Abstract
Human nondisjunction errors in oocytes are the leading cause of pregnancy loss, and for pregnancies that continue to term, the leading cause of intellectual disabilities and birth defects. For the first time, we have conducted a candidate gene and genome-wide association study to identify genes associated with maternal nondisjunction of chromosome 21 as a first step to understand predisposing factors. A total of 2,186 study participants were genotyped on the HumanOmniExpressExome-8v1-2 array. These participants included 749 live birth offspring with standard trisomy 21 and 1,437 parents. Genotypes from the parents and child were then used to identify mothers with nondisjunction errors derived in the oocyte and to establish the type of error (meiosis I or meiosis II). We performed a unique set of subgroup comparisons designed to leverage our previous work suggesting that the etiologies of meiosis I and meiosis II nondisjunction differ for trisomy 21. For the candidate gene analysis, we selected genes associated with chromosome dynamics early in meiosis and genes associated with human global recombination counts. Several candidate genes showed strong associations with maternal nondisjunction of chromosome 21, demonstrating that genetic variants associated with normal variation in meiotic processes can be risk factors for nondisjunction. The genome-wide analysis also suggested several new potentially associated loci, although follow-up studies using independent samples are required.
Author summary
Approximately one of every 700 babies is born with trisomy 21—an extra copy of chromosome 21. Trisomy 21 is caused by the failure of chromosomes to segregate properly during meiosis, generally in the mother. Past studies have defined altered patterns of recombination along nondisjoined chromosomes as risk factors for human nondisjunction and model systems have clearly shown that specific genes involved recombination and other early meiotic processes play a role in the fidelity of chromosome segregation. However, no genome-wide genetic study (GWAS) has ever been conducted using maternal human nondisjunction as the disease phenotype. This study takes the first step to understand predisposing factors. We used chromosome 21 genotypes from the parents and child to identify mothers with nondisjunction errors derived in the oocyte and to establish the type of error (meiosis I or meiosis II). We then conducted a unique set of subgroup comparisons designed to leverage our previous work that shows that the etiologies of meiosis I and meiosis II nondisjunction differ for trisomy 21. Both the candidate gene study and the GWAS provide evidence that meiotic-specific structures and processes are vulnerable to genetic variants that lead to increased risk of human chromosome nondisjunction.
Introduction
Correct segregation of chromosomes during the two successive meiotic divisions is essential for the formation of haploid gametes. At least 10% of human pregnancies produce aneuploid embryos with too many or too few chromosomes, the majority of which are lost during pregnancy. If they survive to term, many have severe congenital defects and developmental and intellectual disability. Thus, meiotic nondisjunction is the leading cause of pregnancy loss and birth defects in humans and an important limiting factor in women’s reproductive life span. (reviewed in [1–4]).
In humans, meiosis in females is highly prone to chromosome segregation errors [i.e., nondisjunction or premature separation of sister chromatids (PSSC)] and these errors increase exponentially with increasing maternal age. The differences between the development of oocytes and sperm clearly influence susceptibility for meiotic nondisjunction. Most importantly, they work on different timelines. In both sexes, meiosis starts with an initial step of DNA replication and the establishment of sister chromatid cohesion, followed by synapsis and recombination between homologous chromosomes. Homologs then separate at the end of meiosis I (MI), whereas sister chromatids separate in meiosis II (MII). Spermatogenesis begins after puberty and cells entering meiosis move from one stage to the other without delay. In contrast, oogenesis begins during fetal development and is arrested in prophase I after chromosomes synapse and recombine. MI resumes in the woman’s adult life just before the ovulation; MI is completed and the first polar body is extruded. MII begins but arrests for a short period as the oocyte travels through the Fallopian tubes, and is only completed if the oocyte is fertilized. Thus, meiosis in females extends over a 10 to 50 year period; the age of the woman at conception reflects the age of the oocyte, and basically the period of arrest in MI. Given the mechanistic differences and temporal separation of maternal MI and MII, it is not surprising that associated risk factors differ for MI and MII nondisjunction errors (reviewed in [5]).
Trisomy 21 has become an important resource to understand meiotic nondisjunction in humans, as it is one of the few aneuploid conditions that survives to term. However, even for trisomy 21, involving the smallest human autosome, about 50–80% conceptions are estimated to be lost during pregnancy [6, 7]. Using chromosome 21 genetic markers to categorize the type of meiotic error among live births with trisomy 21, over 90% are derived from errors in the oocyte, of which at least 75% are estimated to be initiated in MI and about 25% in MII (e.g., [8]).
In this study, our goal was to discover genetic variants that increase the risk for maternal nondisjunction of chromosome 21 using both a candidate gene approach and a genome-wide association study. We focused on candidate genes that have been associated with chromosome dynamics early in meiosis. Accurate segregation depends on the coordinated control of sister chromatid cohesion with chromosome synapsis and the assembly of the synaptonemal complex (SC) and, within these structures, meiotic recombination [9, 10]. Below we provide a brief overview of the role of some of the important meiotic genes that mediate these processes, and that we have examined in the present study. Bolcun-Filas and Schimenti [9] have summarized the meiotic defects that are observed in the associated mutant mouse models.
In a meiotic cell, DNA is organized as an array of loops along a proteinaceous axis. The axes are composed of the meiosis-specific synaptonemal complex, in association with condensin/cohesin complexes. Several of the components of meiotic cohesin are meiosis-specific, including those encoded by SMC1β, REC8, RAD21L, and STAG3. The SC brings homologous chromosomes into close proximity and promotes recombination and chiasmata formation [11]. The mature SC is a tripartite structure, composed of two parallel axial/lateral elements that bind to each homolog and a central element, with transverse filaments joining the individual axial/lateral elements [12, 13]. SYCP2 and SYCP3 are components of the axial/lateral elements. SYCP1 is a component of the transverse filaments and components of the central element are encoded by SYCE1, SYCE2, SYCE3, and TEX12. In addition to these structural sub-units, HORMAD1 and HORMAD2 code for proteins that load onto axes of meiotic chromosomes throughout early prophase I but are removed upon synapsis, a process that depends on the presence of TRIP13 [14]. In general, HORMAD1 and HORMAD2 play a role in coordinating progression of chromosome synapsis with meiotic recombination [15].
Meiotic recombination is initiated by programmed DNA double-strand breaks (DSBs) that occur as the meiotic chromosome axes develop early in prophase I. These breaks are generated by the SPO11 protein and its interacting partners MEI1, MEI4 and REC114 (reviewed in Cole et al. [16]). The DSBs are processed to generate single-stranded DNA that is bound by strand-exchange proteins DMC1 (meiosis specific) and RAD51 (ubiquitously expressed). The single-stranded DNA then engages in homology search. Proper function of DMC1 requires interactions with several meiotic accessory proteins, one of which is MND1. MND1, complexed with HOP2, stabilizes the DMC1 filaments on the resected end of the DSBs. This complex also increases the ability of the pre-synaptic filament to capture the double-stranded DNA (reviewed in Sansam and Pezza [17]).
As homologs synapse, so-called early recombination nodules transiently associate with ZMM proteins, including DNA mismatch repair proteins MSH4 and MSH5. Subsequently, a proportion of these are converted into late recombination nodules, detected by the mismatch repair proteins MLH1 and MLH3, and representing the vast majority of crossovers [18–22]. In addition, EXO1 and BLM function in crossover regulation, and with MLH1 and MLH3, appear to play a role in the crossover pathway that is subject to crossover interference (reviewed in Manhart and Alani [23]).
In addition to these candidate genes, we chose another group of genes that have been associated with the amount of global meiotic recombination in humans. The motivation for these candidates is based on the altered recombination patterns observed along nondisjoined chromosomes, a well-established predisposing factor for maternal nondisjunction of almost all human chromosomes studied to date (reviewed in [24]). Specifically for maternal chromosome 21 nondisjunction, altered meiotic recombination patterns are associated with both MI and MII error types [25–29]. For maternal MI-derived trisomy 21, about 40–47% of MI cases are derived from oocytes with no meiotic exchange [25, 27, 30]. Among those with a single exchange, the majority of exchanges occur in the telomeric region of chromosome 21. MII errors are associated with pericentromeric exchanges [25, 27, 29, 30]. This apparent effect of an MI process–recombination–on MII nondisjunction suggests that at least a portion of so-called MII errors may have their origin in MI. In addition, there is evidence that genome-wide recombination counts in oocytes with a MI nondisjunction error of chromosome 21 are reduced compared to oocytes with normal meiosis [31, 32]. Also, previous studies indicate that oocyte-specific dysregulation of global recombination may contribute to the nondisjunction event [31]. Thus, we chose candidate genes identified in the largest GWAS study of meiotic recombination conducted on humans, a study based on 71,929 parent-offspring pairs from Iceland [33]. They found evidence for 13 variants in eight regions that were associated with genome-wide recombination counts.
For both the candidate gene and genome-wide association studies, we took a unique approach by using several different GWAS group comparisons (Table 1). These comparisons were crafted to address the likelihood that there are both distinct genetic factors influencing MI and MII nondisjunction and common factors affecting both. In addition, some of our analyses target the conflated phenotype of nondisjunction with survival to term. Study design issues are discussed in more detail below.
Table 1. Description of primary analyses and associated sample sizes.
| Analysis | Analysis groups | Sample size | Contrast able to detect: |
|---|---|---|---|
| Logistic regression | Mothers vs. fathers | 705 vs. 645 | Maternal NDJ and survival to term |
| . . . | MI mothers vs. fathers | 535 vs. 645 | MI-specific effects and survival to term |
| . . . | MII mothers vs. fathers | 157 vs. 645 | MII- specific effects and survival to term |
| . . . | MI mothers vs. MII mothers | 535 vs. 157 | MI- or MII-specific effects |
| TDT | All complete case trios | 615 trios | Survival to term |
NDJ: nondisjunction; MI: maternal MI nondisjunction error; MII: maternal MII nondisjunction error.
Methods
Study sample
Our study participants included 749 live born offspring with free (non-translocation), maternally-derived trisomy 21 (both full and mosaic trisomy 21 were included) and their available biological parents. In almost all instances, the trisomy was confirmed by karyotype, although in some it was confirmed by birth record or parent report. Recruitment occurred in the U.S. by multiple sites since 1989, when the first population-based study was initiated. Recruitment for these population-based studies used birth surveillance systems to identify infants born with Down syndrome (details are provided in Freeman et al. [8]). Later, our recruitment strategy was not population-based, but instead a convenience sample of families with Down syndrome identified through our network of assessment sites, social and website media, and parent groups. Using self-reported race/ethnicity, 72% reported as White, 4% as Hispanic descent, 2% as African/African-American or Asian descent and about 23% with other or unknown descent.
Ethics statement
Participants were recruited from several geographic areas with the collaboration of several institutions, including Arkansas (University of Arkansas for Medical Sciences, Arkansas Center for Birth Defects Research and Prevention, Arkansas Children’s Hospital, Arkansas Reproductive Health Monitoring Systems), California (California Birth Defects Monitoring Program, Public Health Institute), Georgia (Department of Human Genetics, Emory University; Centers for Disease Control and Prevention), Iowa (University of Iowa, Registry for Congenital and Inherited Disorders), New Jersey (New Jersey Department of Health and Senior Services; Special Child Health Services Registry; Eagleton Institute), and New York (New York State Department of Health Congenital Malformations Registry). Each recruitment site obtained IRB approval for their protocol, consent forms, and data sharing during the project period from their respective institutions. All samples were collected under written consent by each participant or their legal guardian. Emory University was the site for the data and biorepository. They obtained IRB approval for all sample processing and de-identified sample submission to the Center for Inherited Disease Research genotyping service (Emory School of Medicine IRB number IRB00005100). IRB approvals for genotyping samples and uploading to dbGaP were approved prior to the initiation of that genotyping project (dbGaP: phs000718).
Genotyping
DNA samples were obtained from lymphoblastoid cell lines (LCLs) (36.8%), saliva (23.7%), buffy coat (15.7%), whole blood (13%), unknown source (i.e., no record available) (8.4%), and buccal cell (0.09%). The remaining 2.2% of genotyped samples were HapMap controls derived from LCLs that were used by the Center for Inherited Disease Research (CIDR) for quality control (QC). The samples were genotyped in batches corresponding to 96-well plates and each plate contained two study duplicates and HapMap controls. Duplicates were randomly selected from all samples with sufficient DNA. Families were randomly distributed across plates with all members of each family on the same plate.
Genotyping was performed on the Illumina HumanOmniExpressExome-8v1-2 array by the Center for Inherited Disease Research (CIDR). The algorithm used for calling genotypes was GenomeStudio version 2011.1, Genotyping Module version 1.9.4 and GenTrain version 1.0. Genotype data that passed initial QC at CIDR were released to the Quality Assurance (QA)/QC) analysis team at the University of Washington Genetics Coordinating Center (UWGCC) for data cleaning and imputation. Details of these procedures can be found in Laurie et al. [34] and all data are available in dbGaP along with specifics of genotyping and QC (dbGaP: phs000718). After QC, genotypes were available for 2,186 unique study participants. We filtered SNPs based a deviation of Hardy-Weinberg equilibrium (HWE) at p < 10−6. Overall, the median call rate was 99.86% and the error rate estimated from 53 pairs of study sample duplicates is 1x10-4. All samples had a missing call rate < 2%. The percent of SNPs with a minor allele frequency (MAF) of < 2% was 30% for the autosomes and 32.1% for the X chromosome. This calculation was based on all study participants for SNPs not located on chromosome 21 and on only study parent samples for SNPs on chromosome 21. Trisomic genotypes for all 749 children in the study were called from raw genotyping data with previously-developed methods [35].
Possible chromosomal abnormalities beyond trisomy 21 were examined as possible artifacts of the use of DNA from LCLs. This was done using Log R Ratio" (LRR) and “B Allele Frequency" (BAF) [36, 37] and applying the methods outlined in Laurie et al. [38]. Regions or chromosomes containing identified anomalies were excluded for genotype imputation purposes (see below). For chromosomes other than chromosome 21, 50 large anomalies were identified, of which 15 were filtered out of the dataset by setting genotypes in the identified region to missing. In addition, Mendelian inconsistencies were examined and one additional family was identified to have a genotype pattern consistent with uniparental chromosome 16 in the offspring. Genotypes at this chromosome were also set to missing.
Seven participants with neither parent genotyped were excluded from subsequent analyses. Thus in the remaining 742 families, genotypes were available for both the child and either the mother only (n = 95), the father only (n = 17), or both parents (n = 630).
Adjustment for population structure
Binary trait analyses using logistic regression are our primary statistical approach in this GWAS study. To adjust for population structure, we first used principal components analysis (PCA) as described by Patterson et al. [39], and implemented in R (SNPRelate package). SNPs used for PCA were selected by LD pruning from an initial pool that included all non-chromosome 21 autosomal SNPs with a missing call rate < 5% and MAF > 5%. In addition, the 2q21 (LCT), HLA, 8p23, and 17q21.31 regions were excluded from the initial pool. The first three eigenvectors were used in subsequent analyses.
Imputation
The UWGCC used IMPUTE2 software [40] to perform genotype imputation. Details of their methods and QC are available at dbGaP:phs000718. The worldwide reference panel of 1,092 samples from the 1000 Genomes Project’s Phase I integrated variant set [41] was used for imputation. We included only imputed variants with a quality metric of ≥ 0.3, as previously recommended [42].
