Abstract
Megakaryopoiesis produces specialized hematopoietic stem cells in the bone marrow that give rise to megakaryocytes which ultimately produce platelets. Defects in megakaryopoiesis can result in altered platelet counts and physiology, leading to dysfunctional hemostasis and thrombosis. Additionally, dysregulated megakaryopoiesis is also associated with myeloid pathologies. Transcription factors play critical roles in cell differentiation by regulating the temporal and spatial patterns of gene expression which ultimately decide cell fate. Several transcription factors have been described as regulating megakaryopoiesis including myocyte enhancer factor 2C (MEF2C), however the genes regulated by MEF2C that influence megakaryopoiesis have not been reported. Using chromatin immunoprecipitation-sequencing (ChIP-Seq) and Gene Ontology (GO) data we identified five candidate genes that are bound by MEF2C and regulate megakaryopoiesis: MOV10, AGO3, HDAC1, RBBP5, and WASF2. To study expression of these genes, we silenced MEF2C gene expression in the Meg01 megakaryocytic cell line and in induced pluripotent stem cells by CRISPR/Cas9 editing. We also knocked down MEF2C expression in cord blood derived hematopoietic stem cells by siRNA. We found that absent or reduced MEF2C expression resulted in defects in megakaryocytic differentiation and reduced levels of the candidate target genes. Luciferase assays confirmed that genomic sequences within the target genes are regulated by MEF2C levels. Finally, we demonstrate that small deletions linked to a platelet count-associated single nucleotide polymorphism alter transcriptional activity, suggesting a mechanism by which genetic variation in MEF2C alters platelet production. These data help elucidate the mechanism behind MEF2C regulation of megakaryopoiesis and genetic variation driving platelet production.
Keywords: MEF2C, megakaryopoiesis, megakaryocytes, hematopoiesis, transcription
Introduction
Genome-wide Association Studies (GWAS) have identified genetic variants associated with a large number of physiological and pathological phenotypic traits in the past several years. One important aspect of these studies, however, is that the associated variants are not necessarily causative of the observed associations. Moreover, no simple experiment is available to identify functional genes and variants. This is primarily due to two factors. First, genetic linkage results in large intervals of DNA containing multiple variants that are co-inherited and jointly associated with phenotypic outcome, leading to a lack of resolution in identifying the functional drivers from among many candidates. Second, some platforms used in in GWAS measure only a subset of the variants in any region, and the true causal variant or variants may be not directly observed.
Several platelet-related phenotypes such as platelet count and volume have been studied by GWAS.1–4 Platelets are anucleate blood cells that are key regulators of hemostasis and thrombosis. Normal platelet count is widely considered 150–400 × 109/L of whole blood and values outside these ranges (thrombocytopenia and thrombocytosis) has been associated with bleeding disorders or indicative of bone marrow disorders. Even within the normal range, there is an association of platelet count with cardiovascular disease risk.5 The large number of genetic associations with platelet count and the high heritability of platelet count suggests there are numerous undiscovered genetic determinants of platelet number.6,7 One such gene associated with platelet count is the transcription factor Myocyte Enhancer Factor 2C (MEF2C).1,4
Transcription factors play a critical role in cellular differentiation by determining temporal sequence of gene expression in the cell by binding to regulatory elements on DNA. Several transcription factors significant to megakaryopoiesis have been identified including RUNX1, GATA1, FLI1, GFIb, ETV6, SCL/TAL1.8,9 The importance of these transcription factors to physiology is underlined by mutations in them that result in defects in platelet production and function.10
MEF2C, a member of the MADS box containing family of transcription factors, is highly expressed in skeletal muscle, heart, brain, and spleen.11 While MEF2C was first identified as a regulator of muscle differentiation, its role in the hematopoietic system was uncovered in a gene expression profile of peripheral CD34+ hematopoietic stem cells (HSCs).12 Enforced constitutive expression of Mef2c in the bone marrow of mice resulted in increased numbers of monocytes and reduced granulocytes, leading to the model of MEF2C as a determinant of myeloid cell fate.13 Later it was shown that Mef2c deficient mice exhibited reduced platelet counts, increased platelet size, and altered platelet shape and granularity.14 Mef2C has also been described as critical regulator of lymphoid versus myeloid differentiation.15 High MEF2C expression is associated with worse outcomes in pediatric acute myeloid leukemia.16 However, the target genes of MEF2C critical for megakaryopoiesis remain unknown.
In this report we utilize publicly available genomic data sources to prioritize and identify variants that are linked to the GWAS-implicated MEF2C variant and alter transcriptional regulation. In addition, by utilizing Gene Ontology information and chromatin immunoprecipitation-deep sequencing (ChIP-Seq) data we identified five potential MEF2C target genes that regulate megakaryopoiesis. We have validated these target genes by developing a megakaryocytic cell line and an induced pluripotent stem cell (iPSC) line that are deficient in MEF2C expression by CRISPR/Cas9 genomic editing. In addition, we find evidence for a potential mechanism by which MEF2C genetic variants alter platelet production.
Methods
Cell Culture.
Meg01 cells were obtained from ATCC (Manassas, VA) and cultured according to protocol in RPMI medium supplemented with 10% fetal bovine serum. CHOP10 iPSCs were obtained from the Human Pluripotent Stem Cell Core at the Children’s Hospital of Philadelphia. The derivation of CHOP10 iPSC cells has be described17 and were cultured and differentiated to megakaryocytes according the published protocol developed by the core facility.18 CHOP10 cells were differentiated into hematopoietic progenitor cells (HPCs) which were then isolated and induced to differentiate into megakaryocytes for 11 days by treatment with SCF, TPO, and IL-3.18 Human umbilical cord blood (CB) CD34+ HSCs were isolated and differentiated as previously described.19 A 10μM ON-TARGET plus SMART pool against human MEF2C siRNA or a negative control (Dharmacon, Lafayette, CO) was transfected into the differentiating cells on days 3, 6, and 9 of the protocol using Lipofectamine (Thermo Fisher, Waltham, MA).
Construction of All-in-One CRISPR vector and MEF2C gene editing.
We generated a vector similar to the All-In-One-GFP vector (AIO-GFP, Addgene #74119, Cambridge, MA) with the difference of it containing a wildtype Cas9 as compared to the Cas9(D10A) nickase which we termed AIO-GFP(Cas9).20 AIO-GFP(Cas9) contains dual U6 promoter-driven sgRNAs and EGFP-coupled Cas9 nuclease.
