Abstract
Bacteria convert changes in sensory inputs into alterations in gene expression, behavior, and lifestyles. A common lifestyle choice that bacteria make is whether to exhibit individual behavior and exist in the free-living planktonic state or to engage in collective behavior and form sessile communities called biofilms. Transitions between individual and collective behaviors are controlled by the chemical cell-to-cell communication process called quorum sensing. Here, we show that quorum sensing represses Pseudomonas aeruginosa biofilm formation and virulence by activating expression of genes encoding the KinB–AlgB two-component system (TCS). Phospho-AlgB represses biofilm and virulence genes, while KinB dephosphorylates and thereby inactivates AlgB. We discover that the photoreceptor BphP is the kinase that, in response to light, phosphorylates and activates AlgB. Indeed, exposing P. aeruginosa to light represses biofilm formation and virulence gene expression. To our knowledge, P. aeruginosa was not previously known to detect and respond to light. The KinB–AlgB–BphP module is present in all pseudomonads, and we demonstrate that AlgB is the partner response regulator for BphP in diverse bacterial phyla. We propose that in the KinB–AlgB–BphP system, AlgB functions as the node at which varied sensory information is integrated. This network architecture provides a mechanism enabling bacteria to integrate at least two different sensory inputs, quorum sensing (via RhlR-driven activation of algB) and light (via BphP–AlgB), into the control of collective behaviors. This study sets the stage for light-mediated control of P. aeruginosa infectivity.
Photosensing and quorum sensing are integrated to control collective behaviors of the pathogenic bacterium Pseudomonas aeruginosa; the information is transduced via a phosphorylation–dephosphorylation sensory system. The study has implications for light-mediated control of P. aeruginosa infectivity.
Introduction
Bacterial responses to self-generated and exogenous stimuli influence their survival, persistence in particular niches, and lifestyle transitions such as alterations between being free-swimming or existing as a member of a biofilm. Biofilms are three-dimensional structured communities of bacterial cells encased in an extracellular matrix [1,2]. Bacteria living in biofilms exhibit superior resilience to environmental stresses such as antimicrobials and host immune responses [2,3]. Frequently, biofilm formation is governed by intracellular signaling molecules such as cyclic di-guanosine monophosphate [4,5] and extracellular signaling molecules such as quorum-sensing autoinducers [6,7]. Quorum sensing is a cell-to-cell communication process that relies on the production, release, and population-wide detection of autoinducers [8,9]. Quorum sensing allows groups of bacteria to synchronously alter behavior in response to changes in the population density and species composition of the vicinal community. Many pathogenic bacteria, including the global pathogen P. aeruginosa, require quorum sensing to establish successful infections [10]. Here, we show that quorum-sensing signaling converges with the detection of and response to another sensory cue, light, to control biofilm formation and virulence factor production in P. aeruginosa. We define the pathway connecting the light and quorum-sensing inputs to the virulence and biofilm outputs.
Light is a common environmental cue that is detected by photoreceptors present in all domains of life [11,12]. Particular photoreceptor photosensory domains are activated by specific wavelengths of light [13]. Photoreceptors fall into 7 families depending on the structure of the light-absorbing chromophore: rhodopsins, xanthopsins, cryptochromes, light–oxygen–voltage (LOV)-domain–containing phototropins, blue-light sensing using flavin (BLUF)-domain proteins, cobalamin (Vitamin B12)-binding proteins, and phytochromes [12–15]. In bacteria, the most abundant photoreceptors are phytochromes [16], typically possessing an amino-terminal chromophore-binding domain and a carboxy-terminal histidine kinase (HK) domain. Bacteriophytochromes assemble with the chromophore called biliverdin (BV) [17]. Surprisingly, very few bacteria encode a partner response regulator (RR) in close proximity to the gene specifying the bacteriophytochrome [18], leaving the systems mostly undefined.
In the human pathogen P. aeruginosa, LuxR-type quorum-sensing receptors, which function as transcriptional activators when they are bound to their partner autoinducers, are required for virulence and biofilm formation [19,20]. In this study, we examine the mechanism by which the P. aeruginosa LuxR-type quorum-sensing receptor called RhlR represses biofilm formation [21,22]. A genetic screen reveals that RhlR activates the expression of the algB–kinB operon (Fig 1). KinB and AlgB are a TCS sensor HK and partner RR, respectively [23,24]. KinB is required for acute infection in zebrafish embryos, in which it controls production of virulence factors such as pyocyanin and promotes motility [24,25]. In the context of our screen, we find that phospho-AlgB (AlgB-P) is a repressor of biofilm formation and KinB is a phosphatase that dephosphorylates and thereby inactivates AlgB. Using genetic suppressor analysis and in vitro phosphorylation assays, we discover that BphP is the HK that phosphorylates and activates AlgB, enabling AlgB to repress biofilm formation and genes encoding virulence factors (Fig 1). BphP is a far-red-light–sensing bacteriophytochrome [26], and indeed, we demonstrate that P. aeruginosa biofilm formation and virulence gene expression are repressed by far-red light. Phylogenetic analyses show that the KinB–AlgB–BphP module is conserved in all pseudomonads, and moreover, AlgB is present in the majority of bacteria that possess BphP orthologs. This final finding suggests that the BphP–AlgB interaction is widespread. As proof of this notion, we show that P. aeruginosa BphP can phosphorylate AlgB orthologs from α-, β-, and γ-Proteobacteria. Thus, we propose that AlgB functions as an integrator that conveys multiple environmental cues, including those specifying population density and the presence or absence of light into the regulation of collective behaviors (Fig 1). We further predict that AlgB functions as the partner RR for BphP in all bacteria that possess BphP as an orphan HK. The downstream signal transduction components and the outputs of photosensory cascades are not known in the majority of nonphotosynthetic bacteria that possess them, making their physiological roles difficult to discern. This study provides the entire cascade—light as the input, BphP as the detector, AlgB as the signal transducer, and biofilm formation and virulence factor production as the outputs—enabling insight into light-driven control of bacterial behaviors.
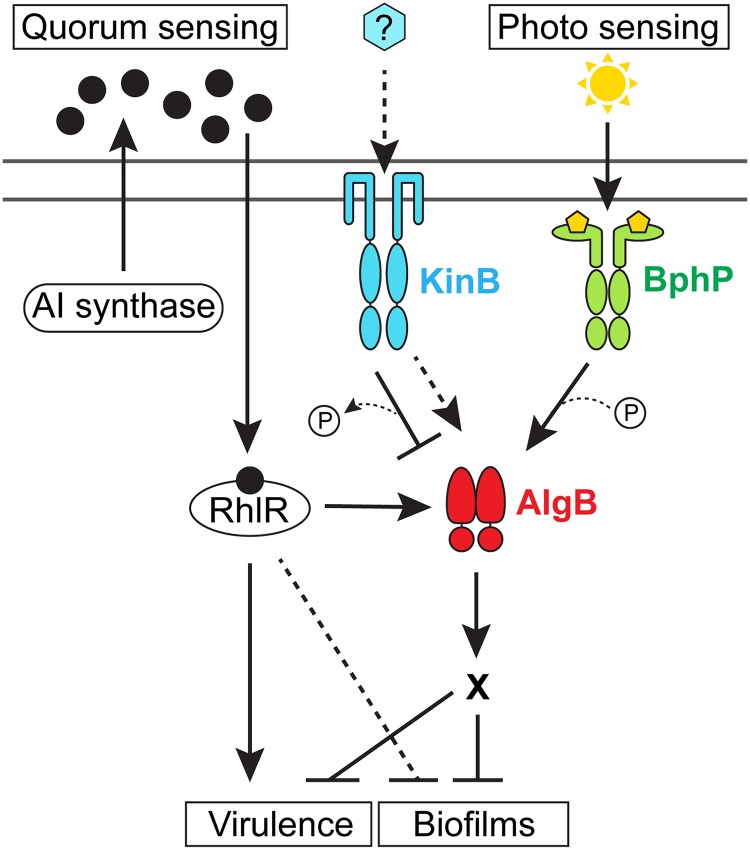
Fig 1. Model for P. aeruginosa integration of quorum-sensing and photosensing information into the control of virulence and biofilm development.
The RhlR quorum-sensing receptor binds its partner AI produced by either the RhlI or PqsE autoinducer synthase (black circles) at high cell density [22]. The RhlR–AI complex represses biofilm formation and virulence gene expression by activating transcription of the algB–kinB operon encoding the KinB HK and the AlgB RR, the latter an indirect repressor of biofilm formation. KinB antagonizes AlgB by dephosphorylation. The stimulus (blue hexagon) for KinB is unknown. In the presence of this putative stimulus, the hypothesis is that KinB functions as a kinase for AlgB (dashed arrow). Photosensing stimulates the BphP HK to autophosphorylate and subsequently transfer the phosphoryl group to AlgB to activate AlgB. AlgB-P activates transcription of genes required for repression of group behaviors such as biofilm formation and virulence. A “P” in a circle denotes addition or removal of a phosphate moiety (dotted line). X denotes that the genes functioning downstream of AlgB in the process are not known. The RhlR–AI complex directly activates virulence gene expression and also represses biofilm formation by additional unknown mechanisms (dashed T-bar). AI, autoinducer; HK, histidine kinase; RR, response regulator.
Results
KinB activates and AlgB represses RhlR-dependent group behaviors
We recently discovered that the P. aeruginosa quorum-sensing receptor RhlR represses biofilm formation [21,22]. Specifically, on Congo red agar biofilm medium, wild-type (WT) P. aeruginosa UCBPP-PA14 (hereafter called PA14) exhibits a rugose-center/smooth-periphery colony biofilm phenotype, while the ΔrhlR mutant forms a larger hyper-rugose colony biofilm (Fig 2A). To determine the mechanism by which RhlR impedes colony biofilm formation, we randomly mutagenized the ΔrhlR strain using the transposon Tn5 IS50L derivative ISlacZ/hah [27] and screened for colonies exhibiting either a WT or a smooth colony biofilm phenotype. Our rationale was that inactivation of a gene encoding a component that functions downstream of RhlR in biofilm formation would sever the connection between RhlR and repression of colony biofilm formation. We screened 5,000 transposon insertion mutants. Strains harboring insertions located in genes encoding hypothetical proteins, proteins involved in twitching motility, and proteins required for Pel polysaccharide synthesis all produced smooth colony biofilms (S1 Table). Most of these genes were already known to play roles in P. aeruginosa biofilm formation [28]. Here, we focus on one transposon insertion mutant that exhibited a smooth colony biofilm phenotype that mapped to the gene PA14_72390 encoding the KinB transmembrane HK (Fig 2A) [23,24,25]. kinB is located immediately downstream of algB in a dicistron that is conserved in all sequenced pseudomonads (S1A Fig; www.pseudomonas.com). To verify that KinB plays a role in colony biofilm formation, we generated an in-frame markerless deletion of kinB in the chromosomes of the WT and ΔrhlR strains. Both the ΔkinB single and ΔrhlR ΔkinB double mutants failed to form colony biofilms and instead exhibited smooth colony phenotypes (Fig 2A). Introduction of a plasmid carrying the kinB gene conferred a hyper-rugose phenotype to the WT and restored colony biofilm formation to the ΔkinB and ΔrhlR ΔkinB mutants (Fig 2A). By contrast, introduction of a plasmid carrying rhlR did not alter the smooth biofilm phenotype of the ΔrhlR ΔkinB double mutant (Fig 2A). We conclude that in P. aeruginosa, KinB is essential for biofilm formation, KinB is an activator of biofilm formation, and KinB functions downstream of RhlR in the biofilm formation process.
