Abstract
Glioblastoma multiforme (GBMs) are recurrent lethal brain tumours. Recurrent GBMs often exhibit mesenchymal, stem-like phenotypes that could explain their resistance to therapy. Analyses revealed that recurrent GBMs have increased tension and express high levels of glycoproteins that increase the bulkiness of the glycocalyx. Studies showed that a bulky glycocalyx potentiates integrin mechanosignalling and tissue tension and promotes a mesenchymal, stem-like phenotype in GBMs. Gain- and loss-of-function studies implicated integrin mechanosignalling as an inducer of GBM growth, survival, invasion and treatment resistance, and a mesenchymal, stem-like phenotype. Mesenchymal-like GBMs were highly contractile and expressed elevated levels of glycoproteins that expanded their glycocalyx, and they were surrounded by a stiff extracellular matrix that potentiated integrin mechanosignalling. Our findings suggest that there is a dynamic and reciprocal link between integrin mechanosignalling and a bulky glycocalyx, implying a causal link towards a mesenchymal, stem-like phenotype in GBMs. Strategies to ameliorate GBM tissue tension offer a therapeutic approach to reduce mortality due to GBM.
GBMs (Grade IV gliomas) are resistant to therapy and have a high rate of recurrence1. Recurrent human GBMs are invariably lethal and often exhibit a mesenchymal-like phenotype2–5. Nevertheless, it remains unclear whether and how factors that foster a mesenchymal-like phenotype also promote GBM recurrence.
GBMs with a mesenchymal-like phenotype show altered extracellular matrix (ECM) proteins, including tenascin, which stiffens the ECM and is increased in recurrent GBMs with mutant IDH-16. GBMs develop within a stiffened ECM that potentiates their growth, survival and invasion6,7. ECM stiffness also compromises vascular integrity to impede macromolecular diffusion and induce hypoxia8. Hypoxia-inducible factor 1α (HIF1α) increases the expression of genes that promote an epithelial-to-mesenchymal (EMT) transition with a stem-like phenotype9,10. Transforming growth factor-β (TGF-β)11 and the Hippo-Yes-associated protein (YAP)/ transcription activator with PDZ binding motif (TAZ) are two additional tension-activated factors that induce a mesenchymal transition and a stem-like phenotype in tumour cells12,13. Thus, brain tumour stiffness could drive treatment resistance and GBM recurrence by stimulating the expansion of pre-existing transformed glioma stem cells (GSCs) or by inducing a de novo EMT transition that fosters the stem-like behaviour of gliomas.
GBMs often express high levels of glycoconjugates such as glycosphingolipids, glycoproteins and proteoglycans that comprise the glycocalyx14–17. A bulky glycocalyx can enhance integrin-dependent cell growth and survival, and compromise immune recognition14,18,19. Primary tumours that express glycoconjugates that increase the bulkiness of the glycocalyx are more metastatic, and circulating tumour cells (CTCs) express bulky glycoproteins20–22. Consistently, therapies that de-bulk the glycocalyx have shown promising therapeutic efficacy20,22–24. Importantly, tumours with a mesenchymal-like phenotype often express high levels of bulky glycoconjugates, including the glycoprotein MUC1 and the polysaccharide hyaluronan25,26. Moreover, stem cells exhibit a unique profile of glycoproteins, including several that increase glycocalyx bulkiness27–29. Thus, there may be a link between a mesenchymal and stem-like phenotype and a bulky glycocalyx. Indeed, recurrent tumours often express high levels of bulky glycoproteins20,22. Here, we determined whether the stiff ECM of gliomas promotes a mesenchymal, stem-like phenotype in GBMs, and whether this was linked to a bulky glycocalyx.
Results
Recurrent GBMs have high myosin activity and exhibit a mesenchymal-like phenotype.
Recurrent human GBMs harbouring IDH-1 mutations are highly contractile (high phosphorylated myosin-like chain kinase (pMLCK)), exhibit elevated integrin mechanosignalling and are surrounded by a stiff, tenascin-rich ECM6. We found that recurrent IDH-1 wild-type GBMs also contain more tenascin (Fig. 1a,b) and that recurrent tumour cells have increased myosin activity (phosphor myosin light chain 2 (pMLC2); Supplementary Fig. 1) that correlates positively with stromal tenascin (Fig. 1c). A bioinformatics analysis of publicly available human data revealed that patients with glioma and high tenascin levels also had poor survival (Fig. 1d), and patients whose tumours are enriched for EMT genes showed a significant reduction in survival5,30 (Fig. 1e). Moreover, many recurrent GBMs exhibit a mesenchymal-like phenotype2–4, and human GBMs with a mesenchymal phenotype express high levels of tenascin (Fig. 1f).
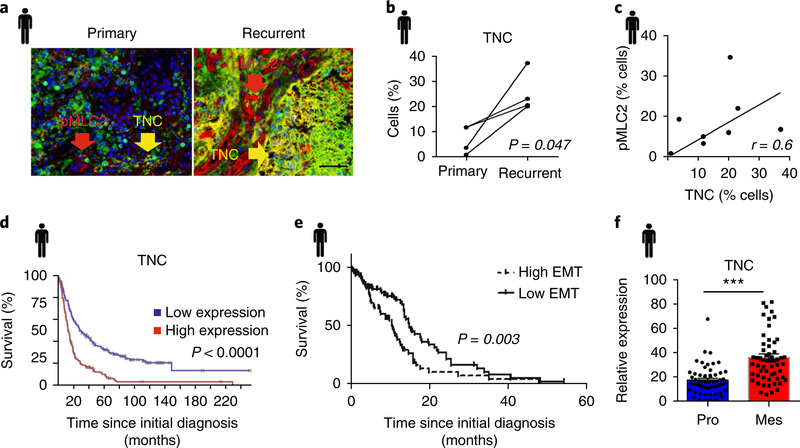
Fig. 1 |. Recurrent wild-type GBM is associated with tenascin C.
a, Representative image of the multiplex immunohistochemistry analysis of primary and recurrent GBM patient tissue samples (four patients) stained for tenascin C (TNC) in yellow and pMLC2 in red (Nam cohort). Scale bar, 50 μm. b, Quantification of cells expressing TNC in matched primary and recurrent GBM patient samples from a (n = 4 patients; P = 0.047 using a two-sided paired t-test). c, Correlation plot of percent cells expressing TNC versus pMLC2 from eight human GBM samples using Spearman correlation. d, Kaplan–Meier survival analysis of Rembrandt data for TNC based on its expression level across all glioma samples (n = 214 GBM, 66 oligodendroglioma and 145 astrocytoma patients; P value calculated using two-sided logrank test). e, Kaplan–Meier analysis of TCGA GBM data (n = 401 patients) showing the survival probability of patients with high versus low EMT gene expression (see gene list in Supplementary Table 1) in glioma. P value calculated using logrank test. f, TCGA data analysed for the expression level of TNC in human proneural (Pro; n = 57 patients) and mesenchymal (Mes; n = 58 patients) GBM. Mean ± s.e.m.; ***P = 4.27 × 10−8 using two-sided Mann–Whitney U-test.
To investigate the relationship between tenascin, GBM tension and a mesenchymal-like GBM phenotype, we analysed GBMs formed 2–10 weeks following the injection of two mesenchymal and two proneural31,32 patient-derived xenograft GBMs (Fig. 2a). Analyses revealed that the stroma of the mesenchymal GBMs contained more fibronectin and tenascin, and the GBM cells had higher integrin-focal adhesion mechanosignalling (pY397-focal adhesion kinase (FAK)) and actomyosin activity (pMLC2; Fig. 2b,g; Supplementary Fig. 2a). PCR with reverse transcription (RT-PCR) analysis confirmed that the mice harbouring the mesenchymal tumours also expressed higher levels of the mesenchymal genes MET, Twist1, MMP9 and WNT5a (Fig. 2c–f; Supplementary Fig. 2b). Furthermore, atomic force microscopy (AFM) revealed that the ECM surrounding the mesenchymal GBMs was stiffer (Fig. 2h). The mice harbouring the mesenchymal GBMs also had a significantly reduced overall survival (Fig. 2i; Supplementary Fig. 2c) and lower microvascular density33 (Fig. 2j). These findings suggest that there is a causal link between mesenchymal-like GBMs, tenascin and tissue tension.
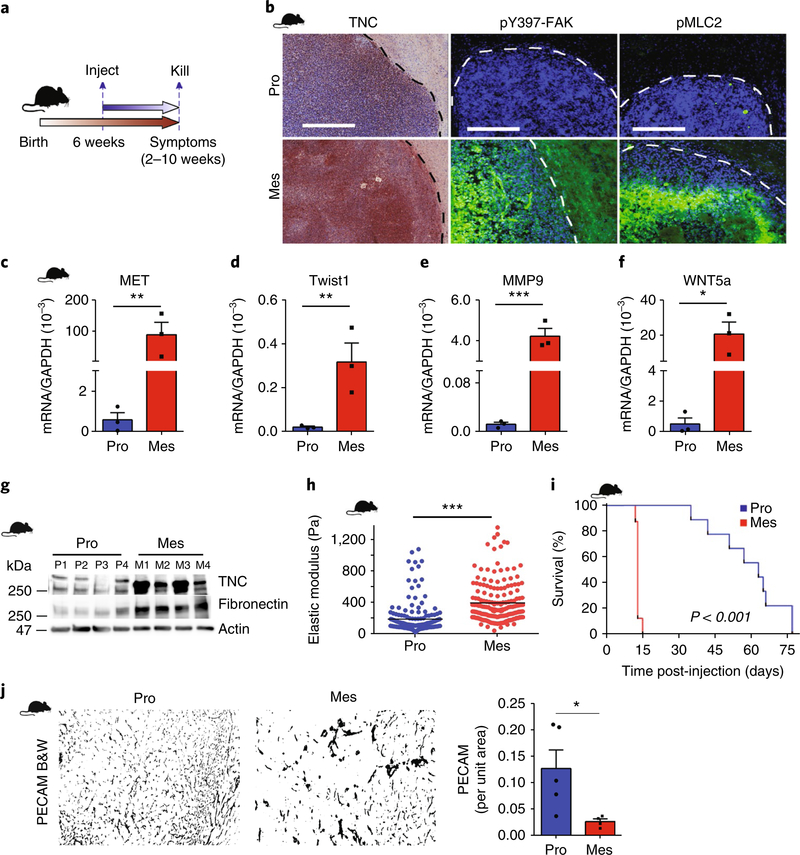
Fig. 2 |. GBM aggression is associated with a mesenchymal-like phenotype and elevated tissue tension.