The design of sgRNA pairs for targeting and prediction of off-target sites were based on online tools: CRISPR Design (http://crispr.mit.edu/) and CRISPOR (http://www.crispor.org). Pairs of complementary DNA oligos containing sgRNA sequences were individually phosphorylated with T4 polynucleotide kinase and then annealed. The two sgRNAs were cloned into the BbsI and BsaI sited in the AIO-GFP(Cas9) vector. Cloning was confirmed by PCR. To target MEF2C gene expression, we used following annealed sgRNAs: sgRNA1 forward: 5′-ACCGACAACGAGCCGCATGAGAGC-3′, reverse: 5′-AAACGCTCTCATGCGGCTCGTTGT-3’ .sgRNA2 forward: 5′-ACCGGAGAAGCACTTTGTCCATGT-3′, reverse: 5′-AAACACATGGACAAAGTGCTTCTC-3’. The AIO-GFP(Cas9) plasmid was transfected into Meg01 and CHOP10 iPSC cells using Nucleofector II (Lonza, Basel, Switzerland) based on manufacturer’s instruction. 24 hours following transfection, individual GFP positive cells were sorted into each well of a 96-well plate. Meg01-MEF2CKO and CHOP10-MEF2CKO cell clones were identified and confirmed by target region DNA PCR sequencing and western blots.
Reverse transcription and quantitative real time PCR.
Total RNA was isolated using Trizol Reagent (Thermo Fisher), from Meg01 or Meg01-MEF2CKO cells treated with 40nM phorbol 12-myristate 13-acetate (PMA) or DMSO for 24 hours; CHOP10 or CHOP10-MEF2CKO HPCs after 11 days culture in megakaryocytic differentiation media,18 and from CB CD34+ derived megakaryocytes after 13 days culture in megakaryocytic differentiation media.19 3ug total RNA was used for first strand cDNA synthesis using Superscript III First-Strand synthesis kit (Thermo Fisher Scientific). To evaluate relative expression levels of mRNAs, we performed qRT-PCR with the Power SYBR Green PCR master mix (Life Technologies, Carlsbad, CA) normalized to GAPDH. Detailed primer information is shown in Supplemental Table 1. We carried out real time PCR reaction and analyses in 384-well optical reaction plates using the CFX384 instrument (Bio-Rad, Hercules, CA).
Western blotting.
Cell were lysed in RIPA buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, and 1× protease inhibitor cocktail (Millipore Sigma, St. Louis, MO). 40μg of cell extracts were resolved by 10% SDS-PAGE and transferred onto PVDF membranes. We immunostained membranes with Rabbit anti-human MEF2C antibody (Cell Signaling, Danvers, MA) and mouse anti-human GAPDH monoclonal antibody (Santa Cruz Biotechnology, Dallas, TX). Detection and densitometric analysis was performed with Odyssey Infrared Imaging System (Li-Cor, Lincoln, NE).
Flow cytometry.
Meg01, Meg01-MEF2CKO, CHOP10, CHOP10-MEF2C-KO, and CB-derived megakaryocytes were treated as described above. Cells were stained with anti-CD34 (Biolegend, San Diego, CA), anti-CD235a, anti-CD45, anti-CD41, or anti-CD42 antibodies (BD Biosciences, San Jose, CA) and then analyzed by flow cytometry using an Accuri C6 flow cytometer (BD Biosciences).
Luciferase assays.
297bp genomic DNA fragments of RBBP5, AGO3, HDAC1, MOV10, and WASF2 genes containing proposed MEF2C binding motifs were amplified by PCR. NheI and HindIII sites were incorporated into the 5’ or 3’ end of primers respectively and ligated into pGL4.28 luciferase vector (Promega, Madison, WI). MEF2C variant luciferase reporter plasmids were constructed as follows: 80bp oligonucleotides containing sequence surrounding the SNPs rs4521516, rs200234178 and rs34845073 were synthesized and annealed (sticky NheI and HindIII sites were formed after annealing) and ligated into pGL4.28 luciferase vector (Promega). All the plasmids were validated by DNA sequencing. Detailed primer information is shown in Supplemental Table 1. For MEF2C target gene luciferase assays, 2 μg of reporter plasmid was co-transfected with 0.5 μg ß-gal expression plasmid into 5×105 Meg01 or Meg01-MEF2CKO cells with Nucleofector II (Lonza, Basel, Switzerland) based on the manufacturer’s instructions. For SNP luciferase assays, 2 μg of reporter plasmid was co-transfected with 0.5 μg ß-gal plasmid into 5×105 K562 cells with Lipofectamine 2000. 24 hours post-transfection, luciferase activity was detected on a FLUOstar OPTIMA plate reader (BMG Labtech, Cary, NC). ß-gal activity was quantified with a ß-gal assay kit (Thermo Fisher Scientific). The data are presented as Luciferase/ß-gal.
Statistical Analysis.
For assays in which two normally distributed means were compared, a two-sample t-test was utilized. For assays in which a normally distributed mean was compared against a normalized ratio of 1, a one-sample t-test was utilized. For correlation analysis, Pearson’s correlation coefficient was calculated, and significance calculated using a t-distribution. Graphpad Prism or Microsoft Excel was used to perform the calculations.
Results
Abrogated Megakaryopoiesis in MEF2C Reduced and Deficient Cells.
