinhabiting species and that the biofilm lifestyle confers protection from antimicrobial insults, the above studies suggest that structural components produced by one microbe can greatly influence the drug sensitivity of a coexisting microbe within a mixed-species biofilm.
Interactions between bacterial species have also been shown to influence biofilm structure. Staphylococcal protein A (SpA), an adhesin known to mediate S. aureus biofilm formation (173), can also bind to the P. aeruginosa exopolysaccharide Psl and inhibit initial P. aeruginosa biofilm formation (174). In a different study, the interaction between these two adhesins led to striking changes in the architecture of mature P. aeruginosa biofilms (67). Specifically, exposure to SpA transformed P. aeruginosa biofilms into densely packed aggregates that occupied less surface area and were much less susceptible to tobramycin than untreated biofilms. While the observed shifts in biofilm structure and drug tolerance were shown to require the presence of both adhesins, it is currently unclear how the interaction between SpA and Psl causes the formation of aggregates. Together, these data indicate that interspecies interactions can modify biofilm architecture in diverse ways and, as a consequence, can greatly influence the drug susceptibility of a polymicrobial community.
INTERSPECIES INTERACTIONS WITHIN POLYMICROBIAL BIOFILMS INDUCE CELL WALL CHANGES
Several recent studies indicate that polymicrobial interactions can influence susceptibility to antibiotics by altering characteristics of their neighbor’s cell wall. One study examined the drug susceptibility profiles within a three-species biofilm community composed of Streptococcus anginosus, P. aeruginosa, and S. aureus. When part of this polymicrobial biofilm, S. anginosus became more tolerant to vancomycin, while S. aureus became less tolerant (175). Additionally, the authors found that exposure to S. aureus culture supernatant protected S. anginosus biofilm cells, but not planktonic cells, from vancomycin (175). In a subsequent study, the same group found that residence within the multispecies biofilm led to the upregulation of S. anginosus cell wall biosynthesis genes and caused an increase in cell wall thickness, which was hypothesized to protect S. anginosus biofilms from the drug (176). These data are consistent with the well-documented association between increased cell wall thickness and decreased susceptibility to vancomycin in S. aureus (177–180). Together, these findings suggest that interspecies interactions can alter properties of bacterial cell walls, which may contribute to changes in antimicrobial susceptibility within biofilm communities.
CONCLUSIONS
As we have reviewed in this article, biofilms are recalcitrant to antimicrobial therapy due to a combination of genetic and phenotypic mechanisms. These same mechanisms operate within polymicrobial biofilms, but with added layers of complexity produced by interspecies interactions. There are many examples of polymicrobial interactions influencing the antibiofilm efficacy of antibiotics. Here, we highlight the few, better-understood mechanisms that have been uncovered (Fig. 1), including those involved in a well-studied interspecies interaction that shift the drug susceptibility of S. aureus biofilms (Fig. 2). Antibiotic recalcitrance mechanisms within polymicrobial biofilms include horizontal gene transfer of antibiotic resistance genes, enzymatic degradation of antibiotics (by β-lactamases), the induction of transcriptional changes by primary metabolites (e.g., indole and 2,3-butanedione) or quorum sensing signals (e.g., DSF and farnesol), inhibition of electron transport (by HQNO), and changes in membrane fluidity (by HQNO). Chronic polymicrobial infections, such as those associated with airway infections in patients with CF, are likely impacted by many of these mechanisms, as has been highlighted in a recent literature review (181).
FIG 1.
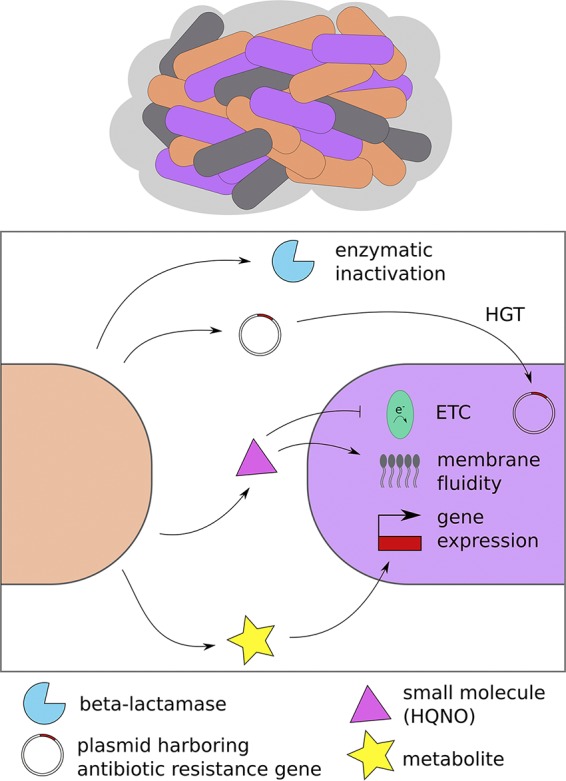
Multiple mechanisms contribute to the recalcitrance of polymicrobial biofilms to antimicrobial treatment. Interspecies interactions can shift the drug sensitivity profiles of microbes within multispecies biofilms, as represented by the differently colored microbes in a biofilm community at the top of the panel, and can do so via several mechanisms, including enzymatic inactivation of antibiotics by β-lactamases, interspecies exchange of antibiotic resistance genes, inhibition of electron transport, altered membrane fluidity, and metabolite-induced transcriptional changes. Abbreviations: HGT, horizontal gene transfer; ETC, electron transport chain; HQNO, 2-heptyl-4-hydroxyquinolone N-oxide.
FIG 2.