a,b, Schematic of the xenograft model experimental setup (a) and immunohistochemistry (TNC) and immunofluorescence (b) for the analysis of pY397-FAK and pMLC2 in xenografts of human proneural (Pro) and mesenchymal (Mes) GBMs injected intracranially into NCR nude mice. Broken lines delineate tumour—haematoxylin or DAPI (4,6-diamidino-2-phenylindole)-dense areas—from normal brain. Image representative of three mice per group. Scale bars, 250 μm. c–f, RNA from these xenografts was analysed by qPCR for the indicated transcripts. Mean ± s.e.m.; n = 3 mice per group. **P = 0.008 (MET), **P = 0.001 (Twist), ***P = 4.64 × 10–5 (MMP9) and *P = 0.01 (WNT5a) by two-sided unpaired t-test. g, Immunoblot of TNC and fibronectin in lysates of proneural (n = 4 samples, P1–4) and mesenchymal (n = 4 samples, M1–4) xenografts. h, ECM stiffness (measured as elastic modulus) measured by AFM in proneural (n = 4 tissue samples; three 90 × 90 μm regions per sample, 191 indentations) and mesenchymal (n = 4 tissue samples, three 90 × 90 μm regions per sample, 192 indentations) xenograft tissue. Each point represents force per indentation. Mean ± s.e.m.; ***P = 1.65 × 10–15 by two-sided Mann–Whitney U-test. i, Kaplan–Meier analysis of nude mice injected with human proneural or mesenchymal GBM cells (n = 9 mice per group). P value calculated using a two-sided logrank test. j, Representative images and quantification of PECAM in proneural and mesenchymal xenograft tissue (n = 5 and 4 mice per group for proneural and mesenchymal, respectively). B&W denotes black and white images post-masking of PECAM-stained blood vessels. Mean ± s.e.m.; *P = 0.03 by two-sided Mann–Whitney U-test.
Tissue tension, integrin mechanosignalling and a GBM mesenchymal-like phenotype.
To determine whether and how elevated tissue tension could induce a mesenchymal-like phenotype, we plated proneural GBM cells on soft (75–400 Pa; gliotic brain) and stiff (60,000 Pa; glioma tissue) fibronectin-coated polyacrylamide hydrogels (PA gels)6,34. In response to the stiffer PA gel, the proneural GBMs spread more (Fig. 3a) and activated more pY397-FAK (Fig. 3b), reflecting enhanced integrin focal adhesion assembly35. Stiffness-induced integrin–focal adhesion signalling also promoted proneural GBM cell proliferation, as revealed by more cyclin D1 protein (Fig. 3b). Proneural GBMs on the stiff PA gels also showed longer migration trajectories and higher migration velocities (Fig. 3c; Supplementary Fig. 3a). Proneural GBM cells expressing the human V737N mutant integrin β136 (V737N proneural GBM), which increases integrin focal adhesion assembly and enhances integrin signalling, showed enhanced transwell invasion and mesenchymal gene expression (Fig. 3d and Supplementary Fig. 3b, respectively) compared to proneural GBMs expressing a human wild-type integrin β1. Quantitative RT-PCR array analysis revealed that proneural GBM cells plated on the stiff PA gels (48 h) exhibited upregulated cell proliferation, motility and mesenchymal transition genes (Fig. 3e). Reducing FAK-dependent integrin mechanosignalling in the mesenchymal GBM cells decreased contractility, as revealed by traction force microscopy and confirmed by FAK and phosphorylated myosin light chain phosphatase (pMYPT) immunoblotting (Fig. 3f,g; Supplementary Fig. 3c). Inhibiting integrin mechanosignalling in the mesenchymal GBMs also reduced tenascin and vimentin expression; two genes that are enriched in mesenchymal GBMs (Fig. 3h; Supplementary Fig. 3d–f). These findings suggest that there is a causal link between tissue tension, integrin mechanosignalling and a mesenchymal phenotype in GBMs.
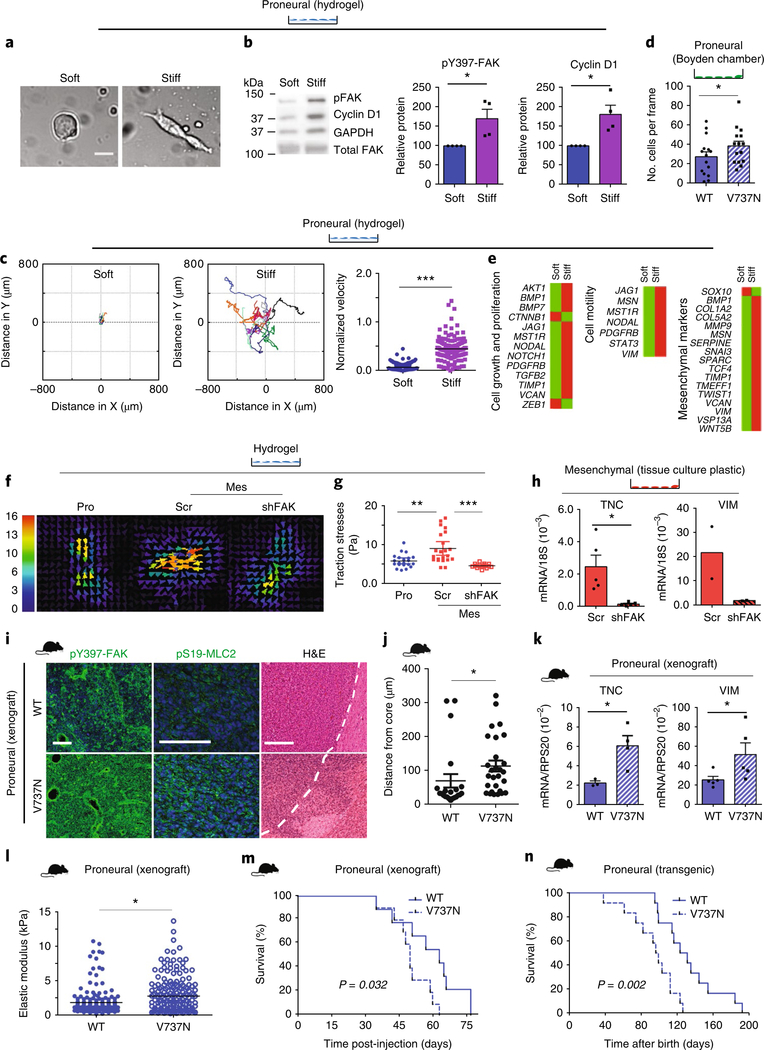
Fig. 3 |. Mechanosignalling drives a mesenchymal phenotype.
a, Proneural cells on soft and stiff hydrogels after 48 h (n = 3 independent experiments). Scale bar, 20 μm. b, pY397-FAK and cyclin D1 expression in proneural cells. Mean ± s.e.m.; n = 4 independent experiments, normalized to GAPDH. *P = 0.032 and 0.04 for pY397-FAK and cyclin D1, respectively, by one- and two-sided paired t-test. c, Migration tracking (left) of proneural cells on hydrogels; n = 10 (soft) and 11 (stiff) cells pooled from 3 independent experiments. Velocity plot (right) represents distance per time per frame; n = 144 frames per condition. Mean ± s.e.m.; ***P = 2.72 × 10–41 by two-sided paired t-test. d, Proneural cells expressing wild-type integrin β1 (WT) or V737N integrin β1 mutant (V737N) assayed for invasion. Each point represents invasive cells per frame; n = 15 frames. Mean ± s.e.m.; *P = 0.011 by two-sided paired t-test. e, qPCR array and pathway analysis of proneural cells on hydrogels; n = 2 independent experiments. f,g, A representative traction map; the colour bar represents increasing magnitude of stress with 0 (blue) denoting low stress and 16 (red) denoting high stress regions of the cell (f) and traction stresses (g) of proneural (Pro), control (Scr) or FAK knockdown (shFAK) mesenchymal cells (Mes) on 2.7 kPa hydrogels analysed by traction force microscopy. Mean ± s.e.m. of traction forces; n = 15 (shFAK) and 21 (all others) measurements, pooled from 3 independent experiments. **P = 0.0015 and ***P = 2.73 × 10–5 by two-sided Mann–Whitney U-test. h, qPCR results for mesenchymal Scr and shFAK cells; n = 5 and 2 independent experiments for TNC and vimentin (VIM), respectively. Mean ± s.e.m.; *P = 0.036 by two-sided paired t-test for TNC. i, Stained xenografted tissue from proneural tumours expressing WT or V737N. Scale bars, 100 μm pY397-FAK and pS19-MLC2; 250 μm H&E (n = 4 mice per group). j, Local invasion was quantified. Mean ± s.e.m.; n= 22 and 28 distance measurements for WT and V737N tumours, respectively, for 4 tissues per group. *P = 0.0012 by two-sided unpaired t-test. k, RNA from these tumours was analysed. Mean ± s.e.m.; n = 4 and 5 mice per group for TNC and vimentin, respectively. *P = 0.002 (TNC) and 0.032 (vimentin) by two-sided unpaired t-test. l, ECM stiffness for n = 3 tissues per group, 90 × 90 μm. Scatter represents 192 indentations per sample. Mean ± s.e.m.; *P = 0.022 by two-sided Mann–Whitney U-test. m,n, Survival of xenografted (m) and transgenic (n) mice overexpressing WT or V737N; n = 9 WT and 10 V737N (xenografts); n = 12 mice per group (transgenic). P value calculated using two-sided logrank test. Source data: Supplementary Table 4; unprocessed blot: Supplementary Fig. 9.
The xenograft tumours generated by the human V737N proneural GBM and by control integrin β1 cells showed more pY397-FAK and pS19-MLC2 immunofluorescence staining, reflecting increased focal adhesion mechanosignalling and tumour cell tension, respectively (Fig. 3i). Elevating integrin mechanosignalling also increased the invasiveness of the proneural GBM xenograft, as revealed by haematoxylin and eosin (H&E) stained tumour cells migrating beyond the tumour core (Fig. 3i,j; Supplementary Fig. 7). The V737N proneural GBMs also expressed more tenascin and vimentin (Fig. 3k) as well as MET, Twist1 and WNT5a (Supplementary Fig. 3g), and developed a stiffer ECM (Fig. 3l). The mice injected with the V737N proneural GBMs were also more hypoxic and had increased HIF1α levels (Supplementary Fig. 2d), accompanied by reduced mouse survival (Fig. 3m). An immunocompetent proneural mouse model of glioma (S100β-vErb:p53+/–)37 engineered to conditionally express the V737N mutant integrin β1 under the control of the GFAP-Cre transgene38 similarly demonstrated reduced survival (Fig. 3n) and increased tumour burden (Supplementary Fig. 3h). Moreover, injecting immunocompetent syngeneic hosts with primary tumour cells derived from the S100β-vErb:p53+/– mouse model, transduced with either V737N or wild-type integrin β1, corroborated the invasive phenotype of both the V737N proneural GBM xenograft and the V737N transgenic murine tumours, as revealed by the human integrin β1 expressed by the invading glioma cells (Supplementary Fig. 3i). These V737N-expressing S100β-vErb:p53+/– integrin β1 syngeneic tumours also expressed significantly higher levels of fibronectin, TGFβ1 and vimentin, consistent with induction of a mesenchymal-like phenotype (Supplementary Fig. 3j). These data provide additional evidence of a causal link between tissue tension, integrin mechanosignalling and a mesenchymal phenotype in GBMs. The findings imply that a stiff tenascin-rich stroma enhances integrin mechanosignalling6 to induce a mesenchymal phenotype in proneural GBMs. Thereafter, GBMs with a mesenchymal-like phenotype are more contractile and secrete more tenascin that further stiffens their stroma to foster their growth, survival and invasion by supporting an integrin-linked tension reinforcement loop.