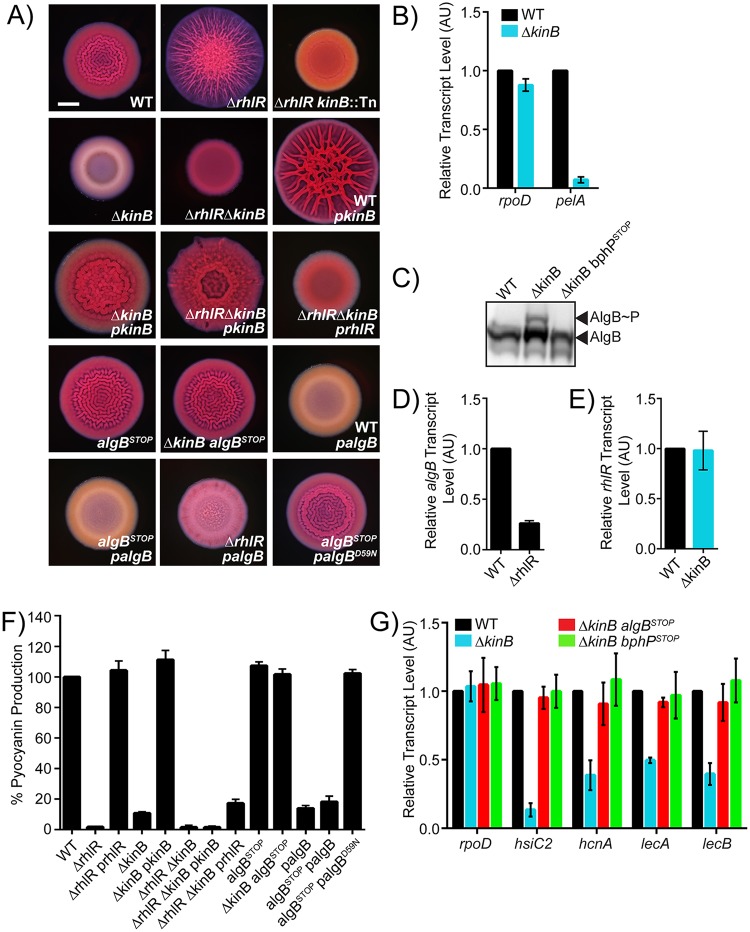
Fig 2. RhlR represses biofilm formation via KinB.
(A) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 72 h of growth. kinB::Tn refers to a mutant identified in a genetic screen harboring a transposon insertion in kinB. pkinB, prhlR, and palgB refer to kinB, rhlR, and algB, respectively, under the Plac promoter on pUCP18. Scale bar for all images is 2 mm. (B) Relative expression levels of rpoD and pelA measured by qRT-PCR in WT and ΔkinB mutant colony biofilms grown as in (A). The pelABCDEFG operon encodes proteins required for the production of the Pel polysaccharide, which is essential for the formation of pellicles, SSA biofilms, and colony biofilms [29]. (C) Phos-tag western blot analysis of the indicated strains probed for 3xFLAG-AlgB. (D) Relative algB transcript levels measured by qRT-PCR in WT PA14 and the ΔrhlR mutant grown planktonically to OD600 = 1.0. (E) Relative rhlR transcript levels measured by qRT-PCR in WT PA14 and the ΔkinB mutant grown planktonically to OD600 = 1.0. (F) Pyocyanin production (OD695) was measured in WT PA14 and the designated mutants. Production from the WT was set to 100%. (G) Relative expression of rpoD, hsiC2, hcnA, lecA, and lecB measured by qRT-PCR in WT PA14 and the designated mutants grown planktonically to OD600 = 1.0. rpoD is used as the control for comparison. For panels B, D, E, and G, data were normalized to 16S RNA levels, and the WT levels were set to 1.0. For data in panels B, D, E, F, and G, error bars represent SEM for 3 biological replicates. Data for panels B, D, E, F, and G can be found in supplemental file S1 Data. The original western blot showing the data for panel C is available in supplemental file S2 Data. AU, arbitrary unit; OD, optical density; qRT-PCR, quantitative Reverse Transcriptase-Polymerase Chain Reaction; SEM, standard error of the mean; SSA, solid-surface–associated; WT, wild type.
PA14 requires Pel, the primary biofilm matrix exopolysaccharide for biofilm formation [29] (Note: PA14 does not produce the Psl exopolysaccharide, and alginate does not contribute significantly to the PA14 biofilm matrix, unlike in P. aeruginosa PAO1 [30]). To examine whether the mechanism by which KinB alters biofilm formation is by changing Pel production, we performed quantitative RT-PCR analyses on WT and ΔkinB colony biofilms, probing for the expression of the housekeeping gene rpoD as a control and the Pel biosynthetic gene pelA (Fig 2B). Expression of rpoD did not change between the WT and the ΔkinB mutant, while transcription of pelA was approximately 14-fold lower in the ΔkinB strain than in the WT. We conclude that KinB activates Pel production, which is why KinB is required for PA14 biofilm formation.
KinB is a transmembrane HK that undergoes autophosphorylation and then transfers the phosphate to its partner RR AlgB [23]. To determine whether AlgB functions downstream of KinB to control colony biofilm formation, we introduced a stop codon in place of the codon specifying residue 10 in the algB gene to obtain an algBSTOP mutant. We note that the algBSTOP mutation does not alter kinB expression (S1B Fig). The algBSTOP mutant had a colony biofilm phenotype indistinguishable from the WT (Fig 2A). However, introduction of the algBSTOP mutation into the ΔkinB strain restored colony biofilm formation (Fig 2A). Furthermore, overexpression of algB repressed colony biofilm formation in the WT, as evidenced by the resulting smooth colony biofilm phenotype (Fig 2A). Overexpression of algB also repressed colony biofilm formation in the algBSTOP and ΔrhlR strains (Fig 2A). Thus, these data suggest that KinB activates while AlgB represses biofilm development.
AlgB has an amino-terminal domain containing the site of phosphorylation (residue D59), a central ATP-binding domain, and a carboxy-terminal helix-turn-helix motif for binding DNA (S1C and S2 Figs) [23,31]. AlgB is a member of the NtrC subfamily of RRs, and it possesses the hallmark GAFTGA motif required for interaction with RpoN (σ54) [32]. Typically, NtrC-type RRs act as transcriptional activators when they are phosphorylated [33]. To investigate whether phosphorylation of AlgB is required for the repression of colony biofilm formation, we substituted the aspartate at residue 59 with an asparagine residue to preclude phosphorylation [32]. We overexpressed the algBD59N allele in the PA14 strain carrying the algBSTOP mutation. Unlike WT AlgB, AlgBD59N failed to repress colony biofilm formation (Fig 2A, S3A, S3B and S4A Figs). To ensure the validity of this result, we generated amino-terminal 3xFLAG-tagged algB and algBD59N fusions and expressed them from a plasmid in the algBSTOP mutant. A western blot showed that both proteins are produced at similar levels (S3A Fig). We also introduced the algBD59N mutation into the algB gene in the chromosome of WT PA14 and the ΔkinB mutant. Identical to the results from the plasmid-based experiment, both the algBD59N single and the ΔkinB algBD59N double mutants produced colony biofilms (S3B Fig). We conclude that the phosphorylated form of AlgB is active and is required for AlgB-mediated repression of biofilm development. We presume that AlgB-P functions indirectly as a transcriptional activator to promote the expression of a gene encoding a negative regulator of biofilm formation (Fig 1).
Our results show that AlgB functions downstream of KinB and that KinB and AlgB have opposing activities with respect to PA14 biofilm formation. In vitro, KinB possesses both kinase and phosphatase activities [25]. One mechanism by which KinB could antagonize AlgB function is by acting as a phosphatase that dephosphorylates AlgB, rendering it inactive. To test this possibility, we integrated the 3xFLAG-tagged algB allele at the native algB locus in the chromosomes of WT PA14 and the ΔkinB mutant. Colony biofilm analyses show that 3xFLAG-AlgB is functional (S3B Fig). Next, we assessed the phosphorylation status of 3xFLAG-AlgB in vivo. Fig 2C shows that AlgB-P accumulates in the ΔkinB mutant compared to in the WT. To verify these claims regarding the signal transduction mechanism, we engineered a missense mutation into KinB at a conserved proline (P390) that is required for phosphatase activity [25]. Specifically, we generated both kinB-SNAP and kinBP390S-SNAP fusions and introduced these alleles at the native kinB locus on the chromosome of PA14. Carboxy-terminal tagging of KinB with SNAP does not interfere with its function because the strain carrying kinB-SNAP forms biofilms that are indistinguishable from those of WT PA14 (S3C and S3D Fig). The KinBP390S-SNAP protein is also produced and stable (S3C Fig); however, identical to the ΔkinB mutant, the strain carrying kinBP390S-SNAP fails to form colony biofilms (S3D Fig). These data demonstrate that KinB acts as a phosphatase to inhibit AlgB function in vivo. We therefore hypothesize—and we come back to this point below—that some other HK must phosphorylate AlgB to activate it and enable it to function as a repressor of biofilm development.
Our data show that the KinB–AlgB TCS functions downstream of RhlR to repress biofilm formation. An obvious mechanism by which RhlR could influence KinB–AlgB activity is by activating transcription of the algB–kinB operon. Indeed, RT-PCR shows that algB transcript levels are approximately 4-fold higher in the WT than in the ΔrhlR mutant (Fig 2D). Thus, RhlR activates expression of the algB–kinB operon. By contrast, deletion of kinB has no effect on rhlR transcript levels (Fig 2E), confirming their epistatic relationship.
KinB has been reported to be required for pyocyanin production [25]. Pyocyanin is a RhlR-dependent virulence factor [21,34]. Our findings of a regulatory connection between KinB and RhlR suggest that KinB and RhlR could jointly regulate pyocyanin production. To test this idea, we measured pyocyanin production in planktonic cultures of WT, ΔrhlR, ΔkinB, and ΔrhlR ΔkinB strains. Similar to what has been reported previously, deletion of rhlR and/or kinB abolished pyocyanin production (Fig 2F). Overexpression of rhlR in the ΔrhlR strain and overproduction of kinB in the ΔkinB strain restored pyocyanin production, demonstrating that our expression constructs are functional (Fig 2F). By contrast, overexpression of either rhlR or kinB in the ΔrhlR ΔkinB double mutant failed to rescue pyocyanin production (Fig 2F). Thus, RhlR and KinB are both required activators of pyocyanin production in PA14. Consistent with AlgB functioning as the RR for KinB, inactivation of AlgB (i.e., algBSTOP) in the ΔkinB background restored WT levels of pyocyanin production, while overexpression of algB in the WT and the algBSTOP mutant reduced pyocyanin levels (Fig 2F). Lastly, unlike overexpression of algB, overexpression of algBD59N failed to repress pyocyanin production, suggesting that phosphorylation of AlgB is required for AlgB activity (Fig 2F).
To further explore the role of the KinB–AlgB TCS on RhlR-driven gene expression, we quantified expression of 4 other RhlR-activated genes [21]—hsiC2 (type-VI secretion), hcnA (hydrogen cyanide synthase), lecA (galactose-binding lectin), and lecB (fucose-binding lectin), all encoding virulence factors—in the WT and the ΔkinB mutant. Expression of all 4 genes was lower in the ΔkinB mutant than in the WT (Fig 2G). Introduction of the algBSTOP mutation into the ΔkinB mutant restored expression of all 4 virulence genes to WT levels (Fig 2G). Thus, both RhlR and KinB activate virulence gene expression in P. aeruginosa. Moreover, we conclude that AlgB is epistatic to KinB for all the phenotypes tested here, and thus, KinB and AlgB function in the same pathway, albeit in opposing manners, to control biofilm formation and virulence factor production.
