Abstract
Background:
Electronic medical record (EMR) systems need functionality that decreases cognitive overload by drawing the clinician’s attention to the right data, at the right time. We developed a Learning EMR (LEMR) system that learns statistical models of clinician information-seeking behavior and applies those models to direct the display of data in future patients. We evaluated the performance of the system in identifying relevant patient data in intensive care unit (ICU) patient cases.
Methods:
To capture information-seeking behavior, we enlisted critical care medicine physicians who reviewed a set of patient cases and selected data items relevant to the task of presenting at morning rounds. Using patient EMR data as predictors, we built machine learning models to predict their relevancy. We prospectively evaluated the predictions of a set of high performing models.
Results:
On an independent evaluation data set, 25 models achieved precision of 0.52, 95% CI [0.49, 0.54] and recall of 0.77, 95% CI [0.75, 0.80] in identifying relevant patient data items. For data items missed by the system, the reviewers rated the effect of not seeing those data from no impact to minor impact on patient care in about 82% of the cases.
Conclusion:
Data-driven approaches for adaptively displaying data in EMR systems, like the LEMR system, show promise in using information-seeking behavior of clinicians to identify and highlight relevant patient data.
Keywords: Electronic medical records, Machine learning, Critical care, Information-seeking behavior
1. Introduction
With increasing amounts of patient data being captured by electronic medical record (EMR) systems, intelligent integration and presentation of data to clinicians is a vital need. In complex and dynamic workplaces, such as the intensive care unit (ICU), decision making is highly reliant on situational awareness of a patient’s condition. In the ICU, clinicians are required to peruse a large amount of continuously changing patient data, determine what data are relevant, synthesize the relevant data, and act on the assessment [1].
In the ICU, and in other settings, patient data accumulate rapidly. A study estimated that about 1348 data points are generated per patient per day in the pediatric ICU [2]. Clinicians face cognitive overload in all three aspects of cognitive processes that underpin situational awareness: observation of relevant patient data, comprehension of the meaning and synthesis of the data, and prediction of future patient states and clinical events. In current EMR systems, data are scattered across many places and clinicians laboriously navigate, view, and filter out irrelevant data. The increased amount of patient data and their location in various places in the EMR may lead to cognitive overload and patient data may be inadvertently missed [3]. Cognitive overload is detrimental to both clinicians and patients because it may prevent a clinician from rapidly identifying, aggregating, and synthesizing relevant data for clinical tasks. Advanced EMR systems should aim to provide clinicians with excellent situational awareness [4]. In particular, it is imperative to improve the usability of EMR systems with functionality that draws the clinician’s attention to the right data for the right patient at the right time [5].
As a step towards developing an EMR system that decreases cognitive overload, we developed a Learning EMR (LEMR) system that focuses on highlighting relevant data for clinical assessment and decision-making. We hypothesized that predictive modeling could be used to automatically highlight relevant information. The purpose of this study was to create those predictive models and evaluate their ability to highlight patient data that are most relevant to clinicians.
2. Background
Investigators have explored several approaches to display patient data for clinical tasks that enable rapid comprehension of the overall state of the patient and characterization of significant changes in the clinical course [6]. Examples of approaches for integrated data display include summarizing using graphs [7], summarizing using temporal trends, and organizing data in organ system-based [8,9] or disease-focused frameworks [10,11]. Graphical summaries of data take advantage of the acute perceptual capabilities of humans. For example, Anders et al. demonstrated that a graphical trending display helped ICU nurses to better synthesize information about patients’ evolving clinical conditions, when compared to a tabular display [12]. Much of patient data are temporal and investigators have explored ways to summarize and display clinical time series data. For example, Klimov et al. developed methods for intelligently visualizing and exploring time-oriented medical records [13], and Post et al. developed temporal abstractions to automatically identify temporal patterns in clinical data [14]. Rind et al. surveyed a comprehensive range of visualization system designs based on timeline exploration of clinical data [15]. Wright et al. published a systematic review of display approaches and design frameworks that are applicable to the ICU [16].