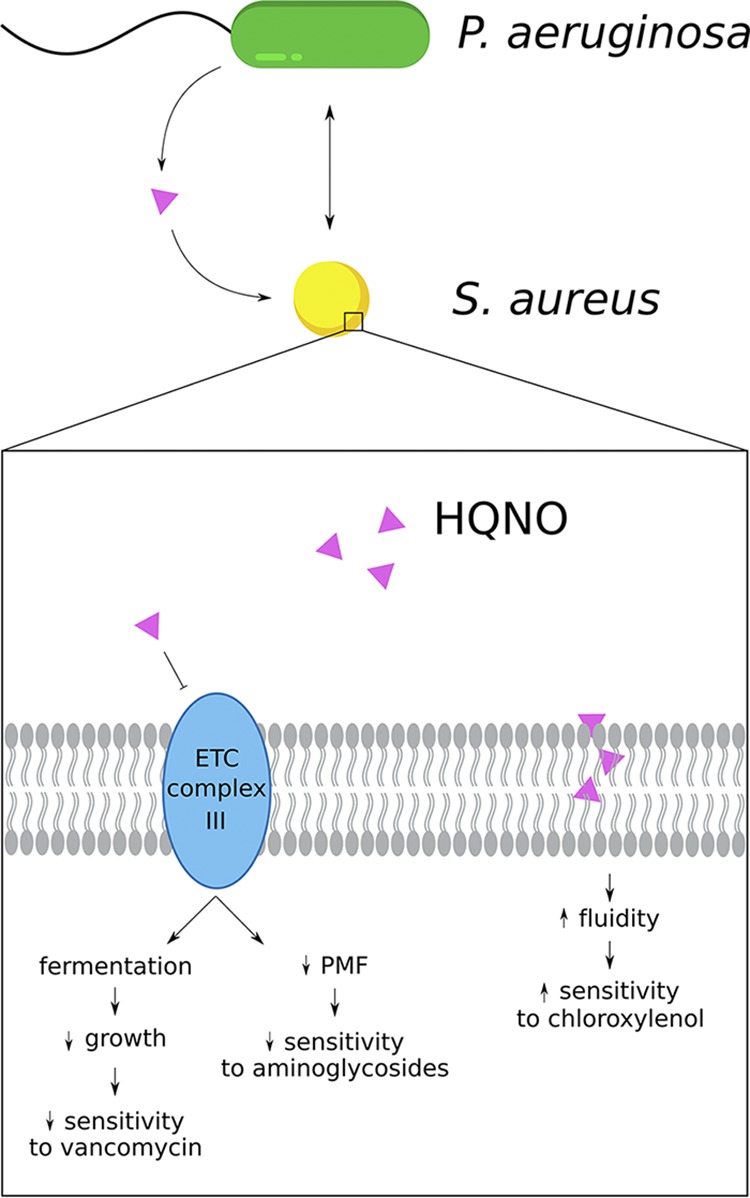
Interspecies interactions between Pseudomonas aeruginosa and Staphylococcus aureus influence the efficacy of antibiotics against S. aureus biofilms. The P. aeruginosa-secreted molecule 2-heptyl-4-hydroxyquinolone N-oxide (HQNO) alters the sensitivity of biofilm-grown S. aureus to multiple antibacterial agents, including cell wall-targeting drugs (e.g., vancomycin), protein synthesis inhibitors (e.g., aminoglycosides), and membrane-active compounds (e.g., chloroxylenol). HQNO-mediated inhibition of the S. aureus electron transport chain (ETC) leads to slow growth and decreases the proton motive force (PMF), which promote tolerance to vancomycin and aminoglycosides, respectively. Additionally, exposure to HQNO increases the fluidity of the S. aureus cell membrane, enhancing the ability of the membrane-targeting antiseptic chloroxylenol (and other hydrophobic compounds) to eradicate S. aureus biofilms.
Based on the studies reviewed above, it is clear that interspecies interactions can modulate the sensitivity of entire polymicrobial communities in dramatic and highly unpredictable ways, often enabling mixed-species biofilms to withstand even greater antibiotic challenge compared to single-species biofilms. Therefore, we believe that the results of monoculture experiments cannot always be extrapolated to predict the behavior of organisms when they are part of a community composed of other pathogens or members of the microbiota. Unfortunately, that is exactly how we currently measure the antimicrobial susceptibility of disease-causing organisms in the clinical setting. Indeed, a recent publication called into question the utility of using MIC testing to guide the treatment of the chronic, polymicrobial communities that are characteristic of CF airway infections (182).
In the past, multiple studies have made the striking observation that a microorganism’s antimicrobial sensitivity profile can be dramatically different when it is grown in mixed culture compared to in pure culture (183–189). As highlighted previously by DeLeon et al., in 1969 Shahidi and Ellner compared the antibiotic sensitivity of an artificial mixed-species community to that of a pure culture (63, 183). Briefly, broth cultures were inoculated either with an individual organism or with different combinations of 10 bacterial species and then spread uniformly onto agar plates. Subsequently, different antibiotic disks were placed on the agar surface, and zones of inhibition were recorded to establish whether an organism was sensitive or resistant to a given drug. To the authors’ surprise, they discovered that the ability of a drug to inhibit an organism was often entirely dependent on whether the organism was part of a pure or mixed culture. Specifically, they found multiple instances in which two organisms that were “sensitive” to an antibiotic in pure culture became “resistant” when mixed together. These results did not occur for all combinations tested, and it was not established in these studies what was driving these different outcomes. From these observations, it was concluded that “since reactions of bacterial mixtures are completely unpredictable, the authors emphasize that antibiotic susceptibility testing be limited to pure cultures” (183). This is not an unreasonable conclusion, but the studies cited above also highlight the limitations of such an approach.
Similar results were observed in later studies that assessed whether performing direct susceptibility testing using patient samples could be a way to expedite the communication of test results to clinicians (184–187, 189). Disk diffusion tests were performed either by directly swabbing clinical specimens (blood, urine, or wound exudates) onto agar plates or by the traditional method of first isolating individual organisms and growing them in broth to a standard inoculum density before plating. Then, the susceptibility results produced by each method were compared. For specimens that contained a single organism, the susceptibility results from the direct method typically closely mirrored those obtained by the traditional method (7.3% discrepancy in one study [186]). In contrast, for specimens that contained multiple microbes, the results often differed greatly between the two methods (42.6% discrepancy [186]). In other words, the sensitivity profiles for an organism were notably different under the mixed-culture compared to pure-culture conditions. The authors of this study echoed those above by saying the results obtained from mixed-culture experiments are “completely unreliable” and “provide clinically misleading information” (183, 189) and reached the consensus that susceptibility testing should be performed only on organisms grown in pure culture. However, these studies did not consider the intriguing hypothesis suggested by their observations that one species can profoundly influence the antimicrobial susceptibility of another, which may have important implications for the treatment of polymicrobial infections in patients.