Phenotyping
Our primary association studies were based on mothers who had a live birth with full or mosaic trisomy 21 as determined by karyotype and then determined to be due to a maternal nondisjunction error based on the characterization of the chromosome 21 genotype contributions from parent to the child with trisomy 21. Genotypes were obtained from the Illumina HumanOmniExpressExome-8v1-2 array and from previously genotyped variants along chromosome 21 using both STRs and SNPs [27, 29] The groups based on maternal nondisjunction errors were compared with fathers of the children with trisomy 21 who represent a random group of individuals from the population.
Methods for defining the type of nondisjunction errors are described in detail in our previous work (e.g.,[27, 29]). Briefly, parental origin of the meiotic error (maternal or paternal) was first determined by establishing the contribution of informative parental chromosome 21 genotypes to the child with trisomy 21. In families with both parents genotyped and where the parent of origin was unambiguously confirmed to be the mother (the vast majority of these families), we defined the meiotic stage of origin. We scored the genotype at each informative SNP and STR marker on chromosome 21q as either reduced to homozygosity (R) or not (N), according to whether the mother transmitted two identical or two different alleles, respectively, to her child at that locus. The meiotic stage of nondisjunction (MI or MII) was called according to the zygosity at the loci most proximal to the centromere (N or R, respectively). In a few cases (n = 7), MII nondisjunction was called on the basis of a single, well-genotyped R SNP nearest the centromere (followed by a series of N SNPs), but because of the dense SNP genotyping on the chip, stage was more typically supported by many markers.
In families with only one parent genotyped, a slightly different approach was required, as missing parental data led to some markers that are partially informative, but not dispositive of zygosity. Briefly, we considered the ratio of information in the SNPs near the centromere, and called each case as MI or MII depending on the ratio. The threshold for this ratio was selected by performing an experiment with the complete trios; for each complete trio, we masked the genotype of one parent, calculated the ratio described above, and found the cutoff that optimized the predictive accuracy.
Lastly, when all informative markers in the parent of origin were reduced to homozygosity along 21q, the origin of error was inferred to be a post-zygotic, mitotic error and the case was excluded from this study, consistent with previous studies [25]. However, we recognize that a proportion of these cases may be MII nondisjunction errors with no recombination.
Analysis
Sample size
As described above, samples from 2,186 participants were genotyped for this study, comprising 749 children and 1,437 parents. Participants with unresolved identity swaps, probands (children) with neither parent genotyped, and mothers in cases of potentially mitotically-arising trisomy were excluded from GWAS. After this filtering, 705 mothers and 645 fathers were retained for analysis, comprising 612 complete parent-child trios. Meiotic stage of origin for trisomy was determined to be MI in 535 cases, MII in 157 cases, and was not determined in 13 cases. Sample sizes for our analysis groups are reported in Table 1.
Association studies
We performed five primary GWAS analyses (summarized in Table 1). The comparison for all mothers vs. fathers can identify maternal genetic factors influencing nondisjunction either in MI or MII (or, more powerfully, in both). As noted in the Introduction, some genetic factors affecting MI nondisjunction may be shared with MII nondisjunction. Comparison of MI-only or MII-only mothers with fathers can identify maternal genetic factors influencing MI nondisjunction or MII nondisjunction, respectively. All three of these comparisons will also detect maternally-derived variants affecting survival of the infant to term. We chose to use fathers within our own study as controls rather than turning to external controls because of the significant problem with confounding (chip and study effects) that is introduced when cases genotyped in one study are compared to controls genotyped in another.
However, one risk of using fathers as controls is that in theory the three analyses that compare mothers to fathers may also identify spurious associations due to comparing females to males. We tested this by running a female vs. male GWAS in a large additional dataset and comparing our results to those. The dataset we used was a subset of the COHRA study [43]; this study targeted dental phenotypes, but participants were selected in a community-based setting without regard to phenotype. We used 456 male and 494 female unrelated self-identified white adults in order to have a sample size comparable to the current study. By using sex as the outcome measure in a sample that was unselected with regard to phenotype, this analysis gave us a set of results to compare to our trisomy dataset in order to determine whether any of our trisomy results might instead be male vs. female artifacts. The female vs. male analysis in the COHRA dataset did not result in any unusually significant differences (lambda = 0.94). None of the GWAS loci or candidate genes described in the results section appeared among the largest differences between males and females in the COHRA dataset. The Manhattan plot and QQ plots are provided in the Supporting Information (S1 Fig), as well as results from the COHRA analysis in our candidate genes (S1 Table).
The fourth comparison involves MI vs. MII mothers. This comparison has the potential to identify unique factors for MI or MII nondisjunction without confounding by trisomy 21 survival; that is, both groups of mothers had a live birth child with trisomy 21.
For the fifth analysis, we conducted a transmission disequilibrium test (TDT) [44]. This test examines the association between the child’s genotype and the dual phenotype of nondisjunction and survival to term. Our prior hypothesis is that this test is best for identifying fetal “survival genes.” If there is association between maternal genotype and either nondisjunction or survival, this test can in theory identify it, but the association would be weakened. We did not perform this test for the candidate genes, since they were chosen as candidates for nondisjunction, not for survival. For the nondisjoined chromosome 21, we used a trisomic TDT, previously developed by our group [45].
For all analyses except the TDT, we used the logistic regression model logit(p) = SNP + PC1 + PC2 + PC3, where SNP is encoded additively and PC1, PC2, and PC3 are the first three principal components of ancestry. The X chromosome was not examined because our primary comparative analyses involved mothers vs. fathers.
For all analyses, we filtered out SNPs with MAF < 1% or with extreme departure from HWE. Imputed SNPs with info score < 0.5 were also excluded, and imputed genotypes called with less than 90% confidence were coded as missing. Analyses were performed with PLINK and R.
Maternal age effect
Because of the strong maternal age effect in maternal chromosome 21 nondisjunction, it is important to consider how maternal age fits into the above analyses. Previous results from our group and others suggest not only different etiologies for MI and MII nondisjunction, but likely different etiologies in different age groups. Statistically, this would suggest a model that includes not only maternal age effect but also an age X genotype interaction term. However, since our primary analyses compare mothers to fathers, it is not possible to fit such a model (since fathers have no “maternal age”). The logical analysis, then, is to stratify by maternal age group, similar to the approach we took for the MI and MII subgroups. We performed several such analyses, but the sample sizes were prohibitively small for interpretation. We elaborate further on this issue in the Discussion.
Candidate gene analyses
For candidate gene analyses, we examined a window of 60kb on each side of the gene or SNP. We used a statistical significance cutoff based on the method of Li and Ji [46], which calculates the equivalent number of independent SNPs in the region and applies a Bonferroni correction based on that number. Thus the candidate gene analyses are fully corrected for multiple testing at the level of each individual gene.
Follow-up analyses to examine top-ranked GWAS signals
For follow-up analyses of signals of p < 10−5 for the GWAS, we performed literature searches on genes within 500kb. For each of those regions, LocusZoom plots were created in all analyses to identify common associations across analyses.
Results
Candidate gene association studies
We focused on two sets of candidate genes/regions: genes that function in early stages of meiosis and that have been associated with accurate chromosome segregation (n = 24) and regions associated with human recombination genome-wide counts (n = 8) [33]. The Bonferroni-corrected statistical significance cutoffs along with all results are shown in Table 2 and LocusZoom plots are provided in Figs 1–5 and in the Supporting Information (S2–S9 Figs). Each row in Table 2 represents one candidate locus. Each column represents an analysis. For each cell in the table, the most significant association at the locus (not always unique) is reported. P-values significant after correcting for multiple testing are marked with an asterisk and highlighted. Note that for each analysis in each gene, Table 2 lists the most statistically significant result, so that the SNP that appears in a given gene is not necessarily the same in each analysis. More detailed results are shown in the Supporting Information (S2 Table).
Table 2. Candidate gene results.
| Locus | All mothers vs. fathers |
MI mothers vs. fathers |
MII mothers vs. fathers |
MI mothers vs. MII mothers |
Significance threshold |
|---|---|---|---|---|---|
| SYCP1 | P = 0.00238 | P = 0.00255 | P = 0.002 | P = 2.69e-05* | 9.43e-04 |
| SYCP2 | P = 0.017 | P = 0.00592 | P = 0.000735* | P = 3.09e-05* | 8.62e-04 |
| SYCP3 | P = 0.0078 | P = 0.00748 | P = 0.00637 | P = 0.00151 | 9.43e-04 |
| SYCE1 | P = 0.0336 | P = 0.022 | P = 0.0227 | P = 0.00222 | 1.35e-03 |
| SYCE2 | P = 0.00146 | P = 0.000425* | P = 0.0154 | P = 0.00655 | 1.14e-03 |
| SYCE3 | P = 0.00324 | P = 0.000764 | P = 0.0053518 | P = 0.00337 | 5.38e-04 |
| TEX12 | P = 0.00558 | P = 0.0219 | P = 0.013871 | P = 0.0271 | 1.19e-03 |
| BLM | P = 0.0225 | P = 0.0378 | P = 0.00601 | P = 0.0257 | 7.04e-04 |
| DMC1 | P = 0.0184 | P = 0.00993 | P = 0.018619 | P = 0.0134 | 1.19e-03 |
| EXO1 | P = 0.00924 | P = 0.0118 | P = 0.00111 | P = 0.00303 | 6.10e-04 |
| HORMAD1 | P = 0.0152 | P = 0.0174 | P = 0.011377 | P = 0.00361 | 1.35e-03 |
| HORMAD2 | P = 0.0113 | P = 0.0156 | P = 0.00155 | P = 0.00838 | 8.93e-04 |
| MEI1 | P = 0.00655 | P = 0.0485 | P = 0.00951 | P = 0.0204 | 9.26e-04 |
| MEI4 | P = 0.0147 | P = 0.0164 | P = 0.0283 | P = 0.021 | 6.33e-04 |
| MLH1 | P = 0.0151 | P = 0.00297 | P = 0.0162 | P = 0.0427 | 1.14e-03 |
| MLH3 | P = 0.00472 | P = 0.0107 | P = 0.0241 | P = 0.0207 | 1.35e-03 |
| MND1 | P = 0.0273 | P = 0.013 | P = 0.000336* | P = 9.6e-05* | 8.77e-04 |
| MSH5 | P = 0.00677 | P = 0.00135 | P = 0.0417 | P = 0.0241 | 9.26e-04 |
| REC114 | P = 0.0187 | P = 0.0272 | P = 0.0886 | P = 0.0118 | 8.77e-04 |
| REC8 | P = 0.00822 | P = 0.00404 | P = 0.00195 | P = 0.00112 | 9.43e-04 |
| SMC1B | P = 0.0115 | P = 0.00407 | P = 0.047 | P = 0.0349 | 7.81e-04 |
| SPO11 | P = 0.0232 | P = 0.0325 | P = 0.0115 | P = 0.0181 | 1.25e-03 |
| STAG3 | P = 0.005 | P = 0.00472 | P = 0.0798 | P = 0.00958 | 1.19e-03 |
| TRIP13 | P = 0.0026 | P = 0.0122 | P = 0.00358 | P = 0.0206 | 1.61e-03 |
|
rs1254319 (C14orf39 missense) |
P = 0.0278 | P = 0.0488 | P = 0.0372 | P = 0.00464 | 1.39e-03 |
|
rs75502650 (CCDC43 intron) |
P = 0.00203 | P = 0.00137* | P = 0.000628* | P = 0.031 | 7.58e-04 |
|
rs1132644 (CCNB1IP1 UTR) |
P = 0.0232 | P = 0.00657 | P = 0.0236 | P = 0.0195 | 1.32e-03 |
|
rs56162163 (chr17 inversion) |
P = 0.00666 | P = 0.00956 | P = 0.00737 | P = 0.0166 | 6.67e-04 |
|
rs74434767 (CPLX1 intron) |
P = 0.00611 | P = 0.0178 | P = 0.0227 | P = 0.0329 | 1.61e-03 |
|
rs5745459 (MSH4 missense) |
P = 0.0389 | P = 0.0283 | P = 0.037 | P = 0.0198 | 1.16e-03 |
|
rs150798754 (PRDM9 intergenic) |
P = 0.00219 | P = 0.006 | P = 0.0123 | P = 0.022 | 1.25e-03 |
|
rs6889665 (PRDM9 upstream) |
P = 0.00219 | P = 0.006 | P = 0.0104 | P = 0.0678 | 8.33e-04 |
|
rs450739 (RAD21L missense) |
P = 2.49e-05* | P = 7.47e-05* | P = 0.00579 | P = 0.00126 | 1.09e-03 |
|
rs4045481 (RNF212 missense) |
P = 0.00475 | P = 0.0136 | P = 0.00514 | P = 0.0131 | 1.02e-03 |
|
rs658846 (RNF212 intron) |
P = 0.00292 | P = 0.0108 | P = 0.00514 | P = 0.0131 | 8.33e-04 |
|
rs12233733 (RNF212 nearby) |
P = 0.00292 | P = 0.0108 | P = 0.00396 | P = 0.031 | 1.52e-03 |
|
rs10135595 (SMEK1 UTR) |
P = 0.0125 | P = 0.007 | P = 0.00306 | P = 0.000183* | 1.39e-03 |
Each row represents one candidate locus (either a gene with a 60kb border on each side or a 60kb window around a SNP). Each column represents an analysis. For each locus-analysis pair, the most significant association at the locus (not always unique) is reported. P-values significant after correcting for multiple testing (i.e., exceeding the Bonferroni-adjusted significance threshold noted in the last column) are marked with an asterisk and highlighted. (MI: meiosis I; MII: meiosis II; P: p-value; OR: odds ratio.) The first 24 loci represent genes selected for their function (above the double line). The latter 13 loci represent SNPs identified by Kong et al. in their GWAS of recombination [33], with annotation in parentheses (below the double line).