Approaches for organized display of patient data in EMR systems can be broadly categorized into two groups: knowledge-based and data-driven. Knowledge-based approaches typically use rules that are derived from general medical knowledge or the experience of clinical experts [17]. For example, easily derived rules from medical knowledge are used to indicate out of range laboratory test values and physiological measurements. More sophisticated approaches, such as the AWARE system, group ICU patient data by organ system, which is a common way for ICU clinicians to summarize data [18]. Knowledge-based approaches have several advantages including that they are likely to be clinically useful and they can be readily implemented in EMR systems. However, such approaches suffer from several disadvantages. Collecting clinical experience data and manually constructing rules is typically tedious and time-consuming [19,20]. Knowledge-based systems are expensive to revise, tune, and update, and furthermore they have limited coverage of the large variety of clinical scenarios.
In contrast to knowledge-based approaches, data-driven approaches can automatically derive patterns from data. Using order-entry data in EMR systems, Klann et al. derived Bayesian network models that can be used to produce patient-specific order menus. Such menus were shown to help physicians in writing complete order sets faster [21]. Gambino et al. created a framework for automatic generation of adaptive and customizable toolbars and menus in medical imaging that is data-driven [22]. Using patient-management activity data in ICU EMR systems, Hauskrecht et al. developed support vector machine classifiers that generate alerts about potential errors in orders entered by clinicians. Such alerts were judged to be clinically useful about 50% of the time [23,24]. A major advantage of such data-driven approaches is that the capacity to learn is limited in scope only by the collective experience that is embodied in the data contained in a health system’s EMR systems. Data-driven approaches can construct sophisticated statistical models that predict clinician information-seeking behavior and update the models rapidly based on new data. Moreover, data-driven approaches are applicable to a wide range of care settings and tasks; hence the coverage is broader than knowledge-based approaches. The LEMR prototype system that we describe in this paper is an example of a data-driven approach for adapting the display of patient data to the needs of the user.
3. Methods
We designed and implemented a prototype LEMR system and used it with past ICU patient cases in our experiments. The LEMR system (1) uses information-seeking behavior of clinicians in viewing past cases to train statistical models to predict which data items are relevant in a current case, and (2) applies these models to automatically highlight data (e.g., vital signs, laboratory test results, and medication orders) that are relevant to a patient’s current clinical condition. To develop a functioning LEMR system, we obtain information-seeking behavior from clinician reviewers who assess past patient cases. However, in a mature LEMR system, such information-seeking behavior would be inferred automatically from clinician user activity in using the EMR.
We created a training data set from past patient cases that were annotated by critical care medicine physicians (i.e., intensivists) for relevant patient data, derived multiple statistical models to predict data relevancy, and evaluated the performance of the models in an independent evaluation data set. We restricted the cases in the training and evaluation data sets to patients with either acute kidney failure or acute respiratory failure that are two common conditions in the ICU.
3.1. The learning EMR system
The prototype LEMR system that we developed can be used to capture information-seeking behavior as well as apply models learned from that data to highlight relevant patient data in an EMR. The current prototype system does not interact with EMR systems that are currently in use in healthcare settings. Our goal was not to replicate an entire modern EMR but to have a useful prototype for displaying patient data that mimics a real EMR in important ways that would support our conducting studies of it in a laboratory setting.
The LEMR system consists of three main components: (1) a user interface implemented in a web browser using HTML, CSS, JavaScript, and Django, (2) a MySQL database for storing patient data, and (3) a module containing statistical models (Fig. 1). The system functions in two modes. In the training mode, the user reviews patient cases and selects data items that are relevant for a specified clinical task. In the evaluation mode, statistical models are applied to new patient cases to identify and highlight data items that are predicted to be sought by the user, and the user reviews highlighted data items in the context of the cases and provides responses to study questions.
Fig. 1.
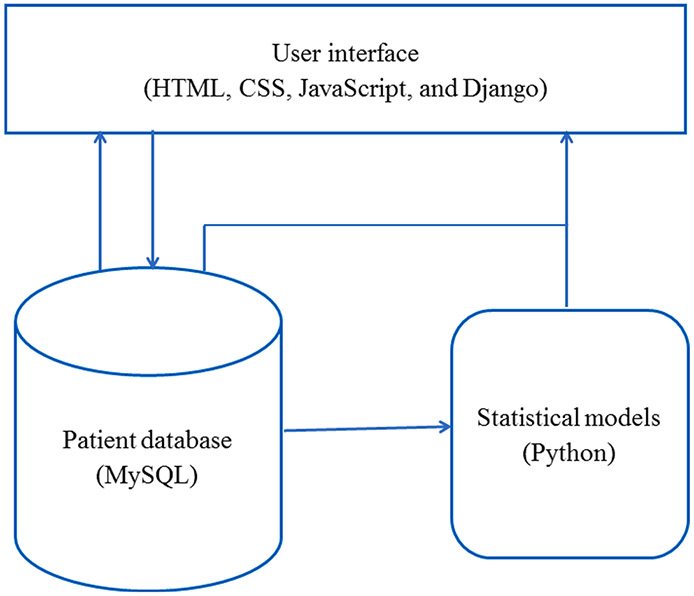
Components of the LEMR system.