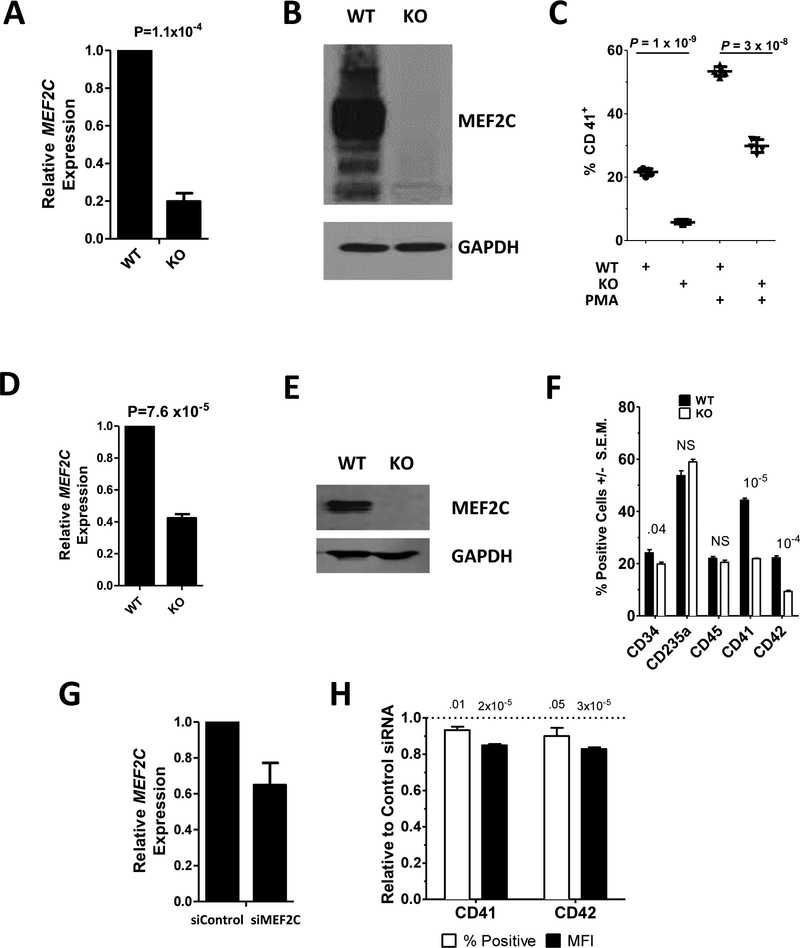
To better understand the role of MEF2C in megakaryopoiesis, we first generated a Meg01 megakaryoblastic cell line that was deficient in MEF2C expression. This was done by utilizing CRISPR-Cas9 guided by two gRNAs that anneal to sequences immediately downstream of the translational start site of the MEF2C gene to excise a segment of genomic DNA. After selection of cells by flow cytometry to isolate cells that express GFP, which is also contained on the Cas9 plasmid, we isolated a clone that contained a 23bp deletion in one copy of MEF2C and a 13bp insertion in the other copy, which were generated by nonhomologous end joining repair. qRT-PCR analysis of this clone (Meg01-MEF2C) with primers located outside the altered region indicated a ~80% reduction of MEF2C mRNA in these cells (Fig. 1A) indicating that while a portion of the mRNA was generated, potentially by alternative splicing, MEF2C mRNA transcription was disrupted by the Cas9-induced alteration. Western blot analysis of the cell line indicated that MEF2C protein expression was lost (Fig. 1B).
Fig. 1. MEF2C deficient cells are defective in megakaryocytic differentiation.
MEF2C (A,D,G) mRNA and (B,E) protein were assayed by qRT-PCR and Western Blot, respectively in: (A,B) Meg01 (WT) and Meg01-MEF2CKO (KO); (D,E) CHOP10 (WT) and CHOP10-MEF2CKO iPSCs; and (G) CB CD34+ derived megakaryocytes transfected with control (siControl) or MEF2C (siMEF2C) targeting siRNA. (C) Surface CD41 levels in Meg01 (WT) and Meg01-MEF2CKO (KO) cells were measured by flow cytometry cells before and after PMA treatment. (F) Percent of cells positive for surface marker expression in CHOP10 (WT) and CHOP10-MEF2CKO (KO) cells measured by flow cytometry. (H) Surface CD41 and CD42 percent positive and mean fluorescent intensity (MFI) of CB CD34+ derived megakaryocytes transfected with anti-MEF2C siRNA. Values are presented as relative to control siRNA transfected cells. P-value above bars. NS=non-significant.
To determine if MEF2C regulates megakaryocytic differentiation in Meg01 cells similar to how it does in murine megakaryocytes, we treated these cells with PMA, which is known to drive the cells toward a more megakaryocyte-like phenotype.21 In wildtype cells treatment of PMA resulted in an increase in CD41 expression, a marker of megakaryocytes, from 20% of cells to 55% of cells (Fig. 1C). However, in MEF2C-KO cells, both basal levels of CD41 (5% vs 20%) and PMA-induced levels (30% vs 55%) was reduced compared to wildtype cells (Fig 1C).
To study the requirement of MEF2C for megakaryopoiesis is a more physiologically relevant cell type, we performed the same CRISPR/Cas9-mediated deletion described above in CHOP10 iPSCs. MEF2C mRNA was reduced ~60% (Fig. 1D) and protein levels were eliminated (Fig. 1E). After 11 days in a megakaryocytic differentiation media, CHOP10 and CHOP10-MEF2CKO derived HPCs were analyzed by flow cytometry for markers of hematopoietic stem cells (CD34), erythrocytes (CD235a), leukocytes (CD45), and megakaryocytes (CD41 and CD42). While the percentage of cells positive for CD235a and CD45 staining was unchanged, the percentage of CD34+ cells was slightly reduced in the CHOP10-MEF2CKO iPSCs compared to wildtype, 20% vs 24% (Fig. 1F). Importantly, cells positive for megakaryocytic markers CD41 and CD42 were considerably reduced in the KO cells: 22% vs 44% for CD41 and 9% vs 22% for CD42 (Fig. 1F). Both the Meg01 and CHOP10 cells were edited independently using the same Cas9 gRNAs to knock out MEF2C expression. Importantly, the similarity of the results indicates that they are not a result of an off-target effect of Cas9, but due to the targeted removal of MEF2C expression.
Finally, to study MEF2C in a primary cell type, we isolated CD34+ hematopoietic stem cells isolated from umbilical cord blood and differentiated them into megakaryocytes. During differentiation, we transfected these cells with a siRNA directed against MEF2C (siMEF2C) or a control siRNA (siControl). Introduction of siMEF2C resulted in a 35% reduction in MEF2C RNA as compared to control (Fig. 1G). After 13 days culture in megakaryocytic differentiation media, siMEF2C treated cells had modest but significant reductions in both the number of cells positive for CD41 and CD42, as well as reduced surface expression of CD41 and CD42 (Fig. 1H). These results in three different cell systems are supportive of a role for MEF2C in megakaryopoiesis.
Lack of MEF2C Alters Expression of Genes Involved in Megakaryocytic Differentiation.