In 1975, Linn and Szabo also observed and considered the problem of the incongruity between monospecies and multispecies antimicrobial sensitivity profiles. Contrary to the previous studies, they argued that “sensitivity testing in mixed cultures may well provide greater clinical relevance since these cultures more closely simulate the situation in the patient than does the pure culture” (188). Interestingly, they found cases where drugs became more effective when certain organisms were combined. They also observed that the differences in antimicrobial sensitivity between pure and mixed cultures were not randomly distributed but that certain organisms or drugs behaved the same way when in combination, knowledge that may offer therapeutic guidance. The literature above suggests that we have rediscovered and extended the findings of Linn and Szabo. Given the polymicrobial etiology of most chronic infections, and the important influence of interspecies interactions on antimicrobial efficacy in vitro, as illustrated by the many findings cited above, we argue that the approaches for measuring the drug susceptibility of mixed-species infections should be reconsidered.
Here, we address several important challenges and propose ideas for testing the antimicrobial susceptibility of microbes within polymicrobial, biofilm infections. A fundamental problem is the lack of effective treatments for polymicrobial infections, such as those of the CF airway or diabetic foot ulcers. Therefore, the first challenge is to direct drug discovery efforts toward identifying antimicrobial agents that can more effectively treat these recalcitrant infections. Furthermore, another key challenge is that we currently do not know how in vitro antimicrobial susceptibilities of mixed-species infections correlate with clinical outcomes. Perhaps, one solution could be to design clinical trials that test whether laboratory susceptibility testing results correlate with treatment outcomes (e.g., does the high in vitro efficacy of a drug against three bacterial species isolated from a wound infection correlate with the successful eradication of that infection following treatment with the same drug). Additionally, it may be possible to mine data retrospectively to assess if particular antibiotic regimens work better against mixed-species infections caused by certain combinations of organisms. Finally, an important challenge is a lack of knowledge about how antimicrobial efficacy changes with the addition of more than two constituent species. A priority moving forward should be to establish new in vitro model systems to interrogate the drug susceptibility profiles of complex polymicrobial communities, as at least one group has begun to do (175).
Once progress has been made in overcoming the above challenges, it may be possible to implement new approaches for susceptibility testing. Here, we propose a few ideas. If and when we reach a stage when we understand which drugs (or drug classes) retain efficacy against particular combinations of microbes, one possible approach could be to develop standardized treatments for particular combinations of microbes within mixed-species infections. As a simple example, if microbes A and B are detected in the same sample, then drug 1 is used, but if microbes A and C are detected together, drug 2 is selected. We envision that these treatments could be used in conjunction with existing MIC testing on pure cultures.
Alternatively, a personalized medicine approach could be a reasonable strategy for evaluating the antimicrobial susceptibility of mixed-species infections. Here, we propose a framework for a potential method. Following the identification of the microbes present in a clinical specimen using rapid, culture-independent methods, it may be possible to select and reconstitute a simplified microbial community (including bacteria and fungi). Perhaps, this community would be composed of the most abundant species, as well as less abundant species that are considered particularly clinically relevant. The selected community would be reconstituted in a microtiter plate and treated with a small number of antimicrobial agents at multiple concentrations, followed by plating onto selective agar for the enumeration of viable counts of each microbial species. This approach would provide a measure of the survival of each species after drug exposure. We recognize that there are many limitations of these proposed approaches for polymicrobial testing in the clinical laboratory. First of all, such strategies might be feasible for sites that are sterile in the absence of infection or sites that contain a relatively small number of resident species. In contrast, this approach could be quite challenging for infections in sites containing a complex microbiota (e.g., the intestine). For example, it is likely that commensals influence the drug sensitivity profiles of pathogens at these infection sites (83, 87); however, including commensals in the reconstituted community could lead to an intractable number of species for testing. Another important limitation for some of the approaches outlined above would be the high labor costs and time-intensive nature of such testing. However, integrating the routine use of culture-independent techniques could reduce the time and laborious culturing protocols that are commonly used for species identification in clinical microbiology laboratories and allow for more time and resources to be available for performing the proposed drug testing methods. Additionally, recent machine learning approaches have utilized the abundant available genome sequence resources to accurately predict MICs of organisms (190, 191); perhaps such approaches could be applied to polymicrobial communities. Additionally, it may be possible to automate several parts of the process using liquid-handling machines and plate-scanning software. Other concerns are the lack of rapid and reliable methods to quantify fungal viability following coculture with other microbes and the problem of how to tackle the antibiotic susceptibility of biofilms, which adds another layer of complexity.
Finally, perhaps the simplest solution to the challenges outlined here is a return to testing the antibiotic susceptibility of direct clinical samples, as described above (186). For example, following species identification, the original clinical specimen could be transferred to microtiter plates and subjected to different antibiotic treatments. Afterward, the treated sample could be plated on selective medium to determine viability of each species, which can be compared to the starting number of cells before antibiotic treatment to evaluate the success of antimicrobial treatment. Alternatively, for the subset of infections not responding to treatment, the clinical sample could be swabbed directly to permissive medium and a suite of antibiotics could be tested using standard MIC assays to empirically determine the antibiotic(s) that most effectively inhibits the organisms in the infection. As reported previously (87), sometimes only the most susceptible member of the community has to be killed to effectively reduce the growth/viability of the entire community. A major concern of this approach is whether testing on direct clinical samples can be standardized. Thus, because mixed-species infections vary widely from one patient to another, it is possible that a personalized approach is the best strategy to measure susceptibility profiles. To move forward, it is essential to promote discussion and collaboration between the clinical setting, academia, and industry to develop new methods to treat and diagnose polymicrobial infections.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R37 AI83256-06 and the Cystic Fibrosis Foundation (OTOOLE16G0) to G.A.O. and the Microbiology and Molecular Pathogenesis Training Grant (T32-AI007519) to G.O.