Mesenchymal-like GBMs exhibit a bulky glycocalyx.
Mesenchymal and stem-like cells frequently express high levels of glycoproteins that enhance the bulkiness of the cellular glycocalyx28,39. Metastatic mesenchymal tumours show a similar increase in bulky glycoproteins14,16,19. An analysis of RNA-sequencing (RNA-seq) data from patients with primary GBM and patients with recurrent GBM (see “Nam cohort” in Methods) showed that transcripts of bulky cell-surface proteins were increased when non-mesenchymal GBMs relapsed with mesenchymal features (Fig. 4a). An analysis of a cohort of 401 patients with GBM (from The Cancer Genome Atlas (TCGA)) revealed that tumours with a high-bulk glycoprotein signature had a significantly shorter overall survival compared to those with a low-bulk signature (Fig. 4b). The data also revealed an increase in the overall bulkiness signature of the glycoproteins in human mesenchymal GBM tumours compared to proneural GBMs (Fig. 4c). A bioinformatics analysis of publicly available human data showed a marked enrichment of many glycoprotein transcripts, such as CD44 and MUC1, in patients with GBM tumours with a mesenchymal subtype (Fig. 4d; Supplementary Fig. 8; Supplementary Table 1). Immunohistochemistry and RT-PCR confirmed that the xenografted mesenchymal GBMs expressed more CD44 and MUC1, and staining with the pan-polysaccharide stain alcian blue suggested that the mesenchymal GBMs had higher overall levels of glycoproteins (Fig. 4e,f). Scanning angle interference microscopy (SAIM), which assesses the thickness of the glycocalyx, revealed that GBMs with a mesenchymal-like phenotype had a thicker glycocalyx, which could be reduced when FAK was knocked down (Fig. 4g). These data reveal a correlation between a bulky glycocalyx, poor patient outcome and GBM tumours with a mesenchymal phenotype, and imply that glycocalyx bulkiness is modulated by integrin mechanosignalling.
Fig. 4 |. A bulky glycocalyx is associated with mesenchymal GBM.
a, Violin plots showing increased expression of genes encoding bulky transmembrane proteins in mesenchymal recurrent GBMs compared to non-mesenchymal primary GBMs (RNA-seq data; Nam cohort). White dots and thick black lines indicate median and interquartile ranges, respectively, of the one-tailed P value distribution of transcripts (n = 39 patients). *P = 0.025 by one-sided Kolmogorov–Smirnov test. Genes encoding proteins with 15 putative N-glycosylation sites and 4 O-glycosylation sites are defined as bulky (N + O). “All” denotes all human transcripts, “Ext” denotes transcripts whose products have extracellular domains, “N” and “O” denotes transcripts with 15 putative N-glycosylation sites and 4 O-glycosylation sites, respectively, and “N+O” denotes a combination of these. b, Kaplan–Meier analysis of TCGA data showing the survival probability of patients (n = 401 patients) with high versus low bulk glycoprotein signatures (Supplementary Table 1) for 22 genes. P value calculated using two-sided logrank test. c, Violin plots showing increased expression of genes encoding bulky transmembrane proteins in human mesenchymal versus proneural GBM (TCGA). White dots and thick black lines indicate the median and interquartile ranges, respectively, of the one-tailed P value distribution of transcripts encoding all transmembrane proteins (Mem) and bulky transmembrane proteins (Bulky). Human proneural n = 57 patients and mesenchymal GBMs n = 58 patients. P = 0.1703 by Mann–Whitney U-test. d, TCGA data analysed for expression levels of indicated transcripts in human proneural (Pro; n = 57 patients) and mesenchymal GBMs (Mes; n = 58 patients). Mean ± s.e.m.; ***P = 4.79 × 10–9 (MUC1) and 6.87 × 10–15 (CD44) by two-sided Mann–Whitney U-test. e, Mouse proneural and mesenchymal xenografts stained for CD44 and total polysaccharides with alcian blue. Scale bar, 250 μm (n = 3 tissue sections per condition). f, RNA from proneural and mesenchymal xenografts was analysed. Mean ± s.e.m.; n = 5 mice per group. **P = 0.006 (MUC1) and *P = 0.014 (CD44) by two-sided unpaired t-test. g, Glycocalyx height was measured in proneural, mesenchymal control (Scr) and mesenchymal shFAK GBM cells by SAIM. Scale bar, 10 μm. Mean ± s.e.m.; each point represents measurement of glycocalyx height per frame (n = 62 measurements for proneural and Scr and 66 for shFAK over 3 independent experiments). *P = 0.04 and 0.03 for proneural versus scr and Scr versus shFAK, respectively, by two-sided paired t-test.
Reciprocal links between integrin mechanosignalling, a bulky glycocalyx and mesenchymal GBMs.
We next investigated the relationship between integrin mechanosignalling, glycocalyx bulkiness and a mesenchymal phenotype in GBMs. Reducing integrin mechanosignalling in mesenchymal GBM cells through short hairpin RNA (shRNA)-mediated knockdown or inhibition of FAK not only decreased cell spreading (data not shown) but also reduced levels of the bulky glycoproteins MUC1, CD44 and/ or Has2 (Fig. 5a; Supplementary Figs. 4a and 3e) and lowered the amount of secreted carbohydrate (Fig. 5b). The proneural xenograft and syngeneic tumours expressing the V737N integrin β1 (Fig. 3i–m; Supplementary Fig. 3g,i,j) also exhibited upregulated glycoprotein expression (Fig. 5c; Supplementary Fig. 4b). The xenografted, syngeneic and transgenic proneural tumours that expressed the V737N mutant additionally stained more robustly with alcian blue, indicating an overall increase in total polysaccharide content (Fig. 5d; Supplementary Fig. 4c). These data show that integrin mechanosignalling not only promotes and sustains a mesenchymal-like phenotype in GBMs but also regulates the expression of glycoproteins that modulate the bulkiness of the glycocalyx.
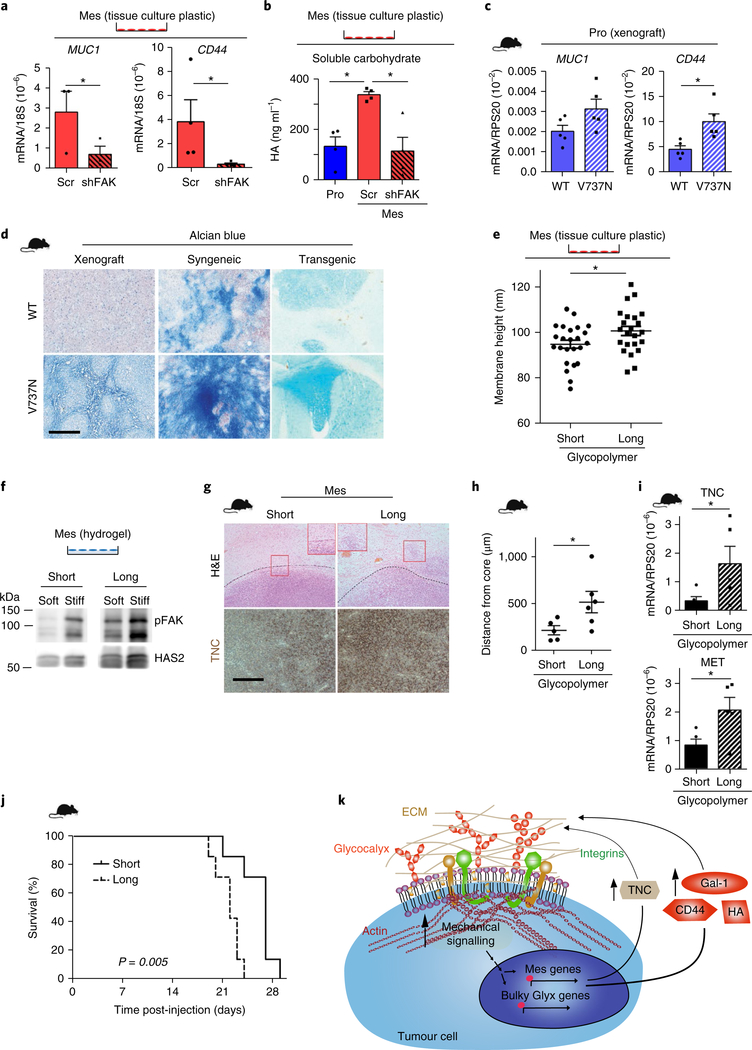
Fig. 5 |. Glycocalyx bulkiness is fostered by enhanced mechanics and is associated with GBM aggression.
a, RNA from control (Scr) and mesenchymal shFAK cells was analysed by qPCR. Mean ± s.e.m. (n = 3 and 4 independent experiments for MUC1 and CD44, respectively). *P = 0.03 and 0.027 for MUC1 and CD44, respectively, by one-sided paired t-test. b, Hyaluronan (HA) was measured from human proneural (Pro), Scr and shFAK cells. Mean ± s.e.m. (n = 4 independent experiments). *P = 0.01 and 0.021 for proneural verus Scr and Scr versus shFAK, respectively, by two-sided paired t-test. c, qPCR of V737N and WT proneural xenografts. Mean ± s.e.m. (n = 5 mice per group). *P = 0.08 and 0.012 for MUC1 and CD44, respectively, by two-sided unpaired t-test. d, Alcian blue staining of WT and V737N xenograft, syngeneic and transgenic proneural tumour models. Scale bar, 250 μm (n = 3 mice per group). e, Mesenchymal cells were loaded with short (3 nm) or long (90 nm) glycopolymers, and glycocalyx height was measured by SAIM. Mean ± s.e.m.; each point represents measurement of glycocalyx height per frame (n = 24 and 23 measurements for short and long glycopolymers, respectively, pooled from two independent experiments). *P = 0.035 by two-sided paired t-test. f, Immunoblot assays were performed for pY397-FAK and HAS2 on soft and stiff hydrogels for long versus short glycopolymer decorated mesenchymal cells (n = 3 independent experiments). g, Cells loaded with long or short glycopolymers were injected into nude mice, and xenografts stained for TNC and by H&E. Insets show differential invasion and black broken lines indicate tumour boundary (n = 7 mice per group). Scale bar, 250 μm. h, Quantification of invasive edge was performed as for Fig. 3j. Mean ± s.e.m. (n = 5 and 6 distance measurements for WT and V737N tumours, respectively, for 4 tissue samples per group). *P = 0.047 by two-sided unpaired t-test. i, qPCR was performed for these tumours. Mean ± s.e.m. (n = 5 tissues per group). *P = 0.048 and 0.028 for TNC and MET, respectively, by one-sided unpaired t-test. j, Survival analysed by Kaplan–Meier test (n = 7 mice per group). P value calculated using two-sided logrank test. k, Schematic showing that enhanced mechanical signalling in tumour cells promotes the upregulation of mesenchymal and bulky glycocalyx (Glyx) genes, which results in the production of ECM proteins such as TNC and bulky glycocalyx components and modulators (CD44, hyaluronan (HA) and Gal-1. Together, these drive glioma aggressiveness in a tension-dependent feedback loop. Unprocessed blot: Supplementary Fig. 9.