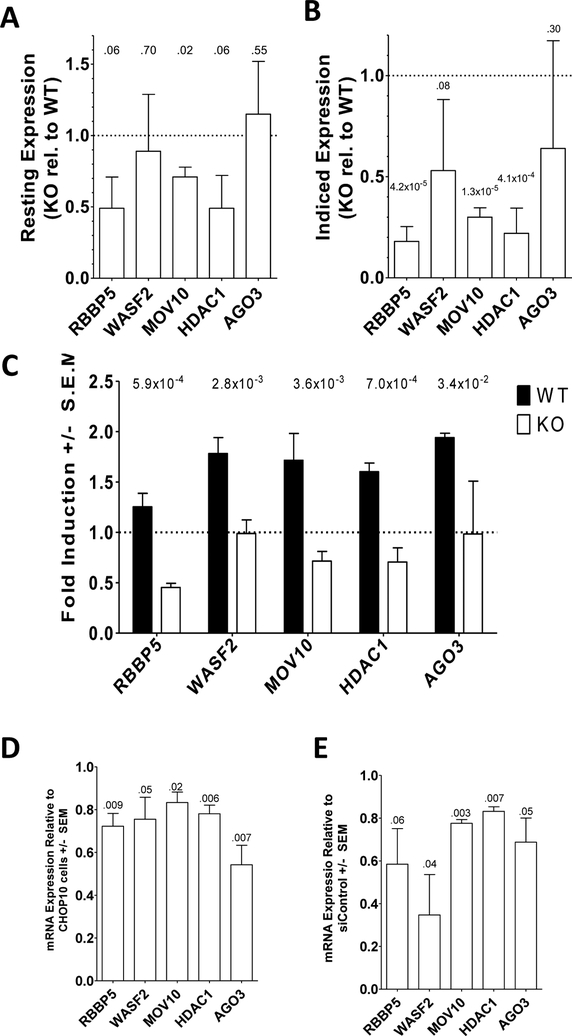
To identify potential targets of MEF2C that regulate megakaryopoiesis we utilized Chromatin Immunoprecipitation - Sequencing (ChIP-Seq) data that identified endogenous binding sites for MEF2C on chromatin-bound genomic DNA in GM12878 lymphoblastoid cell lines conducted as part of the ENCODE project.22 We looked for MEF2C ChIP-Seq peaks, indicating the binding of MEF2C, in genes that are members of Gene Ontology (GO) groups that are related to megakaryopoiesis and platelet production. These peaks were defined by an analysis performed by the ENCODE project using a method of peak calling known as the Irreproducible Discovery Rate (IDR).23 We combined the following GO groups: megakaryocyte development (GO:0035855), megakaryocyte differentiation (GO:0030219), positive regulation of megakaryocyte differentiation (GO:0045654), negative regulation of megakaryocyte differentiation (GO:0045653), regulation of megakaryocyte differentiation (GO:0045652), platelet formation (GO:0030220), platelet morphogenesis (GO:0036344), and platelet maturation (GO:0036345). This resulted in a list of 80 unique genes. Of these, five contained MEF2C ChIP-Seq peaks: Wiskott-Aldrich syndrome protein family member 2 (WASF2), Histone deacetylase 1 (HDAC1), Argonaute-3 (AGO3), Retinoblastoma-binding protein 5 (RBBP5), and Mov10 RISC Complex RNA Helicase (MOV10). To confirm these finding, we also studied the MEF2C ChIP-Seq data obtained in MOLM13 Acute Myeloid Leukemia cells and found binding peaks in this data set as well (Supplementary Fig. S1).24 These sources of ChIP-Seq data provide evidence that these genes are endogenous MEF2C targets.
We measured mRNA levels for RBBP5, WASF2, MOV10, HDAC1, and AGO3 in Meg01 and Meg01-MEF2CKO cells under DMSO- or PMA-treated conditions; in CHOP10 and CHOP10-MEF2CKO iPSCs undergoing megakaryocytic differentiation; and in CB CD34+ HSCs transfected with siRNA targeting MEF2C or control siRNA undergoing megakaryocytic differentiation. Compared to Meg01 cells, Meg01-MEF2CKO MOV10 levels were 30% lower (P=0.02) while the 50% reduction in RBBP5 and HDAC1 mRNA levels trended towards significance (P=0.06) (Fig. 2A). When compared to PMA-stimulated Meg01 cells, PMA-treated Meg01-MEF2CKO cells contained significantly less 82% less RBBP5 (P=4.2×10−5), 70% less MOV10 (P=1.3×10−5), and 78% less HDAC1(P=4.1×10−4) mRNA (Fig. 2B). When comparing PMA-treated to DMSO-treated conditions, all measured mRNAs increased 20–70% in Meg01 cells when treated with PMA. However, no increase in any of the mRNAs was observed in PMA-treated Meg01-MEF2CKO cells. In fact, PMA-treatment resulted in a decrease in mRNA content for RBBP5, MOV10, and HDAC1 (Fig. 2C).
Fig. 2. MEF2C deficient cells are defective in candidate target gene expression.
Expression of WASF2, MOV10, HDAC1, AGO3, and RBBP5 was measured by qRT-PCR in Meg01 and Meg01-MEF2CKO cells under (A) untreated and (B) PMA-exposed conditions. (C) Fold induction values for target genes in Meg01 and Meg01-MEF2CKO cells treated with PMA. (D) Target gene expression in CHOP10-MEF2CKO iPSC derived megakaryocytes relative to control CHOP10 iPSC derived megakaryocytes. (E) Target gene expression in CB CD34+ derived megakaryocytes transfected with siRNA directed against MEF2C relative to control cells. All qRT-PCR values were normalized to GAPDH expression. P-value above bars. NS=non-significant.
When we measured our putative MEF2C target genes in CHOP10 and CHOP10-MEF2CKO iPSCs induced to differentiate into megakaryocytes, we found a reduction of 22% - 46% of the mRNA levels (Fig. 2D). Likewise, when the target gene mRNA levels were measured in CB CD34+ derived megakaryocytes transfected with siRNA targeting MEF2C, reductions of 17%−65% were observed (Fig. 2E). Differences in the extent of reduction between the different cell systems may represent differences in transcription factor repertoire, chromatin state, or epigenetic marks between the different cell types. Moreover, these cells represent very different models of megakaryopoiesis (transformed cell line, reprogrammed iPSC, primary fetal HSC). Nevertheless, the data presented here supports a model in which MEF2C regulated expression of these genes.
Validation of MEF2C-Regulation of Target Gene ChIP-Seq Regions.