REFERENCES
- 1.Henrici AT. 1933. Studies of freshwater bacteria. I. A direct microscopic technique. J Bacteriol 25:277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. 1995. Microbial biofilms. Annu Rev Microbiol 49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Mah T-FC, O’Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 6.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Lebeaux D, Ghigo J-M, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hennequin C, Aumeran C, Robin F, Traore O, Forestier C. 2012. Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J Antimicrob Chemother 67:2123–2130. doi: 10.1093/jac/dks169. [DOI] [PubMed] [Google Scholar]
- 9.Savage VJ, Chopra I, O’Neill AJ. 2013. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother 57:1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maheshwari M, Ahmad I, Althubiani AS. 2016. Multidrug resistance and transferability of bla CTX-M among extended-spectrum β-lactamase-producing enteric bacteria in biofilm. J Glob Antimicrob Resist 6:142–149. doi: 10.1016/j.jgar.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Brown MR, Allison DG, Gilbert P. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother 22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- 12.Evans DJ, Allison DG, Brown MRW, Gilbert P. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J Antimicrob Chemother 27:177–184. doi: 10.1093/jac/27.2.177. [DOI] [PubMed] [Google Scholar]
- 13.Wentland EJ, Stewart PS, Huang CT, McFeters GA. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol Prog 12:316–321. doi: 10.1021/bp9600243. [DOI] [PubMed] [Google Scholar]
- 14.Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother 47:317–323. doi: 10.1128/aac.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodionov DG, Ishiguro EE. 1995. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J Bacteriol 177:4224–4229. doi: 10.1128/jb.177.15.4224-4229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppée J-Y, Ghigo J-M, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e1003144. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon CA, Hodges NA, Marriott C. 1988. Antibiotic interaction and diffusion through alginate and exopolysaccharide of cystic fibrosis-derived Pseudomonas aeruginosa. J Antimicrob Chemother 22:667–674. doi: 10.1093/jac/22.5.667. [DOI] [PubMed] [Google Scholar]
- 19.Hatch RA, Schiller NL. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:974–977. doi: 10.1128/AAC.42.4.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lynch SV, Dixon L, Benoit MR, Brodie EL, Keyhan M, Hu P, Ackerley DF, Andersen GL, Matin A. 2007. Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob Agents Chemother 51:3650–3658. doi: 10.1128/AAC.00601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Mah TF. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J Bacteriol 190:4447–4452. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 24.Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, Kaess H, Deterding RR, Accurso FJ, Pace NR. 2007. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci U S A 104:20529–20533. doi: 10.1073/pnas.0709804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filkins LM, Hampton TH, Gifford AH, Gross MJ, Hogan DA, Sogin ML, Morrison HG, Paster BJ, O’Toole GA. 2012. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol 194:4709–4717. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, Wolfgang MC. 2012. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stressmann FA, Rogers GB, van der Gast CJ, Marsh P, Vermeer LS, Carroll MP, Hoffman L, Daniels TWV, Patel N, Forbes B, Bruce KD. 2012. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax 67:867–873. doi: 10.1136/thoraxjnl-2011-200932. [DOI] [PubMed] [Google Scholar]
- 28.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim YW, Schmieder R, Haynes M, Willner D, Furlan M, Youle M, Abbott K, Edwards R, Evangelista J, Conrad D, Rohwer F. 2013. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J Cyst Fibros 12:154–164. doi: 10.1016/j.jcf.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filkins LM, O’Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boisvert A-A, Cheng MP, Sheppard DC, Nguyen D. 2016. Microbial biofilms in pulmonary and critical care diseases. Ann Am Thorac Soc 13:1615–1623. doi: 10.1513/AnnalsATS.201603-194FR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol 54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 33.Schaudinn C, Gorur A, Keller D, Sedghizadeh PP, Costerton JW. 2009. Periodontitis: an archetypical biofilm disease. J Am Dent Assoc 140:978–986. doi: 10.14219/jada.archive.2009.0307. [DOI] [PubMed] [Google Scholar]
- 34.Post JC, Preston RA, Aul JJ, Larkins-Pettigrew M, Rydquist-White J, Anderson KW, Wadowsky RM, Reagan DR, Walker ES, Kingsley LA, Magit AE, Ehrlich GD. 1995. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA 273:1598–1604. doi: 10.1001/jama.1995.03520440052036. [DOI] [PubMed] [Google Scholar]
- 35.Hendolin PH, Markkanen A, Ylikoski J, Wahlfors JJ. 1997. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol 35:2854–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Post JC. 2015. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 125:2003–2014. doi: 10.1002/lary.25291. [DOI] [PubMed] [Google Scholar]
- 37.Ronald A. 2003. The etiology of urinary tract infection: traditional and emerging pathogens. Dis Mon 49:71–82. doi: 10.1067/mda.2003.8. [DOI] [PubMed] [Google Scholar]
- 38.Kline KA, Lewis AL. 2016. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol Spectr 4(2):UTI-0012-2012. doi: 10.1128/microbiolspec.UTI-0012-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soto SM. 2014. Importance of biofilms in urinary tract infections: new therapeutic approaches. Adv Biol 2014:543974. doi: 10.1155/2014/543974. [DOI] [Google Scholar]
- 40.Giacometti A, Cirioni O, Schimizzi AM, Del Prete MS, Barchiesi F, D’Errico MM, Petrelli E, Scalise G. 2000. Epidemiology and microbiology of surgical wound infections. J Clin Microbiol 38:918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Citron DM, Goldstein EJC, Merriam CV, Lipsky BA, Abramson MA. 2007. Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J Clin Microbiol 45:2819–2828. doi: 10.1128/JCM.00551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen B, Klein BM, Krogfelt KA. 2006. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 3:225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Körber A, Schmid EN, Buer J, Klode J, Schadendorf D, Dissemond J. 2010. Bacterial colonization of chronic leg ulcers: current results compared with data 5 years ago in a specialized dermatology department. J Eur Acad Dermatol Venereol 24:1017–1025. doi: 10.1111/j.1468-3083.2010.03570.x. [DOI] [PubMed] [Google Scholar]
- 45.Percival SL, Hill KE, Williams DW, Hooper SJ, Thomas DW, Costerton JW. 2012. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen 20:647–657. doi: 10.1111/j.1524-475X.2012.00836.x. [DOI] [PubMed] [Google Scholar]