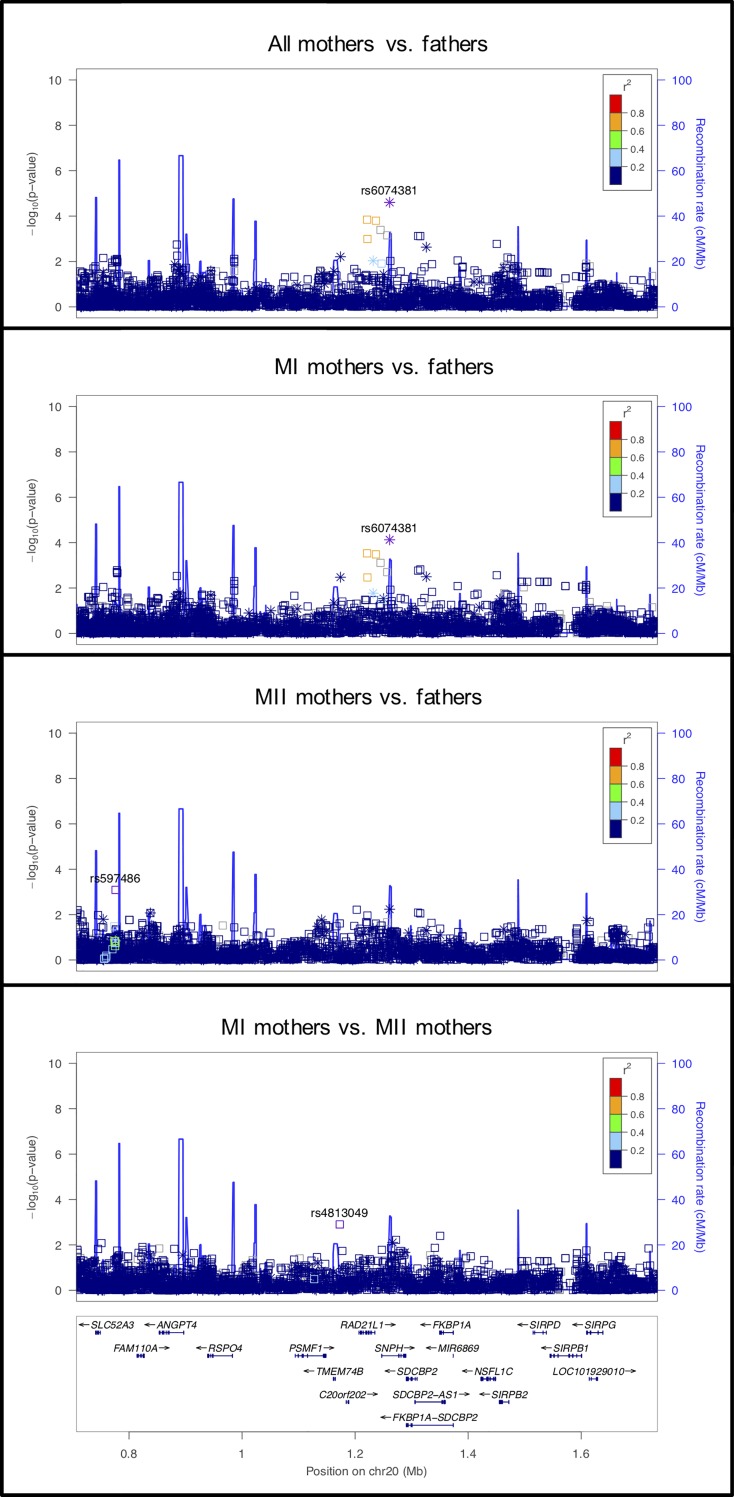
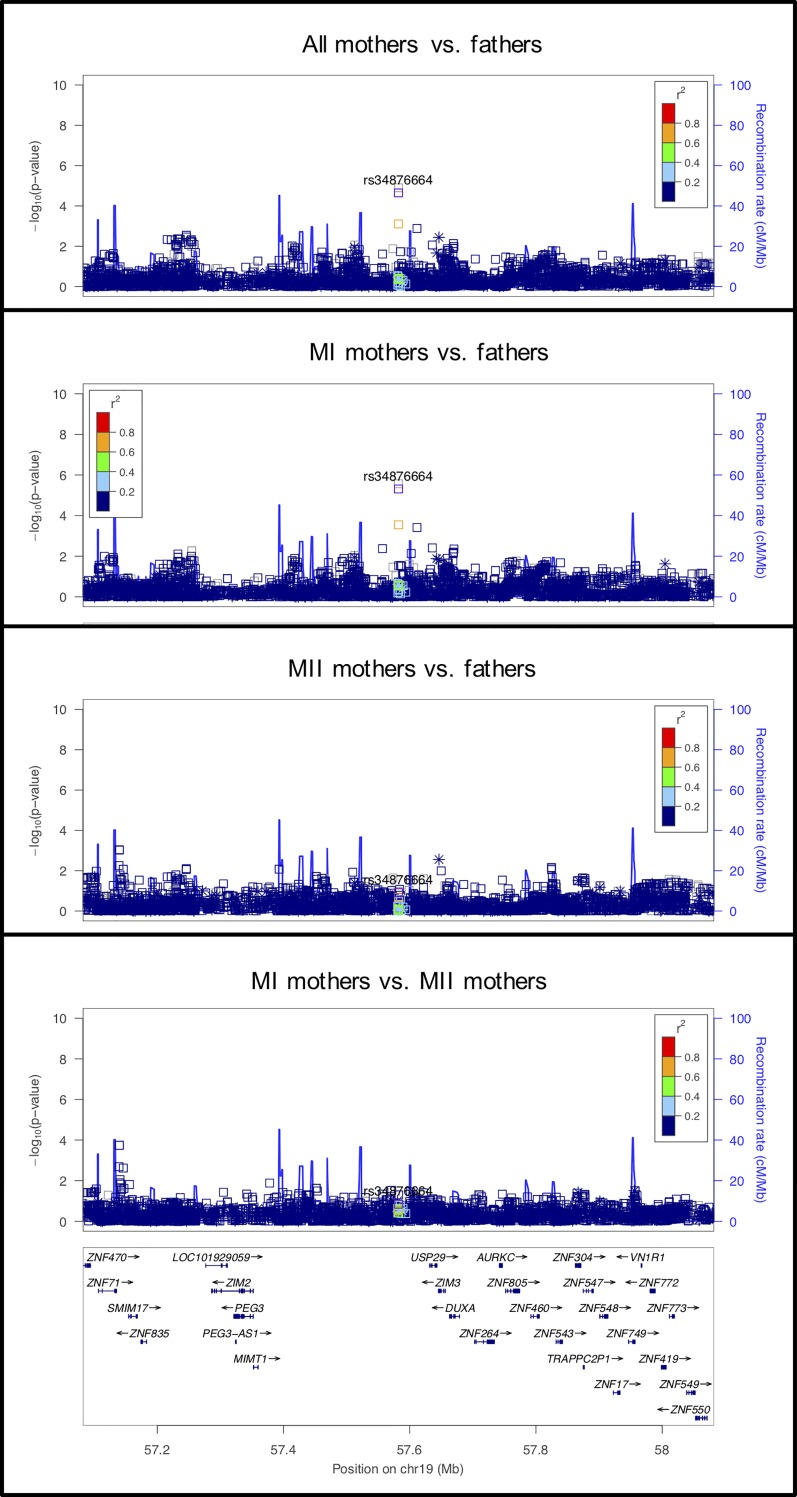
Fig 1. LocusZoom plot for RAD21L.
In this Figure (as in Figs 2–12) four LocusZoom plots show the results at one locus across all four analyses. Each point is one variant, with the x- and y-axes representing physical position on the chromosome and -log10(p-value), respectively. Open squares and asterisks represent genotyped and imputed variants, respectively. Coloring represents linkage disequilibrium (red = stronger, blue = weaker) with the tagging SNP (which is purple). The overlaid blue curve shows the recombination rate.
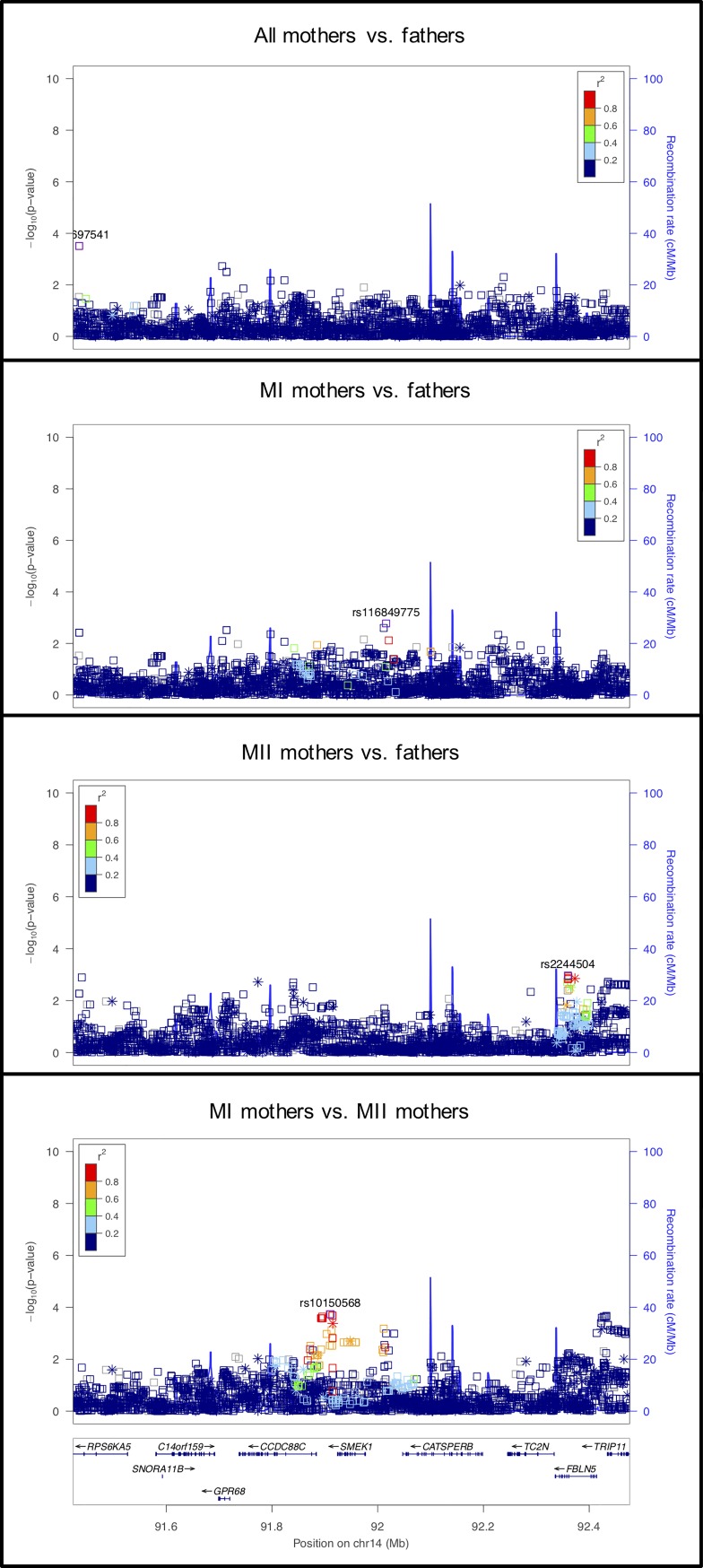
Fig 5. LocusZoom plot for SMEK1.
Candidate genes associated with chromosome segregation
These genes are shown in the top half (above the double line) of Table 2. Examination of genes involved in the meiotic cohesion complex showed a statistically significant association with RAD21L, a meiosis-specific member of the α-kleisin protein family [47–50]. This was significant in both the mothers vs. fathers and the MI mothers vs. fathers comparisons, and has a similar effect (odds ratio) in the MII cases at the same SNP (Fig 1). Meiotic cohesins are essential for sister chromatid cohesion, but also have an effect on other prophase I processes, including formation of the axial/lateral elements, assembly of the SC, and crossing-over (e.g., [51, 52]). Gene disruption of RAD21L leads to sexually dimorphic phenotypes in mice. Male mice are infertile, whereas female mice show age-related infertility, reminiscent of primary ovarian insufficiency. The reduced efficiency in synapsis in fetal oocytes may result in a lower ovarian reserve to be established [50] In human males, variants in RAD21L have been implicated in meiotic arrest and Sertoli cell-only syndrome [53].
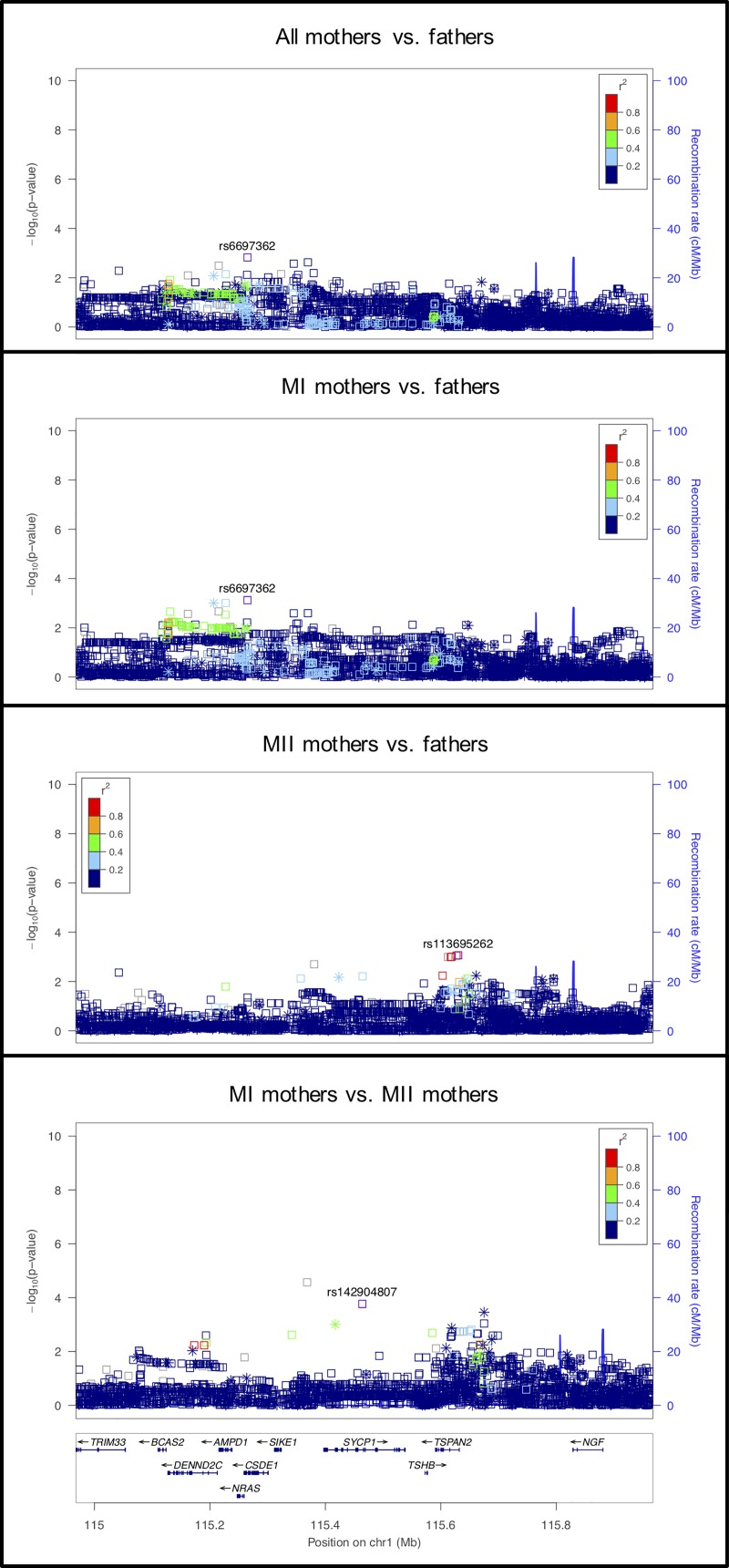
Variants in seven genes coding for components of the SC were also investigated in this candidate gene group. Of the genes coding for components of the central element of the SC (i.e., SYCE1, SYCE2, SYCE3, TEX12), SYCE2 showed a statistically significant association (in the MI mothers vs. fathers) (Fig 2), although the association with SYCE3 was close to the cutoff for significance (also in MI mothers vs. fathers).
Fig 2. LocusZoom plot for SYCE2.
The other SC genes we examined code for the transverse filament (SYCP1) and components of the axial/lateral elements (SYCP2 and SYCP3). SYCP1 showed significant association in the MI vs. MII analysis, but not in the other analyses (Fig 3). The signal in SYCP1 was primarily located at an imputed SNP, at rs35401563, so this result requires confirmation by further genotyping. SYCP2 showed highly significant associations in both the MII mothers vs. fathers and the MI vs. MII comparisons (Fig 4), suggesting the potential for an effect specific to MII. SYCP3 was nearly significant in the MI vs. MII comparison.
Fig 3. LocusZoom plot for SYCP1.
Fig 4. LocusZoom plot for SYCP2.