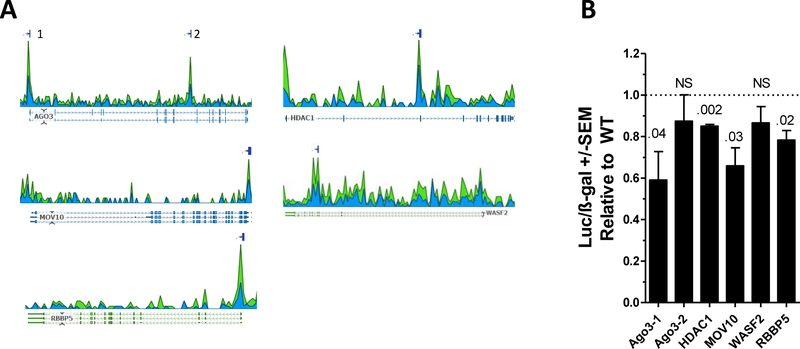
We next identified potential MEF2C binding sites within the genomic regions delineated by MEF2C ChIP-Seq peaks defined by the ENCODE project in GM12878 cells. All candidate genes contained one identified ChIP-Seq peak except for AGO3 which contained two (designated AGO3–1 and AGO3–2, Fig 3A). These sequences contained in these peaks were analyzed using the SCAN tool on the JASPAR 2018 (http://jaspar.genereg.net) website to identify the putative MEF2C binding sites.25 The highest scoring predicted MEF2C sites for each peak are shown in Table 1. These sites and the surrounding genomic sequence were cloned 5′ to a luciferase reporter cassette and transfected into Meg01 and Meg01-MEF2CKO cells. Transcriptional activity driven by the RBBP5, AGO3–1, HDAC1, and MOV10 ChIP-Seq peaks was significantly lower in the KO cells as compared to the wildtype, indicating a contribution of MEF2C to the regulation of these sequences (Fig. 3B).
Fig. 3. Identification of MEF2-C responsive genomic sequences in target genes.
(A) Read depth of MEF2C ChIP-Seq in GM12878 B-cells. Reads are aligned either to the forward (blue) or reverse (green) strands. Lines above the coverage plot indicate peaks identified using the optimal Irreproducible Discovery Rate (IDR).23 The AGO3 gene contains two peaks, labeled 1 and 2. (B) Reporter constructs containing the putative MEF2C binding sites were transfected into WT and MEF2C KO cells and assayed for luciferase activity. Results are normalized to ß-gal activity. P-value above bars. NS=non-significant.
Table 1.
Putative MEF2C binding sites found in ChIP-Seq peaks
| Peak | Sequence | Relative Score |
|---|---|---|
| AGO3–1 | GGACGGAAAACGGCA | 0.69 |
| AGO3–2 | GGTACAAGAATAGGT | 0.85 |
| HDAC1 | TGGTCAAAAATAACT | 0.92 |
| MOV10 | GGGCTAAACTTAGAA | 0.83 |
| WASF2 | TTTGCTCAAATAGCC | 0.81 |
| RBBP5 | GGACTTTAAATAGCG | 0.87 |
HDAC1 and RBBP5 expression are correlated with MEF2C expression during hematopoiesis.
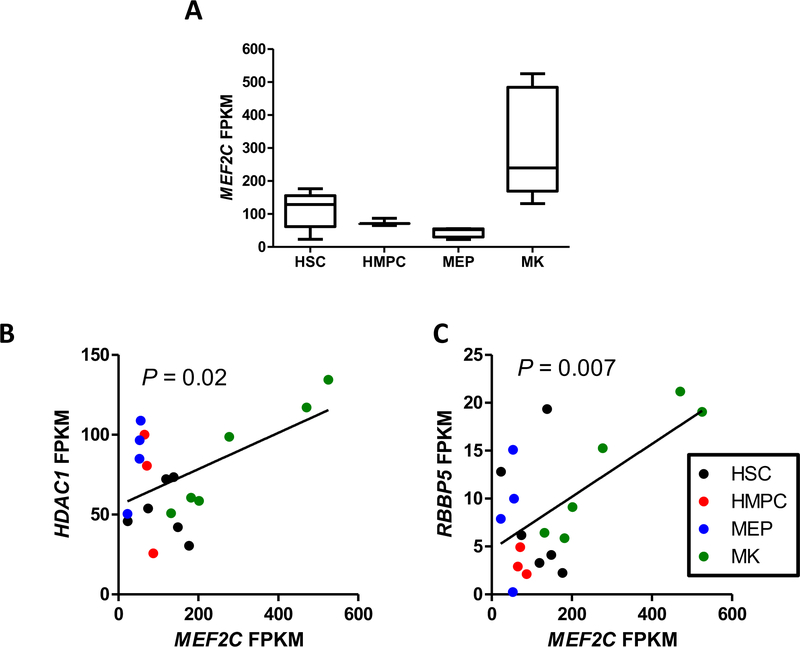
In addition to observing the changes in the target genes expression level in the modified Meg01 cells, we wanted to determine if the target gene levels were correlated with MEF2C levels during megakaryopoiesis. To do so, we accessed RNA-Seq data from the BLUEPRINT hematopoietic epigenome project.26 RNA-Seq FPKM (fragments per kilobase of exon model per million reads mapped) values for MEF2Cwere downloaded for 6 hematopoietic stem cell (HSC), 3 hematopoietic multipotent precursor cell (HMPC), 4 megakaryocyte-erythroid precursor cell (MEP), and 6 megakaryocyte (MK) samples (Fig. 4A). Utilizing this data, we uncovered significant correlations of MEF2C mRNA levels with HDAC1 and RBBP5mRNA (Fig. 4B). However, this correlation appears to be a result of the megakaryocytic expression levels and may not by related to differentiation status.
Fig 4. MEF2C expression levels correlate with HDAC1 and RBBP5 mRNA levels during hematopoiesis.
(A) MEF2C FPKM values from RNA-Seq data in hematopoietic stem cells (HSC), hematopoietic multipotent precursor cells (HMPC), megakaryocyte-erythrocyte precursors (MEP, and megakaryocytes (MK) from the BLUEPRINT project. Correlations between MEF2C levels and (B) HDAC1 and (C) RBBP5 levels.
GWAS associated MEF2CSNPs regulate MEF2C Expression.