The bacteriophytochrome BphP is the HK required to activate AlgB to mediate repression of quorum-sensing–controlled behaviors
We have invoked the existence of a putative HK to activate AlgB via phosphorylation. To identify this component, we used genetic suppressor analysis, reasoning that mutants with defects in the upstream component required to phosphorylate AlgB would render AlgB nonfunctional. We further reasoned that such suppressor mutants would transform the ΔkinB smooth colony biofilm phenotype back to the rugose phenotype because in such mutants, AlgB could not act as a repressor of biofilm formation. We isolated 12 spontaneously arising rugose mutants from ΔkinB smooth colony biofilms and analyzed them by whole-genome sequencing (Fig 3A). Eight suppressors contained deletions or missense mutations in the algB gene, while the remaining 4 suppressors harbored mutations in the bphP gene (Fig 3B, S2 Table). bphP is located in a dicistron immediately downstream of bphO (Fig 3B, S1 Fig) [35]. We discuss bphO below; here we focus on bphP. Exactly analogous to mutation of algB, mutation of bphP was epistatic to kinB for all of the phenotypes tested. Specifically, engineering a stop codon in place of the codon specifying residue 50 in the bphP gene showed no effect in WT PA14, but it restored colony biofilm formation, pyocyanin production, and virulence gene expression to the ΔkinB mutant (Figs 3C–3D and 2G). Consistent with BphP being required to activate AlgB, unlike in the WT, in the bphPSTOP mutant, overexpression of algB failed to repress colony biofilm formation and pyocyanin production (Fig 3C and 3D). Furthermore, while overexpression of bphP in the WT reduced pyocyanin production to the levels of the ΔkinB mutant, overexpression of bphP had no effect in the algBSTOP mutant (Fig 3D). There is a severe growth defect associated with the overexpression of bphP. For this reason, in Fig 3D, rather than using plasmid pUCP18, we expressed bphP from the low-copy–number plasmid pBBR1-MCS5. Unfortunately, the presence of the empty pBBR-MCS5 plasmid in WT and mutant PA14 strains abrogates colony biofilm formation, so we could not perform the companion colony biofilm assay to test overexpression of bphP. Nonetheless, we can conclude from Fig 3C and 3D that BphP is necessary and sufficient to activate AlgB.
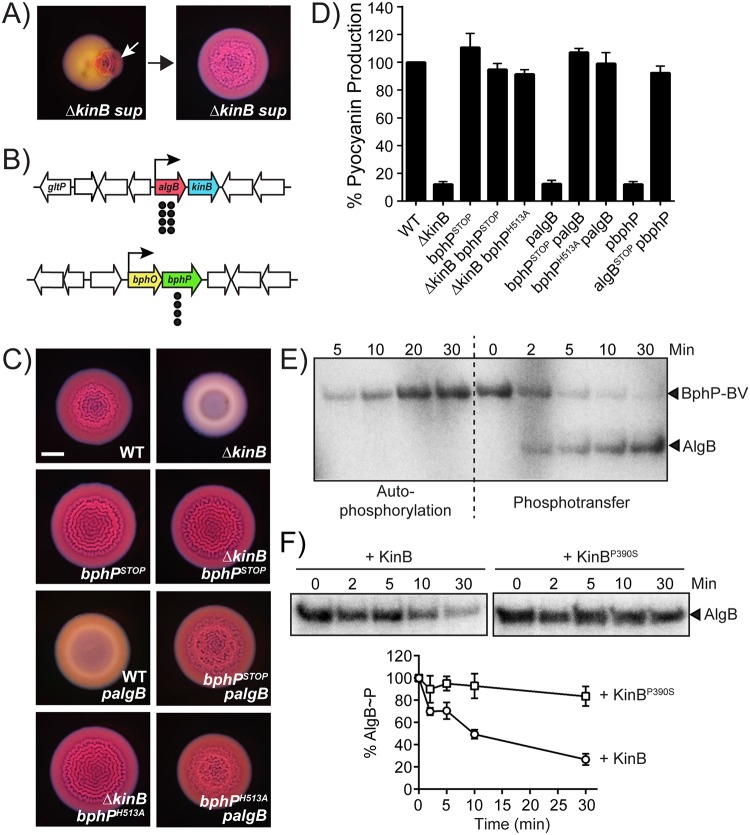
Fig 3. BphP and AlgB are a two-component HK–RR pair.
(A) Shown is a representative isolation of a suppressor mutation of the ΔkinB smooth colony biofilm phenotype. The white arrow in the left panel indicates a region of rugose sectoring in the ΔkinB smooth colony biofilm that is diagnostic of the emergence of a suppressor mutation. The right panel shows the colony biofilm phenotype of a mutant following isolation. (B) Chromosomal arrangements of the algB (red), kinB (blue), bphO (yellow), and bphP (green) genes. Large white arrows represent open reading frames (lengths not to scale), black bent arrows indicate promoters, and black circles indicate the locations of suppressor mutations. (C) Colony biofilm phenotypes of WT PA14 and the designated mutants on Congo red agar medium after 72 h of growth. palgB refers to algB under the Plac promoter on the pUCP18 plasmid. Scale bar is 2 mm for all images. (D) Pyocyanin production (OD695) was measured in WT PA14 and the designated mutants. pbphP refers to bphP under the Plac promoter on the pBBR-MCS5 plasmid. Error bars represent SEM for 3 biological replicates. (E) Autophosphorylation of BphP–BV and phosphotransfer to AlgB. (Left) Autophosphorylation of BphP–BV was carried out for 30 min and samples were removed at the indicated times for electrophoresis. (Right) An equimolar amount of AlgB was added to phospho-BphP-BV for 30 min and samples were removed at the indicated times for electrophoresis. (F) Dephosphorylation of AlgB-P by KinB or KinBP390S. Phosphotransfer to AlgB from phospho-BphP-BV was carried out for 30 min. ATP was removed from the reaction, and either KinB or KinBP390S was added. Samples were removed at the indicated times for electrophoresis. The top panel shows representative images of gels. The bottom graph shows percent AlgB-P levels at each time point with SEM for 3 independent replicates. Band intensities for AlgB-P when KinB was added (circles) and when KinBP390S was added (squares) were normalized to the levels at time 0. To assess the quality of protein preparations used in panels E and F, see S4B Fig. Data for the graphs in panels D and F can be found in supplemental file S1 Data. The original autoradiographs with the data for panels E and F are available in supplemental file S2 Data. BV, biliverdin; OD, optical density; SEM, standard error of the mean; WT, wild type.
BphP is a bacteriophytochrome that assembles with its chromophore BV, which is produced by the heme oxygenase BphO (Fig 3B, S1A and S1D Fig) to generate a photosensing HK that is activated by light [35]. P. aeruginosa BphP contains the HDLRNPL motif, which often contains the histidine residue that undergoes autophosphorylation in transmembrane HKs [36]. In P. aeruginosa BphP, this histidine is residue 513 (S1D Fig). To determine whether BphP kinase activity is required for AlgB activation, we generated the bphPH513A mutation, fused it to 3xFLAG, and introduced it onto the chromosome of the ΔkinB mutant. The BphPH513A-3xFLAG protein is produced and stable (S3E Fig), and identically to the bphPSTOP allele, the bphPH513A mutation restored colony biofilm formation and pyocyanin production to the ΔkinB mutant (Fig 3C and 3D). Moreover, overexpression of algB in the bphPH513A mutant failed to repress colony biofilm formation and pyocyanin production (Fig 3C and 3D). These results show that BphP H513 and AlgB D59 are required for signal transmission, and the signal is presumably phosphorylation.
To assess phosphorelay between BphP and AlgB, we used our 3xFLAG-AlgB in vivo construct. In addition to introducing it into the chromosome of WT PA14, we engineered it onto the chromosome of the bphPSTOP mutant. Consistent with BphP being the kinase for AlgB, Fig 2C shows that the ΔkinB bphPSTOP mutant lacks the band corresponding to AlgB-P. These data suggest that BphP transfers phosphate to AlgB. To verify this finding, we performed in vitro phosphotransfer assays. We purified recombinant BphP and formed a complex with it and commercially available BV to obtain the BphP–BV chromoprotein. Upon incubation with radiolabeled ATP under ambient light, BphP–BV underwent autophosphorylation (Fig 3E). BphP–BV readily transferred radiolabeled phosphate to purified AlgB but not to AlgBD59N (Fig 3E, S4A Fig). Purified apo-BphP and purified apo-BphPH513A complexed with BV both failed to autophosphorylate and thus could not transfer phosphate to AlgB (S4A Fig). Together, these data show that BphP–BV phosphorylates and thereby activates AlgB.
Our data suggest that KinB dephosphorylates AlgB, while BphP phosphorylates AlgB. To directly test this hypothesis, we reconstituted the BphP–AlgB–KinB phosphorelay in vitro. We purified the recombinant KinB and KinBP390S proteins and added them separately, at equimolar concentration, to AlgB-P prephosphorylated by BphP–BV. Fig 3F shows that over time, KinB dephosphorylates AlgB while AlgB-P levels remain unchanged in the presence of KinBP390S. As control experiments, we added either KinB or KinBP390S to AlgB in the presence of ATP but in the absence of BphP–BV. Both KinB and KinBP390S underwent autophosphorylation and transferred phosphate to AlgB in vitro, but only WT KinB acted as a phosphatase to dephosphorylate AlgB (S5A–S5D Fig). Although our findings show that KinB is a dual kinase/phosphatase, under our in vivo conditions, only KinB phosphatase activity was detected. Perhaps KinB can function as a kinase for AlgB when its stimulus is present (Fig 1). Identifying the natural signal that drives the KinB kinase activity is the subject of our future work. We conclude that BphP–AlgB–KinB forms a regulatory module in which the RR AlgB is activated by the kinase activity of the HK BphP and inhibited by the phosphatase activity of the HK KinB.
BphP-mediated photosensing represses P. aeruginosa quorum-sensing–controlled behaviors
The P. aeruginosa BphP bacteriophytochrome has been studied in vitro, and its kinase activity is reported to be activated by light [35]. To explore whether BphP photosensing has any effect on AlgB-controlled group behaviors in vivo, we compared colony biofilm formation by WT, ΔkinB, ΔkinB bphPSTOP, and ΔkinB algBSTOP PA14 strains in the dark and under different light conditions. We note that all of the colony biofilm experiments in the previous sections were performed under ambient light. First, we consider WT PA14 and the ΔkinB mutant in the no light condition. Fig 4A shows that in the dark, both strains formed colony biofilms that were indistinguishable from one another. We interpret these results to mean that in the absence of light, the BphP kinase is inactive in both WT PA14 and the ΔkinB mutant, and AlgB is not phosphorylated, so it too is inactive. Thus, no repression of colony biofilm formation occurs (Fig 1). Now, we address the results under ambient light. WT PA14 formed colony biofilms, but the ΔkinB strain did not (Fig 4A). Our interpretation is that in the WT, ambient light activates the BphP kinase, and phosphotransfer to AlgB occurs. However, the opposing KinB phosphatase activity strips the phosphate from AlgB, thereby eliminating AlgB-dependent repression of colony biofilm formation. Thus, WT PA14 forms colony biofilms under ambient light. In the case of the ΔkinB mutant, since there is no KinB phosphatase present, ambient light is sufficient to drive BphP-mediated phosphorylation of AlgB, AlgB-P accumulates, and it represses colony biofilm formation. Based on these results, we infer that the presence or absence of light can alter group behaviors such as colony biofilm formation in P. aeruginosa.