Among the other candidate genes in this group, the only statistically significant result was for MND1. The observed significant association was strongest in the MI vs. MII comparison, was also strong in the MII vs. fathers comparison, and was much weaker in the MI vs. fathers comparison. This pattern suggests that this locus may be associated with MII nondisjunction. Based on genetic and cellular analysis of deletion mutants, MND1, acting with HOP2, plays a role in the initial processing of DSBs. Specifically, the HOP2-MND1 complex is involved in two separate stages of the DMC1-promoted recombination process: first, in the stabilization of DMC1 filaments on the resected end of the DSBs, and second, in the promotion of the subsequent strand invasion steps. In higher eukaryotes (mouse [54] and Arabidopsis thaliana [55, 56]), MND1 appears to be required for normal male and female fertility. Mutations result in normal recombination initiation, but meiotic DSBs are abnormally repaired and chromosome synapsis is aberrant [17]. The HOP2-MND1 complex has also been implicated in ovarian dysfunction and biochemically, is capable of driving RAD51-mediated alternative lengthening of telomeres in somatic cells [57]. If this association is confirmed, understanding why the effect of the variant is stronger in MII errors vs. MI errors may shed more light on its function.
Genes associated with human genome-wide recombination counts (shown in the bottom half of Table 2, below the double line of Table 2)
We also examined the eight regions identified in Kong et al. that were highly associated with genome-wide recombination counts in a large Icelandic study of 71,929 parent-offspring pairs [33]. Of the eight regions, three showed associations with maternal nondisjunction that were statistically significant according to the cutoffs shown in Table 2. The first region included RAD21L for which results are discussed above, as it was also in the group of candidate genes for meiotic processes. The second statistical signal was in the region of SMEK1 (also known as protein phosphatase 4 regulatory subunit 3 (PPP4R3A)) and was strongest in the MI vs. MII mothers comparison (Fig 5). SMEK1 is known as a regulator of cellular functions, including apoptosis, cell growth, microtubule organization, cell cycle arrest, and TNF and PI3K/Akt signaling (e.g., [58, 59]). It is also known to play a role in endothelial cell function and subsequent angiogenesis [60]. However, its role in meiosis is unknown, although it is known to be expressed in the ovary.
The third signal is in the region of CCDC43, and was evident in both the MI mothers vs. fathers analysis and the MII mothers vs. fathers analysis (see S7 Fig). There is no known function of CCDC43 in meiosis. In the study of Kong et al. [33], the SNP associated with recombination (rs75502650) was located in an intron of CCDC43. It was estimated to increase the global recombination rate by 76 cM and this effect was limited to females.
Strongest results from the genome-wide association study
Because of the limited sample size in this study, the full GWAS produced only suggestive results, though a few of those top results have strong support in the literature for the relevance of the gene functions to meiosis or fetal survival. Manhattan plots and Q-Q plots for each GWAS analysis are included in the Supporting Information (S10 Fig). Tables 3–7 show the most statistically significant results from each of the comparisons in the genome-wide association study. For each result for a given comparison, the corresponding table also gives the smallest p-value within 20kb in each of the other comparisons. Detailed results are included in the Supporting Information (S3–S7 Tables).
Table 3. Top hits from the all mothers vs. fathers genome-wide association study.
| Locus | All mothers vs. fathers | MI mothers vs. fathers | MII mothers vs. fathers | MI mothers vs. MII mothers |
TDT |
|---|---|---|---|---|---|
| rs10948101 | P = 7.65e-07 | P = 1.58e-06 | P = 0.000897 | P = 0.0634 | P = 0.00514 |
| rs35141718 | P = 2.02e-06 | P = 1.92e-05 | P = 0.0109 | P = 0.127 | P = 0.0499 |
| rs11535058 | P = 2.42e-06 | P = 2.1e-05 | P = 7.69e-05 | P = 0.0411 | P = 0.015 |
| rs62086686 | P = 4.45e-06 | P = 0.000292 | P = 8.53e-05 | P = 0.0171 | P = 0.0133 |
| rs75733466 | P = 5.09e-06 | P = 0.00023 | P = 1.24e-05 | P = 0.00189 | P = 0.0212 |
| rs117746305 | P = 5.14e-06 | P = 0.000168 | P = 3.93e-06 | P = 0.056 | P = 0.0881 |
| rs12947774 | P = 5.62e-06 | P = 0.000199 | P = 0.000949 | P = 0.00229 | P = 0.0339 |
| rs1612273 | P = 5.93e-06 | P = 7.22e-05 | P = 1.62e-05 | P = 0.000934 | P = 0.00432 |
| rs12652455 | P = 6.43e-06 | P = 2.43e-05 | P = 0.000437 | P = 0.0429 | P = 0.0518 |
| rs148846406 | P = 6.79e-06 | P = 3.85e-05 | P = 0.00673 | P = 0.0562 | P = 0.00883 |
| rs11026040 | P = 8.22e-06 | P = 0.000238 | P = 3.1e-05 | P = 0.00505 | P = 0.00794 |
| rs7010571 | P = 8.27e-06 | P = 3.41e-05 | P = 0.0185 | P = 0.0796 | P = 0.0477 |
| rs35816728 | P = 9.96e-06 | P = 6.09e-05 | P = 0.00101 | P = 0.00124 | P = 0.00815 |
Suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value.)
Table 7. Top hits from the TDT (transmission disequilibrium test) genome-wide association study.
| Locus | All mothers vs. fathers | MI mothers vs. fathers | MII mothers vs. fathers | MI mothers vs. MII mothers | TDT |
|---|---|---|---|---|---|
| rs3802065 | P = 0.0144 | P = 0.0066 | P = 0.0318 | P = 0.00859 | P = 4.84e-07 |
| rs2867076 | P = 0.0164 | P = 0.0227 | P = 0.0081 | P = 0.00904 | P = 1.05e-06 |
| rs7451700 | P = 0.0186 | P = 0.0169 | P = 0.0544 | P = 0.0195 | P = 1.71e-06 |
| rs7389783 | P = 0.0421 | P = 0.0397 | P = 0.00488 | P = 0.00741 | P = 1.93e-06 |
| rs17769147 | P = 0.182 | P = 0.152 | P = 0.0738 | P = 0.0624 | P = 2.06e-06 |
| rs201634098 | P = 0.00592 | P = 0.00385 | P = 0.0722 | P = 0.015 | P = 3.52e-06 |
| chr23:154539980 | NA | NA | NA | NA | P = 3.63e-06 |
| rs74615884 | P = 0.0801 | P = 0.037 | P = 0.0167 | P = 0.00236 | P = 3.73e-06 |
| rs158866 | P = 0.036 | P = 0.00582 | P = 0.0103 | P = 0.0102 | P = 3.77e-06 |
| rs1187600 | P = 0.0553 | P = 0.0636 | P = 0.204 | P = 0.156 | P = 6.25e-06 |
| rs55743346 | P = 0.0514 | P = 0.0371 | P = 0.0834 | P = 0.0234 | P = 7.34e-06 |
| rs183199067 | P = 0.0131 | P = 0.00285 | P = 0.0396 | P = 0.0254 | P = 9.55e-06 |
| rs140022090 | P = 0.0163 | P = 0.00733 | P = 0.0538 | P = 0.00493 | P = 9.55e-06 |
| rs34518363 | P = 0.00538 | P = 0.00186 | P = 0.049 | P = 0.0394 | P = 9.55e-06 |
| rs6681167 | P = 0.0358 | P = 0.0505 | P = 0.0648 | P = 0.0377 | P = 9.58e-06 |
Suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value.)
Table 4. Top hits from the MI mothers vs. fathers genome-wide association study.
| Locus | All mothers vs. fathers | MI mothers vs. fathers | MII mothers vs. fathers | MI mothers vs. MII mothers | TDT |
|---|---|---|---|---|---|
| rs10948100 | P = 7.65e-07 | P = 1.58e-06 | P = 0.000897 | P = 0.0634 | P = 0.00514 |
| rs35288347 | P = 1.26e-05 | P = 2.72e-06 | P = 0.023 | P = 0.0298 | P = 0.0339 |
| rs4649043 | P = 0.000148 | P = 3.1e-06 | P = 0.00388 | P = 0.000408 | P = 0.00556 |
| rs437933 | P = 1.79e-05 | P = 3.29e-06 | P = 0.0683 | P = 0.05 | P = 0.0186 |
| rs16847735 | P = 2.44e-05 | P = 3.77e-06 | P = 0.0173 | P = 0.0135 | P = 0.0617 |
| rs2467011 | P = 5.02e-05 | P = 4.29e-06 | P = 0.0902 | P = 0.00786 | P = 0.158 |
| rs9442389 | P = 1.88e-05 | P = 5.27e-06 | P = 0.0701 | P = 0.0164 | P = 0.0241 |
| rs731245 | P = 0.000148 | P = 6.91e-06 | P = 0.00772 | P = 0.000809 | P = 0.00654 |
| rs984968 | P = 1.48e-05 | P = 6.95e-06 | P = 0.0741 | P = 0.0425 | P = 0.0833 |
| rs9984132 | P = 5.86e-05 | P = 9.87e-06 | P = 0.0539 | P = 0.00662 | NA |
Suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value.)
Table 5. Top hits from the MII mothers vs. fathers genome-wide association study.
| Locus | All mothers vs. fathers | MI mothers vs. fathers | MII mothers vs. fathers | MI mothers vs. MII mothers | TDT |
|---|---|---|---|---|---|
| rs1855111 | P = 6.48e-05 | P = 0.00213 | P = 2.2e-06 | P = 0.00698 | P = 0.00494 |
| rs76740710 | P = 7.19e-06 | P = 0.000236 | P = 3.93e-06 | P = 0.0341 | P = 0.00648 |
| rs12981234 | P = 0.0013 | P = 0.00911 | P = 4.28e-06 | P = 8.8e-05 | P = 0.0854 |
| rs200216460 | P = 0.000209 | P = 0.0104 | P = 4.99e-06 | P = 0.00373 | P = 0.00284 |
| rs11668205 | P = 0.00588 | P = 0.00218 | P = 5.49e-06 | P = 3.34e-05 | P = 0.0233 |
| rs146838878 | P = 0.0197 | P = 0.0249 | P = 6.16e-06 | P = 0.000328 | P = 0.000463 |
| rs115281615 | P = 0.047 | P = 0.0792 | P = 6.34e-06 | P = 7.33e-07 | P = 0.0712 |
| rs62359711 | P = 0.0024 | P = 0.00108 | P = 6.91e-06 | P = 0.000191 | P = 0.0112 |
| rs73178888 | P = 0.00754 | P = 0.0041 | P = 7.05e-06 | P = 1.72e-05 | P = 0.0122 |
| rs9966603 | P = 0.00967 | P = 0.0639 | P = 7.05e-06 | P = 0.00208 | P = 0.00759 |
| rs13020106 | P = 0.00149 | P = 0.0335 | P = 7.12e-06 | P = 0.00336 | P = 0.0162 |
| rs2560850 | P = 0.0016 | P = 0.00365 | P = 7.69e-06 | P = 0.000332 | P = 0.00604 |
| rs1191234 | P = 0.00176 | P = 0.0137 | P = 9.98e-06 | P = 0.00157 | P = 0.0411 |
Suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value.)
Table 6. Top hits from the MI mothers vs. MII mothers genome-wide association study.
| Locus | All mothers vs. fathers | MI mothers vs. fathers | MII mothers vs. fathers | MI mothers vs. MII mothers | TDT |
|---|---|---|---|---|---|
| rs115281615 | P = 0.047 | P = 0.0792 | P = 6.34e-06 | P = 7.33e-07 | P = 0.0712 |
| rs6440985 | P = 0.0958 | P = 0.0112 | P = 0.000198 | P = 1.31e-06 | P = 0.00796 |
| rs2806747 | P = 0.0934 | P = 0.0656 | P = 0.00037 | P = 1.35e-06 | P = 0.00284 |
| rs9319652 | P = 0.0225 | P = 0.0267 | P = 0.00243 | P = 4.09e-06 | P = 0.039 |
| rs11977478 | P = 0.00474 | P = 0.00902 | P = 0.00019 | P = 4.18e-06 | P = 0.00232 |
| rs11034351 | P = 0.0124 | P = 0.00284 | P = 7e-04 | P = 4.3e-06 | P = 0.0295 |
| rs7685548 | P = 0.00618 | P = 0.000955 | P = 0.00884 | P = 4.82e-06 | P = 0.00226 |
| rs71967233 | P = 0.044 | P = 0.0785 | P = 0.000135 | P = 5.24e-06 | P = 0.0163 |
| rs34282937 | P = 0.0334 | P = 0.0368 | P = 5.95e-05 | P = 5.48e-06 | P = 0.0298 |
| rs77525287 | P = 0.0707 | P = 0.000444 | P = 0.0124 | P = 5.9e-06 | P = 0.099 |
| rs4818884 | P = 0.0454 | P = 0.0312 | P = 0.00354 | P = 9.57e-06 | NA |
| rs61999085 | P = 0.00851 | P = 0.000125 | P = 0.0449 | P = 9.84e-06 | P = 0.00243 |
Suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value.)
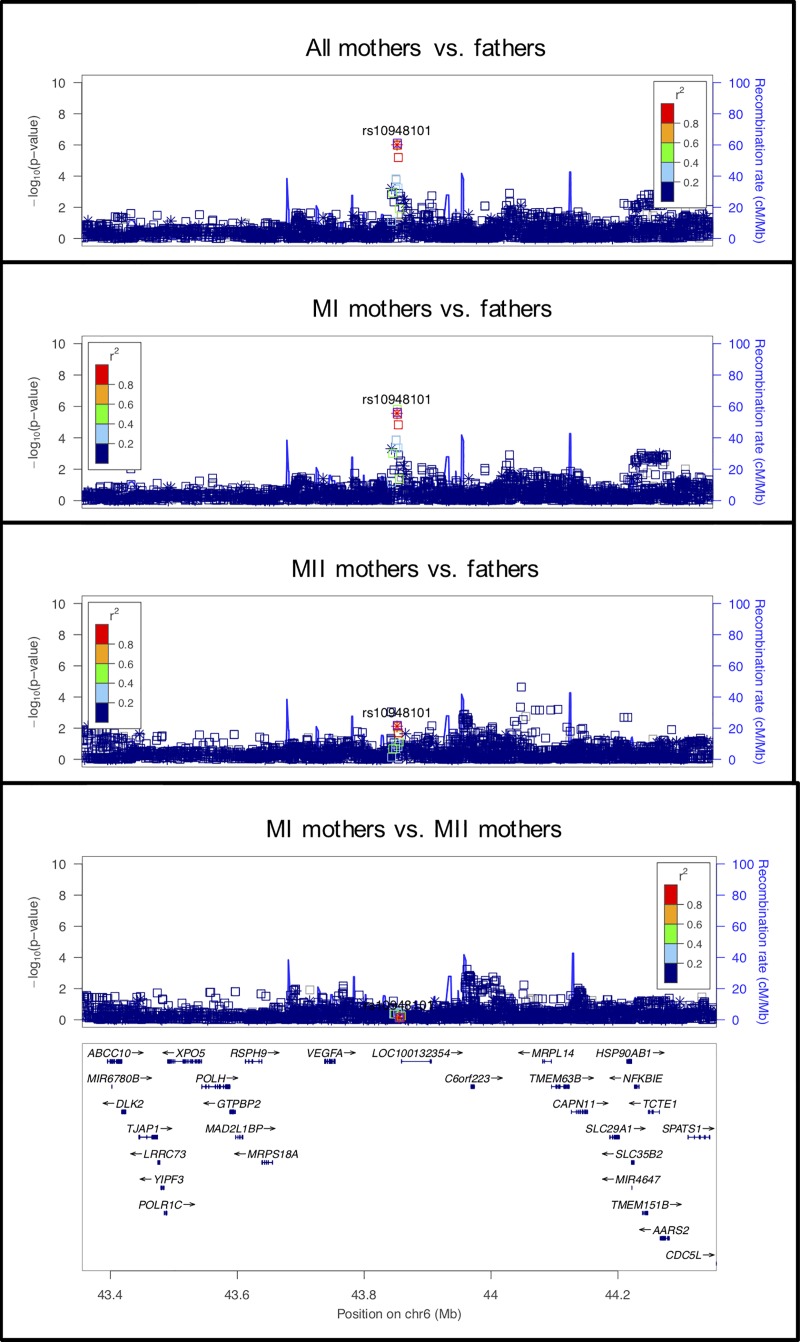
rs10948101 on chromosome 6 near VEGFA
The observed signal for this locus was strongest for the analysis of all mothers vs. fathers and was located within LOC100132354, a long non-coding RNA (lncRNA) (Fig 6). Upstream of this intergenic lncRNA is VEGFA, the gene encoding vascular endothelial growth factor A (VEGFA). In a recent meta-analysis, LOC100132354 was confirmed to be highly associated with VEGF circulating levels in serum [61]. VEGFA plays multiple roles in ovarian development and function (reviewed in McFee and Cupp [62]). Vascularization plays a role in the formation of early ovarian structures, primordial follicle assembly, and follicle activation. Further, ovarian function is highly dependent on the development and continual remodelling of a complex vascular system. This allows the follicle and the corpus luteum to receive the needed oxygen, nutrients, and systemic hormones and the release of ovarian hormones (reviewed in Robinson et al. [63]). If angiogenesis is disrupted, follicular growth is reduced, ovulation is perturbed, and development and function of the corpus luteum is significantly altered. The action of VEGFA is necessary at all these stages of development. We did not see any significant effect in the TDT analysis, so there is no suggestion that this locus is associated with survival to term.