We have previously shown that a bulky glycocalyx enhances integrin activation and integrin focal adhesion assembly14. So, we explored whether a bulky glycocalyx reciprocally increases integrin mechanosignalling to promote a mesenchymal-like phenotype in GBMs. We employed a set of short (3 nm) and long (90 nm) synthetic mucin mimetic glycopolymers to increase the thickness of the glycocalyx in the GBM cells. These mucin mimetic glycopolymers were synthesized to rapidly intercalate into the plasma membrane via a cholesterylamine tail and project perpendicularly to the cell surface where they are retained over multiple cell generations40. SAIM imaging revealed that the GBM cells decorated with the longer glycopolymers had a significantly thicker glycocalyx compared to those decorated with the shorter glycopolymers (Fig. 5e; Supplementary Fig. 4d). The immunoblot analysis confirmed that the cells with the longer glycopolymer had more integrin mechanosignalling, as indicated by increased pY397-FAK compared to cells decorated with the shorter glycopolymers (Fig. 5f ). Immunostaining of the intracranially injected GBM cells decorated with the longer glycopolymers revealed that the xenografted tumours expressed more tenascin, and staining also indicated that the tumours invaded more (Fig. 5g,h; Supplementary Fig. 4e,f ). The RT-PCR analysis of the tumours generated by the cells decorated with the longer glycopolymers showed increased levels of tenascin and MET (Fig. 5i). The mice injected with the GBM cells decorated with the longer glycopolymers also had significantly reduced survival (Fig. 5j). Thus, there appears to be a dynamic and reciprocal link between integrin mechanosignalling and a bulky glycocalyx that associates with and could promote a mesenchymal-like phenotype in GBMs (Fig. 5k).
Reciprocal interplay between the glycocalyx, integrin-mechanosignalling and the mesenchymal phenotype in GBMs.
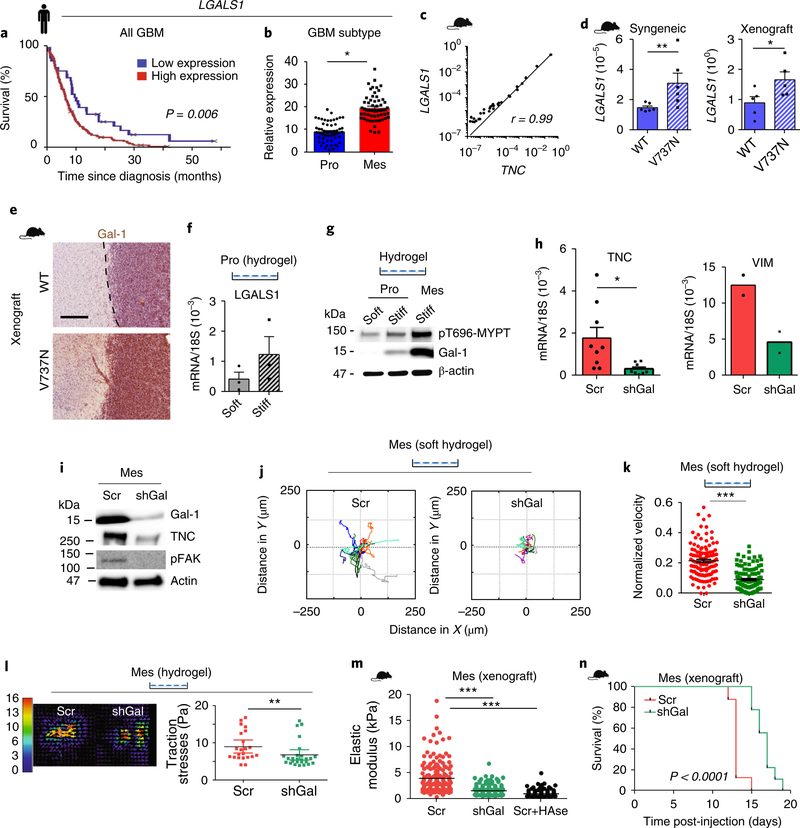
Glycoprotein structure and function are tightly regulated by lectins, which are sugar-binding proteins41. Lectins such as galectin-1 (Gal-1) bind glycoproteins implicated in cell–cell and cell–ECM interactions and modify the organization of transmembrane receptors that regulate cell signalling and morphogenesis41–43. Gal-1 in particular crosslinks surface glycoproteins to modulate the activity of cell signalling receptors implicated in cell growth, survival and motility44. Lectins, including Gal-1, are frequently upregulated in malignancies, including glioma45,46. A bioinformatics analysis of patient data revealed that patients with GBM and patients with glioma expressing higher levels of Gal-1 have a significantly poorer prognosis than those with low Gal-1 levels (Fig. 6a; Supplementary Fig. 5a). Further analysis revealed that Gal-1 is disproportionately elevated in the tumours of patients with mesenchymal GBMs (Fig. 6b). In our mesenchymal xenograft models, Gal-1 levels correlated positively and significantly with tenascin (Fig. 6c). Galectins can modify the activity of cell–ECM adhesion receptors42–44, and we detected a significant increase in the messenger RNA and protein levels of Gal-1 in both the syngeneic and xenograft V737N proneural GBM tumours (Fig. 6d,e; Supplementary Fig. 5b). Proneural GBM tumour cells plated on stiff PA hydrogels showed an increase in Gal-1 mRNA and protein levels (Fig. 6f,g), which was accompanied by elevated ROCK activity, as revealed by higher phospho-MYPT (pT696-MYPT; Fig. 6g). shRNA-mediated knockdown of Gal-1 in mesenchymal GBM cells reduced the height of the glycocalyx (Supplementary Fig. 5c,d), while simultaneously decreasing integrin-dependent mechanosignalling, as indicated by lower pY397-FAK (Fig. 6i; Supplementary Fig. 5e). Decreasing the glycocalyx bulkiness through Gal-1 knockdown also reduced the expression of several mesenchymal genes, including tenascin, vimentin, MET, Twist1 and WNT5a. Hyaluronan production by these cells was also decreased (Fig. 6h,i; Supplementary Fig. 5f,h), as was GBM cell motility, as indicated by decreased migration trajectories and migration velocity (Fig. 6j,k; Supplementary Fig. 5g). Gal-1 knockdown also reduced cell contractility, as illustrated by traction force microscopy (Fig. 6l). The loss of the mesenchymal-like phenotype reduced integrin mechanosignalling in the Gal-1-depleted GBMs that translated into a reduced ECM stiffening following intracranial injection (Fig. 6m). This effect was similar to that exhibited by mouse GBM tumours treated with hyaluronidase (Fig. 6m). Gal-1 knockdown in mesenchymal cells also significantly enhanced mouse survival post-injection (Fig. 6n). These data provide evidence for a reciprocal and dynamic link between a Gal-1-regulated glycocalyx, integrin mechanosignalling and a mesenchymal phenotype in GBMs.
Fig. 6 |. Galectins demonstrate mechanical reciprocity and drive a mesenchymal phenotype.
a, Survival probability versus expression of Gal-1 (LGALS1). n = 214 patients with GBM (TCGA). P value calculated using two-sided logrank test. b, LGALS1 expression in proneural (Pro; n = 57 patients) and mesenchymal (Mes; n = 58 patients) GBM (TCGA). Mean ± s.e.m.; *P = 1.46 × 10–19 by two-sided Mann–Whitney U-test. c, Correlation between LGALS1 and TNC from mesenchymal xenografts (n = 30 cDNA samples). Pearson’s correlation; log values of expression. d, WT and V737N proneural xenograft and syngeneic tumours were analysed for LGALS1 expression. Mean ± s.e.m. (n = 6 (syngeneic) and 5 (xenograft) mice per group). P = **0.009 and *0.032 by two-sided unpaired t-test. e, Xenografts were stained for Gal-1. Scale bar, 250 μm (n = 3 tissue sections). f, Proneural cells were plated on soft and stiff PA gels for LGALS1 expression. Mean ± s.e.m. (n = 3 independent experiments). g, Immunoblotting for Gal-1 and pT696-MYPT in proneural and mesenchymal cells on hydrogels (n = 2 independent experiments). h, RNA from control (Scr) or cells expressing Gal-1-silencing shRNA (shGal) was analysed. Mean ± s.e.m. (n = 9 and 2 independent experiments for TNC and vimentin, respectively). *P = 0.03 for TNC by two-sided paired t-test. i, Scr and shGal cells were analysed via immunoblotting (n = 3 independent experiments). j, Scr and shGal cells on soft hydrogels were imaged over 36 h (n = 12 cells per condition from 3 independent experiments). k, Velocity plot was derived from calculating the distance per time per frame, where n = 119 frames per condition averaged over 12 cells. Mean ± s.e.m.; ***P = 7.2 × 10–20 by two-sided paired t-test. l, Scr and shGal on 2.7 kPa hydrogels were imaged using traction force microscopy. A representative traction map is shown (left); the colour bar represents increasing magnitude of stress with 0 (blue) denoting low stress and 16 (red) denoting high stress regions of the cell. The scatter plot (right) shows mean ± s.e.m. of n = 22 and 26 measurements for shGal and scr, respectively, over 3 independent experiments. **P = 0.004 by two-sided Mann–Whitney U-test. m, ECM stiffness of Scr, shGal and hyaluronidase-treated mesenchymal Scr (Scr+HAse) xenografts was analysed by AFM. Scatter plot represents 192 indentations per sample. Mean ± s.e.m. (n = 3 tumours analysed per group with four 90 × 90 μm regions mapped). ***F (2,573) = 144.3, P = 1.76 × 10–51 verus Scr by one-way ANOVA and Bonferroni correction. n, Survival analysis of mice injected with Scr (n = 8 mice) and shGal-1 (n = 9 mice) GBM cells. P value calculated using two-sided logrank test. Source data: Supplementary Table 4; unprocessed blots: Supplementary Fig. 9.