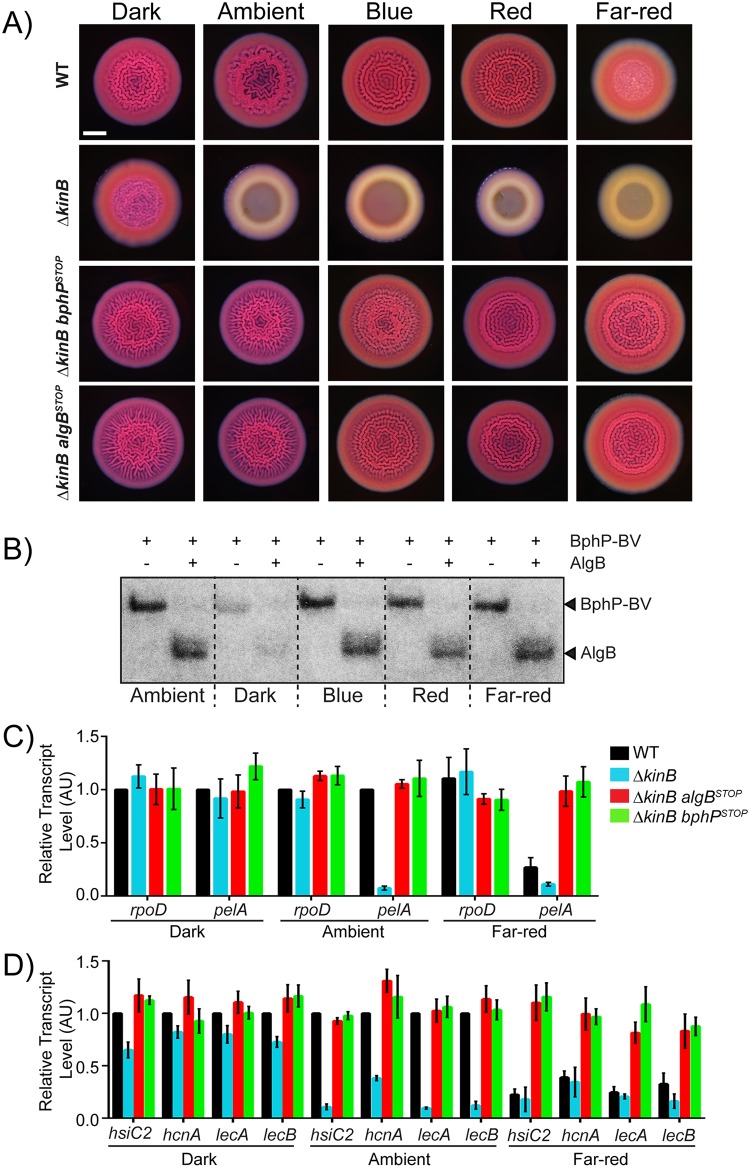
Fig 4. Photosensing, via the BphP–AlgB phosphorelay, represses group behaviors in P. aeruginosa.
(A) Colony biofilm phenotypes are shown for WT PA14 and the designated mutants on Congo red agar medium after 72 h of growth under the indicated light conditions. Scale bar is 2 mm for all images. (B) Autophosphorylation of the BphP–BV complex was carried out for 30 min (left lane in each pair), followed by addition of AlgB (right lane in each pair) for an additional 30 min under the indicated light conditions. (C) Relative expression of rpoD and pelA as measured by qRT-PCR in WT PA14 and the designated mutant strains grown as colony biofilms as in (A) in darkness, ambient light, and far-red light. (D) Relative expression of hsiC2, hcnA, lecA, and lecB measured by qRT-PCR in WT PA14 and the designated mutants grown as colony biofilms as in (A) and light conditions as in (B). For panels B and C, data were normalized to 16S RNA levels, and the WT levels were set to 1.0. Error bars represent SEM for 3 biological replicates. Data for panels C and D can be found in supplemental file S1 Data. The original autoradiograph with the data for panel B is available in supplemental file S2 Data. AU, arbitrary unit; BV, biliverdin; qRT-PCR, quantitative Reverse Transcriptase-Polymerase Chain Reaction; SEM, standard error of the mean; WT, wild type.
Ambient light is a composite of different wavelengths of light. The PA14 BphP bacteriophytochrome is reported to be a far-red-light–sensing HK in vitro [26]. We wondered whether a particular wavelength of light could maximally activate the BphP kinase activity in vivo, and if so, perhaps, under that condition, the BphP kinase activity could override the KinB phosphatase, enabling light to repress biofilm formation in WT PA14. To test this notion, we exposed PA14 strains to blue, red, and far-red light and monitored colony biofilm formation. In contrast to WT PA14, the ΔkinB mutant failed to form colony biofilms under blue, red, and far-red light, suggesting that BphP is a promiscuous photoreceptor that is activated by far-red light and also by blue and red light (Fig 4A). Indeed, BphP phosphorylates AlgB in vitro under ambient (as above), blue, red, and far-red light, but it does not do so in the absence of light (Fig 4B). Importantly, when WT PA14 was exposed to far-red light, it failed to make colony biofilms, but rather, exhibited the smooth colony phenotype, identical to the ΔkinB mutant (Fig 4A). We conclude that far-red light is the preferred wavelength for BphP and is sufficient to repress biofilm formation in WT P. aeruginosa. Finally, we show that light-mediated repression of biofilm formation requires functional BphP and AlgB because the bphPSTOP and algBSTOP single mutants and the ΔkinB bphPSTOP and ΔkinB algBSTOP double mutants did not repress colony biofilm formation under the conditions tested (Fig 4A, S6A Fig). To extend our results concerning light-mediated repression of biofiolm formation, we assessed the effects of light on solid-surface–associated (SSA) biofilm formation [29] by the WT, ΔkinB, ΔkinB algBSTOP, and ΔkinB bphPSTOP PA14 strains (S6B Fig). The results exactly mirror our findings for colony biofilms: far-red light repressed SSA biofilm formation in the WT. The ΔkinB mutant exhibited a severe decrease in SSA biofilm formation under ambient and far-red light, but it produced SSA biofilms in the dark. Lastly, both the ΔkinB algBSTOP and ΔkinB bphPSTOP mutants formed SSA biofilms under all conditions.
One mechanism by which light could suppress biofilm formation via BphP–AlgB is by decreasing the production of the Pel exopolysaccharide. To quantify the effect of light on pel expression and, in turn, on biofilm formation, we measured pelA transcript levels using quantitative RT-PCR analyses of WT, ΔkinB, ΔkinB algBSTOP, and ΔkinB bphPSTOP colony biofilms in darkness and under ambient and far-red light (Fig 4C). We used rpoD transcription as the control. Expression of rpoD did not change under any condition tested. Regarding pelA, analogous to what occurred for colony biofilm formation, there was no significant difference in pelA expression between the WT and the ΔkinB strains in the dark, whereas transcription of pelA was approximately 14-fold lower in the ΔkinB strain than in the WT under ambient light. Repression of pelA expression depended on functional BphP and AlgB because the ΔkinB bphPSTOP and ΔkinB algBSTOP mutants transcribed pelA at high levels under both conditions. We conclude that dephosphorylation of AlgB does not occur in the ΔkinB mutant under ambient light. In this condition, BphP phosphorylates AlgB, and AlgB-P represses colony biofilm formation via down-regulation of pelA expression. Lastly, in the WT, pelA transcript levels were approximately 4-fold lower under far-red light than in darkness. Therefore, far-red light is the strongest activator of BphP such that under far-red light, but not ambient light, the kinase activity of BphP overrides the phosphatase activity of KinB in the WT to drive AlgB-P accumulation, repression of pelA expression, and consequently, repression of biofilm formation.
In Fig 2, we showed that BphP is required for AlgB-dependent repression of virulence gene expression. Our results in Fig 4 suggest that light, by controlling BphP-dependent phosphorylation of AlgB, could control virulence in P. aeruginosa. To explore this idea further, we quantified the expression of the virulence-associated genes hsiC2, hcnA, lecA, and lecB in colony biofilms of WT PA14 and in the ΔkinB, ΔkinB algBSTOP, and ΔkinB bphPSTOP strains under darkness, ambient light, and far-red light. The results mimic those for biofilm formation and pelA transcription. Only in the absence of the opposing KinB phosphatase activity is ambient light sufficient to activate BphP, whereas far-red-light–driven BphP kinase activity can override the KinB phosphatase activity, allowing accumulation of AlgB-P to levels that repress virulence gene expression. Again, light-mediated repression of virulence genes requires functional BphP and AlgB (Fig 4D). We conclude that BphP-dependent photosensing represses virulence gene expression in P. aeruginosa.
Light possesses both color (wavelength) and intensity properties. Above, we demonstrated that BphP can detect blue, red, and far-red light. To explore the possibility that P. aeruginosa BphP is also capable of detecting light intensity, we varied the intensity of far-red light since it has the most dramatic effect on PA14 phenotypes. We used repression of colony biofilm formation as the readout. Colony biofilm formation decreased with increasing intensity of far-red light in the WT and ΔkinB mutant but remained unaltered in the bphPSTOP mutant (Fig 5A). The highest intensity of light we tested (bottom-most panel in Fig 5A) is similar to that present in natural sunlight (5.5 W/m2 in a 5-nm window around 730 nm; ASTM G173-03 Reference Solar Spectra, www.astm.org). At this intensity, WT colony biofilm formation was maximally repressed, showing that BphP kinase dominates over KinB phosphatase. The ΔkinB mutant generates suppressor flares under this condition, suggesting that one role of the KinB phosphatase is to keep the BphP kinase activity in check. To verify that far-red light specifically altered colony biofilm behavior without affecting general physiology, we quantified rpoD and pelA transcript levels in the WT and bphPSTOP mutant colony biofilm samples grown under the different light intensities (Fig 5B). Expression of rpoD did not change under any condition tested, while transcription of pelA decreased progressively in the WT with increasing intensity of far-red light. At the highest intensity of far-red light tested, expression of pelA was approximately 12-fold lower than that in the bphPSTOP mutant that cannot convey the light cue internally to AlgB. These results demonstrate that P. aeruginosa colony biofilm formation can be modulated simply by tuning the intensity of far-red light in which the strain is grown.
Fig 5. Far-red light intensity controls biofilm formation.