- 46.Weinstein MP, Murphy JR, Reller LB, Lichtenstein KA. 1983. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations, with special reference to factors influencing prognosis. Clin Infect Dis 5:54–70. doi: 10.1093/clinids/5.1.54. [DOI] [PubMed] [Google Scholar]
- 47.Faix RG, Kovarik SM. 1989. Polymicrobial sepsis among intensive care nursery infants. J Perinatol 9:131–136. [PubMed] [Google Scholar]
- 48.Roberts FJ, Geere IW, Coldman A. 1991. A three-year study of positive blood cultures, with emphasis on prognosis. Rev Infect Dis 13:34–46. doi: 10.1093/clinids/13.1.34. [DOI] [PubMed] [Google Scholar]
- 49.Pittet D, Li N, Wenzel RP. 1993. Association of secondary and polymicrobial nosocomial bloodstream infections with higher mortality. Eur J Clin Microbiol Infect Dis 12:813–819. doi: 10.1007/bf02000400. [DOI] [PubMed] [Google Scholar]
- 50.Pammi M, Zhong D, Johnson Y, Revell P, Versalovic J. 2014. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect Dis 14:390. doi: 10.1186/1471-2334-14-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim NCS, Lim DKA, Ray M. 2013. Polymicrobial versus monomicrobial keratitis: a retrospective comparative study. Eye Contact Lens 39:348–354. doi: 10.1097/ICL.0b013e3182a3024e. [DOI] [PubMed] [Google Scholar]
- 52.Trifilio S, Zhou Z, Fong JL, Zomas A, Liu D, Zhao C, Zhang J, Mehta J. 2015. Polymicrobial bacterial or fungal infections: incidence, spectrum of infection, risk factors, and clinical outcomes from a large hematopoietic stem cell transplant center. Transpl Infect Dis 17:267–274. doi: 10.1111/tid.12363. [DOI] [PubMed] [Google Scholar]
- 53.Tan TL, Kheir MM, Tan DD, Parvizi J. 2016. Polymicrobial periprosthetic joint infections: outcome of treatment and identification of risk factors. J Bone Joint Surg Am 98:2082–2088. doi: 10.2106/JBJS.15.01450. [DOI] [PubMed] [Google Scholar]
- 54.Jorge LS, Fucuta PS, Oliveira MGL, Nakazone MA, de Matos JA, Chueire AG, Salles M. 2018. Outcomes and risk factors for polymicrobial posttraumatic osteomyelitis. J Bone Jt Infect 3:20–26. doi: 10.7150/jbji.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudson VL, Wielinski CL, Regelmann WE. 1993. Prognostic implications of initial oropharyngeal bacterial flora in patients with cystic fibrosis diagnosed before the age of two years. J Pediatr 122:854–860. doi: 10.1016/s0022-3476(09)90007-5. [DOI] [PubMed] [Google Scholar]
- 56.Rosenbluth DB, Wilson K, Ferkol T, Schuster DP. 2004. Lung function decline in cystic fibrosis patients and timing for lung transplantation referral. Chest 126:412–419. doi: 10.1378/chest.126.2.412. [DOI] [PubMed] [Google Scholar]
- 57.Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, Goldberg JB. 2016. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis 35:947–953. doi: 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 58.Maliniak ML, Stecenko AA, McCarty NA. 2016. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: a single-center study. J Cyst Fibros 15:350–356. doi: 10.1016/j.jcf.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 59.Limoli DH, Hoffman LR. 2019. Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 74:684–692. doi: 10.1136/thoraxjnl-2018-212616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton BA, Morrison HG, Sogin ML, Czum J, Ashare A. 2016. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakeman CA, Moore JL, Noto MJ, Zhang Y, Singleton MD, Prentice BM, Gilston BA, Doster RS, Gaddy JA, Chazin WJ, Caprioli RM, Skaar EP. 2016. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun 7:11951. doi: 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. 2014. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O’Toole GA. 2015. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michelsen CF, Christensen A-M, Bojer MS, Høiby N, Ingmer H, Jelsbak L. 2014. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J Bacteriol 196:3903–3911. doi: 10.1128/JB.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tognon M, Köhler T, Gdaniec BG, Hao Y, Lam JS, Beaume M, Luscher A, Buckling A, van Delden C. 2017. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J 11:2233–2243. doi: 10.1038/ismej.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beaudoin T, Yau YCW, Stapleton PJ, Gong Y, Wang PW, Guttman DS, Waters V. 2017. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 3:25. doi: 10.1038/s41522-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR, Grahl N, Hogan DA, Rahme LG, Howell PL, O’Toole GA, Goldberg JB. 2017. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan K, Dammel C, Stein J, Rabin H, Surette MG. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 70.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog 4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korgaonkar AK, Whiteley M. 2011. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 193:909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC. 2013. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. 2008. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol 68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 75.Armbruster CE, Hong W, Pang B, Weimer KED, Juneau RA, Turner J, Swords WE. 2010. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1:e00102-10. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bragonzi A, Farulla I, Paroni M, Twomey KB, Pirone L, Lorè NI, Bianconi I, Dalmastri C, Ryan RP, Bevivino A. 2012. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS One 7:e52330. doi: 10.1371/journal.pone.0052330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. 2011. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lightbown JW. 1954. An antagonist of streptomycin and dihydrostreptomycin produced by Pseudomonas aeruginosa. J Gen Microbiol 11:477–492. doi: 10.1099/00221287-11-3-477. [DOI] [PubMed] [Google Scholar]
- 79.Lebrun M, de Repentigny J, Mathieu LG. 1978. Diminution of the antibacterial activity of antibiotics in cultures and in experimental mixed infections. Can J Microbiol 24:154–161. doi: 10.1139/m78-028. [DOI] [PubMed] [Google Scholar]
- 80.Hoffman LR, Deziel E, D’Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harriott MM, Noverr MC. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother 53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bernier SP, Létoffé S, Delepierre M, Ghigo J-M. 2011. Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol Microbiol 81:705–716. doi: 10.1111/j.1365-2958.2011.07724.x. [DOI] [PubMed] [Google Scholar]