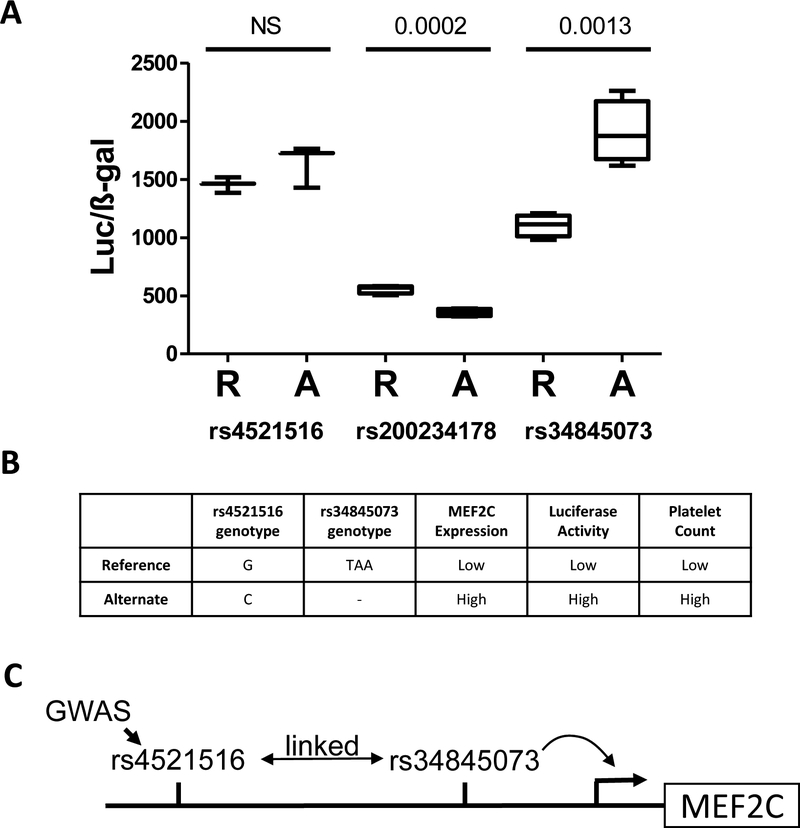
Variants in the MEF2Cgene have previously been identified as associated with platelet phenotypes by GWAS.1,2,4 Given that we have observed that alteration of MEF2C levels could alter megakaryopoiesis, we were interested if GWAS-implicated SNPs or variant linked to these SNPs, functioned by altering MEF2C levels. Intronic MEF2C SNP rs4521516 has been associated by GWAS with platelet count (P=6.75 × 10−40), platelet volume (P=3.31×10−38), platelet distribution width (P=1.26×10−27), and plateletcrit (P=1.17×10−16).1,2 Importantly, rs4521516 has also been associated with the expression level of MEF2C in a whole blood expression Quantitative Trait Loci (eQTL) study.27 We cloned the genomic sequences surrounding both the reference (R) and alternate (A) alleles of rs4521516 in front of a basic promoter and luciferase vector and found no significant difference in transcriptional activity between the variants (Fig. 5A). We then looked for variants linked to rs4521516 (R2 > 0.8) and found rs34845073 (R2=0.97), a 3bp deletion, which has promoter epigenetic marks in whole blood,28 and rs200234178, a 9bp deletion, which has previously been identified as an RNA polymerase II binding site by ChIP-Seq by ENCODE. We again performed luciferase assays to quantify the transcriptional activity of the reference and alternate alleles of these variants and found significant allelic differences for both variants (Fig 5A). Notably, the 3bp deletion of rs34845073 resulted in a 70% increase in reporter gene activity.
Fig 5. Deletion variants linked to GWAS-sentinel MEF2C SNP rs4521516 alter transcription.
(A) (A) Reporter vectors containing either the reference (R) or alternate alleles (A) of rs4521516, rs200234178, or rs34845073 were generated, transfected into K562 cells, and then assayed for luciferase activity. Values are normalized to ß-gal activity. P-value above bars. NS=non-significant. (B) Summary of associations between variants, MEF2C expression, luciferase activity, and platelet count. (C) Model of mechanism by which MEF2C genetic variants influence platelet count.
The ‘C’ allele of rs4521516 has been associated with both higher MEF2C expression and higher platelet count (Fig 5B). The ‘C’ allele of rs4521516 is commonly found in haplotypes with the 3bp deletion at rs34845073, which is our assay, leads to higher transcriptional activity (Fig. 5B). This allows us to propose a model in which the GWAS-identified SNP is linked to the 3bp deletion rs34845073. This 3bp deletion results in higher levels of MEF2C expression and thus higher platelet counts (Fig 5C).
Discussion
In this report, we have integrated genomic data and gene ontology content together with direct experimental evidence to identify target genes of the transcription factor MEF2C that may regulate megakaryopoiesis. These target genes: MOV10, AGO3, RBBP5, HDAC1, and WASF2 have all been identified as MEF2C target genes in ChIP-Seq experiments conducted by the ENCODE project. We demonstrated that abrogation of MEF2C expression in megakaryocytic cell lines, iPSC-derived megakaryocytes, and CB CD34+ HSC-derived megakaryocytes results in defects in megakaryocytic differentiation, altered target gene expression, and altered transcriptional regulation of the genomic region containing the MEF2C ChIP-Seq peak of these target genes. In addition, we have identified deletion variants linked to a GWAS sentinel SNP platelet count that alters MEF2C gene expression.
MEF2C was initially identified as a myocyte differentiation factor and has since become recognized for its role in healthy and pathological hematopoiesis.12–16,24,29,30 MEF2C was first identified as a potential megakaryopoiesis regulator by the observation that its level increased as peripheral blood CD34+ HSCs were induced to differentiate megakaryocytes.12 MEF2C was later shown to be a regulator of the cell fate decisions of lymphoid vs myeloid and monocyte vs granulocyte made during hematopoiesis.13,15 Gekas et al. reported that Mef2C null mice had reduced platelet counts and increased platelet volume and granularity while genomic studies have indicated MEF2C as being associated with platelet counts in human.1,4,14 MEF2C has also been implicated in pathologic hematopoiesis. MEF2C expression was induced in granulocyte-macrophage progenitor cells by the oncogenic fusion protein MLL-AF9 and in primary myelofibrosis cells.31,32 High levels of MEF2C have been associated with worse outcomes in pediatric AML.16
One strength of the approach we have taken here is the use of MEF2C ChIP-Seq data; under this approach the candidate genes we tested have strong evidence that they are in vivo genomic targets of MEF2C regulation. To narrow the list of 423 genes which contain a MEF2C ChIP-Seq signal peak to a reasonable number which could analyzed, we looked for overlap of these genes with those that have been categorized by gene ontology as regulating megakaryocyte or platelet development. This resulted in the list of five genes we tested in our experimental studies MOV10, AGO3, HDAC1, RBBP5, and WASF2.