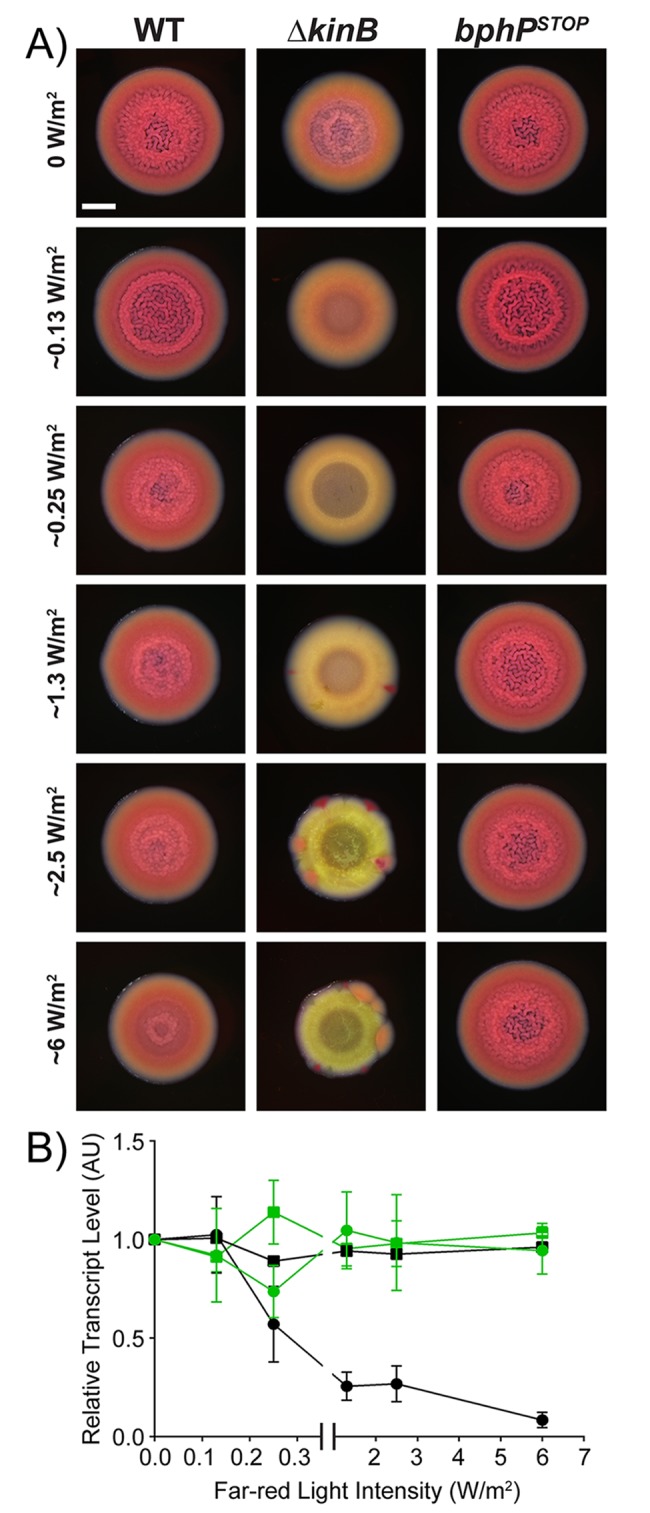
(A) Colony biofilm phenotypes are shown for WT PA14 and the designated mutants on Congo red agar medium after 72 h of growth under the indicated far-red light intensities. Scale bar is 2 mm for all images. (B) Relative expression of rpoD (squares) and pelA (circles) measured by qRT-PCR in WT PA14 (black) and in the bphPSTOP mutant (green) grown as colony biofilms as in (A). Data were normalized to 16S RNA levels, and the WT levels at 0 mW/m2 far-red light were set to 1.0. Error bars represent SEM for 3 biological replicates. Data for panel B can be found in supplemental file S1 Data. AU, arbitrary unit; qRT-PCR, quantitative Reverse Transcriptase-Polymerase Chain Reaction; SEM, standard error of the mean; WT, wild type.
The BphP–AlgB phosphorelay is conserved in diverse bacteria
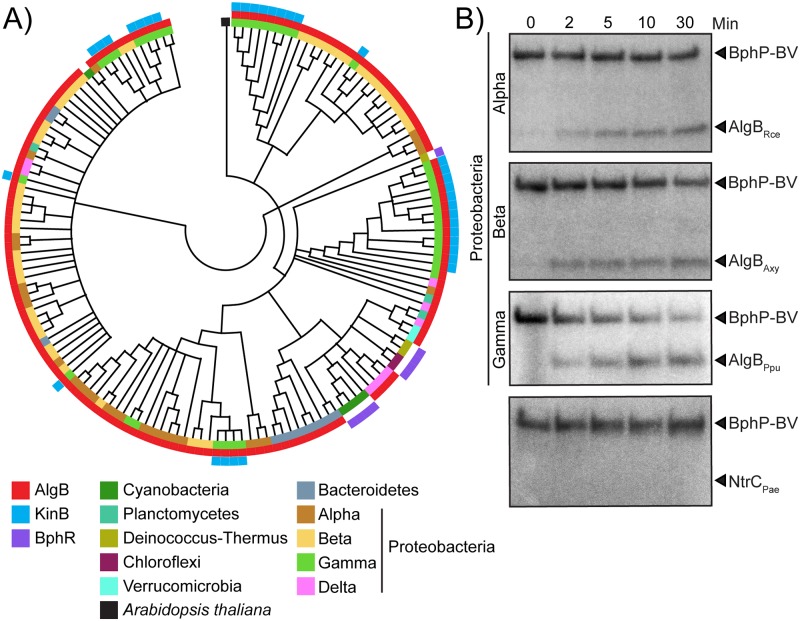
BphP bacteriophytochromes are a major class of photoreceptors widely distributed in nonphotosynthetic bacteria [16]. These BphP HKs either lack a partner RR or, when they are co-transcribed with a partner RR gene, the physiological output of the circuit is unknown. Thus, the downstream signaling components and consequences of photosensing in nonphotosynthetic bacteria are not understood. Our discovery of AlgB as the partner RR for the orphan light-sensing BphP HK in a nonphotosynthetic bacterium, coupled with our demonstration of the biofilm and virulence outputs of photosensing, puts us in a position to test the generality of our findings. As a first step, we generated a phylogenetic tree containing 150 BphP orthologs that are the closest homologs to P. aeruginosa BphP (Fig 6A, S7 Fig). The majority of these BphP orthologs are present in nonphotosynthetic bacteria from diverse phyla. The pseudomonads fall into discrete clusters, hinting at acquisition of BphP via horizontal gene transfer. With respect to AlgB and KinB, we find that while KinB is present only in the pseudomonads, Acinetobacter baumannii, and Enterobacter cloacae (Fig 6A, S1 and S7 Figs), AlgB is present in approximately 93% of the bacterial species in our BphP-based phylogenetic tree (Fig 6A, S7 Fig). We note that in all of the bacteria that do not encode AlgB, for example, Deinococcus spp., BphR is the partner RR for BphP (Fig 6A, S1, S2 and S7 Figs; [35]). None of these bphP-encoding bacteria possess both BphR and AlgB. Therefore, the pattern that emerges is that BphB is widely distributed in nonphotosynthetic bacteria and the partner RR is either AlgB or BphR.
Fig 6. The BphP–AlgB phosphotransfer relay is conserved in diverse bacteria.
(A) Maximum likelihood based phylogenetic tree for BphP showing the 150 closest orthologs to P. aeruginosa BphP. Co-occurrences of AlgB and KinB are depicted in red and blue, respectively. BphR is shown in purple. The other colors indicate bacterial phyla. The black square indicates Arabidopsis thaliana as the root of the tree. (B) In vitro phosphorylation of AlgB orthologs from the α-Proteobacterium Rce, the β-Proteobacterium Axy, and the γ-Proteobacterium Ppu by P. aeruginosa BphP–BV that had been autophosphorylated for 30 min. The bottom panel shows that phosphotransfer from P. aeruginosa phospho-BphP-BV to P. aeruginosa NtrC does not occur. To assess the quality of the protein preparations used in panel B, see S4B Fig. The original autoradiographs with the data for panel B are available in supplemental file S2 Data. Axy, Achromobacter xylosocidans; BV, biliverdin; Ppu, P. putida; Rce, Rhodospirillum centenum.
To test whether BphP can interact with and phosphorylate AlgB in bacteria other than P. aeruginosa, we purified AlgB orthologs from diverse Proteobacteria: Rhodospirillum centenum (α), Achromobacter xylosoxidans (β), and P. putida (γ) (S2 Fig). We incubated these AlgB proteins with an equimolar concentration of autophosphorylated P. aeruginosa BphP–BV. Phosphotransfer from BphP–BV to the AlgB orthologs occurred in all cases, albeit to varying degrees (Fig 6B). To eliminate the possibility that phospho-BphP-BV is a promiscuous kinase for NtrC family RRs, we purified NtrC from PA14 and incubated it with autophosphorylated BphP–BV. Phospho-BphP-BV failed to phosphorylate NtrC (Fig 6B). We conclude that BphP is the specific HK for AlgB, and AlgB appears to have a conserved function in photosensory signal transduction in diverse bacteria.
Discussion
Our study reveals that the nonphotosynthetic pathogenic bacterium P. aeruginosa detects and responds to light to repress group behaviors, including virulence factor production and biofilm formation. The photoreceptor BphP functions as a light-activated HK that phosphorylates the AlgB RR. AlgB-P represses group behaviors but is antagonized by its partner HK KinB. Specifically, KinB dephosphorylates AlgB, and thus, KinB functions as an activator of group behaviors. Our work shows that AlgB functions as a hub protein that has 3 inputs—quorum sensing via RhlR, photosensing via BphP, and an unknown signal via KinB. We note that bphP expression has been reported to be controlled by LasR-LasI quorum sensing and the stationary-phase alternative sigma factor RpoS [37], suggesting that other sensory cues can be integrated into the AlgB–KinB–BphP circuit. While quorum sensing activates algB expression, photosensing activates AlgB function, and thus, the presence or absence of light can override the quorum-sensing input from RhlR. We reason that at high cell density, RhlR will drive AlgB production. However, if there is no light, BphP will not phosphorylate and activate AlgB. In turn, AlgB will not repress group behaviors. To our knowledge, the BphP–AlgB photosensory signal transduction cascade represents the first example of light-mediated control of group behaviors in the global pathogen P. aeruginosa.
Light is a ubiquitous source of energy that drives the anabolic process of photosynthesis in photosynthetic organisms. However, the wide distribution of photoreceptors in all domains of life suggests roles for photosensing in behaviors far beyond photosynthesis. Plants, for example, use light cues to regulate activities such as seed germination [38], stomatal opening [39], and defenses against microbes [40–42]. Furthermore, plant vascular systems can function as bundles of optical fibers to efficiently transmit light, particularly far-red light, that is not absorbed by plant pigments, allowing opportunities for photosensing in roots and possibly in the rhizosphere [43]. Many of the bphP-encoding bacteria from the phylogenetic tree in Fig 6A that also possess AlgB are members of the rhizosphere microbiome [44]. Perhaps these nonphotosynthetic bacteria exploit light cues to colonize and/or to fine-tune their mutualistic or pathogenic interactions with their plant hosts as well as adjust their physiology in the rhizosphere. While we do not know the evolutionary forces that drove P. aeruginosa to become a photosensing bacterium, we speculate below on possible advantages P. aeruginosa could accrue by detecting light in the environment and in the human host.
Light provides spatial and temporal information to higher organisms. Does light serve a similar purpose in bacteria? Recent studies have reported that BphP plays a role in multiple stages of infection by the foliar plant pathogens Xanthomonas campestris pv. campestris and P. syringae pv. syringae [45,46], in each case via an unknown but putative downstream RR. Based on our phylogenetic analysis, we speculate that AlgB fulfils this role. We further speculate that P. aeruginosa, which is a plant pathogen [47], responds to light cues via the BphP–AlgB TCS to appropriately modulate its biofilm and virulence programs, particularly to inhibit virulence during daylight, enabling avoidance of plant defense mechanisms. For instance, during the day, chlorophyll in leaves removes most of the red wavelength from sunlight but little of the far-red spectrum [48]. Thus, far-red light is readily available, and based on our work here, could signal to P. aeruginosa to tamp down virulence factor production and biofilm formation, allowing it to optimize those programs in line with host conditions because shaded leaves are more susceptible to infection than leaves exposed to direct light [49]. In the same vein, our work suggests the possibility that inside the mammalian host, the lack of light would drive P. aeruginosa to form biofilms and produce virulence factors because, in the absence of light, BphP–AlgB-mediated repression of biofilms and virulence would not occur.