- 83.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. 2013. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc Natl Acad Sci U S A 110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, Burgess KE, Lang S, Millington O, Mackay WG, Williams C, Ramage G. 2017. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol 8:258. doi: 10.3389/fmicb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Orazi G, O’Toole GA. 2017. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8:e00873-17. doi: 10.1128/mBio.00873-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Radlinski L, Rowe SE, Kartchner LB, Maile R, Cairns BA, Vitko NP, Gode CJ, Lachiewicz AM, Wolfgang MC, Conlon BP. 2017. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol 15:e2003981. doi: 10.1371/journal.pbio.2003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Adamowicz EM, Flynn J, Hunter RC, Harcombe WR. 2018. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J 12:2723–2735. doi: 10.1038/s41396-018-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Orazi G, Ruoff KL, O’Toole GA. 2019. Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio 10:e01501-19. doi: 10.1128/mBio.01501-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kline KA, Schwartz DJ, Gilbert NM, Hultgren SJ, Lewis AL. 2012. Immune modulation by group B Streptococcus influences host susceptibility to urinary tract infection by uropathogenic Escherichia coli. Infect Immun 80:4186–4194. doi: 10.1128/IAI.00684-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chekabab SM, Silverman RJ, Lafayette SL, Luo Y, Rousseau S, Nguyen D. 2015. Staphylococcus aureus inhibits IL-8 responses induced by Pseudomonas aeruginosa in airway epithelial cells. PLoS One 10:e0137753. doi: 10.1371/journal.pone.0137753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Radlinski L, Conlon BP. 2018. Antibiotic efficacy in the complex infection environment. Curr Opin Microbiol 42:19–24. doi: 10.1016/j.mib.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gerber AU, Craig WA. 1981. Worldwide clinical experience with cefoperazone. Drugs 22(Suppl 1):108–118. doi: 10.2165/00003495-198100221-00022. [DOI] [PubMed] [Google Scholar]
- 94.Hilf M, Yu VL, Sharp J, Zuravleff JJ, Korvick JA, Muder RR. 1989. Antibiotic therapy for Pseudomonas aeruginosa bacteremia: outcome correlations in a prospective study of 200 patients. Am J Med 87:540–546. doi: 10.1016/0002-9343(89)90695-5. [DOI] [PubMed] [Google Scholar]
- 95.Doern G. 1992. In vitro activity of loracarbef and effects of susceptibility test methods. Pediatr Infect Dis J 11:6A–16S. [DOI] [PubMed] [Google Scholar]
- 96.Doern GV. 1995. Interpretive criteria for in vitro antimicrobial susceptibility tests. Rev Med Microbiol 6:126–136. doi: 10.1097/00013542-199504000-00006. [DOI] [Google Scholar]
- 97.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. 2003. Susceptibility testing of Pseudomonas aeruginosa isolates and clinical response to parenteral antibiotic administration: lack of association in cystic fibrosis. Chest 123:1495–1502. doi: 10.1378/chest.123.5.1495. [DOI] [PubMed] [Google Scholar]
- 98.Doern GV, Brecher SM. 2011. The clinical predictive value (or lack thereof) of the results of in vitro antimicrobial susceptibility tests. J Clin Microbiol 49:S11–S14. doi: 10.1128/JCM.00580-11. [DOI] [Google Scholar]
- 99.Rex JH, Pfaller MA, Barry AL, Nelson PW, Webb CD. 1995. Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B as treatment of nonneutropenic patients with candidemia. NIAID Mycoses Study Group and the Candidemia Study Group. Antimicrob Agents Chemother 39:40–44. doi: 10.1128/AAC.39.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kartsonis N, Killar J, Mixson L, Hoe C-M, Sable C, Bartizal K, Motyl M. 2005. Caspofungin susceptibility testing of isolates from patients with esophageal candidiasis or invasive candidiasis: relationship of MIC to treatment outcome. Antimicrob Agents Chemother 49:3616–3623. doi: 10.1128/AAC.49.9.3616-3623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Greenwood D. 1981. In vitro veritas? Antimicrobial susceptibility tests and their clinical relevance. J Infect Dis 144:380–385. doi: 10.1093/infdis/144.4.380. [DOI] [PubMed] [Google Scholar]
- 102.Miller TE, North DK. 1981. Clinical infections, antibiotics and immunosuppression: a puzzling relationship. Am J Med 71:334–336. doi: 10.1016/0002-9343(81)90145-5. [DOI] [PubMed] [Google Scholar]
- 103.Pai MP, Momary KM, Rodvold KA. 2006. Antibiotic drug interactions. Med Clin North Am 90:1223–1255. doi: 10.1016/j.mcna.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 104.Hausner M, Wuertz S. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl Environ Microbiol 65:3710–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. 2005. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol 3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 106.Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, Takahashi H, Matsuda A, Sawamura A, Kato S. 2006. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol Lett 255:115–120. doi: 10.1111/j.1574-6968.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- 107.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 108.Christensen BB, Sternberg C, Molin S. 1996. Bacterial plasmid conjugation on semi-solid surfaces monitored with the green fluorescent protein (GFP) from Aequorea victoria as a marker. Gene 173:59–65. doi: 10.1016/0378-1119(95)00707-5. [DOI] [PubMed] [Google Scholar]
- 109.Stalder T, Top E. 2016. Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2:16022. doi: 10.1038/npjbiofilms.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 111.Reisner A, Haagensen JAJ, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 112.Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cvitkovitch DG, Li Y-H, Ellen RP. 2003. Quorum sensing and biofilm formation in streptococcal infections. J Clin Invest 112:1626–1632. doi: 10.1172/JCI20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scrudato Lo M, Blokesch M. 2012. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet 8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 116.Moreno-Gámez S, Sorg RA, Domenech A, Kjos M, Weissing FJ, van Doorn GS, Veening J-W. 2017. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat Commun 8:854–812. doi: 10.1038/s41467-017-00903-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Y-H, Tian X. 2012. Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel) 12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 119.Tanner WD, Atkinson RM, Goel RK, Toleman MA, Benson LS, Porucznik CA, VanDerslice JA. 2017. Horizontal transfer of the blaNDM-1 gene to Pseudomonas aeruginosa and Acinetobacter baumannii in biofilms. FEMS Microbiol Lett 364:fnx048. doi: 10.1093/femsle/fnx048. [DOI] [PubMed] [Google Scholar]
- 120.Potron A, Poirel L, Nordmann P. 2011. Plasmid-mediated transfer of the bla(NDM-1) gene in Gram-negative rods. FEMS Microbiol Lett 324:111–116. doi: 10.1111/j.1574-6968.2011.02392.x. [DOI] [PubMed] [Google Scholar]