Integrin-mechanosignalling, the glycocalyx and a GBM mesenchymal, stem-like phenotype.
GBM relapse has been attributed to an increase in the frequency of treatment-resistant, stem-like glioma cells47,48. Many bulky glycoproteins are also stem cell markers27,29,49. Consistently, the glycocalyx modulator Gal-1 correlated positively with the glycoprotein CD44, a marker often used to identify GSCs50, in human samples of GBM (Fig. 7a). A multiplex immunofluorescence analysis of a cohort of paired primary and recurrent GBMs showed increased levels of another stem-like cancer cell marker as well as a bulky glycoprotein, podoplanin (PDPN)51,52 (Supplementary Fig. 6a,b). These findings are consistent with an analysis of patient datasets that indicated that there is a reduced overall patient survival in patients with GBM whose tumours express high levels of PDPN (Fig. 7b). A bioinformatics analysis of GBM patient data further revealed that PDPN, which is a bulky glycoprotein, is enriched in the mesenchymal compared to the proneural GBM subtype (Fig. 7c). CD44 is both a bulky glycoprotein and a common marker of tumour initiation potential53,54. Fluorescence activated cell sorting (FACS) identified a CD44:PDPN double-positive (DP) cell population present predominantly in the patient-derived mesenchymal GBMs (Fig. 7d). Compared to tumour cells only expressing CD44 (single positive (SP)), the sorted DP cells not only expressed higher levels of the stem cell genes HES1 and SOX2 but also showed enrichment for tenascin and Gal-1 (Fig. 7e; Supplementary Fig. 6e). The DP tumour cells also had elevated integrin mechanosignalling and were more contractile, as indicated by higher pY397-FAK and pS19-MLC2 (Supplementary Fig. 6e). The DP tumour cells exhibited heightened resistance to the chemotherapy agent temozolomide compared to the SP GBM tumours (Fig. 7f ). Further evidence of stem-like properties in the DP cells was the ability of these cells to give rise to SP cells, eventually reconstituting the parental CD44:PDPN distribution (Supplementary Fig. 6h,i). Knocking down Gal-1 to reduce glycocalyx bulkiness and integrin mechanosignalling, or directly reducing integrin mechanosignalling by knocking down FAK, reduced the number of DP cells and sensitized the mesenchymal GBMs to temozolomide (Fig. 7g,h). Thus, a bulky glycocalyx can modulate the stem-like phenotype of some GBMs, at least in culture, and does so by regulating integrin mechanosignalling.
Fig. 7 |. Tissue mechanics fosters GBM stemness by regulating the glycocalyx.
a, Correlation between LGALS1 and CD44 levels in patient samples of primary and recurrent GBM (UCSF cohort; n = 10 patients per group). r from Pearson’s correlation. b, Survival probability versus expression level of PDPN (n = 214 GBM patients (TCGA)). P value calculated using two-sided logrank test. c, PDPN expression in proneural (Pro; n = 57 patients) and mesenchymal (Mes; n = 58 patients) GBM (TCGA). Mean ± s.e.m.; *P = 1.78 × 10–14 by two-sided Mann–Whitney U-test. d, CD44 and PDPN expression was analysed in proneural and mesenchymal cultures to distinguish a DP population (n = 3 independent sorting experiments). e, RNA from these populations was analysed. Mean ± s.e.m. (n = 4, 5, 3 and 6 independent experiments for SOX2, HES1, TNC and LGALS1, respectively). *P = 0.12, 0.06, 0.12 and 0.03, respectively, by two-sided paired t-test. f, CD44:PDPN DP and CD44 SP cells were treated with temozolomide (TMZ) for 72 h (n = 4 per dose per group). Mean ± s.e.m.; **P = 0.0001 and ***P = 1.31 × 10–18 by two-way ANOVA and Bonferroni post-test. g, The DP population was quantified from n = 2 for proneural and n = 3 independent experiments for others. Mean ± s.e.m.; F (3,7) = 149.0252, ***P = 1.05 × 10–6 by one-way ANOVA and Bonferroni post-test. h, Mesenchymal Scr, shGal and shFAK cells were treated with 50% growth inhibitory (GI50) dose of temozolomide at 7.5 μM for 72 h (n = 3 independent experiments per dose per group). *P = 0.01 and 0.04 for shGal and shFAK, respectively, verus Scr by one-way ANOVA. i, Secondary sphere colony formation was performed using primary neural stem cells (n = 4 independent experiments). *P = 0.03 by two-sided Mann–Whitney U-test. RNA from these spheres was assayed for HES1 expression (n = 3 independent experiments). Mean ± s.e.m.; **P = 0.002 by two-sided paired t-test. j, RNA from Scr, shGal and shFAK cells was analysed. Mean ± s.e.m. (n = 5, 4 and 3 independent experiments for SOX2, POU3F2 and HES1, respectively). P = *0.04, 0.2 and *0.03 by one-way ANOVA. k, Limiting dilution assays were performed using mesenchymal Scr and shGal-1 cells (n = 3 independent experiments). Mean ± s.e.m.; P = *0.02, *0.04 and **0.008 for 1,000, 100 and 10 cells per well, respectively, by two-sided paired t-test. l,m, In vivo limiting dilution studies using 100 and 1,000 cells per mouse (n = 5 mice per group). Tumour-bearing mice shown at study endpoint (l) and quantified for indicated dilutions (m).
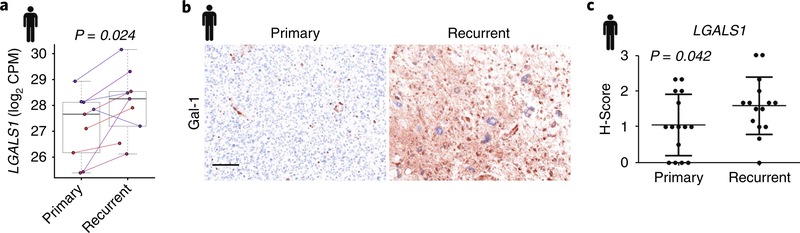
Next, we determined whether a causal link exists between integrin mechanosignalling, glycocalyx bulkiness and a stem-like phenotype in GBMs. We performed secondary sphere formation assays using primary neural stem cells isolated from the subventricular zone of normal mouse brains in which FAK activity was enhanced through targeted expression (GFAP-Cre) of the mutant V737N integrin β1. The neural cells expressing the V737N integrin β1 expressed higher levels of the stem cell marker HES1, and their isolated neurospheres showed enhanced self-renewal capacity (Fig. 7i). Mesenchymal GBMs in which mechanosignalling was reduced either through reduction in the glycocalyx bulkiness following Gal-1 knockdown or via shRNA knockdown of FAK exhibited reduced expression of stem cell markers, including SOX2, POU3F2 and HES1 (Fig. 7j). Moreover, the brain tumours generated by GBM cells decorated with long (bulky) mimetic glycopolymers showed increased SOX2 protein and higher mRNA levels of PDPN and HES1 (Supplementary Fig. 6c,d). These findings suggest that integrin mechanosignalling mediated by a bulky glycocalyx engages a reinforcing feedback loop that promotes a stem-like phenotype in GBMs. Consistently, limiting dilution assays showed that reducing the glycocalyx and integrin mechanosignalling through Gal-1 knockdown reduced colony formation in vitro (Fig. 7k; Supplementary Fig. 6f) and tumour initiation in vivo (Fig. 7l,m; Supplementary Fig. 6g). The clinical relevance of this phenotype was underscored by an analysis of RNA-seq data of paired primary and recurrent patient samples from the Nam cohort. An increase in Gal-1 expression was evident in the recurrent GBMs compared to primary GBMs (Fig. 8a). Immunohistochemistry analyses of these paired primary and recurrent samples (Nam cohort) confirmed an increase in Gal-1 protein expression (Fig. 8b). Moreover, an immunohistochemistry analysis and quantification of paired primary and recurrent GBMs (UCSF cohort; Fig. 8c), similarly revealed that there is an increase in Gal-1 in recurrent GBMs, and further showed that this correlated with the stem cell marker CD44 (Fig. 7a). These data demonstrate how a bulky glycocalyx can potentiate integrin mechanosignalling to promote a mesenchymal and stem-like phenotype in GBMs and could explain the enrichment for this phenotype in recurrent, treatment-resistant GBMs.
Fig. 8 |. Gal-1 is upregulated in recurrent wild-type GBM.
a, Before-and-after plot of LGALS1 expression in paired primary and recurrent GBM patient samples from the Nam cohort (RNA-seq data; n = 9 patients per group). Individual dots represent log2 expression of LGALS1 transcript levels. Boxes indicate the group median and interquartile range, and the whiskers extend to the most extreme observed value. P value calculated using two-sided paired t-test. b, Representative images of paired human primary and recurrent GBM patient samples (Nam cohort) stained for Gal-1 using multiplex immunohistochemistry. Scale bar, 100 μm. c, LGALS1 expression in paired primary versus recurrent GBM human patient samples (UCSF cohort; n = 15 patients). Mean ± s.e.m.; P value calculated using one-tailed paired t-test.
Discussion and conclusion
GBM recurrence has been attributed to a glioma population with a mesenchymal, stem-like phenotype that is resistant to treatment47,48. GSCs could arise through transdifferentiation or via the expansion of resident, oncogenically modified stem cells induced by tumour-associated microenvironmental factors52,53,55,56. GSC frequency could also be increased by treatment-induced changes in the microenvironment, such as by radiation-induced TGFβ activation or by changes in the vascular niche57. Our findings suggest that the mesenchymal, stem-like phenotype of some recurrent GBMs could also be promoted by elevated tissue tension and an increased expression of glycoproteins that increase the bulkiness of the glycocalyx. Indeed, we showed that there is a consistent upregulation of glycoproteins in recurrent human GBMs and presented experimental evidence of their functional promotion of a mesenchymal- and stem-like phenotype4,20,22,58.