In addition to providing spatial and temporal information, light can also reveal other key parameters to which bacteria respond. Detection of blue light via LOV- and BLUF-domain proteins modulates general stress responses in some nonphotosynthetic bacteria such as Bacillus subtilis and Caulobacter crescentus [50,51]. Light, through the LOV-HK of the mammalian pathogen Brucella abortus, is crucial for virulence in a macrophage infection model, although the components connecting light to the virulence response remain undefined. It is also proposed that B. abortus uses light as an indicator of whether it is inside or outside of its animal host [52]. The P. aeruginosa genome does not encode LOV or BLUF domain proteins [11]. P. aeruginosa possesses only one identifiable photoreceptor, BphP. Nonetheless, we showed that P. aeruginosa is capable of detecting blue, red, and far-red light via BphP (Fig 4A and 4B). Perhaps an advantage of BphP promiscuity is that it enables detection of higher energy and therefore phototoxic blue light, in addition to the lower-energy but highly penetrative far-red light. Such a scenario would endow P. aeruginosa with the plasticity to diversify its physiological outputs in response to particular wavelengths of light without the necessity of a distinct photoreceptor for each wavelength.
Bacteriophytochromes are thought to function as reversible photoswitches that convert between red-absorbing and far-red–absorbing states [17]. The P. aeruginosa BphP kinase has been reported to be activated by far-red light in vitro [26]. Interestingly, however, the published BV absorption spectrum possesses peaks at blue, red, and far-red light [53], hinting at promiscuity in light detection. Based on our in vitro and in vivo data, we suggest that P. aeruginosa BphP is active as a kinase in both the red and far-red states as well as under blue light. We are currently investigating the mechanism by which the BphP–BV holocomplex is activated by different wavelengths of light.
An advantage P. aeruginosa could gain by sensing light on or within a mammalian host would be the ability to tune into the host circadian rhythm and its associated responses. Circadian clocks influence various aspects of health and disease such as sleep/wake cycles and metabolism [54,55]. Disruption of circadian rhythms are associated with fitness costs [55]. In mammals, both innate and adaptive immune responses are controlled by the circadian clock such that the immune system is primed to combat pathogens during the host active phase while immune functions undergo regeneration and repair during the resting phase of the daily cycle. Parasites such as Plasmodium spp., which cause malaria, synchronize their replication cycle with host circadian rhythms for optimized infection and dissemination [56]. Likewise, viruses such as herpes and influenza A have been shown to exploit the mammalian circadian clock for their own gain, i.e., to successfully avoid host immune responses, enabling maximal replication [57,58]. Perhaps P. aeruginosa uses light as a signal that reveals when the host immune response is at peak function, and accordingly, at that time, P. aeruginosa represses biofilm formation and virulence factor expression as a mechanism that enhances evasion of host defenses. If so, a human host infected with P. aeruginosa during the night would be colonized to higher levels compared to a host acquiring an infection during the day. Synchronizing infectivity with light/dark cues to enable optimal infection could be a common feature of nonphotosynthetic photoreceptor-harboring pathogens.
P. aeruginosa is a priority pathogen on the Centers for Disease Control and Prevention (CDC) ESKAPE pathogen list (a set of bacteria including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp. that are designated as multidrug-resistant pathogens requiring new antimicrobials for treatment) and a critical pathogen on the World Health Organization (WHO) priority list [59,60]. Our phylogenetic analysis suggests that the KinB–AlgB–BphP module is conserved in the genomes of A. baumannii and E. cloacae, perhaps acquired from P. aeruginosa via horizontal gene transfer because the AlgB primary sequence is nearly identical between the 3 species. We speculate that beyond P. aeruginosa, BphP–AlgB-dependent photosensing also affects the physiology and possibly the virulence of these ESKAPE pathogens. Collectively, the results from this study provide unanticipated insight into P. aeruginosa physiology and a surprising possibility for therapeutic intervention—shining light on a deadly and actively studied pathogen, P. aeruginosa, to attenuate virulence and biofilm formation. One can imagine such a strategy could be deployed in external infections such as burns, which are highly susceptible to P. aeruginosa and could, moreover, be subjected to precise light regimes.
Materials and methods
Bacterial strains and growth conditions
All strains and plasmids used in this study are listed in S3 and S4 Tables, respectively. P. aeruginosa PA14 and mutants were grown at 37 °C in lysogeny broth (LB) (10 g tryptone, 5 g yeast extract, 5 g NaCl per L), in 1% Tryptone broth (TB) (10 g tryptone per L), or on LB plates fortified with 1.5% Bacto agar. When appropriate, antimicrobials were included at the following concentrations: 400 μg/mL carbenicillin, 30 μg/mL gentamycin, and 100 μg/mL irgasan. Escherichia coli was grown at 37 °C in LB or on LB plates fortified with 1.5% Bacto agar and the following concentrations of antimicrobials as appropriate: 15 μg/mL gentamycin, 50 μg/mL kanamycin, and 100 μg/mL ampicillin. IPTG (Sigma-Aldrich, St. Louis, MO, USA) was added to the medium at the indicated concentrations when appropriate.
Mutant strain and plasmid construction
Strains and plasmids were constructed as described previously [22]. Briefly, to construct markerless in-frame chromosomal deletions and substitutions in PA14, DNA fragments flanking the gene of interest were amplified, assembled by the Gibson method [61], and cloned into suicide vector pEXG2 [62]. The resulting plasmids were used to transform E. coli SM10λpir and subsequently mobilized into PA14 strains via biparental mating. Exconjugants were selected on LB containing gentamycin and irgasan, followed by recovery of deletion mutants on LB medium containing 5% sucrose. Candidate mutants were confirmed by PCR and Sanger sequencing. Transposon insertions in the PA14 chromosome were generated by mating the PA14 ΔrhlR parent strain with E. coli SM10λpir harboring pIT2 (ISlacZ/hah) [27]. Insertion mutants were selected on LB agar containing 60 μg/mL tetracycline, and 100 μg/mL irgasan was included in the agar to counterselect against the E. coli donor. Transposon insertion locations were determined by arbitrary PCR and sequencing as described previously [27].
Protein production constructs were generated by amplifying the algB, kinB, and bphP coding regions and cloning them in pET28b or pET21b expression vectors (Addgene, Watertown, MA, USA) to obtain pET28b-His6-AlgB, pET21b-KinB-His6, and pET21b-BphP-His6, respectively. To generate the AlgBD59N, KinBP390S, and BphPH513A variants, the corresponding mutations were engineered onto the pET28b-His6-AlgB, pET21b-KinB-His6, and pET21b-BphP-His6 plasmids, respectively, via Gibson assembly. AlgB orthologs from R. centenum and A. xylosoxidans were amplified from gene fragments obtained from Integrated DNA Technologies (Coralville, IA, USA), and that from P. putida was amplified from the P. putida KT2440 genome. All of the gene orthologs were cloned into the pET28b plasmid.
Pyocyanin assay
PA14 strains were grown overnight in LB liquid medium at 37 °C with shaking at 250 rotations per minute (rpm). The cells were pelleted by centrifugation at 21,130 × g for 2 min, and the clarified supernatants were passed through 0.22-μm filters (Millipore, Burlington, MA, USA) into clear plastic cuvettes. The OD695 of each sample was measured on a spectrophotometer (Beckman Coulter DV 730; Brea, CA, USA) and normalized to the culture cell density, which was determined by OD600.
Colony biofilm assay
The procedure for establishing colony biofilms has been described [22]. Briefly, 1 μL of overnight cultures of PA14 strains were spotted onto 60 × 15 mm Petri plates containing 10 mL 1% TB medium fortified with 40 mg/L Congo red and 20 mg/L Coomassie brilliant blue dyes and solidified with 1% agar. Colony biofilms were grown at 25 °C for 72 h in an incubator (Benchmark Scientific, Sayreville, NJ, USA), and images were acquired using a Leica stereomicroscope M125 (Wetzlar, Germany) mounted with a Leica MC170 HD camera at 7.78× zoom.
For colony biofilms exposed to specific wavelengths of light, the following light-emitting diodes (LEDs) were used: blue, 430 nm (Diffused RGB LED, #159; Adafruit, New York, NY, USA); red, 630 nm (Diffused RGB LED, #159; Adafruit); and far-red, 730 nm (LST1-01G01-FRD1-00; Opulent Americas, Raleigh, NC, USA). Ambient light exposure refers to biofilms grown under laboratory light conditions. For the colony biofilms shown in Fig 4A, light intensity was normalized by photon flux, and the following intensities were used: blue (0.7 W/m2), red (1 W/m2), and far-red (1.1 W/m2). Light intensity was calibrated using a laser power meter (Ophir, North Logan, UT, USA) in a 5-nm window at the appropriate wavelength. Colony biofilm samples were grown in custom laser-cut acrylic chambers. Each chamber housed a single LED light source and an individual Petri plate containing 4 technical replicates. Samples exposed to darkness were housed in the same chambers as the light-exposed samples, but with no current applied to the LEDs.
SSA biofilm formation assay
The procedure for establishing SSA biofilms has been described [29]. Briefly, overnight cultures of PA14 strains were diluted to a final OD600 of 0.01 in 1% TB. These samples were used to make standing 3-mL cultures in 18 × 150 mm borosilicate glass tubes that were incubated (Benchmark Scientific) at 25 °C for 72 h under the light conditions described above for the colony biofilm assay. The cultures were poured out of the tubes, and the tubes were washed vigorously with tap water. The remaining biofilms were stained by the addition of 5 mL 0.1% crystal violet solution into each tube. After 30 min, tubes were washed twice with tap water and left to dry overnight. Subsequently, 5 mL 33% glacial acetic acid solution was added to each tube. The crystal violet stain was quantified at OD550 using a spectrophotometer (Beckman Coulter DV 730).
qRT-PCR
WT PA14 and mutant strains were harvested from planktonic cultures (OD600 = 1.0) or from colony biofilms grown for 72 h. RNA was purified using the Zymo Research kit, and the preparations were subsequently treated with DNAse (TURBO DNA-free; Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and quantified using PerfeCTa SYBR Green FastMix Low ROX (Quanta Biosciences, Beverly, MA, USA).
Protein purification
His6-AlgB. The pET28b-His6-AlgB protein production vector was transformed into E. coli BL21 (DE3) and the culture grown to approximately 0.8 OD600 in 1 L of LB supplemented with 50 μg/mL kanamycin at 37 °C with shaking at 220 rpm. Protein production was induced by the addition of 1 mM IPTG, followed by incubation of the culture for another 3 h at 25 °C with shaking. The cells were pelleted by centrifugation at 16,100 × g for 20 min and resuspended in AlgB-lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 0.1% Triton X-100, 10 mM imidazole, and protease inhibitor cocktail [Roche, Basel, Switzerland]). The preparation was frozen at −80 °C overnight. The frozen cell pellet was thawed on ice, and the cells were lysed by sonication (1-s pulses for 15 s). The sample was subjected to centrifugation at 32,000 × g for 30 min at 4 °C. The resulting clarified supernatant was combined with Ni-NTA resin (Novagen) and incubated for 3 h at 4 °C. The bead/lysate mixture was loaded onto a 1-cm separation column (Bio-Rad, Hercules, CA, USA), the resin was allowed to pack, and then it was washed with AlgB-wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 0.1% Triton X-100, 30 mM imidazole, and protease inhibitor cocktail [Roche]). Resin-bound His6-AlgB was eluted twice with 1 mL AlgB-wash buffer containing 250 mM imidazole. Fractions were analyzed by SDS-PAGE, and the gel was stained with Coomassie brilliant blue to assess His6-AlgB purity. Purified protein was dialyzed in AlgB-storage buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1 mM MgCl2, 1 mM DTT, 5% glycerol, and 0.1% Triton X-100) and stored at −80 °C.