- 121.Ready D, Pratten J, Roberts AP, Bedi R, Mullany P, Wilson M. 2006. Potential role of Veillonella spp. as a reservoir of transferable tetracycline resistance in the oral cavity. Antimicrob Agents Chemother 50:2866–2868. doi: 10.1128/AAC.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hannan S, Ready D, Jasni AS, Rogers M, Pratten J, Roberts AP. 2010. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol Med Microbiol 59:345–349. doi: 10.1111/j.1574-695X.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 123.Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, Zhu W, Musser KA, Thompson J, Kohlerschmidt D, Dumas N, Limberger RJ, Patel JB. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother 51:231–238. doi: 10.1128/AAC.00576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bush K. 2010. Bench-to-bedside review: the role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit Care 14:224. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Høiby N. 2000. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 126.Schaar V, Nordström T, Mörgelin M, Riesbeck K. 2011. Moraxella catarrhalis outer membrane vesicles carry β-lactamase and promote survival of Streptococcus pneumoniae and Haemophilus influenzae by inactivating amoxicillin. Antimicrob Agents Chemother 55:3845–3853. doi: 10.1128/AAC.01772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee J, Lee E-Y, Kim S-H, Kim D-K, Park K-S, Kim KP, Kim Y-K, Roh T-Y, Gho YS. 2013. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother 57:2589–2595. doi: 10.1128/AAC.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sorg RA, Lin L, van Doorn GS, Sorg M, Olson J, Nizet V, Veening J-W. 2016. Collective resistance in microbial communities by intracellular antibiotic deactivation. PLoS Biol 14:e2000631. doi: 10.1371/journal.pbio.2000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kim SW, Park SB, Im SP, Lee JS, Jung JW, Gong TW, Lazarte JMS, Kim J, Seo J-S, Kim J-H, Song J-W, Jung HS, Kim GJ, Lee YJ, Lim S-K, Jung TS. 2018. Outer membrane vesicles from β-lactam-resistant Escherichia coli enable the survival of β-lactam-susceptible E. coli in the presence of β-lactam antibiotics. Sci Rep 8:5402. doi: 10.1038/s41598-018-23656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Budhani RK, Struthers JK. 1998. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob Agents Chemother 42:2521–2526. doi: 10.1128/AAC.42.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weimer KED, Juneau RA, Murrah KA, Pang B, Armbruster CE, Richardson SH, Swords WE. 2011. Divergent mechanisms for passive pneumococcal resistance to β-lactam antibiotics in the presence of Haemophilus influenzae. J Infect Dis 203:549–555. doi: 10.1093/infdis/jiq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Perez AC, Pang B, King LB, Tan L, Murrah KA, Reimche JL, Wren JT, Richardson SH, Ghandi U, Swords WE. 2014. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog Dis 70:280–288. doi: 10.1111/2049-632X.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giwercman B, Meyer C, Lambert PA, Reinert C, Høiby N. 1992. High-level beta-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob Agents Chemother 36:71–76. doi: 10.1128/aac.36.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brook I, Yocum P. 1995. Bacteriology and beta-lactamase activity in ear aspirates of acute otitis media that failed amoxicillin therapy. Pediatr Infect Dis J 14:805–807. [PubMed] [Google Scholar]
- 135.Brook I. 2009. The role of beta-lactamase-producing-bacteria in mixed infections. BMC Infect Dis 9:202. doi: 10.1186/1471-2334-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Flynn JM, Niccum D, Dunitz JM, Hunter RC. 2016. Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog 12:e1005846. doi: 10.1371/journal.ppat.1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 138.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. 1985. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 109:135–141. doi: 10.1007/bf00391888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Whiteson KL, Meinardi S, Lim YW, Schmieder R, Maughan H, Quinn R, Blake DR, Conrad D, Rohwer F. 2014. Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. ISME J 8:1247–1258. doi: 10.1038/ismej.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim K-S, Lee S, Ryu C-M. 2013. Interspecific bacterial sensing through airborne signals modulates locomotion and drug resistance. Nat Commun 4:1809. doi: 10.1038/ncomms2789. [DOI] [PubMed] [Google Scholar]
- 141.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Iyer R, Wu Z, Woster PM, Delcour AH. 2000. Molecular basis for the polyamine-OmpF porin interactions: inhibitor and mutant studies. J Mol Biol 297:933–945. doi: 10.1006/jmbi.2000.3599. [DOI] [PubMed] [Google Scholar]
- 143.Tkachenko AG, Pozhidaeva ON, Shumkov MS. 2006. Role of polyamines in formation of multiple antibiotic resistance of Escherichia coli under stress conditions. Biochemistry (Mosc) 71:1042–1049. doi: 10.1134/S0006297906090148. [DOI] [PubMed] [Google Scholar]
- 144.Kwon DH, Lu CD. 2007. Polyamine effects on antibiotic susceptibility in bacteria. Antimicrob Agents Chemother 51:2070–2077. doi: 10.1128/AAC.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cepl J, Blahůšková A, Cvrčková F, Markoš A. 2014. Ammonia produced by bacterial colonies promotes growth of ampicillin-sensitive Serratia sp. by means of antibiotic inactivation. FEMS Microbiol Lett 354:126–132. doi: 10.1111/1574-6968.12442. [DOI] [PubMed] [Google Scholar]
- 146.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. 2009. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 148.Bernier SP, Surette MG. 2013. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol 4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. 2010. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem 285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nijland R, Burgess JG. 2010. Bacterial olfaction. Biotechnol J 5:974–977. doi: 10.1002/biot.201000174. [DOI] [PubMed] [Google Scholar]
- 151.Arora DP, Hossain S, Xu Y, Boon EM. 2015. Nitric oxide regulation of bacterial biofilms. Biochemistry 54:3717–3728. doi: 10.1021/bi501476n. [DOI] [PubMed] [Google Scholar]
- 152.Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol 49:443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- 153.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. 2009. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol 7:e1000068. doi: 10.1371/journal.pbio.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pereira CS, Thompson JA, Xavier KB. 2013. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev 37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 156.Schertzer JW, Boulette ML, Whiteley M. 2009. More than a signal: non-signaling properties of quorum sensing molecules. Trends Microbiol 17:189–195. doi: 10.1016/j.tim.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 157.Kong EF, Tsui C, Kucharíková S, Van Dijck P, Jabra-Rizk MA. 2017. Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob Agents Chemother 61:e01573-17. doi: 10.1128/AAC.01573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lightbown JW, Jackson FL. 1956. Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem J 63:130–137. doi: 10.1042/bj0630130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mates SM, Patel L, Kaback HR, Miller MH. 1983. Membrane potential in anaerobically growing Staphylococcus aureus and its relationship to gentamicin uptake. Antimicrob Agents Chemother 23:526–530. doi: 10.1128/aac.23.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Eiff von C, Heilmann C, Proctor RA, Woltz C, Peters G, Götz F. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J Bacteriol 179:4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kalani M, Brismar K, Fagrell B, Ostergren J, Jorneskog G. 1999. Transcutaneous oxygen tension and toe blood pressure as predictors for outcome of diabetic foot ulcers. Diabetes Care 22:147–151. doi: 10.2337/diacare.22.1.147. [DOI] [PubMed] [Google Scholar]