Fig 6. LocusZoom plot for VEGFA.
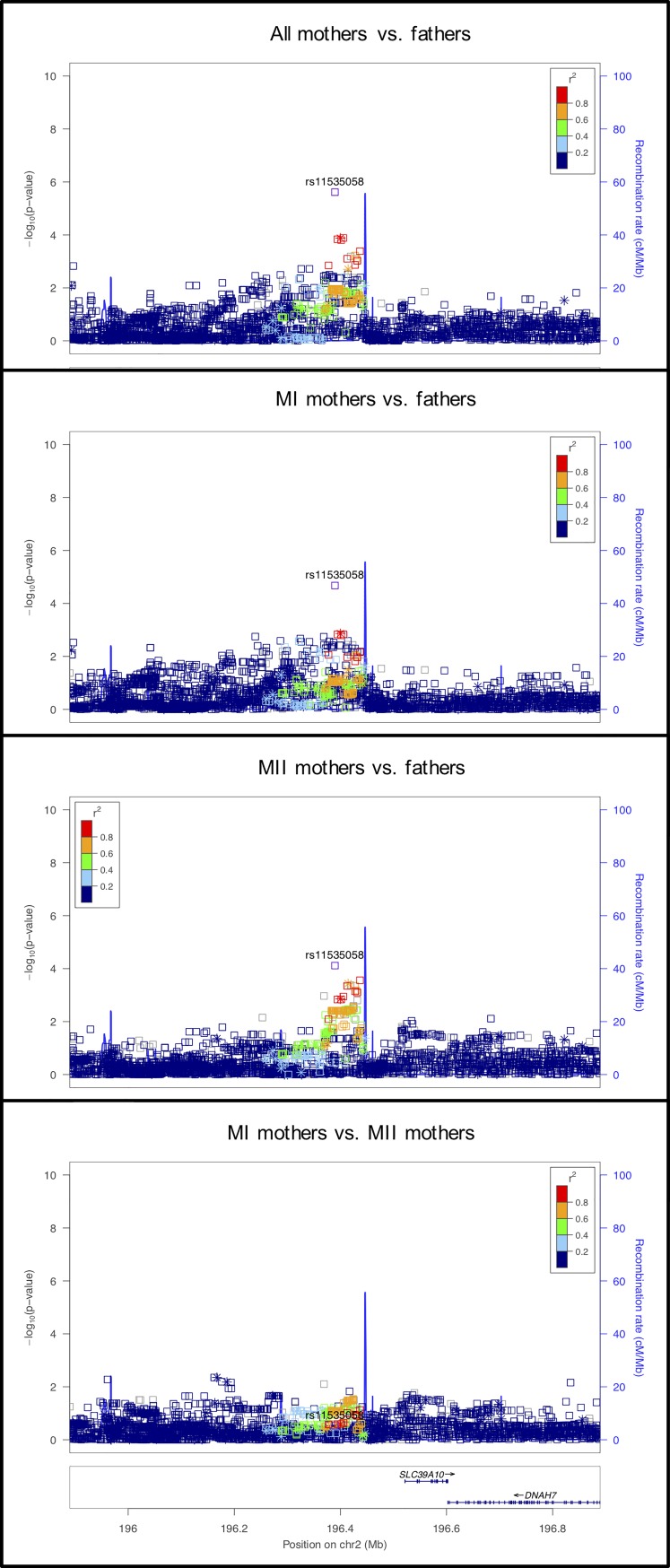
rs11535058 on chromosome 2 near SLC39A10
The observed signal in this region, strongest in the mothers vs. fathers comparison, was located 132kb downstream of SLC39A10 (Fig 7), with a similar, but non-significant, effect size for both MI mothers vs. fathers and MII mothers vs. fathers. SLC39A10 is involved in the zinc transport network. Regulation of intracellular zinc is essential for oocyte maturation and activation. In mouse, progression of the oocyte from a cell arrested in prophase of MI into a mature egg arrested at metaphase of MII is accompanied by an increase in total zinc content. This increase is required for proper meiotic progression [64]. Also, exit from MII during oocyte activation requires decreasing cellular zinc through the rapid export of zinc from the oocyte. These ‘zinc sparks’ are required for oocyte activation and resumption of the cell cycle [65].
Fig 7. LocusZoom plot for SLC39A10.
Specific to SLC39A10, Lisle et al. [66] found a complex zinc transport network present in the cumulus-oocyte complex in mouse oocytes. They found that mRNA transcripts for specific zinc transporter proteins (SLC family), including Slc39a10 were higher in oocytes, while another unique set of zinc transporter protein transcripts were higher in cumulus cells. Thus, zinc homeostasis, regulated in the cumulus-oocyte complex, may affect both MI and MII processes.
rs35288347 on chromosome 19 near AURKC
This signal occurs primarily in the MI mothers vs. fathers analysis (Fig 8), with a similar but less significant effect in the mothers vs. fathers analysis. Consistent with the possibility of an effect specific to MI, mutations in the AURKC gene in this region (160kb away) cause tetraploidy in human sperm and MI arrest in mouse oocytes [67, 68].
Fig 8. LocusZoom plot for AURKC.
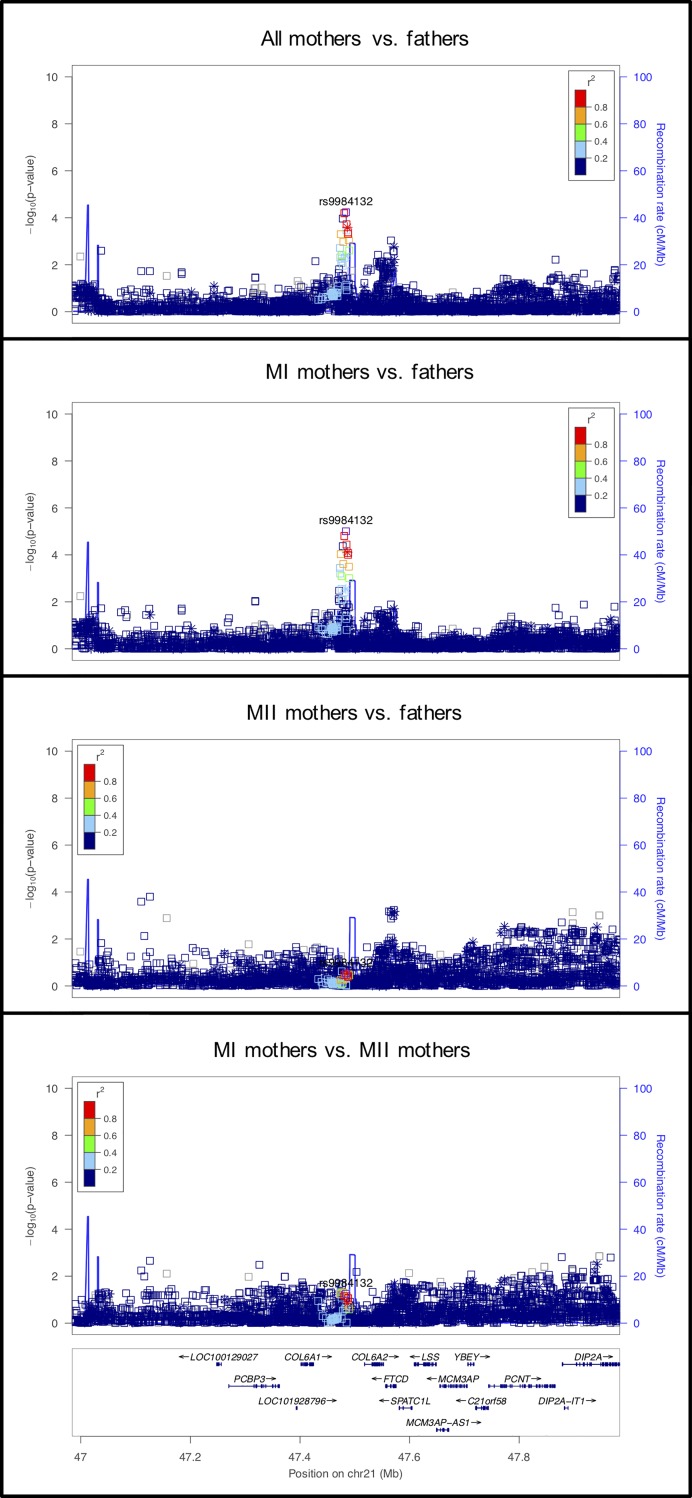
rs9984132 on chromosome 21 located in a gene rich region
The signal at rs9984132 on chromosome 21 is located in a gene-rich region (Fig 9). The strongest signal at this locus was identified in the comparison of MI mothers with fathers. Two genes stand out as possible candidates for involvement in chromosome segregation. COL6A2, located 34kb upstream of the signal, codes for one of the components of collagen that is part of the extracellular matrix (ECM) formed by cumulus cells. This ovarian follicular ECM is related to proliferation, steroidogenesis, and luteinization [69]. As the formation of this ECM is involved in fertilization and embryo quality, the observation that this signal appears to be only related to MI nondisjunction reduces the support of COL6A2 as a candidate.
Fig 9. LocusZoom plot for rs9984132 on chromosome 21.
PCNT is a gene located about 260kb upstream from the signal. Pericentrin, coded by PCNT, is a highly conserved component of the acentriolar microtubule-organizing centers (aMTOCs) in mouse oocytes. aMTOCs play a vital role in meiotic spindle assembly and stability. Depletion of pericentrin in mouse oocytes leads to increased rates of aneuploidy [70]. Human oocytes differ from mouse oocytes in that they lack PNCT and aMTOCs in MI, where spindle assembly is mediated from chromosomes by the small guanosine triphosphates [71]. Thus, more work is needed to confirm this signal and its underlying genetic association.
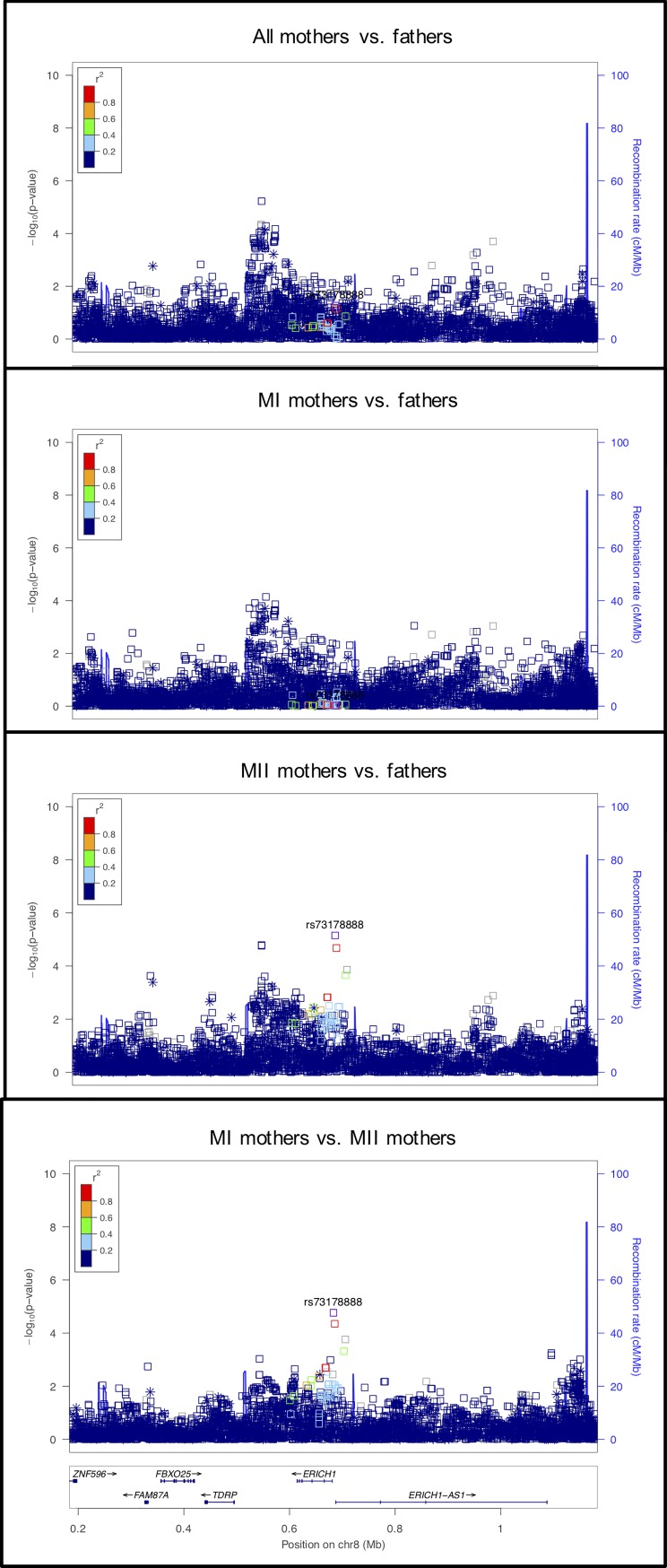
rs73178888 on chromosome 8 near a region associated with meiotic recombination
This signal, located in an intron of ERICH1, is primarily observed in the MII mothers vs. fathers analysis and in the MI vs. MII analysis, suggesting that it might be an MII risk locus (Fig 10). The location is noteworthy because Begum et al. [72] identified a variant 59kb away in this region as potentially associated with meiotic recombination (specifically recombination outside of hotspots) in a euploid population. There is no evidence to suggest that ERICH1 or DLGAP2 (also known as ERICH1-AS1), 3kb from the signal, is involved in recombination or meiosis. The next closest gene is TDRP. The deficiency of TDRP in mice is suggested to be involved in sperm motility and may play a role in spermatogenesis [73], but there is no evidence for involvement is oogenesis.