MOV10 is a 5’ to 3’ ATP dependent RNA helicase that is a component of the Argonaute-containing RNA induced silencing complex (RISC).33 RISC incorporates single stranded small noncoding microRNAs which then allows it to recognize mRNA targets. Once bound to the mRNA, protein expression is attenuated by inhibition of translation or degradation of the RNA.34 The importance of the miRNA pathway to megakaryopoiesis has been well characterized.35 Of the four human argonaute proteins (Ago1–4), only Ago2 has mRNA degrading slicer activity.36 MEF2C target Ago3 plays a role in non-catalytic RISC complex, inhibiting translation while not affecting mRNA stability.37 The non-degredory mechanism of RISC has shown to be important to hematopoiesis. Over expression of miR-376a in K562 erythroleukemia cells prevented erythroid differentiation and resulted in the decrease of Ago2 and CDK2 protein while having minimal effect on RNA levels.38 Likewise, miR-27a inhibits RUNX protein translation while having no effect on mRNA levels in a 70/Z pre-B cell line.39
Chromatin modifying complexes are critical regulators of gene expression.40 HDAC1 is a histone deacetylase which negatively regulates transcription by deacetylating histones. Deletion of HDAC1 together with HDAC2 in mice resulted in loss hematopoietic stem cells and bone marrow failure.41 Overexpression of HDAC1 in murine bone marrow cells resulted in defective myeloid differentiation.42 HDAC1, in complex with Sin3A, interacts with numerous transcription factors important to megakaryopoiesis including TAL1 and RUNX1.43,44 RBBP5 is a member of the WRAD complex (WDR5, RbBP5, Ash2L, and DPY-30) which is required for dimethylation of histone H3–K4 as part of the mixed lineage leukemia (MLL) core complex.45 MLL physically interacts with RUNX1 where it regulates AML1 target gene PU.1.46 This suggests that MEF2C may regulate multiple epigenetic regulators that play critical roles in healthy and pathogenic hematopoiesis. Interestingly, regulation of RUNX1 expression and function are characteristics of MOV10, AGO3, HDAC1, and RBBP5.
The WASF2 gene encodes the protein Wiskott-Aldrich syndrome protein (WASp) family verprolin-homologous protein 2 (WAVE2). WAVE2 is an actin-polymerization regulator which is a key feature of filopodia and lamellipodia formation. WAVE2−/−embryonic stem cells were impaired in terminal megakaryocytic differentiation and platelet production. The megakaryocytes that did form in these cells were defective in spreading on fibrinogen.47 HSCs in which WAVE2 has been knocked down were defective in bone marrow reconstitution.48 Altogether, the five candidate MEF2C target genes studied here have highly plausible roles in megakaryopoiesis and require further study to determine the effect of MEF2C expression levels on their ability to regulate megakaryopoiesis.
There is a current push in genomics to move beyond genetic associations to identify functional variants that drive observed associations. Most GWAS-identified SNPs are not themselves functional, but instead are sentinel variants located in non-coding regions of the genome that mark the region in which causal variants reside; therefore, much of the focus has been on variants that alter regulation of the proximal gene. MEF2C has been identified as a gene associated with platelet count by multiple groups.1,4 We found that while one of the sentinel variants, rs4521516, had no effect of transcriptional regulation, two linked small deletions did, rs3485073 and rs200234178. Given the concordance of rs4521516 genotype, rs3485073 genotype, MEF2Cexpression, platelet count, and luciferase activity, we were able generate a model by which MEF2C genetic variation affects platelet production (Fig. 5B–C). More study is warranted, particularly because of the prevalence of rare variation in MEF2C and its impact on diverse disease phenotypes.49 Future studies may also consider the potential that SNP associations may be synthetic markers for additional functional rare variation in MEF2C.50
We have identified in this paper target genes of the transcription factor, MEF2C, that may regulate megakaryopoiesis. MEF2C has increasingly been appreciated as a regulator hematopoietic cells, however there is a lack of data as to its actual function. Work remains to be done to better characterize the function of these target genes and their contribution to megakaryopoiesis, including quantifying the effect on proplatelet formation and ploidy. This work establishes a clear direction to follow to better understand MEF2C in both normal and oncogenic hematopoiesis.
Supplementary Material
Summary Table.
| What is known on this topic | What this paper adds |
|---|---|
|
|
Acknowledgements
This work was funded by grant HL128234 from the National Institute of Health.
Footnotes
Conflict of Interest
None.