- 162.Wattel F, Mathieu D, Coget JM, Billard V. 1990. Hyperbaric oxygen therapy in chronic vascular wound management. Angiology 41:59–65. doi: 10.1177/000331979004100109. [DOI] [PubMed] [Google Scholar]
- 163.Kopf SH, Sessions AL, Cowley ES, Reyes C, Van Sambeek L, Hu Y, Orphan VJ, Kato R, Newman DK. 2016. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A 113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jabra-Rizk MA, Meiller TF, James CE, Shirtliff ME. 2006. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother 50:1463–1469. doi: 10.1128/AAC.50.4.1463-1469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Inoue Y, Togashi N, Hamashima H. 2016. Farnesol-induced disruption of the Staphylococcus aureus cytoplasmic membrane. Biol Pharm Bull 39:653–656. doi: 10.1248/bpb.b15-00416. [DOI] [PubMed] [Google Scholar]
- 166.Radlinski LC, Rowe SE, Brzozowski R, Wilkinson AD, Huang R, Eswara P, Conlon BP. 2019. Chemical induction of aminoglycoside uptake overcomes antibiotic tolerance and resistance in Staphylococcus aureus. Cell Chem Biol. doi: 10.1016/j.chembiol.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hogan DA, Kolter R. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 168.Shirtliff ME, Peters BM, Jabra-Rizk MA. 2009. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett 299:1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Morales DK, Hogan DA. 2010. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6:e1000886. doi: 10.1371/journal.ppat.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. 2016. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio 7:e01365-16. doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.De Brucker K, Tan Y, Vints K, De Cremer K, Braem A, Verstraeten N, Michiels J, Vleugels J, Cammue BPA, Thevissen K. 2015. Fungal β-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob Agents Chemother 59:3052–3058. doi: 10.1128/AAC.04650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Adam B, Baillie GS, Douglas LJ. 2002. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol 51:344–349. doi: 10.1099/0022-1317-51-4-344. [DOI] [PubMed] [Google Scholar]
- 173.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penadés JR, Lasa I. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Armbruster CR, Wolter DJ, Mishra M, Hayden HS, Radey MC, Merrihew G, MacCoss MJ, Burns J, Wozniak DJ, Parsek MR, Hoffman LR. 2016. Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio 7:e00538-16. doi: 10.1128/mBio.00538-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tavernier S, Crabbé A, Tuysuz M, Stuer L, Henry S, Rigole P, Dhondt I, Coenye T. 2017. Community composition determines activity of antibiotics against multispecies biofilms. Antimicrob Agents Chemother 61:e00302-17. doi: 10.1128/AAC.00302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tavernier S, Sass A, De Bruyne M, Baeke F, De Rycke R, Crabbé A, Vandecandelaere I, Van Nieuwerburgh F, Coenye T. 2018. Decreased susceptibility of Streptococcus anginosus to vancomycin in a multispecies biofilm is due to increased thickness of the cell wall. J Antimicrob Chemother 73:2323–2330. doi: 10.1093/jac/dky216. [DOI] [PubMed] [Google Scholar]
- 177.Cui L, Ma X, Sato K, Okuma K, Tenover FC, Mamizuka EM, Gemmell CG, Kim M-N, Ploy M-C, El-Solh N, Ferraz V, Hiramatsu K. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 41:5–14. doi: 10.1128/jcm.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cui L, Iwamoto A, Lian J-Q, Neoh H-M, Maruyama T, Horikawa Y, Hiramatsu K. 2006. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cázares-Domínguez V, Cruz-Córdova A, Ochoa SA, Escalona G, Arellano-Galindo J, Rodríguez-Leviz A, Hernández-Castro R, López-Villegas EO, Xicohtencatl-Cortes J. 2015. Vancomycin tolerant, methicillin-resistant Staphylococcus aureus reveals the effects of vancomycin on cell wall thickening. PLoS One 10:e0118791. doi: 10.1371/journal.pone.0118791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Vandeplassche E, Tavernier S, Coenye T, Crabbé A. 2019. Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens. Eur Respir Rev 28:190041. doi: 10.1183/16000617.0041-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Waters VJ, Kidd TJ, Canton R, Ekkelenkamp MB, Johansen HK, LiPuma JJ, Bell SC, Elborn JS, Flume PA, VanDevanter DR, Gilligan P, Antimicrobial Resistance International Working Group in Cystic Fibrosis . 2019. Reconciling antimicrobial susceptibility testing and clinical response in antimicrobial treatment of chronic cystic fibrosis lung infections. Clin Infect Dis doi: 10.1093/cid/ciz364. [DOI] [PubMed] [Google Scholar]
- 183.Shahidi A, Ellner PD. 1969. Effect of mixed cultures on antibiotic susceptibility testing. Appl Microbiol 18:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Barry AL, Joyce LJ, Adams AP, Benner EJ. 1973. Rapid determination of antimicrobial susceptibility for urgent clinical situations. Am J Clin Pathol 59:693–699. doi: 10.1093/ajcp/59.5.693. [DOI] [PubMed] [Google Scholar]
- 185.Ellner PD, Johnson E. 1976. Unreliability of direct antibiotic susceptibility testing on wound exudates. Antimicrob Agents Chemother 9:355–356. doi: 10.1128/aac.9.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hollick GE, Washington JA. 1976. Comparison of direct and standardized disk diffusion susceptibility testing of urine cultures. Antimicrob Agents Chemother 9:804–809. doi: 10.1128/aac.9.5.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Johnson JE, Washington JA. 1976. Comparison of direct and standardized antimicrobial susceptibility testing of positive blood cultures. Antimicrob Agents Chemother 10:211–214. doi: 10.1128/aac.10.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Linn BS, Szabo S. 1975. The varying sensitivity to antibacterial agents of micro-organisms in pure vs. mixed cultures. Surgery 77:780–785. [PubMed] [Google Scholar]
- 189.Mirrett S, Reller LB. 1979. Comparison of direct and standard antimicrobial disk susceptibility testing for bacteria isolated from blood. J Clin Microbiol 10:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nguyen M, Brettin T, Long SW, Musser JM, Olsen RJ, Olson R, Shukla M, Stevens RL, Xia F, Yoo H, Davis JJ. 2018. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci Rep 8:421. doi: 10.1038/s41598-017-18972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nguyen M, Long SW, McDermott PF, Olsen RJ, Olson R, Stevens RL, Tyson GH, Zhao S, Davis JJ. 2019. Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J Clin Microbiol 57:e01260-18. doi: 10.1128/JCM.01260-18. [DOI] [PMC free article] [PubMed] [Google Scholar]