Fig 10. LocusZoom plot for DLGAP2.
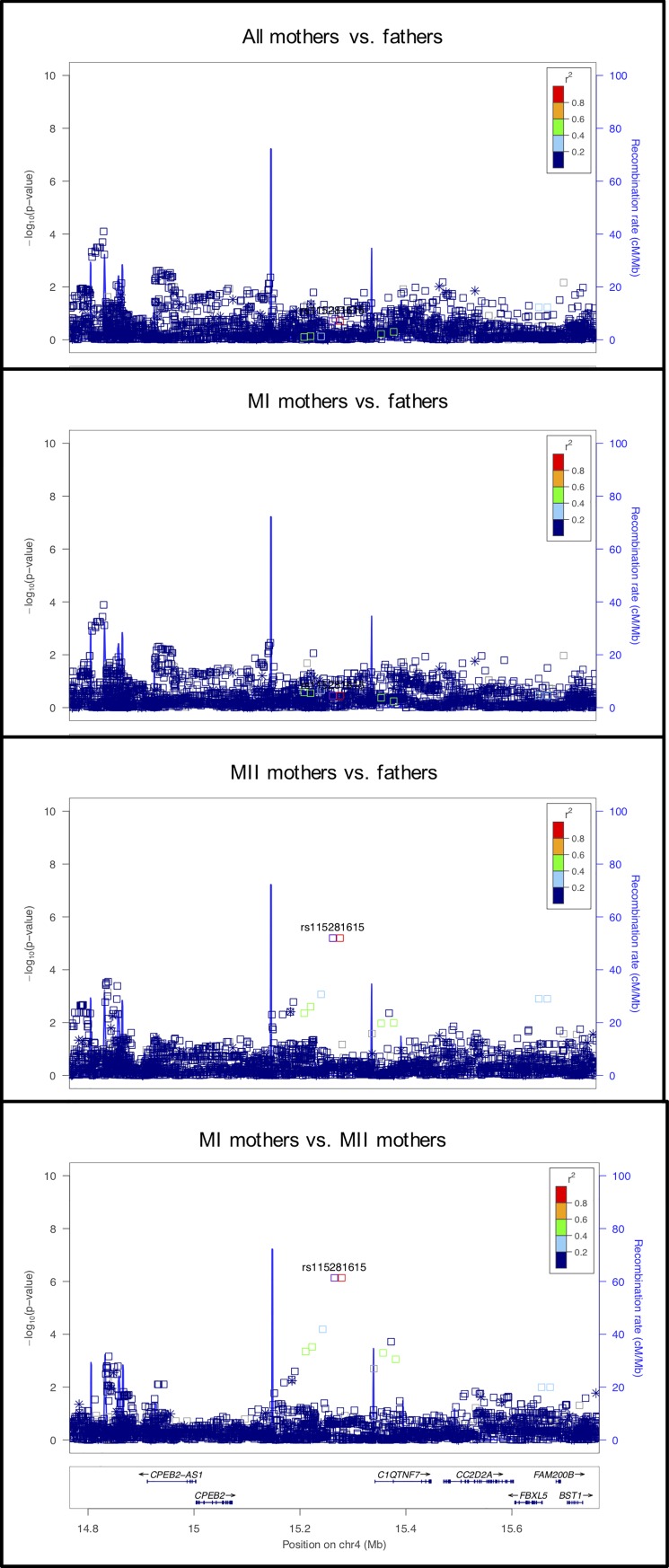
rs115281615 on chromosome 4 near CPEB2
The signal at this locus is primarily observed in the MII mothers vs. fathers comparison (Fig 11). The genes in the region, C1QTNF7, and CC2D2A, do not have evidence for a role in meiosis. CPEB2, located 193kb from the signal, encodes an RNA-binding protein, cytoplasmic polyadenylation element binding protein and is thought to be involved in regulated translation, a system that allows the rapid production of selective proteins in response to a physiological signal. CPEB2 interacts with the elongation factor, eEF2, to slow down peptide elongation of CPEB2-bound RNA [74]. In mice, this protein is highly similar to the family of CPEBs that regulate cytoplasmic polyadenylation of mRNA as a trans-factor in oogenesis and spermatogenesis. CPEB2 is expressed post-meiotically in mouse spermatogenesis, which suggests a possible role in translational regulation of stored mRNAs in transcriptionally inactive haploid spermatids [75].
Fig 11. LocusZoom plot for CPEB2.
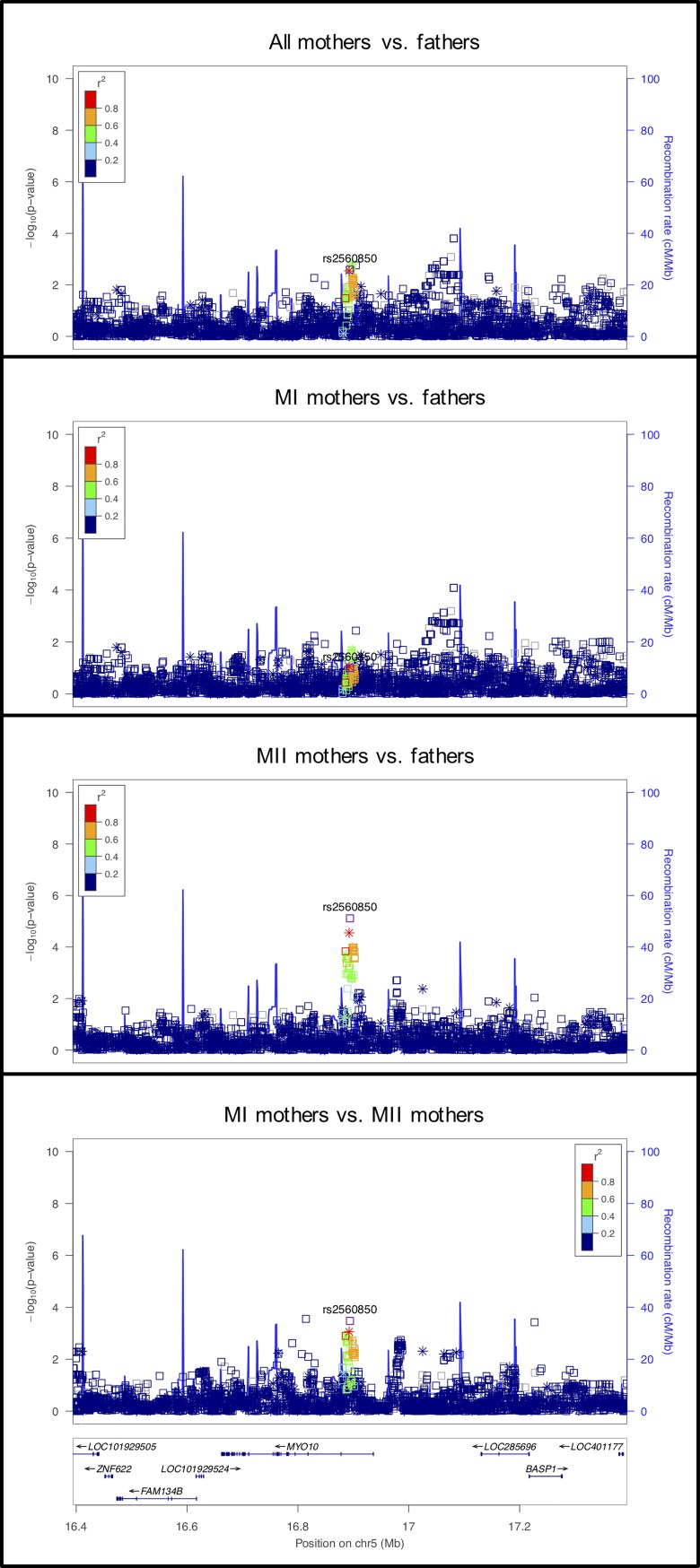
rs2560850 on chromosome 5 in an intron of MYO10
The comparison between MII mothers vs. fathers shows the strongest statistical significance for this locus which includes MYO10 (Fig 12). Myosin-10 proteins are phosphoinositide-binding, actin-based motors that play an important role during meiosis in the integration of the F-actin and microtubule cytoskeletons. Proper spindle positioning and orientation are essential for asymmetric cell division and these functions are particularly important in meiosis. In Xenopus oocytes, Weber et al. [76] showed that myosin-10 is associated with microtubules and is concentrated where the meiotic spindle contacts the F-actin-rich cortex. This observation and others suggest that myosin-10, the microtubule-binding myosin, is required for anchoring the spindle and an actin-binding kinesin is required for meiotic cytokinesis [76, 77]. Recently F-actin was shown to permeate the microtubule spindles in oocytes of many mammals, including human, where it prevents lagging chromosomes and thus segregation errors, including during anaphase I [78].
Fig 12. LocusZoom plot for MYO10.
Strongest results from the TDT analyses
Together with the other case/control association tests to identify genes associated with nondisjunction, we also had the opportunity to use the TDT. We assumed that the TDT might help tease apart genes associated with nondisjunction from those associated with fetal “survival genes.” If there is association between maternal genotype and either nondisjunction or survival, this test can theoretically identify it. We only performed this test in our GWAS approach, not our candidate genes, since the candidates were chosen for possible involvement in nondisjunction per se. Here we highlight two genes where the statistical signal was relatively strong, although not genome-wide significant.
The first is located at rs17769147, 24kb upstream of the gene RNF182, which encodes a RING-finger-containing transmembrane protein that includes an E3 ubiquitin ligase activity. There was no statistical signal for any of the case/control comparisons, only for the TDT. Studies have shown that this gene is expressed preferentially in the brain and is up-regulated in Alzheimer’s disease brain and in neuronal cell cultures that are subject to stress-induced cell death [79]. In another line of study, RNF182 was found to be one of the gene targets of MeCP2, the gene involved in Rett syndrome. The group of identified gene targets are involved in the regulation of the cell growth and survival of neuronal cells [80].
Another strong TDT signal was found at rs158866, with no statistical signal for any of the case/control comparisons. This signal is located within NEDD4L (also referred to as NEDD4-2), a gene encoding a ubiquitin ligase. This protein binds and regulates membrane-associated proteins (although not exclusively), particularly ion channels and transporters (reviewed in Goel et al. [81]). NEDD4L interacts with several other proteins and may regulate other important substrates as well (e.g., [82, 83]). Based on current evidence, this ligase is essential for the maintenance of cellular homeostasis.
Discussion
We present, for the first time, a candidate gene study and GWAS of chromosome 21 maternal nondisjunction. The goal of this project was to gain insight into factors that may predispose a woman to this common chromosomal error.
Genes associated with cohesin complex
The meiosis-specific cohesin subunits are encoded by SMC1β, REC8, RAD21L, and STAG3. Of these genes, we found that variation in RAD21L was associated with nondisjunction, with the strongest signal with MI nondisjunction. As part of the cohesion complex [47–50], RAD21L plays a role in the structural maintenance of chromosomes (SMC) complex. The SMC complex includes cohesin, condensin and SMC5/6, and is an important regulator of chromosome dynamics and structure during both mitosis and meiosis. In female mice, mutations in meiosis-specific cohesins and in the SMC complex increase the frequency of oocyte aneuploidy and primary ovarian insufficiency [50, 84–86]. The study of Kong et al. [33] found a highly significant association of RAD21L with male recombination and a much weaker signal in females; Begum et al. [72] did not replicate this finding, although the genomic region had poor coverage in two of the three datasets of their study. Together, this suggests a sex-specific role of RAD21L and one in females that maybe more directly related to segregation of bivalents than recombination counts per se.
There are several possible explanations for why genetic variation in RAD21L was associated with maternal nondisjunction whereas variation in REC8 was not, although both are α-kleisin subunits that are part of the cohesin complex. One possible explanation is that there is reduced power to detect a signal based on allele frequencies of variants in REC8 compared with RAD21L. Alternatively one gene may play a more essential role in meiosis, where variation is not tolerated or not compatible with the oocyte surviving to fertilization. It is known that these genes play unique roles during meiosis and thus have differential effects on the meiotic process [87–89].
Genes associated with the synaptonemal complex (SC)
The general structure of the SC is highly conserved across yeast and mammals, although the genes and proteins involved are not always conserved (reviewed in Cahoon and Hawley [10]). The tripartite protein structure extends along the entire length of the synapsed homologues and assembles alongside cohesin and cohesin-like proteins that hold the sister chromatids of the homologues together (e.g., [90, 91]). Mutations in genes coding for SC components have been identified previously among women with infertility or recurrent miscarriages (reviewed in Geisinger and Benavente [92]). At this time, only mutations in SYCP3 and SYCE1 have been identified, but most studies had <100 women available for study. In our study, we found evidence for an association of variants in genes of all three SC components with maternal nondisjunction. We had supporting evidence for SYCP3 (although not statistically significant), with the strongest statistical signal being found in the comparison of MI vs. MII. In addition, we found a statistically significant association of SYCE2 in MI vs. fathers and with SYCP2 in the comparison of MI vs. MII. Thus, with an increased sample size and a more homogeneous reproductive outcome, we were able to confirm the importance of the SC structure for proper segregation of chromosome during human oogenesis.
Association with recombination-related variants
It is now well established that there is both significant sex-specific and individual variation in genome-wide recombination counts and location of events, in spite of the need for the recombination process to be tightly controlled [93–95]. When there are alterations in the number of recombinants (reduced or no recombination) or their location (pericentromeric or telomeric), there is a high risk for human chromosome nondisjunction [96–98]. Variation in genes that play a role in recombination has been identified and we examined those that were identified in a large Icelandic study using linkage analyses of live births [33]. Begum et al. [72] also attempted to replicate the association of these variants in a population of primarily Northern European ancestry. Their GWAS meta-analyses were extended to the study of recombination phenotypes, including the average recombination count along with those related to placement relative to historical recombination hotspots. Here, we asked whether these variants would also explain susceptibility to maternal nondisjunction. Our results are interesting both with respect the identification of associated regions and to the lack of evidence in the others.
Both Kong et al. [33] and Begum et al. [72] found a strong association of SMEK1 (also known as PPP4R3A) with recombination in females only. Our results identified the strongest association in the comparison of MI vs. MII nondisjunction, which suggests a stage-specific role of this protein, as well as a sex-specific role identified in the recombination studies. At this time, there is no known role of SMEK1 in meiosis. It is a member of the PP2A subfamily. PP2A is involved in de-protection of centromere cohesin in MII of mammals, a process that is essential for proper sister chromatid segregation (reviewed in Wassmann [99]). Although it is intriguing to think that variation in SMEK1 may alter this MII-associated process, this is no direct link at this time.
Another locus that deserves follow-up is on chromosome 8 near rs73178888, one of the top hits in our genome-wide analyses. Begum et al. [72] identified this same region as potentially associated with meiotic recombination in a euploid population. Although the genes in the region do not appear to be linked to recombination, further investigation is warranted.
With respect to the two most well-established genes associated with recombination, RNF212 and PRDM9, our data showed no association with nondisjunction. Variation in RNF212 is sex-specific, some variants being associated with increased recombination in males and others in females [33, 70, 95, 100–102]. RNF212 is known to form many discrete foci along chromosomes early in meiotic prophase I; these foci are then reduced to a few sites where crossovers are formed [103]. For small chromosomes such as chromosome 21, perhaps female-specific variation in this process is less evident compared with genome-wide alterations.