Many of the glycoproteins that increase the bulkiness of the cancer cell glycocalyx are also expressed by stem-like tumour cells and are enriched in mesenchymal-like GBMs20,22,27,29,49,59. A bulky glycocalyx can act as a steric modulator of integrin mechanosignalling and as an inducer of cell and stromal tension14,42,44. We confirmed this finding by showing that reducing glycocalyx bulkiness represses integrin signalling, while enhancing glycocalyx bulkiness promotes integrin adhesion assembly and signalling. We also noted that increasing integrin-dependent mechanosignalling, through expression of a mutant V737N integrin β1 that enhances FAK activity, induced the expression of several bulky glycoproteins, and that inhibiting integrin signalling decreased their expression. These findings imply that there is a functional link between integrin mechanosignalling and the expression of bulky glycoproteins, and further indicate that this mechanism is self-reinforcing. Whether the overall structure and function of the glycocalyx in GBM, which is influenced by the nature of glycosylation, including the level of sialylation, fucosylation, N- or O-linked glycosylation60,61, would influence integrin mechanosignalling to alter this relationship, and how, remains unclear. Nevertheless, our observations that increased expression of many of the glycoproteins that enhance glycocalyx bulkiness also induces a mesenchymal, stem-like phenotype in GBM suggests that there is an integrin mechanosignalling-linked glycocalyx-mediated regulatory circuit that, once engaged, drives and sustains this treatment-resistant, tumour-initiating phenotype. Accordingly, inhibiting regulatory hubs of integrin–ECM signalling or glycocalyx bulkiness should disrupt this regulatory circuit and constitute attractive targets to treat GBM24,62.
Methods
Methods, including statements of data availability and any associated accession codes and references, are available at https://doi.org/10.1038/s41556–018-0183–3.
Methods
Human samples.
Human tissue samples, lacking any patient-identifying information, were obtained from the University of California, San Francisco Brain Tissue Bank (UCSF cohort). De-identified biospecimens for multiplex immunohistochemistry were imaged and analysed by D.-H.N.’s group at the Samsung Medical Center BioBank, Republic of Korea (Nam cohort). The study is compliant with all relevant ethical regulations regarding research involving human participants, and all human tissue samples were collected in compliance with the informed consent policy under the institutional review boards of the University of California and the Samsung Research Center.
Mouse studies.
All mice were maintained in accordance with University of California Institutional Animal Care and Use Committee (IACUC) guidelines under protocol number AN109372, and the study was compliant with all relevant ethical regulations regarding animal research. Animals were randomly distributed among the different conditions since the animals did not exhibit differences in appearance at the onset of the experiments. Cells were washed with PBS, collected by trypsinization (0.05%, Gibco), counted and re-suspended in PBS at 100,000 cells per μl. For intracranial injections, 6-week-old (NCR nude for xenograft experiments, FVB/n for syngeneic experiments) female mice were anaesthetized with 2% isoflurane, injected subcutaneously with buprenorphine (0.03 mg per kg) and slowly injected with a 2 μl tumour cell suspension (200,000 cells per injection, unless otherwise noted) into the striatum. Transgenic mice were generated by crossing the proneural oligodendroglioma S100β-vErb:p53+/– model (provided by W. Weiss, UCSF) with GFAP-Cre mice (Jax Labs FVB-Tg(GFAP-cre)25Mes/J) and LSL-V737N mice (described previously63), all on a pure FVB/n background. GFAP-Cre:LSL-V737N mice were used for neural stem cell studies. Both transgenic and transplant tumour-bearing mice were sacrificed upon exhibiting 15% weight loss or signs of tumour burden, including ataxia and laboured breathing. No animals were excluded.
Immunoblotting.
Immunoblotting was performed as described previously6. For all immunoblotting, cells were lysed in 2% SDS in Tris buffer saline (TBS), pH 8. Antibodies used for the immunoblotting analysis are listed in Supplementary Table 3.
Antibodies and reagents.
Comprehensive information on the primary antibodies used for immunoblotting, immunohistochemistry, immunofluorescence and flow cytometry is included in Supplementary Table 3 “Antibody Information”. Alexa Fluor-conjugated anti-mouse IgG and anti-rabbit IgG secondary antibodies (Invitrogen, A11012 and A11005) and HRP-conjugated rabbit and rat secondary antibodies (Vector Labs, PI-1000 and PI-9400), Hypoxyprobe was purchased from HPI (Hypoxyprobe-1, lot no. 06242014), alcian blue stain (Diagnostic BioSystems, ANC250), propidium iodide (AcrosOrganics, no. 440300250), temozolomide (Selleck Chem, no. S1237) and FAK inhibitor (Selleck Chem, no. PND-1186). shRNAs were purchased from Sigma (LGALS1 clone no. TRCN0000057423–27; FAK clone no. TRCN0000196310). The sequence 5′-GCAAGCTGACCCTGAAGTTCAT-3′ was used as a nonspecific control cloned into the lentiviral pLKO.1 puro vector (Sigma-Aldrich). Lentiviral constructs encoding wild-type and V737N mutant β1 integrin (used to transduce proneural GBM cells and wild-type S100β-vErb:p53+/– derived tumour cells) have been described previously6. Hyaluronic acid detection was performed using a Quantikine ELISA (DHYAL0).
Primary cell culture conditions.
All primary GBM patient-derived cells were maintained at 37 °C and 5% CO2. These primary GBM cells (mesenchymal GBM43 and GBM10, and proneural GBM5 and GBM12), described previously31,32, were authenticated based on morphology and on the validated markers tenascin and vimentin. The cells were not cultured beyond ten passages to avoid drift. Cells were cultured in neurobasal media (Invitrogen) supplemented with B27 supplement (Invitrogen), epidermal growth factor (20 ng ml–1; Peprotech), fibroblast growth factor (20 ng ml–1; Peprotech) and penicillin–streptomycin (100 units per ml). Cells were tested for mycoplasma contamination using a commercially available kit (PCR-Mycoplasma Test Kit I/C (Promokine, PK-CA91–1024)) according to the manufacturer’s instructions. Knockdown cells were generated via transduction with a lentiviral vector-encoding shRNA targeting LGALS1, FAK or a control (Scr) sequence (all clones listed above). HEK 293T cells (ATCC) were used for lentiviral transduction. For PA gel studies, cells were plated on fibronectin-conjugated gels for 18–48 h before collecting cells for quantitative PCR (qPCR) or western blotting.
Quantitative real-time PCR and in situ hybridization.
PCR reactions were performed in triplicate with LightCycler Fast Start DNA Master SYBR Green Mix (Roche, catalogue no. 03752186001) using a Realplex2 epGradient S Mastercycler (Eppendorf), and the relative amount of complementary DNA was calculated using the comparative CT method based on the following: (1) human 18S RNA for cell culture studies; (2) human GAPDH and/or RPS20 for xenograft studies (RPS20 does not exist in the mouse, thus this approach normalizes to the input human tumour cells); (3) vERB for syngeneic studies (this transgene is only expressed in the Ep53 tumour cells, so this approach normalizes to input mouse tumour cells). Primer sequences for all qPCR reactions are listed in Supplementary Table 2 “Primer Sequences”.
qPCR arrays were purchased from Qiagen (epithelial-to-mesenchymal transition array no. 330231), and pathway analysis was performed using the freely available Qiagen data analysis centre, available at http://saweb2.sabiosciences.com/pcr/arrayanalysis.php.
AFM measurements.
AFM and analyses were performed using an MFP3D-BIO inverted optical atomic force microscope mounted on a Nikon TE2000-U inverted fluorescence microscope (Asylum Research) as described previously6. Freshly dissected brains were OCT-embedded and flash-frozen in liquid nitrogen. Sections were cut at 20 μm on Fisher Superfrost Plus Gold slides. Immediately before AFM, sections were thawed at room temperature, and optimal cutting temperature compound (OCT) was rinsed off using room temperature PBS, then covered in HEPES-buffered PBS containing complete protease inhibitor cocktail (Roche) and 1 μg ml–1 propidium iodide (to allow visualization of nuclei). Hyaluronidase-treated mesenchymal Scr xenografts were incubated with hyaluronidase for 30 min at 37 °C, and control and Gal-1 samples were treated with PBS for 30 min at 37 °C. Slides were then magnet-anchored to the stage of the microscope. All samples were measured in liquid media in contact mode using Novascan cantilevers (2.5-μm radius beaded silica glass tip, k = 0.06 N per m), which were calibrated using the thermal tune method. Force measurements were collected over a 90 × 90 μm grid at a resolution of 16 × 16 μm at a trigger force of 1.5 nN. The resulting force data were converted to elastic modulus values using the Hertz Model program (tissue samples were assumed to be noncompressible, and a Poisson’s ratio of 0.5 was used in the calculation of the Young’s elastic modulus values) in IgorPro v.6.22, supplied by Asylum Research.
Preparation of PA gel substrates.
Glass and silicon (SAIM) substrates were prepared by glutaraldehyde activation followed by conjugation with 10 μg ml−1 (glass) or 20 μg ml−1 (silicon) fibronectin as previously described64. PA gel substrates (soft: 2.5% acrylamide, 0.03% bis-acrylamide; stiff: 10% acrylamide, 0.5% bis-acrylamide) were conjugated overnight with 20 μg ml−1 fibronectin at 4 °C and rinsed twice with PBS and DMEM before cell plating. A stiffness of 75–400 Pa was considered soft (comparable to gliotic tissue) and a stiffness of 6,000–60,000 Pa was considered stiff (comparable to GBM tissue).
Traction force microscopy.
PA gels were prepared as described above and red fluorescent beads (0.5 μm; Molecular Probes) were incorporated as described previously64. Cells were seeded overnight on gels and images were taken of the cells and beads before 0.2% SDS was added to lyse the cells, after which cell-free images were taken. Images were registered using ImageJ, and a particle image velocimetry program was implemented65. Then, the traction force of individual cells was calculated using the Fourier transform traction cytometry method (with a Poisson ratio of 0.5) using the freely available software at https://sites.google.com/site/qingzongtseng/tfm66.
Immunohistochemistry, immunofluorescence and imaging.
Tissues were fixed and labelled, and multiple independent fields imaged on a Zeiss LSM 510 microscope system with either ×20 Apo NA 0.75 air or ×40 Apo LWD NA 1.15 water objective, and 488 nm Argon, 543 nm HeNe and 633 nm HeNe excitation lines. Analyses of tissue immunofluorescence and immunohistochemistry (not including multiplex immunohistochemistry) were done using ImageJ/FIJI software. For immunohistochemistry, slides were deparaffinized in xylene and rehydrated in ethanol. Antigen retrieval was performed in Tris-buffered saline buffer (pH 9.0) or citric acid (pH 6.0) using a pressure cooker. Slides were blocked with 5% serum–2% BSA solution, followed by incubation in primary antibody overnight at 4 °C. Sections were incubated in biotinylated secondary antibodies for 1 h at room temperature followed by 30 min in Vectastain ABC (Vector Labs, no. PK-6100) and DAB treatment (Vector Labs, no. SK-4100). Slides were stained with haematoxylin, dehydrated, mounted and imaged using a standard brightfield microscope. For alcian blue staining, slides were incubated at 37 °C in pH 2.5 alcian blue (Vector Labs) for 10 min after deparaffinization, washed twice in 3% acetic acid, followed by xylene dehydration and mounting.