References
- 1.Gieger C, Radhakrishnan A, Cvejic A, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480(7376):201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astle WJ, Elding H, Jiang T, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167(5):1415–1429 e1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eicher JD, Chami N, Kacprowski T, et al. Platelet-Related Variants Identified by Exomechip Metaanalysis in 157,293 Individuals. Am J Hum Genet. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schick UM, Jain D, Hodonsky CJ, et al. Genome-wide Association Study of Platelet Count Identifies Ancestry-Specific Loci in Hispanic/Latino Americans. Am J Hum Genet. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vannucchi AM, Barbui T. Thrombocytosis and thrombosis. Hematology Am Soc Hematol Educ Program. 2007:363–370 [DOI] [PubMed] [Google Scholar]
- 6.Pujol-Moix N, Vazquez-Santiago M, Morera A, et al. Genetic determinants of platelet large-cell ratio, immature platelet fraction, and other platelet-related phenotypes. Thromb Res. 2015;136(2):361–366 [DOI] [PubMed] [Google Scholar]
- 7.Buckley MF, James JW, Brown DE, et al. A novel approach to the assessment of variations in the human platelet count. Thromb Haemost. 2000;83(3):480–484 [PubMed] [Google Scholar]
- 8.Daly ME. Transcription factor defects causing platelet disorders. Blood Rev. 2016 [DOI] [PubMed] [Google Scholar]
- 9.Tijssen MR, Ghevaert C. Transcription factors in late megakaryopoiesis and related platelet disorders. J Thromb Haemost. 2013;11(4):593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Songdej N, Rao AK. Hematopoietic Transcription Factors Mutations - Important Players in Inherited Platelet Defects. Blood. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cante-Barrett K, Pieters R, Meijerink JP. Myocyte enhancer factor 2C in hematopoiesis and leukemia. Oncogene. 2014;33(4):403–410 [DOI] [PubMed] [Google Scholar]
- 12.Fuhrken PG, Chen C, Apostolidis PA, Wang M, Miller WM, Papoutsakis ET. Gene Ontology-driven transcriptional analysis of CD34+ cell-initiated megakaryocytic cultures identifies new transcriptional regulators of megakaryopoiesis. Physiol Genomics. 2008;33(2):159–169 [DOI] [PubMed] [Google Scholar]
- 13.Schuler A, Schwieger M, Engelmann A, et al. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood. 2008;111(9):4532–4541 [DOI] [PubMed] [Google Scholar]
- 14.Gekas C, Rhodes KE, Gereige LM, et al. Mef2C is a lineage-restricted target of Scl/Tal1 and regulates megakaryopoiesis and B-cell homeostasis. Blood. 2009;113(15):3461–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stehling-Sun S, Dade J, Nutt SL, DeKoter RP, Camargo FD. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat Immunol. 2009;10(3):289–296 [DOI] [PubMed] [Google Scholar]
- 16.Laszlo GS, Alonzo TA, Gudgeon CJ, et al. High expression of myocyte enhancer factor 2C (MEF2C) is associated with adverse-risk features and poor outcome in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. J Hematol Oncol. 2015;8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguire JA, Gagne AL, Jobaliya CD, Gandre-Babbe S, Gadue P, French DL. Generation of human control iPS cell line CHOPWT10 from healthy adult peripheral blood mononuclear cells. Stem Cell Res. 2016;16(2):338–341 [DOI] [PubMed] [Google Scholar]
- 18.Mills JA, Paluru P, Weiss MJ, Gadue P, French DL. Hematopoietic differentiation of pluripotent stem cells in culture. Methods in molecular biology. 2014;1185:181–194 [DOI] [PubMed] [Google Scholar]
- 19.Edelstein LC, Simon LM, Montoya RT, et al. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat Med. 2013;19(12):1609–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang TW, le Sage C, Larrieu D, Demir M, Jackson SP. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Scientific reports. 2016;6:24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogura M, Morishima Y, Okumura M, et al. Functional and morphological differentiation induction of a human megakaryoblastic leukemia cell line (MEG-01s) by phorbol diesters. Blood. 1988;72(1):49–60 [PubMed] [Google Scholar]
- 22.Becker PB. A User’s Guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biology. 2011;9(4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. The Annals of Applied Statistics. 2011;5(3):1752–1779 [Google Scholar]
- 24.Tarumoto Y, Lu B, Somerville TDD, et al. LKB1, Salt-Inducible Kinases, and MEF2C Are Linked Dependencies in Acute Myeloid Leukemia. Mol Cell. 2018;69(6):1017–1027 e1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan A, Fornes O, Stigliani A, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Research. 2018;46(D1):D260–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez JM, de la Torre V, Richardson D, et al. The BLUEPRINT Data Analysis Portal. Cell Syst. 2016;3(5):491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G. Epigenomics: Roadmap for regulation. Nature. 2015;518(7539):314–316 [DOI] [PubMed] [Google Scholar]
- 29.Schwieger M, Schuler A, Forster M, et al. Homing and invasiveness of MLL/ENL leukemic cells is regulated by MEF2C. Blood. 2009;114(12):2476–2488 [DOI] [PubMed] [Google Scholar]
- 30.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annual review of cell and developmental biology. 1998;14:167–196 [DOI] [PubMed] [Google Scholar]
- 31.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–822 [DOI] [PubMed] [Google Scholar]
- 32.Norfo R, Zini R, Pennucci V, et al. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014;124(13):e21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meister G, Landthaler M, Peters L, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15(23):2149–2155 [DOI] [PubMed] [Google Scholar]
- 34.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379 [DOI] [PubMed] [Google Scholar]
- 35.Edelstein LC, McKenzie SE, Shaw C, Holinstat MA, Kunapuli SP, Bray PF. MicroRNAs in platelet production and activation. J Thromb Haemost. 2013;11 Suppl 1:340–350 [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441 [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol. 2008;18(17):1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, Yu J, Yang GH, Wang XS, Zhang JW. Regulation of erythroid differentiation by miR-376a and its targets. Cell Res. 2011;21(8):1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106(1):238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier K, Brehm A. Chromatin regulation: how complex does it get? Epigenetics. 2014;9(11):1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heideman MR, Lancini C, Proost N, Yanover E, Jacobs H, Dannenberg JH. Sin3a-associated Hdac1 and Hdac2 are essential for hematopoietic stem cell homeostasis and contribute differentially to hematopoiesis. Haematologica. 2014;99(8):1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada T, Kikuchi J, Nishimura N, Shimizu R, Kitamura T, Furukawa Y. Expression levels of histone deacetylases determine the cell fate of hematopoietic progenitors. J Biol Chem. 2009;284(44):30673–30683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X, Li X, Valverde K, et al. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc Natl Acad Sci U S A. 2009;106(25):10141–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo H, Friedman AD. Phosphorylation of RUNX1 by cyclin-dependent kinase reduces direct interaction with HDAC1 and HDAC3. J Biol Chem. 2011;286(1):208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shinsky SA, Hu M, Vought VE, et al. A non-active-site SET domain surface crucial for the interaction of MLL1 and the RbBP5/Ash2L heterodimer within MLL family core complexes. Journal of molecular biology. 2014;426(12):2283–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang G, Zhao X, Wang L, et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood. 2011;118(25):6544–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eto K, Nishikii H, Ogaeri T, et al. The WAVE2/Abi1 complex differentially regulates megakaryocyte development and spreading: implications for platelet biogenesis and spreading machinery. Blood. 2007;110(10):3637–3647 [DOI] [PubMed] [Google Scholar]
- 48.Ogaeri T, Eto K, Otsu M, Ema H, Nakauchi H. The actin polymerization regulator WAVE2 is required for early bone marrow repopulation by hematopoietic stem cells. Stem Cells. 2009;27(5):1120–1129 [DOI] [PubMed] [Google Scholar]
- 49.Lambert L, Bienvenu T, Allou L, et al. MEF2C mutations are a rare cause of Rett or severe Rett-like encephalopathies. Clin Genet. 2012;82(5):499–501 [DOI] [PubMed] [Google Scholar]
- 50.Orozco G, Barrett JC, Zeggini E. Synthetic associations in the context of genome-wide association scan signals. Hum Mol Genet. 2010;19(R2):R137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.