Variation in PRDM9 is known to be associated with recombination hotspots in both males and females. Kong et al. [33] showed that variants were also associated with total recombination counts in males, but not females. Begum et al. [72] provided further evidence that in females, variants were associated with both recombinant counts within and outside of historical hotspots, in opposite directions. They suggested that females might have a compensatory mechanism, such that increased recombination in hotspots is balanced by decreased recombination elsewhere; thereby not altering the overall recombination count. In males, variants were only associated with recombinants within historical hotspots. In our previous study, we found that historical hotspot usage along maternally-derived nondisjoined chromosomes 21 was similar to that in controls, particularly among MI errors, indicating that the observed altered telomeric placement probably does not involve differential hotspot usage [28]. Subsequently, Oliver et al. [104] studied sequence variation in the zinc finger-binding domain (ZFBD) of PRDM9 in a subset of the study sample presented here. They found that the frequency of the PRDM9 ZFBD minor alleles was significantly increased among women who had a chromosome 21 nondisjunction event and no observed recombination on 21q. Further, when these PRDM9 minor alleles were compared with the major A-allele, fewer predicted binding sites on 21q were found. Together, these observations suggest that allelic variation in PRDM9 may play a role in nondisjunction, but that the effect size may be small and it may be limited to nondisjunction of achiasmate chromosomes.
Gene discovery
When we conducted a GWAS, no variants were genome-wide significant; thus, the marginally significant signals need replication prior to additional speculation. We highlighted a few findings in Results for signals in genes that are known to be involved in oogenesis. If these are true signals, our data are consistent with the idea that the underlying susceptibility for chromosome nondisjunction involves different components of oogenesis.
Conclusion and future direction
Our candidate gene study was successful in detecting statistically significant associations of maternal nondisjunction of chromosome 21 and variation in genes that are essential for proper chromosome segregation during meiosis. Future studies are needed to investigate other known risk factors and their interaction with the genetic variation. For example, stratification by maternal age at the time of birth of the infant with trisomy 21 could provide insight into mechanism of the identified genetic variants. In our exploratory analyses, we did not observe unique age-associated variants; however, our sample sizes were limited. Thus, expansion of the study sample with enrichment of the youngest and oldest maternal age groups would be valuable. Also, studies that further stratify meiotic errors by recombination risk patterns known to increase susceptibility of nondisjunction, namely lack of observed recombination, a single telomeric recombination event or a pericentromeric event, may provide further insight into the function of the genetic variant.
Another possible approach is to examine other sources of samples from which information on aneuploidy may be drawn to obtain larger sample sizes. For example McCoy et al. [105] studied day-3 embryos obtained from in vitro fertilization cycles and parents to identify both meiotic and mitotic segregation errors. Irrespective, we have begun to gain insight into which meiotic proteins may be more susceptible to genetic variation, leading to abnormal chromosome segregation. Independent studies are needed to replicate findings from our GWAS study to further identify novel susceptibility genes.
Supporting information
In each Manhattan plot in S1 (and S10), each point is one variant, with the the x-axis representing chromosome number and the y-axis representing -log10(p-value). In each QQ plot, the observed vs. expected quantiles of -log10(p-value) are plotted, with the genomic inflation factor lambda shown below.
(TIFF)
In this Figure (and in S3–S9 Figs) the four LocusZoom plots in a row show the results at one locus across all four analyses. Each point is one variant, with the x- and y-axes representing physical position on the chromosome and -log10(p-value), respectively. Open squares and asterisks represent genotyped and imputed variants, respectively. Coloring represents linkage disequilibrium (red = stronger, blue = weaker) with the tagging SNP (which is purple). The overlaid blue curve shows the recombination rate.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Each row represents one candidate locus (either a gene with a 60kb border on each side or a 60kb window around a SNP). Each column represents an analysis. For each locus-analysis pair, the most significant association at the locus (not always unique) is reported. P-values significant after correcting for multiple testing (i.e., exceeding the Bonferroni-adjusted significance threshold noted in the last column) are marked with an asterisk and highlighted. (MI: meiosis I; MII: meiosis II; P: p-value; OR: odds ratio.) The first 24 loci represent genes selected for their function (above the double line). The latter 13 loci represent SNPs identified by Kong et al. in their GWAS of recombination [33], with annotation in parentheses (below the double line). Note that for each analysis in each gene, S2 Table lists the most statistically significant result within a window, so that the SNP that appears in a given gene is not necessarily the same in each analysis. This also means that some odds ratios appear to “flip” between analyses; for example, a result that shows an odds ratio of 2.0 for one SNP in the MI vs. fathers analysis may be represented as an odds ratio of approximately 0.5 (i.e. 1/2.0) for a nearby SNP in the MII vs. fathers analysis.
(TIFF)
In this table (and in S4–S7 Tables) suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value; OR: odds ratio).
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Acknowledgments
The authors thank Karen Schindler and the Origins of Aneuploidy Research Consortium for helpful discussions of aneuploidy and candidate genes.
Data Availability
Genetic data are available in dbGaP along with specifics of genotyping and QC (dbGaP: phs000718 - https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000718.v1.p1).
Funding Statement
Support for this work was provided by NIH grants R01 HD38979 (PI: S.S.; National Institute of Child Health and Development, National Institutes of Health, Bethesda, MD, USA) and X01 HG007493 (PIs: E.F. and S.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16 Spec No. 2:R203-8. Epub 2007/10/04. 10.1093/hmg/ddm243 . [DOI] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91. Epub 2001/04/03. 10.1038/35066065 . [DOI] [PubMed] [Google Scholar]
- 3.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13(7):493–504. Epub 2012/06/19. 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, et al. Aneuploidy across individual chromosomes at the embryonic level in trophectoderm biopsies: changes with patient age and chromosome structure. J Assist Reprod Genet. 2014;31(11):1501–9. Epub 2014/09/23. 10.1007/s10815-014-0333-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherman SL, Allen EG, Bean LJH. Maternal age and oocyte aneuploidy: lessons Learned from trisomy 21 In: Schlegel P, Fauser B, Carrell D, Racowsky C, editors. Biennial Review of Infertility. New York, NY: Springer; 2013. pp. 69–85 [Google Scholar]
- 6.Hook EB, Mutton DE, Ide R, Alberman E, Bobrow M. The natural history of Down syndrome conceptuses diagnosed prenatally that are not electively terminated. Am J Hum Genet. 1995;57(4):875–81. Epub 1995/10/01. [PMC free article] [PubMed] [Google Scholar]
- 7.Hassold T, Chiu D, Yamane JA. Parental origin of autosomal trisomies. Ann Hum Genet. 1984;48(2):129–44. Epub 1984/05/01. 10.1111/j.1469-1809.1984.tb01008.x . [DOI] [PubMed] [Google Scholar]
- 8.Freeman SB, Allen EG, Oxford-Wright CL, Tinker SW, Druschel C, Hobbs CA, et al. The National Down Syndrome Project: design and implementation. Public Health Rep. 2007;122(1):62–72. Epub 2007/01/24. 10.1177/003335490712200109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolcun-Filas E, Schimenti JC. Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol. 2012;298:179–227. Epub 2012/08/11. 10.1016/B978-0-12-394309-5.00005-5 . [DOI] [PubMed] [Google Scholar]
- 10.Cahoon CK, Hawley RS. Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol. 2016;23(5):369–77. Epub 2016/05/05. 10.1038/nsmb.3208 . [DOI] [PubMed] [Google Scholar]
- 11.Zickler D, Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol. 2015;7(6). Epub 2015/05/20. 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moses MJ. Chromosomal structures in crayfish spermatocytes. J Biophys Biochem Cytol. 1956;2(2):215–8. Epub 1956/03/25. 10.1083/jcb.2.2.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fawcett DW. The fine structure of chromosomes in the meiotic prophase of vertebrate spermatocytes. J Biophys Biochem Cytol. 1956;2(4):403–6. Epub 1956/07/25. 10.1083/jcb.2.4.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5(10):e1000702 Epub 2009/10/24. 10.1371/journal.pgen.1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol. 2011;13(5):599–610. Epub 2011/04/12. 10.1038/ncb2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole F, Keeney S, Jasin M. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 2010;24(12):1201–7. Epub 2010/06/17. 10.1101/gad.1944710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sansam CL, Pezza RJ. Connecting by breaking and repairing: mechanisms of DNA strand exchange in meiotic recombination. FEBS J. 2015;282(13):2444–57. Epub 2015/05/09. 10.1111/febs.13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P, et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 1999;13(5):523–31. Epub 1999/03/11. 10.1101/gad.13.5.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85(7):1125–34. Epub 1996/06/28. 10.1016/s0092-8674(00)81312-4 . [DOI] [PubMed] [Google Scholar]
- 20.Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H Jr., et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14(9):1085–97. Epub 2000/05/16. [PMC free article] [PubMed] [Google Scholar]
- 21.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31(4):385–90. Epub 2002/07/02. 10.1038/ng931 . [DOI] [PubMed] [Google Scholar]
- 22.Plug AW, Peters AH, Keegan KS, Hoekstra MF, de Boer P, Ashley T. Changes in protein composition of meiotic nodules during mammalian meiosis. J Cell Sci. 1998;111 (Pt 4):413–23. Epub 1998/04/04. . [DOI] [PubMed] [Google Scholar]
- 23.Manhart CM, Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst). 2016;38:84–93. Epub 2015/12/22. 10.1016/j.dnarep.2015.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb NE, Sherman SL, Hassold TJ. Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenet Genome Res. 2005;111(3–4):250–5. Epub 2005/09/30. 10.1159/000086896 . [DOI] [PubMed] [Google Scholar]
- 25.Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14(4):400–5. Epub 1996/12/01. 10.1038/ng1296-400 . [DOI] [PubMed] [Google Scholar]
- 26.Lamb NE, Yu K, Shaffer J, Feingold E, Sherman SL. Association between maternal age and meiotic recombination for trisomy 21. Am J Hum Genet. 2005;76(1):91–9. Epub 2004/11/20. 10.1086/427266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver TR, Feingold E, Yu K, Cheung V, Tinker S, Yadav-Shah M, et al. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet. 2008;4(3):e1000033 Epub 2008/03/29. 10.1371/journal.pgen.1000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver TR, Middlebrooks CD, Tinker SW, Allen EG, Bean LJ, Begum F, et al. An examination of the relationship between hotspots and recombination associated with chromosome 21 nondisjunction. PLoS One. 2014;9(6):e99560 Epub 2014/06/14. 10.1371/journal.pone.0099560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver TR, Tinker SW, Allen EG, Hollis N, Locke AE, Bean LJ, et al. Altered patterns of multiple recombinant events are associated with nondisjunction of chromosome 21. Hum Genet. 2012;131(7):1039–46. Epub 2011/12/14. 10.1007/s00439-011-1121-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb NE, Feingold E, Savage A, Avramopoulos D, Freeman S, Gu Y, et al. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum Mol Genet. 1997;6(9):1391–9. Epub 1997/09/01. 10.1093/hmg/6.9.1391 . [DOI] [PubMed] [Google Scholar]
- 31.Middlebrooks CD, Mukhopadhyay N, Tinker SW, Allen EG, Bean LJ, Begum F, et al. Evidence for dysregulation of genome-wide recombination in oocytes with nondisjoined chromosomes 21. Hum Mol Genet. 2014;23(2):408–17. Epub 2013/09/10. 10.1093/hmg/ddt433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown AS, Feingold E, Broman KW, Sherman SL. Genome-wide variation in recombination in female meiosis: a risk factor for non-disjunction of chromosome 21. Hum Mol Genet. 2000;9(4):515–23. Epub 2000/03/04. 10.1093/hmg/9.4.515 . [DOI] [PubMed] [Google Scholar]
- 33.Kong A, Thorleifsson G, Frigge ML, Masson G, Gudbjartsson DF, Villemoes R, et al. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet. 2014;46(1):11–6. Epub 2013/11/26. 10.1038/ng.2833 . [DOI] [PubMed] [Google Scholar]
- 34.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34(6):591–602. Epub 2010/08/19. 10.1002/gepi.20516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Tseng GC, Cheong SY, Bean LJ, Sherman SL, Feingold E. Smarter clustering methods for SNP genotype calling. Bioinformatics. 2008;24(23):2665–71. Epub 2008/10/02. 10.1093/bioinformatics/btn509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19(7):1263–75. Epub 2010/01/08. 10.1093/hmg/ddq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–48. Epub 2006/08/11. 10.1101/gr.5402306 PubMed Central PMCID: PMC1557768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44(6):642–50. Epub 2012/05/09. 10.1038/ng.2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190 Epub 2006/12/30. 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529 Epub 2009/06/23. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. Epub 2012/11/07. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17(R2):R122–8. Epub 2008/10/15. 10.1093/hmg/ddn288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaffer JR, Wang X, Feingold E, Lee M, Begum F, Weeks DE, et al. Genome-wide association scan for childhood caries implicates novel genes. J Dent Res. 2011;90(12):1457–62. Epub 2011/09/24. 10.1177/0022034511422910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993;52(3):506–16. Epub 1993/03/01. [PMC free article] [PubMed] [Google Scholar]
- 45.Kerstann KF, Feingold E, Freeman SB, Bean LJ, Pyatt R, Tinker S, et al. Linkage disequilibrium mapping in trisomic populations: analytical approaches and an application to congenital heart defects in Down syndrome. Genet Epidemiol. 2004;27(3):240–51. Epub 2004/09/25. 10.1002/gepi.20019 . [DOI] [PubMed] [Google Scholar]
- 46.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb). 2005;95(3):221–7. Epub 2005/08/04. 10.1038/sj.hdy.6800717 . [DOI] [PubMed] [Google Scholar]
- 47.Lee J, Hirano T. RAD21L, a novel cohesin subunit implicated in linking homologous chromosomes in mammalian meiosis. J Cell Biol. 2011;192(2):263–76. Epub 2011/01/19. 10.1083/jcb.201008005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011;12(3):267–75. Epub 2011/01/29. 10.1038/embor.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez-Caballero C, Herran Y, Sanchez-Martin M, Suja JA, Barbero JL, Llano E, et al. Identification and molecular characterization of the mammalian alpha-kleisin RAD21L. Cell Cycle. 2011;10(9):1477–87. Epub 2011/04/30. 10.4161/cc.10.9.15515 . [DOI] [PubMed] [Google Scholar]