Primary neural stem cell isolation.
For neural stem cell assays, GFAP-Cre:LSL-V737N or control (LSL-V737N) mice were killed at weaning age (day 21), and subventricular microdissections were performed and digested for neurosphere generation as previously described67. Subventricular dissections were digested at 37 °C for 10 min in papain (1 mg ml–1; Worthington Chemical), washed and filtered. Single cells were cultured at 1 × 105 cells per ml in neurobasal media (Invitrogen) supplemented with B27 supplement, FGF (fibroblast growth factor; 20 ng ml–1; Peprotech), EGF (epidermal growth factor; 20 ng ml–1; Sigma-Aldrich) and L-glutamine (2 mM), and incubated at 37 °C in 5% CO2. After primary spheres developed (7–14 days), they were mechanically dissociated with a pipette and replated at 1 × 105cells per ml, and secondary spheres were enumerated with a low-power objective after 7–14 days.
Cell motility and invasion assays.
Cells were plated on soft and stiff substrates for 48 h before imaging. Live cell imaging was performed using a Nikon TiE microscope, and images were captured using NIS Elements software AR v.4.30.02. For tracking and quantifying migrating cells, the Manual Tracking function in ImageJ and GraphPad Prism v.6.0 were employed. For the invasion assay, 8 μm Boyden chambers were used (BD, no. 353097). The membrane was coated with fibronectin (20 μg ml–1; Millipore). Cells resuspended in neurobasal media (without EGF or bFGF) were seeded in the top chamber. The bottom chamber contained neurobasal media containing 5% FBS (Gibco). The membrane was fixed with paraformaldehyde after 12 h, stained with 0.1% crystal violet, allowed to dry overnight and imaged at ×10 magnification using a brightfield microscope. For the quantification of local invasion in tissue samples, H&E stained sections were imaged at ×20 and ×40 magnification using a brightfield microscope. Quantification of the invasive edge was performed by measuring the distance between the edge of the tumour core to the point where tumour cells were seen migrating into the normal parenchyma.
Limiting dilution assays.
Cells were trypsinized (0.05% trypsin, Gibco), counted and diluted to 1,000 cells per 3 μl for in vivo studies and 1,000 cells per 100 μl for in vitro studies. The cells were then serially diluted to 100, 10 or 1 cell and injected into mice or plated into 96 well plates. For in vitro assays, cells were grown for 10 days, fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and imaged using a brightfield microscope. For in vivo studies, mice injected with cells were imaged starting at day 7 of injection, 1–2 times per week until tumour initiation was observed. Live mice imaging was performed using bioluminescence imaging on a Xenogen IVIS spectrum imaging system.
SAIM.
Cells ectopically expressing farnesylated GFP were plated overnight on fibronectin-coated reflective silicon substrates, fixed and imaged randomly as previously described68, scanning the incident angle of excitation light from 0° to 48° with a one-degree sampling rate. Z-positions were localized with custom algorithms68.
Glycomimetic studies.
Glycopolymers were synthesized as previously described40. Lipid-conjugated RAFT agents were synthesized, from which methyl vinyl ketone was polymerized, to generate polymers of various lengths with low polydispersities. The ketone pendant groups were functionalized with alkoxy-amine containing N-acetylgalactosamine monomers and purified to give the final lipid-terminated glycopolymers. Glycopolymers were loaded onto cell surfaces40. Cells in suspension were mixed with stock concentrates of aqueous solutions of glycopolymers, and mixtures were incubated for 1 h at room temperature for labelling to occur. The cells were then pelleted, the labelling solution aspirated, and the excess glycopolymers washed away by an additional dilution–pelleting step. The cells were then used for setting up xenograft studies and/or for SAIM analysis. Short glycopolymers were 3 nm long and long glycopolymers were 90 nm long.
Flow cytometry.
Cells were suspended in 0.05% trypsin-EDTA (Gibco) at a concentration of 1 × 105 cells per 200 μl and incubated with the fluorochrome-conjugated antibodies as indicated (listed above) for 30 min on ice. Cells were pelleted, washed and suspended in PBS, strained through 40-μm filters, and analysed using a Becton Dickinson LSR II or Fortessa with the channels corresponding to the fluorochromes. For sorting, cells were similarly prepared, but at a concentration of 1 × 106 cells per 500 μl, and run on a FACSAria and sorted with a 100-μm nozzle. All flow cytometry data were collected using FACS Diva and analysed and plotted using FloJo and Graphpad Prism v.6.0.
Peptide spectral mapping.
Peptide mapping was performed as previously described69. Tissue solubilization and enrichment of membrane proteins and membrane-associated proteins was accomplished according to the Thermo Mem-PER Plus Extraction Kit (Thermo Fisher Scientific). Following the final membrane extraction step, the remaining pellet was solubilized in 100 mM ammonium bicarbonate, 6 M Guanidium-HCl pH 8.0 for 2 h at room temperature. Samples were analysed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nanoLC 1000 system through a nanoelectrospray source. Data acquisition was performed using the Xcalibur (v.2.1) software. Peptide spectral matching was performed with Mascot (v.2.5) against the Uniprot mouse database (release 201701). Scaffold (v.4.4.6, Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as determined by the Trans-Proteomic Pipeline and the Protein Prophet algorithms. Relative quantification was performed based on peptide spectral matches determined in Scaffold.
Multiplex immunohistochemistry.
Multiplex staining was performed using an Opal Manual kit (NEL811001KT, PerkinElmer) according to the manufacturer’s protocol. The slides were deparaffinized in xylene and rehydrated in ethanol. Antigen retrieval was performed in Tris-buffered saline buffer (pH 9.0) using microwave treatment. Details of primary antibodies, including dilutions, are included in Supplementary Table 3. In a serial fashion, primary antibodies were incubated for 30 min in a humidified chamber at room temperature, followed by detection using a mouse/rabbit SuperPicture Polymer Detection HRP Kit. Visualization of the primary antibody was accomplished using Opal Fluorophore Working Solution (TSA, 1:50), after which the slide was placed in Tris-buffered saline buffer (pH 9.0) and repeated using microwave treatment. The slides were examined using a VECTRA 3.0 Automated Quantitative Pathology Imaging System (PerkinElmer). We used InForm image analysis software (PerkinElmer) to analyse each marker expression. Spearman correlation was used to calculate the correlation coefficient, r, for multiplex imaging data.
Bioinformatics analysis of gene expression datasets.
Recurrent versus primary GBM analyses (Nam cohort).
To determine whether bulky cell-surface proteins are systematically upregulated when non-mesenchymal GBMs relapse with mesenchymal-like features, we used a paired primary and recurrent GBM patient dataset from D.-H.N.’s group at the Samsung Medical Center, Republic of Korea, to identify genes whose expression levels increase during this transition. We used voom/limma70 to fit a gene-wise mixed-effects linear model to quantify the evidence for upregulation of each gene during the acquisition of the mesenchymal-like state (for example, P values derived from a one-sided test on the coefficient t-statistics estimated in the framework of the linear model). Gene expression level changes that correlated with the relapsed mesenchymal state were estimated as a fixed effect, and we used a random effect to capture and control for baseline patient-level variation in gene expression. Raw RNA-seq data from the Nam cohort were normalized to log2 CPM (count-per-million) and a before–after plot was generated for assessing expression of LGALS1 in paired primary and recurrent patient samples.
Estimation of bulky glycocalyx genes.
To determine whether genes whose products were likely to contribute to membrane bulkiness were disproportionately transcriptionally upregulated with respect to GBM subtype, we compared summarized expression levels in relevant publicly available datasets (TCGA) as previously described14. We separated genes into classes on the basis of their estimated extramembrane bulk. Genes encoding proteins with ten or more putative N-glycosylation sites and four or more putative O-glycosylation sites are defined as bulky (see genes in Supplementary Table 1), but similar trends were observed across a range of bulk cut-offs. Then we determined the evidence that they were upregulated in tumours of different grade or subtype using a gene-wise one-sided t-test. The resulting P value distributions across bulkiness classes were then compared using a Kolmogorov–Smirnov test. See previous study14 for additional details.
Statistics and reproducibility.
All quantitative results were assessed with the test indicated in the figure legends, after confirming that the data met appropriate assumptions (normality, homogeneous variance and independent sampling), t-tests, non-parametric Mann–Whitney exact test (using the normal approximation for the U score), Kaplan–Meier (survival). Unless otherwise stated, all indicated P values are two-tailed and all data are plotted with standard error bars, indicating standard error of the mean. All the experiments were performed using sample sizes based on standard protocols in the field. For in vitro and mouse studies, as well as human patient sample analysis, sample sizes were chosen to provide 85% power to detect an effect size of 2.5 with a two-sided error of less than or equal 5%, wherever pertinent. All results were reproduced at least twice in the laboratory. The figure legends indicate the number of experiments that were repeated individually with similar results. Microsoft Excel and Graphpad Prism v.5.0 software were used to conduct statistical analyses of the data. P values less than or equal to 0.05 were considered significant. Animals were randomly distributed and no animals were excluded. For mouse studies and assessment of clinical specimens, mechanical testing was performed blinded, and immunostaining intensity of tissue sections was scored blinded.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability.