BphP-His6. The pET21b-BphP-His6 protein production vector was transformed into E. coli BL21-CodonPlus (DE3)-RIPL (Agilent Technologies, Santa Clara, CA, USA). BphP-His6 was purified as described for His6-AlgB with the following changes in buffers: BphP-lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1% Triton X-100, 0.1% β-mercaptoethanol, 10 mM imidazole, and protease inhibitor cocktail [Roche]), BphP-wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1% Triton X-100, 0.1% β-mercaptoethanol, 30 mM imidazole, and protease inhibitor cocktail [Roche]), and BphP-storage buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 1% Triton X-100, 0.1% β-mercaptoethanol, 5% glycerol).
KinB-His6. The pET21b-KinB-His6 protein production vector was transformed into E. coli BL21 (DE3). KinB-His6 protein was purified exactly as described above for BphP-His6.
Phosphorylation assays
Autophosphorylation assays were performed with purified WT BphP and the BphPH513A variant or with KinB and the KinBP390S variant. 100 μM BphP or BphPH513A was incubated under ambient light with 10-fold molar excess of BV (Sigma-Aldrich) for 1 h prior to the assay to form the light-activated BphP–BV stocks. Reactions were carried out in phosphorylation buffer (50 mM Tris [pH 8.0], 100 mM KCl, 5 mM MgCl2, and 10% (v/v) glycerol) and were initiated with the addition of 100 μM ATP and 2 μCi [γ-32P]-ATP (PerkinElmer, Waltham, MA, USA). Reactions were incubated at room temperature and terminated by the addition of SDS-PAGE loading buffer. Reaction products were separated using SDS-PAGE. Gels were dried at 80 °C on filter paper under vacuum, exposed to a phosphoscreen overnight, and subsequently analyzed using a Typhoon 9400 scanner (GE Healthcare, Chicago, IL, USA) and ImageQuant software. For phosphotransfer to AlgB, an equimolar concentration of AlgB was added to the phosphorylated BphP–BV or phosphorylated KinB proteins (all proteins assayed at 5 μM). Reactions were incubated at room temperature for the indicated times and terminated by the addition of SDS-PAGE loading buffer. For the BphP–AlgB phosphorelay shown in S4B Fig, BphP–BV was incubated under specific light wavelengths using the same devices as described above for colony biofilm assay.
Dephosphorylation of AlgB-P: 10 μM AlgB was phosphorylated for 30 min in reactions containing 10 μM BphP–BV, 100 μM ATP, and 2 μCi [γ-32P]-ATP in phosphorylation buffer. Subsequently, the reactions containing AlgB-P were applied to gel filtration spin columns (Probe Quant G-50, GE Healthcare) to remove ATP. Dephosphorylation reactions were initiated by adding 10 μM KinB or KinBP390S. Aliquots were taken at the indicated times and analyzed as described above.
Phos-tag SDS-PAGE and western blotting
WT PA14 and mutant strains were harvested from planktonic cultures (OD600 = 1.0). Cells were resuspended in 100 μl of ice-cold BugBuster reagent (Novagen) containing EDTA-free Protease Inhibitor Cocktail (Roche), followed by end-over-end rotation on a nutator at room temperature for 30 min. Cell debris was removed by centrifugation (4 °C at 10,000 rpm for 1 min). 50 μL 4× SDS-PAGE loading buffer (Thermo Fisher Scientific) containing 15% β-mercaptoethanol was combined with 50 μL of the sample supernatant. Ten μL samples were loaded onto a 12.5% SuperSepTM Phos-tag gel (Wako Pure Chemical Industries, Osaka, Japan). Samples were subjected to electrophoresis at 4 °C for 3 h. Gels were incubated for 20 min on a shaking platform in 1× transfer buffer containing 1 mM EDTA and re-equilibrated for 20 min in 1× transfer buffer lacking EDTA. Proteins were transferred to nitrocellulose membranes, blocked with 5% skim milk in TBS at room temperature for 1 h, and incubated with primary anti-FLAG antibody (Sigma-Aldrich) at 1:5,000 dilution in 5% skim milk in TBS overnight at 4 °C on a rocking platform. Membranes were washed 3 times with TBS-Tween 20 at room temperature for 10 min on a rocking platform and subsequently developed with a SuperSignal West Femto Kit (Thermo Scientific) and captured with an LAS-4000 Imager (GE Healthcare).
Whole-genome sequencing
P. aeruginosa strains were harvested from planktonic cultures (OD600 = 2.0), and DNA was purified using DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The Nextera DNA Library Prep kit (Illumina, San Diego, CA, USA) was employed with 2 ng of genomic DNA to prepare the library. Unique barcodes were added to each sample to enable multiplexing. The libraries were examined for quality using Bioanalyzer DNA High Sensitivity chips (Agilent Technologies) and quantified using a Qubit fluorometer (Invitrogen). DNA libraries from the different strains were pooled at equal molar amounts and sequenced using an Illumina MiSeq as pair-end 2 × 100 nt reads. Only the Pass-Filter (PF) reads were used for further analysis.
Whole-genome sequencing data were processed with breseq version 0.33.2 to identify mutations relative to the reference P. aeruginosa UCBPP-PA14 genome (www.pseudomonas.com; [63]). All high-confidence and putative SNPs and deletion events were confirmed by a manual examination of the read pileups with GenomeViewer IGV 2.4.8. A sample collected prior to the suppressor mutation screen was aligned against the reference genome of PA14, yielding a manually curated list of 25 differences acquired by our laboratory strain prior to the experiment (19 SNPs, 6 single-nucleotide indels). Applying these differences to PA14 using gdtools (part of the breseq package) yielded an updated reference genome against which all other samples were compared. S2 Table reports all high-confidence mutations identified in this analysis.
Phylogenetic analysis
The maximum likelihood tree for BphP orthologs was generated using MEGA-X software as described previously [64].
Supporting information
(A) The genes flanking kinB, algB, bphO, and bphP are diagrammed for the indicated genomes. The relative gene positions and orientations are accurate, but gene lengths are not to scale. (B) Relative expression of kinB measured by qRT-PCR in WT PA14 and the algBSTOP mutant grown planktonically to OD600 = 1.0. Data were normalized to 16S RNA levels, and the WT levels were set to 1.0. Error bars represent SEM for 3 biological replicates. (C) The domain architecture of the AlgB monomer is shown. Residue 59 is required for phosphorylation; the GAFTGA motif, indicated by the magenta shading, is required for interaction with σ54; and HTH refers to the helix-turn-helix DNA binding domain. Adapted from [31]. (D) Domain organization of the BphP monomer consisting of the PAS, GAF, PHY, and HK domains is shown. BV binds to the GAF domain, and residue H513 is required for autophosphorylation. Adapted from [17]. Data for panel B can be found in supplemental file S1 Data. AU, arbitrary unit; BV, biliverdin; GAF, cGMP-specific phosphodiesterases, adenylate cyclases, and FhlA; HK, histidine kinase; PAS, Per-Arnt-Sim; PHY, phytochrome; qRT-PCR, quantitative Reverse Transcriptase-Polymerase Chain Reaction; SEM, standard error of the mean.
(TIF)
Primary sequence alignment of NtrC (first line) and AlgB (second line) from Pae and AlgB orthologs (third through twelfth lines) from Pfl, Psy, Ppr, Pst, Pen, Ppu, Aba, Ecl, Axy, Rce, and BphR (thirteenth line) from Deinococcus radiodurans. Highly conserved amino acids are highlighted in black. Residue 59 is shown by the green asterisk. The GAFTGA motif required for interaction with σ54 is indicated by the magenta line. Aba, Acinetobacter baumanii; Axy, Achromobacter xylosoxidans; Ecl, E. cloacae; Pae, P. aeruginosa; Pen, P. entomophila; Pfl, P. fluorescens; Ppr, P. protegens; Ppu, P. putida; Pst, P. stutzeri; Psy, P. syringae; Rce, R. centenum.
(TIF)
(A) Western blot analysis of whole cell lysates from the indicated strains, all of which have the algBSTOP allele at the native locus in the genome and carry an empty vector or 3xFLAG-algB or 3xFLAG-algBD59N on the pBBR1-MCS5 plasmid under the Plac promoter. The same cell lysates were probed for RNAP as the loading control. (B) Colony biofilm phenotypes of WT PA14 and the designated mutants. Scale bar is 2 mm. (C) SDS-PAGE analysis of whole cell lysates from the indicated strains. The gel was stained for SNAP using SNAP-Cell 647-SiR fluorescent substrate (New England Biolabs, Ipswich, MA, USA). Lysozyme was added as the loading control. (D) Colony biofilm phenotypes of the kinB-SNAP and kinBP390S-SNAP strains. Scale bar is 2 mm. (E) Western blot analysis of whole cell lysates from the indicated strains. The same cell lysates were probed for RNAP as the loading control. The original western blots showing the data for panels A, C, and E are available in supplemental file S2 Data. RNAP, RNA Polymerase; WT, wild type.
(TIF)
(A) Autophosphorylation of the BphP–BV complex was carried out for 30 min (leftmost lane), followed by addition of AlgB (second lane) or AlgBD59N (third lane) for an additional 30 min. The kinase-defective BphPH513A-BV complex was incubated with radiolabeled ATP for 30 min (fourth lane), followed by addition of AlgB (fifth lane) for an additional 30 min. The apo-BphP protein was incubated with radiolabeled ATP for 30 min (sixth lane). (B) SDS-PAGE gel stained with Coomassie brilliant blue showing the indicated purified proteins. Ten μL of a 20 μM stock of each protein was loaded. The original autoradiograph showing the data for panel A is available in the supplemental file S2 Data. BV, biliverdin.
(TIF)
(A) Autophosphorylation of KinB was carried out for 30 min, and samples were removed at the indicated times. (B) An equimolar amount of AlgB was added to KinB that had been autophosphorylated for 30 min as in (A). Samples were taken at the indicated times. (C and D) As in A and B, respectively, but for the phosphatase-deficient protein KinBP390S. The original autoradiographs with the data for this figure are available in supplemental file S2 Data.
(TIF)
(A) Colony biofilm phenotypes are shown for WT PA14 and the designated mutants on Congo red agar medium after 72 h of growth under the indicated light conditions. Scale bar is 2 mm for all images. (B) SSA biofilm phenotypes assessed by crystal violet staining are shown for WT PA14 and the designated mutants after 72 h of growth under the indicated light conditions. Data can be found in supplemental file S1 Data. SSA, solid-surface–associated; WT, wild type.
(TIF)
Enlarged maximum-likelihood–based phylogenetic tree for BphP from Fig 6A showing the 150 closest orthologs to P. aeruginosa BphP. Co-occurrences of AlgB and KinB are depicted using red and blue dots, respectively. The presence of BphR is shown by purple dots. The colored squares indicate the corresponding bacterial phyla. The black square indicates A. thaliana as the root of the tree.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(PDF)
Acknowledgments
We thank Wei Wang and the Genomics Core Facility at Princeton University for help with whole-genome sequencing. We thank Ned Wingreen, Anne-Florence Bitbol, Joseph E. Sanfillipo, and all members of the Bassler group for thoughtful discussions.
Abbreviations
- AU
arbitrary unit
- BLUF
blue-light sensing using flavin
- BV
biliverdin
- ESKAPE
Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.