- 50.Herran Y, Gutierrez-Caballero C, Sanchez-Martin M, Hernandez T, Viera A, Barbero JL, et al. The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. EMBO J. 2011;30(15):3091–105. Epub 2011/07/12. 10.1038/emboj.2011.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400(6743):461–4. Epub 1999/08/10. 10.1038/22774 . [DOI] [PubMed] [Google Scholar]
- 52.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98(1):91–103. Epub 1999/07/21. 10.1016/S0092-8674(00)80609-1 . [DOI] [PubMed] [Google Scholar]
- 53.Minase G, Miyamoto T, Miyagawa Y, Iijima M, Ueda H, Saijo Y, et al. Single-nucleotide polymorphisms in the human RAD21L gene may be a genetic risk factor for Japanese patients with azoospermia caused by meiotic arrest and Sertoli cell-only syndrome. Hum Fertil (Camb). 2017;20(3):217–20. Epub 2017/06/22. 10.1080/14647273.2017.1292004 . [DOI] [PubMed] [Google Scholar]
- 54.Pezza RJ, Voloshin ON, Volodin AA, Boateng KA, Bellani MA, Mazin AV, et al. The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res. 2014;42(4):2346–57. Epub 2013/12/07. 10.1093/nar/gkt1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerzendorfer C, Vignard J, Pedrosa-Harand A, Siwiec T, Akimcheva S, Jolivet S, et al. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J Cell Sci. 2006;119(Pt 12):2486–96. Epub 2006/06/10. 10.1242/jcs.02967 . [DOI] [PubMed] [Google Scholar]
- 56.Domenichini S, Raynaud C, Ni DA, Henry Y, Bergounioux C. Atmnd1-delta1 is sensitive to gamma-irradiation and defective in meiotic DNA repair. DNA Repair (Amst). 2006;5(4):455–64. Epub 2006/01/31. 10.1016/j.dnarep.2005.12.007 . [DOI] [PubMed] [Google Scholar]
- 57.Zhao W, Sung P. Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res. 2015;43(8):4055–66. Epub 2015/03/31. 10.1093/nar/gkv259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee DH, Goodarzi AA, Adelmant GO, Pan Y, Jeggo PA, Marto JA, et al. Phosphoproteomic analysis reveals that PP4 dephosphorylates KAP-1 impacting the DNA damage response. EMBO J. 2012;31(10):2403–15. Epub 2012/04/12. 10.1038/emboj.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoon YS, Lee MW, Ryu D, Kim JH, Ma H, Seo WY, et al. Suppressor of MEK null (SMEK)/protein phosphatase 4 catalytic subunit (PP4C) is a key regulator of hepatic gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107(41):17704–9. Epub 2010/09/30. 10.1073/pnas.1012665107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim BR, Seo SH, Park MS, Lee SH, Kwon Y, Rho SB. sMEK1 inhibits endothelial cell proliferation by attenuating VEGFR-2-dependent-Akt/eNOS/HIF-1alpha signaling pathways. Oncotarget. 2015;6(31):31830–43. Epub 2015/09/18. 10.18632/oncotarget.5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi SH, Ruggiero D, Sorice R, Song C, Nutile T, Vernon Smith A, et al. Six Novel Loci Associated with Circulating VEGF Levels Identified by a Meta-analysis of Genome-Wide Association Studies. PLoS Genet. 2016;12(2):e1005874 Epub 2016/02/26. 10.1371/journal.pgen.1005874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McFee RM, Cupp AS. Vascular contributions to early ovarian development: potential roles of VEGFA isoforms. Reprod Fertil Dev. 2013;25(2):333–42. Epub 2012/10/02. 10.1071/RD12134 . [DOI] [PubMed] [Google Scholar]
- 63.Robinson RS, Woad KJ, Hammond AJ, Laird M, Hunter MG, Mann GE. Angiogenesis and vascular function in the ovary. Reproduction. 2009;138(6):869–81. Epub 2009/09/30. 10.1530/REP-09-0283 . [DOI] [PubMed] [Google Scholar]
- 64.Kim AM, Vogt S, O'Halloran TV, Woodruff TK. Zinc availability regulates exit from meiosis in maturing mammalian oocytes. Nat Chem Biol. 2010;6(9):674–81. Epub 2010/08/10. 10.1038/nchembio.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim AM, Bernhardt ML, Kong BY, Ahn RW, Vogt S, Woodruff TK, et al. Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem Biol. 2011;6(7):716–23. Epub 2011/04/30. 10.1021/cb200084y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lisle RS, Anthony K, Randall MA, Diaz FJ. Oocyte-cumulus cell interactions regulate free intracellular zinc in mouse oocytes. Reproduction. 2013;145(4):381–90. Epub 2013/02/14. 10.1530/REP-12-0338 . [DOI] [PubMed] [Google Scholar]
- 67.Dieterich K, Zouari R, Harbuz R, Vialard F, Martinez D, Bellayou H, et al. The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet. 2009;18(7):1301–9. Epub 2009/01/17. 10.1093/hmg/ddp029 . [DOI] [PubMed] [Google Scholar]
- 68.Fellmeth JE, Ghanaim EM, Schindler K. Characterization of macrozoospermia-associated AURKC mutations in a mammalian meiotic system. Hum Mol Genet. 2016;25(13):2698–711. Epub 2016/04/24. 10.1093/hmg/ddw128 . [DOI] [PubMed] [Google Scholar]
- 69.Adriaenssens T, Mazoyer C, Segers I, Wathlet S, Smitz J. Differences in collagen expression in cumulus cells after exposure to highly purified menotropin or recombinant follicle-stimulating hormone in a mouse follicle culture model. Biol Reprod. 2009;80(5):1015–25. Epub 2009/01/24. 10.1095/biolreprod.107.067462 . [DOI] [PubMed] [Google Scholar]
- 70.Baumann C, Wang X, Yang L, Viveiros MM. Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J Cell Sci. 2017;130(7):1251–62. Epub 2017/02/15. 10.1242/jcs.196188 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holubcova Z, Blayney M, Elder K, Schuh M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science. 2015;348(6239):1143–7. Epub 2015/06/06. 10.1126/science.aaa9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Begum F, Chowdhury R, Cheung VG, Sherman SL, Feingold E. Genome-Wide Association Study of Meiotic Recombination Phenotypes. G3 (Bethesda). 2016;6(12):3995–4007. Epub 2016/10/14. 10.1534/g3.116.035766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mao S, Wu F, Cao X, He M, Liu N, Wu H, et al. TDRP deficiency contributes to low sperm motility and is a potential risk factor for male infertility. Am J Transl Res. 2016;8(1):177–87. Epub 2016/04/14. [PMC free article] [PubMed] [Google Scholar]
- 74.Chen PJ, Huang YS. CPEB2-eEF2 interaction impedes HIF-1alpha RNA translation. EMBO J. 2012;31(4):959–71. Epub 2011/12/14. 10.1038/emboj.2011.448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurihara Y, Tokuriki M, Myojin R, Hori T, Kuroiwa A, Matsuda Y, et al. CPEB2, a novel putative translational regulator in mouse haploid germ cells. Biol Reprod. 2003;69(1):261–8. Epub 2003/04/04. 10.1095/biolreprod.103.015677 . [DOI] [PubMed] [Google Scholar]
- 76.Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431(7006):325–9. Epub 2004/09/17. 10.1038/nature02834 . [DOI] [PubMed] [Google Scholar]
- 77.Samwer M, Dehne HJ, Spira F, Kollmar M, Gerlich DW, Urlaub H, et al. The nuclear F-actin interactome of Xenopus oocytes reveals an actin-bundling kinesin that is essential for meiotic cytokinesis. EMBO J. 2013;32(13):1886–902. Epub 2013/06/04. 10.1038/emboj.2013.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mogessie B, Schuh M. Actin protects mammalian eggs against chromosome segregation errors. Science. 2017;357(6353). Epub 2017/08/26. 10.1126/science.aal1647 . [DOI] [PubMed] [Google Scholar]
- 79.Liu QY, Lei JX, Sikorska M, Liu R. A novel brain-enriched E3 ubiquitin ligase RNF182 is up regulated in the brains of Alzheimer's patients and targets ATP6V0C for degradation. Mol Neurodegener. 2008;3:4 Epub 2008/02/27. 10.1186/1750-1326-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nectoux J, Fichou Y, Rosas-Vargas H, Cagnard N, Bahi-Buisson N, Nusbaum P, et al. Cell cloning-based transcriptome analysis in Rett patients: relevance to the pathogenesis of Rett syndrome of new human MeCP2 target genes. J Cell Mol Med. 2010;14(7):1962–74. Epub 2010/06/24. 10.1111/j.1582-4934.2010.01107.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557(1):1–10. Epub 2014/11/30. 10.1016/j.gene.2014.11.051 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Low LH, Chow YL, Li Y, Goh CP, Putz U, Silke J, et al. Nedd4 family interacting protein 1 (Ndfip1) is required for ubiquitination and nuclear trafficking of BRCA1-associated ATM activator 1 (BRAT1) during the DNA damage response. J Biol Chem. 2015;290(11):7141–50. Epub 2015/01/30. 10.1074/jbc.M114.613687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu C, Fan CD, Wang X. Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase. Oncogene. 2015;34(3):281–9. Epub 2014/01/15. 10.1038/onc.2013.557 . [DOI] [PubMed] [Google Scholar]
- 84.Hodges CA, Revenkova E, Jessberger R, Hassold TJ, Hunt PA. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat Genet. 2005;37(12):1351–5. Epub 2005/11/01. 10.1038/ng1672 . [DOI] [PubMed] [Google Scholar]
- 85.Murdoch B, Owen N, Stevense M, Smith H, Nagaoka S, Hassold T, et al. Altered cohesin gene dosage affects Mammalian meiotic chromosome structure and behavior. PLoS Genet. 2013;9(2):e1003241 Epub 2013/02/15. 10.1371/journal.pgen.1003241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hwang G, Sun F, O'Brien M, Eppig JJ, Handel MA, Jordan PW. SMC5/6 is required for the formation of segregation-competent bivalent chromosomes during meiosis I in mouse oocytes. Development. 2017;144(9):1648–60. Epub 2017/03/18. 10.1242/dev.145607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward A, Hopkins J, McKay M, Murray S, Jordan PW. Genetic Interactions Between the Meiosis-Specific Cohesin Components, STAG3, REC8, and RAD21L. G3 (Bethesda). 2016;6(6):1713–24. Epub 2016/05/14. 10.1534/g3.116.029462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Biswas U, Hempel K, Llano E, Pendas A, Jessberger R. Distinct Roles of Meiosis-Specific Cohesin Complexes in Mammalian Spermatogenesis. PLoS Genet. 2016;12(10):e1006389 Epub 2016/10/30. 10.1371/journal.pgen.1006389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agostinho A, Manneberg O, van Schendel R, Hernandez-Hernandez A, Kouznetsova A, Blom H, et al. High density of REC8 constrains sister chromatid axes and prevents illegitimate synaptonemal complex formation. EMBO Rep. 2016;17(6):901–13. Epub 2016/05/14. 10.15252/embr.201642030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McNicoll F, Stevense M, Jessberger R. Cohesin in gametogenesis. Curr Top Dev Biol. 2013;102:1–34. Epub 2013/01/05. 10.1016/B978-0-12-416024-8.00001-5 . [DOI] [PubMed] [Google Scholar]
- 91.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. Epub 2000/02/26. 10.1146/annurev.genet.33.1.603 . [DOI] [PubMed] [Google Scholar]
- 92.Geisinger A, Benavente R. Mutations in Genes Coding for Synaptonemal Complex Proteins and Their Impact on Human Fertility. Cytogenet Genome Res. 2016;150(2):77–85. Epub 2016/12/21. 10.1159/000453344 . [DOI] [PubMed] [Google Scholar]
- 93.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63(3):861–9. Epub 1998/08/27. 10.1086/302011 PubMed Central PMCID: PMC1377399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheung VG, Burdick JT, Hirschmann D, Morley M. Polymorphic variation in human meiotic recombination. Am J Hum Genet. 2007;80(3):526–30. Epub 2007/02/03. 10.1086/512131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fledel-Alon A, Leffler EM, Guan Y, Stephens M, Coop G, Przeworski M. Variation in human recombination rates and its genetic determinants. PLoS One. 2011;6(6):e20321 Epub 2011/06/24. 10.1371/journal.pone.0020321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hou Y, Fan W, Yan L, Li R, Lian Y, Huang J, et al. Genome analyses of single human oocytes. Cell. 2013;155(7):1492–506. Epub 2013/12/24. 10.1016/j.cell.2013.11.040 . [DOI] [PubMed] [Google Scholar]
- 97.Brieno-Enriquez MA, Cohen PE. Double trouble in human aneuploidy. Nat Genet. 2015;47(7):696–8. Epub 2015/06/27. 10.1038/ng.3344 . [DOI] [PubMed] [Google Scholar]
- 98.MacLennan M, Crichton JH, Playfoot CJ, Adams IR. Oocyte development, meiosis and aneuploidy. Semin Cell Dev Biol. 2015;45:68–76. Epub 2015/10/11. 10.1016/j.semcdb.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wassmann K. Sister chromatid segregation in meiosis II: deprotection through phosphorylation. Cell Cycle. 2013;12(9):1352–9. Epub 2013/04/12. 10.4161/cc.24600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319(5868):1395–8. Epub 2008/02/02. 10.1126/science.1151851 . [DOI] [PubMed] [Google Scholar]
- 101.Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5(9):e1000648 Epub 2009/09/19. 10.1371/journal.pgen.1000648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A, Gudbjartsson DF, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319(5868):1398–401. Epub 2008/02/02. 10.1126/science.1152422 . [DOI] [PubMed] [Google Scholar]
- 103.Qiao H, Prasada Rao HB, Yang Y, Fong JH, Cloutier JM, Deacon DC, et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet. 2014;46(2):194–9. Epub 2014/01/07. 10.1038/ng.2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oliver TR, Middlebrooks C, Harden A, Scott N, Johnson B, Jones J, et al. Variation in the Zinc Finger of PRDM9 is Associated with the Absence of Recombination along Nondisjoined Chromosomes 21 of Maternal Origin. J Down Syndr Chromosom Abnorm. 2016;2(2). Epub 2017/07/14. 10.4172/2472-1115.1000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, et al. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science. 2015;348(6231):235–8. Epub 2015/04/11. 10.1126/science.aaa3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In each Manhattan plot in S1 (and S10), each point is one variant, with the the x-axis representing chromosome number and the y-axis representing -log10(p-value). In each QQ plot, the observed vs. expected quantiles of -log10(p-value) are plotted, with the genomic inflation factor lambda shown below.
(TIFF)
In this Figure (and in S3–S9 Figs) the four LocusZoom plots in a row show the results at one locus across all four analyses. Each point is one variant, with the x- and y-axes representing physical position on the chromosome and -log10(p-value), respectively. Open squares and asterisks represent genotyped and imputed variants, respectively. Coloring represents linkage disequilibrium (red = stronger, blue = weaker) with the tagging SNP (which is purple). The overlaid blue curve shows the recombination rate.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Each row represents one candidate locus (either a gene with a 60kb border on each side or a 60kb window around a SNP). Each column represents an analysis. For each locus-analysis pair, the most significant association at the locus (not always unique) is reported. P-values significant after correcting for multiple testing (i.e., exceeding the Bonferroni-adjusted significance threshold noted in the last column) are marked with an asterisk and highlighted. (MI: meiosis I; MII: meiosis II; P: p-value; OR: odds ratio.) The first 24 loci represent genes selected for their function (above the double line). The latter 13 loci represent SNPs identified by Kong et al. in their GWAS of recombination [33], with annotation in parentheses (below the double line). Note that for each analysis in each gene, S2 Table lists the most statistically significant result within a window, so that the SNP that appears in a given gene is not necessarily the same in each analysis. This also means that some odds ratios appear to “flip” between analyses; for example, a result that shows an odds ratio of 2.0 for one SNP in the MI vs. fathers analysis may be represented as an odds ratio of approximately 0.5 (i.e. 1/2.0) for a nearby SNP in the MII vs. fathers analysis.
(TIFF)
In this table (and in S4–S7 Tables) suggestive associations (p < 10−5) are recorded (highlighted cells). For each such locus, the most significant association within 20kb is recorded for each of the other four genome-wide analyses. Rows are ordered by significance. (MI: meiosis I; MII: meiosis II; P: p-value; OR: odds ratio).
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Data Availability Statement
Genetic data are available in dbGaP along with specifics of genotyping and QC (dbGaP: phs000718 - https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000718.v1.p1).