Previously published paired primary and recurrent patient RNA-seq data (Nam cohort) that were re-analysed here are available on EGA under the accession codes EGAS00001001041, EGAS00001001800 and EGAS00001001880. Data from TCGA (http://cancergenome.nih.gov) and Repository for Molecular Brain Neoplasia Data (Rembrandt) were downloaded from the CBioPortal for Cancer Genomics (http://www.cbioportal.org) and Project Betastasis 6 (www.betastasis.com), and the mean-normalized expression scores for genes of interest determined. The datasets assessed in TCGA were Classical, Mesenchymal, Neural, and Proneural GBM. In REMBRANDT, the datasets assessed were Oligodendroglioma, Astrocytoma, and GBM. Analyses contributing to Fig. 4a were performed based on previously published literature14,70. R scripts for these can be provided by R.O.B. or the corresponding author upon reasonable request. Source data for Fig. 4b,c and Supplementary Fig. 8 have been provided as Supplementary Table 1, “Bulky genes list”. Source data for figures that were repeated twice (Figs. 3e,h, 5e, 6h and 7g, and Supplementary Figs. 3b,g, 5h and 6f) are provided as Supplementary Table 4 “Statistics Source Data”. All other data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
We thank W. Weiss (UCSF) for the S100β-vErb:p53+/− mice and M. Paszek (Cornell University) for helpful discussions. Animal handling and tissue preparation was supported by L. Korets and N. Korets (UCSF). This work was supported by the following grants: US National Institutes of Health NCI R01 grants CA138818-01A1, CA192914-01, CA174929-01 and CA085482 (V.M.W.); U01 grant CA202241 (V.M.W. and F.K.); U54 grant CA163155 (V.M.W.); Ruth Kirschstein NRSA F32 CA174319 (J.M.B.); UCSF Brain Tumor Research Program T32 CA151022 (J.M.B.); NHLBI T32 HL007544 (J.C.T.); UCSF Brain Tumor SPORE Tissue Core NIH/NCI P50 CA097257 (J.J.P.); NIH 1U01CA168878 (J.J.P); NIH/NINDS R01 NS081117 (J.J.P.); CIRM grant RB5-07409 (V.M.W.); CIRM training grant TG2-01153 (L.P.); and NCI R01 CA227942 (C.R.B. and V.M.W.). This work was also supported by a grant from the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3418). The biospecimens for multiplex immunohistochemistry were provided by Samsung Medical Center BioBank.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41556-018-0183-3.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Louis DN et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Yoon SJ et al. Tumor mesenchymal stem-like cell as a prognostic marker in primary glioblastoma. Stem Cells Int. 2016, 6756983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon SM et al. Recurrent glioblastomas reveal molecular subtypes associated with mechanistic implications of drug-resistance. PLoS ONE 10, e0140528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood MD, Reis GF, Reuss DE & Phillips JJ Protein analysis of glioblastoma primary and posttreatment pairs suggests a mesenchymal shift at recurrence. J. Neuropathol. Exp. Neurol. 75, 925–935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips HS et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Miroshnikova YA et al. Tissue mechanics promote IDH1-dependent HIF1α-tenascin C feedback to regulate glioblastoma aggression. Nat. Cell Biol. 18, 1336–1345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim Y. & Kumar S. CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility. Mol. Cancer Res. 12, 1416–1429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butcher DT, Alliston T. & Weaver VM A tense situation: forcing tumour progression. Nat. Rev. Cancer 9, 108–122 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L. et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor-1alpha in hepatocellular carcinoma. BMC Cancer 13, 108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S. et al. Hypoxia induces TWIST-activated epithelial-mesenchymal transition and proliferation of pancreatic cancer cells in vitro and in nude mice. Cancer Lett. 383, 73–84 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Leight JL, Wozniak MA, Chen S, Lynch ML & Chen CS Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol. Biol. Cell 23, 781–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordenonsi M. et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147, 759–772 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Ikushima H. et al. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 5, 504–514 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Paszek MJ et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511, 319–325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinho SS & Reis CA Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Wade A. et al. Proteoglycans and their roles in brain cancer. FEBS J. 280, 2399–2417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varki A, Kannagi R. & Toole BP in Essentials of Glycobiology 2nd edn (eds Varki Aet al.) Ch. 44 (Cold Spring Harbor Laboratory Press, New York, 2009). [Google Scholar]
- 18.Tinder TL et al. MUC1 enhances tumor progression and contributes toward immunosuppression in a mouse model of spontaneous pancreatic adenocarcinoma. J. Immunol. 181, 3116–3125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woods EC et al. A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. eLife 6, e25752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kufe DW Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer 9, 874–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horm TM & Schroeder JA MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh. Migr. 7, 187–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollingsworth MA & Swanson BJ Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 4, 45–60 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Thompson CB et al. Enzymatic depletion of tumor hyaluronan induces antitumor responses in preclinical animal models. Mol. Cancer Ther. 9, 3052–3064 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Hingorani SR et al. Phase Ib study of PEGylated recombinant human hyaluronidase and gemcitabine in patients with advanced pancreatic cancer. Clin. Cancer Res. 22, 2848–2854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy LD et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene 30, 1449–1459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porsch H. et al. Efficient TGFbeta-induced epithelial-mesenchymal transition depends on hyaluronan synthase HAS2. Oncogene 32, 4355–4365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J. et al. Identification of cell surface glycoprotein markers for glioblastoma-derived stem-like cells using a lectin microarray and LC-MS/MS approach. J. Proteome Res. 9, 2565–2572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muramatsu T. & Muramatsu H. Carbohydrate antigens expressed on stem cells and early embryonic cells. Glycoconj. J. 21, 41–45 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Boheler KR & Gundry RL Concise review: cell surface N-linked glycoproteins as potential stem cell markers and drug targets. Stem Cells Transl Med. 6, 131–138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhaak RG et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandita A, Aldape KD, Zadeh G, Guha A. & James CD Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer 39, 29–36 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Kitange GJ et al. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro. Oncol. 11, 281–291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engler JR et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS ONE 7, e43339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lakins JN, Chin AR & Weaver VM Exploring the link between human embryonic stem cell organization and fate using tension-calibrated extracellular matrix functionalized polyacrylamide gels. Methods Mol. Biol. 916, 317–350 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Barnes JM, Przybyla L. & Weaver VM Tissue mechanics regulate brain development, homeostasis and disease. J. Cell Sci. 130, 71–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paszek MJ et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Weiss WA et al. Genetic determinants of malignancy in a mouse model for oligodendroglioma. Cancer Res. 63, 1589–1595 (2003). [PubMed] [Google Scholar]
- 38.Zhuo L. et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Nishihara S. Glycans define the stemness of naive and primed pluripotent stem cells. Glycoconj. J. 34, 737–747 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Woods EC, Yee NA, Shen J. & Bertozzi CR Glycocalyx engineering with a recycling glycopolymer that increases cell survival in vivo. Angewandte Chemie 54, 15782–15788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu FT & Rabinovich GA Galectins as modulators of tumour progression. Nat. Rev. Cancer 5, 29–41 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Bhat R. et al. Nuclear repartitioning of galectin-1 by an extracellular glycan switch regulates mammary morphogenesis. Proc. Natl Acad. Sci. USA 113, E4820–E4827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camby I, Le Mercier M, Lefranc F. & Kiss R. Galectin-1: a small protein with major functions. Glycobiology 16, 137R–157R (2006). [DOI] [PubMed] [Google Scholar]
- 44.Belardi B, O’Donoghue GP, Smith AW, Groves JT & Bertozzi CR Investigating cell surface galectin-mediated cross-linking on glycoengineered cells. J. Am. Chem. Soc. 134, 9549–9552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Mercier M. et al. Evidence of galectin-1 involvement in glioma chemoresistance. Toxicol. Appl. Pharmacol. 229, 172–183 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Le Mercier M, Fortin S, Mathieu V, Kiss R. & Lefranc F. Galectins and gliomas. Brain Pathol. 20, 17–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J. et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu G. et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 5, 67 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visvader JE & Lindeman GJ Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer 8, 755–768 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Yoshida T, Matsuda Y, Naito Z. & Ishiwata T. CD44 in human glioma correlates with histopathological grade and cell migration. Pathol. Int. 62, 463–470 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Atsumi N. et al. Podoplanin, a novel marker of tumor-initiating cells in human squamous cell carcinoma A431. Biochem. Biophys. Res. Commun. 373, 36–41 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Dahlrot RH, Hermansen SK, Hansen S. & Kristensen BW What is the clinical value of cancer stem cell markers in gliomas? Int. J. Clin. Exp. Pathol. 6, 334–348 (2013). [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ & Clarke MF Prospective identification of tumorigenic breast cancer cells. Proc. Natl Acad. Sci. USA 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Underhill C. CD44: the hyaluronan receptor. J. Cell. Sci. 103, 293–298 (1992). [DOI] [PubMed] [Google Scholar]
- 55.Soeda A. et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene 28, 3949–3959 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Gilbertson RJ & Rich JN Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer 7, 733–736 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Lu KV et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell 22, 21–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubelt C, Hattermann K, Sebens S, Mehdorn HM & Held-Feindt J. Epithelial-to-mesenchymal transition in paired human primary and recurrent glioblastomas. Int. J. Oncol. 46, 2515–2525 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Dai L. et al. Differential profiling studies of N-linked glycoproteins in glioblastoma cancer stem cells upon treatment with gamma-secretase inhibitor. Proteomics 11, 4021–4028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Autelitano F. et al. Identification of novel tumor-associated cell surface sialoglycoproteins in human glioblastoma tumors using quantitative proteomics. PLoS ONE 9, e110316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furukawa J. et al. Comprehensive glycomics of a multistep human brain tumor Model reveals specific glycosylation patterns related to malignancy. PLoS ONE 10, e0128300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolev VN et al. Inhibition of FAK kinase activity preferentially targets cancer stem cells. Oncotarget 8, 51733–51747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laklai H. et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 22, 497–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Przybyla L, Lakins JN, Sunyer R, Trepat X. & Weaver VM Monitoring developmental force distributions in reconstituted embryonic epithelia. Methods 94, 101–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mekhdjian AH et al. Integrin-mediated traction force enhances paxillin molecular associations and adhesion dynamics that increase the invasiveness of tumor cells into a three-dimensional extracellular matrix. Mol. Biol. Cell 28, 1467–1488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tseng Q. et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc. Natl Acad. Sci. USA 109, 1506–1511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM & Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Paszek MJ et al. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nat. Methods 9, 825–827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrett AS et al. Hydroxylamine chemical digestion for insoluble extracellular matrix characterization. J. Proteome Res. 16, 4177–4184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law CW, Chen Y, Shi W. & Smyth GK voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published paired primary and recurrent patient RNA-seq data (Nam cohort) that were re-analysed here are available on EGA under the accession codes EGAS00001001041, EGAS00001001800 and EGAS00001001880. Data from TCGA (http://cancergenome.nih.gov) and Repository for Molecular Brain Neoplasia Data (Rembrandt) were downloaded from the CBioPortal for Cancer Genomics (http://www.cbioportal.org) and Project Betastasis 6 (www.betastasis.com), and the mean-normalized expression scores for genes of interest determined. The datasets assessed in TCGA were Classical, Mesenchymal, Neural, and Proneural GBM. In REMBRANDT, the datasets assessed were Oligodendroglioma, Astrocytoma, and GBM. Analyses contributing to Fig. 4a were performed based on previously published literature14,70. R scripts for these can be provided by R.O.B. or the corresponding author upon reasonable request. Source data for Fig. 4b,c and Supplementary Fig. 8 have been provided as Supplementary Table 1, “Bulky genes list”. Source data for figures that were repeated twice (Figs. 3e,h, 5e, 6h and 7g, and Supplementary Figs. 3b,g, 5h and 6f) are provided as Supplementary Table 4 “Statistics Source Data”. All other data that support the findings of this study are available from the corresponding author upon reasonable request.