- HK
histidine kinase
- LB
lysogeny broth
- LED
light-emitting diode
- LOV
light–oxygen–voltage
- PF
Pass-Filter
- qRT-PCR
quantitative Reverse Transcriptase-Polymerase Chain Reaction
- rpm
rotations per minute
- RR
response regulator
- SEM
standard error of the mean
- SSA
solid-surface–associated
- TB
Tryptone broth
- TCS
two-component system
- WT
wild type
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Howard Hughes Medical Institute, NIH Grant 5R37GM065859, and National Science foundation Grant MCB-1713731 to BLB, and NIH Grant 1K99GM129424-01 to SM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Flemming H, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8: 623–33. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: An emergent form of bacterial life. Nat Rev Microbiol. 2016;14: 563–575. 10.1038/nrmicro.2016.94 [DOI] [PubMed] [Google Scholar]
- 3.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15: 740–755. 10.1038/nrmicro.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahlstrom KM, O’Toole GA. A Symphony of Cyclases: Specificity in Diguanylate Cyclase Signaling. Annu Rev Microbiol. 2017;71: 179–195. 10.1146/annurev-micro-090816-093325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giacalone D, Smith TJ, Collins AJ, Sondermann H, Koziol LJ, O’Toole GA. Ligand-mediated biofilm formation via enhanced physical interaction between a diguanylate cyclase and its receptor. MBio. 2018;9: e01254–18. 10.1128/mBio.01254-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50: 101–114. 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17: 371–382. 10.1038/s41579-019-0186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14: 576–88. 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110: 303–314. 10.1016/s0092-8674(02)00829-2 [DOI] [PubMed] [Google Scholar]
- 10.Rutherford ST, Bassler BL. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb Perspect Med. 2012;2: a012427 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Der Horst MA, Key J, Hellingwerf KJ. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol. 2007;15: 554–562. 10.1016/j.tim.2007.09.009 [DOI] [PubMed] [Google Scholar]
- 12.Kottke T, Xie A, Larsen DS, Hoff WD. Photoreceptors Take Charge: Emerging Principles for Light Sensing. Annu Rev Biophys. 2018;47: 291–313. [DOI] [PubMed] [Google Scholar]
- 13.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha V V. Natural Photoreceptors as a Source of Fluorescent Proteins, Biosensors, and Optogenetic Tools. Annu Rev Biochem. 2015;84: 519–50. 10.1146/annurev-biochem-060614-034411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Z, Yamamoto H, Bauer CE. Cobalamin’s (Vitamin B12) Surprising Function as a Photoreceptor. Trends Biochem Sci. 2016;41(8):647–650. 10.1016/j.tibs.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto H, Fang M, Dragnea V, Bauer CE. Differing isoforms of the cobalamin binding photoreceptor AerR oppositely regulate photosystem expression. Elife. 2018;7: e39028 10.7554/eLife.39028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomelsky M, Hoff WD. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 2011;19: 441–448. 10.1016/j.tim.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 17.Gourinchas G, Etzl S, Winkler A. Bacteriophytochromes–from informative model systems of phytochrome function to powerful tools in cell biology. Curr Opin Struct Biol. 2019;57: 72–83. 10.1016/j.sbi.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beattie GA, Hatfield BM, Dong H, McGrane RS. Seeing the Light: The Roles of Red- and Blue-Light Sensing in Plant Microbes. Annu Rev Phytopathol. 2018;56: 41–66. 10.1146/annurev-phyto-080417-045931 [DOI] [PubMed] [Google Scholar]
- 19.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science. 1998;280: 295–298. 10.1126/science.280.5361.295 [DOI] [PubMed] [Google Scholar]
- 20.Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000;2: 1721–1731. 10.1016/s1286-4579(00)01327-7 [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee S, Moustafa D, Smith CD, Goldberg JB, Bassler BL. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017;13(7): e1006504 10.1371/journal.ppat.1006504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S, Moustafa DA, Stergioula V, Smith CD, Goldberg JB. The PqsE and RhlR proteins are an autoinducer synthase–receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2018;115: E9411–E9418. 10.1073/pnas.1814023115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma S, Wozniak DJ, Ohman DE. Identification of the histidine protein kinase KinB in Pseudomonas aeruginosa and its phosphorylation of the alginate regulator AlgB. J Biol. Chem. 1997;272: 17952–17960. 10.1074/jbc.272.29.17952 [DOI] [PubMed] [Google Scholar]
- 24.Chand NS, Lee JSW, Clatworthy AE, Golas AJ, Smith RS, Hung DT. The sensor kinase KinB regulates virulence in acute Pseudomonas aeruginosa infection. J Bacteriol. 2011;193: 2989–2999. 10.1128/JB.01546-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chand NS, Clatworthy AE, Hung DT. The two-component sensor KinB acts as a phosphatase to regulate Pseudomonas aeruginosa Virulence. J Bacteriol. 2012;194: 6537–6547. 10.1128/JB.01168-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasler R, Moises T, Frankenberg-Dinkel N. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 2005;272: 1927–1936. 10.1111/j.1742-4658.2005.04623.x [DOI] [PubMed] [Google Scholar]
- 27.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100: 14339–14344. 10.1073/pnas.2036282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. Regulation of biofilm formation in Pseudomonas and Burkholderia species.Environ Microbiol. 2014;16(7):1961–81. 10.1111/1462-2920.12448 [DOI] [PubMed] [Google Scholar]
- 29.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51: 675–690. 10.1046/j.1365-2958.2003.03877.x [DOI] [PubMed] [Google Scholar]
- 30.Wozniak D, Wyckoff T, Starkey M, Keyser R, Azadi P, O’Toole G, et al. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2003;100: 7907–12. 10.1073/pnas.1231792100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma S, Selvaraj U, Ohman DE, Quarless R, Hassett DJ, Wozniak DJ. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1998;180: 956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wozniak DJ, Ohman DE. Pseudomonas aeruginosa AlgB, a Two-Component Response Regulator of the NtrC Family, Is Required for algD Transcription. J Bacteriol. 1991;173: 1406–1413. 10.1128/jb.173.4.1406-1413.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bush M, Dixon R. The Role of Bacterial Enhancer Binding Proteins as Specialized Activators of σ54-Dependent Transcription. Microbiol Mol Biol Rev. 2012;76: 497–529. 10.1128/MMBR.00006-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brint JM, Ohman DE. Synthesis of Multiple Exoproducts in Pseudomonas aeruginosa Is under the Control of RhlR-RhlI, Another Set of Regulators in Strain PAO1 with Homology to the Autoinducer-Responsive LuxR-LuxI Family. J Bacteriol. 1995;177: 7155–7163. 10.1128/jb.177.24.7155-7163.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature. 2001;414: 776–779. 10.1038/414776a [DOI] [PubMed] [Google Scholar]
- 36.Bhate MP, Molnar KS, Goulian M, Degrado WF. Signal Transduction in Histidine Kinases: Insights from New Structures. Struct Des. 2015;23: 981–994. 10.1016/j.str.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barkovits K, Schubert B, Heine S, Scheer M, Frankenberg-Dinkel N. Function of the bacteriophytochrome BphP in the RpoS/Las quorum-sensing network of Pseudomonas aeruginosa. Microbiology. 2011;157(Pt 6):1651–64. 10.1099/mic.0.049007-0 [DOI] [PubMed] [Google Scholar]
- 38.Mathews S. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol. 2006; 3483–3503. 10.1111/j.1365-294X.2006.03051.x [DOI] [PubMed] [Google Scholar]
- 39.Shimazaki K, Doi M, Assmann SM, Kinoshita T. Light Regulation of Stomatal Movement. Annu Rev Plant Biol. 2007;58: 219–47. 10.1146/annurev.arplant.57.032905.105434 [DOI] [PubMed] [Google Scholar]
- 40.Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. Defence Responses of Arabidopsis thaliana to Infection by Pseudomonas syringae Are Regulated by the Circadian Clock. PLoS ONE. 2011;6(10): e26968 10.1371/journal.pone.0026968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neukermans J, Li S, Aro E, Noctor G, Kangasja S. Photosynthesis, photorespiration, and light signalling in defence responses. J Exp Bot. 2012; 63:1619–1636. 10.1093/jxb/err402 [DOI] [PubMed] [Google Scholar]
- 42.Roden LC, Ingle RA. Lights, Rhythms, Infection: The Role of Light and the Circadian Clock in Determining the Outcome of Plant–Pathogen Interactions. Plant Cell. 2009;21: 2546–2552. 10.1105/tpc.109.069922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HJ, Park CM, Oh KH, Shin K, Choi HK, Hyeon T, et al. Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci Signal. 2016;9: ra106 10.1126/scisignal.aaf6530 [DOI] [PubMed] [Google Scholar]
- 44.Duran P, Thiergart T, Garrido-oter R, Agler M, Kemen E, Schulze-lefert P, et al. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell. 2018; 973–983. 10.1016/j.cell.2018.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonomi HR, Toum L, Sycz G, Sieira R, Toscani AM, Gudesblat GE, et al. Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Rep. 2016;17: 1565–1577. 10.15252/embr.201541691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, McGrane RS, Beattie GA. Light Regulation of Swarming Motility in Pseudomonas syringae Integrates Signaling Pathways Mediated by a Bacteriophytochrome and a LOV Protein. MBio. 2013;4: e00334–13. 10.1128/mBio.00334-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starkey M, Rahme LG. Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat Protoc. 2009;4: 117–124. 10.1038/nprot.2008.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith H. Phytochromes and light signal perception by plants—an emerging synthesis. Nature. 2000;407: 585–591. 10.1038/35036500 [DOI] [PubMed] [Google Scholar]
- 49.de Wit M, Spoel SH, Sanchez-Perez GF, Gommers CM, Pieterse CMJ, Voesenek LACJ, et al. Perception of low red: far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 2013; 90–103. 10.1111/tpj.12203 [DOI] [PubMed] [Google Scholar]
- 50.Ávila-Pérez Marcela, Hellingwerf KJ, Kort R. Blue Light Activates the σB-Dependent Stress Response of Bacillus subtilis via YtvA. J Bacteriol. 2006;188: 6411–6414. 10.1128/JB.00716-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci. 2007;104: 18241–18246. 10.1073/pnas.0705887104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swartz TE, Tseng T, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, et al. Blue-Light–Activated Histidine Kinases: Two-Component Sensors in Bacteria. Science. 2007;317: 1090–1094. 10.1126/science.1144306 [DOI] [PubMed] [Google Scholar]
- 53.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci U S A. 2008;105(38):14715–20. 10.1073/pnas.0806718105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Curtis AM, Bellet MM, Sassone-corsi P, Neill LAJO. Review Circadian Clock Proteins and Immunity. Immunity. 2014;40: 178–186. [DOI] [PubMed] [Google Scholar]
- 55.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol. 2013;13: 190–198. 10.1038/nri3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donnell AJO, Schneider P, Mcwatters HG, Reece SE. Fitness costs of disrupting circadian rhythms in malaria parasites. Proc Biol Sci. 2011; 2429–2436. 10.1098/rspb.2010.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RS, Stangherlin A, Nagy AD, Nicoll MP, Efstathiou S, O’Neill JS, et al. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci. 2016;113: 10085–10090. 10.1073/pnas.1601895113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundar IK, Ahmad T, Yao H, Hwang J, Gerloff J, Lawrence BP, et al. Influenza A virus-dependent remodeling of pulmonary clock function in a mouse model of COPD. Sci Rep. 2015;5: 1–14. 10.1038/srep09927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice LB. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J Infect Dis. 2008;197: 1079–1081. 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- 60.Tacconelli E, Carmell Y, Harbarth S, Kahlmeter G, Kluytmans J, Mendelson M, et al. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO Press Release [Internet]. 2017 [cited 2019 May 28]. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/.
- 61.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison C a, Smith HO, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6: 343–5. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 62.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10: 1820–41. 10.1038/nprot.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44: D646–D653. 10.1093/nar/gkv1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018; 35:1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]