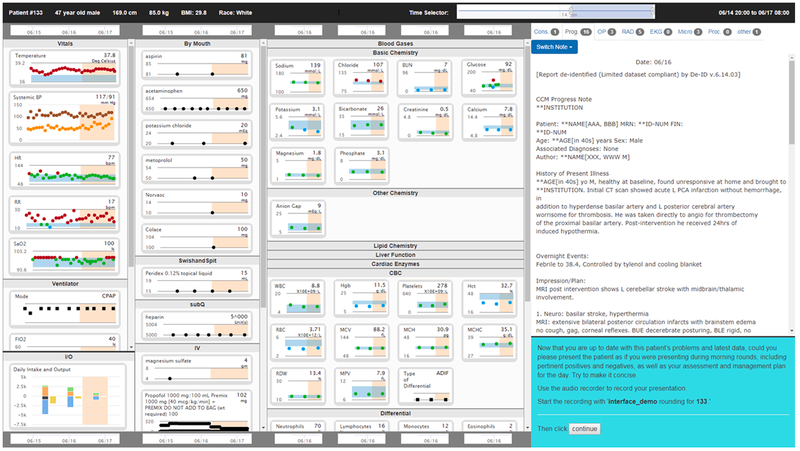
The LEMR user interface displays a patient case in a compact manner. Structured data are shown in graphical time series plots and free-text notes are shown in a separate area in the interface (see Fig. 2). In addition to display of patient data, the interface can record selections made by the user (in training mode) and can highlight patient data that are predicted to be sought by the user through the application of statistical models (in evaluation mode). The LEMR user interface does not support all functionalities and accessibility standards of a commercial EMR system since it was developed to support research studies.
Fig. 2.
The LEMR user interface displays patient data in scrollable columns. Patient data are arranged in four vertical columns: the left-most column displays vital sign measurements, ventilator settings, and fluid intake and output; the second column shows medication administrations; the third column displays laboratory test results; and the right-most column displays free-text notes and reports. Study-related instructions are shown in the lower right-hand corner.
3.2. Clinical task
During ICU morning rounds, a multidisciplinary clinical team evaluates and makes decisions for each patient. For each patient, one member of the team typically reviews relevant data in the EMR system and prepares a summary of the patient’s clinical status. During rounds, this team member delivers an oral presentation of the summary to the team. The task of preparing the summary is often time-consuming, with the clinician laboriously combing the EMR system to identify and retrieve relevant data. To evaluate the LEMR system, we chose the task of identifying relevant patient data for presenting a summary of the patient’s clinical status at morning rounds.
3.3. Training data set
Using the LEMR system in training mode, we showed a set of ICU patient cases to critical care medicine physicians who reviewed the cases and selected data items that were relevant to the task of preparing a summary of each case for presentation at morning rounds. One-hundred seventy-eight patient cases were selected randomly from patients who were admitted between June 2010 and May 2012 to an ICU at the University of Pittsburgh Medical Center, with a diagnosis of either acute kidney failure (AKF; ICD-9 584.9 or 584.5; 93 cases) or acute respiratory failure (ARF; ICD-9 518.81; 85 cases). EMR data were extracted from a research database [25] and a clinical data warehouse [26]. The procedure for case review and selection of data items included three tasks:
Task 1 (familiarization): A random day between day two of admission to the ICU and the day before discharge from the ICU was selected as the “past patient stay.” All available EMR data up until 8:00 am on the day selected for the past patient stay were displayed to the reviewer. The reviewer was instructed to “use the available information to become familiar with the patient case as if they are one of your own patients.” After becoming familiar with the case, the reviewer clicked on a button to advance to Task 2.
Task 2 (preparation): An additional day (from 8:00 am on the day selected for the past patient stay to 8:00 am on the next day i.e., “current time”) of the patient’s EMR data were added to the display. The reviewer was prompted with a dialog box that stated “24-hours have passed” and directed to “use the available information to prepare to present the case during morning rounds.” After preparation was complete, the reviewer clicked on a button to advance to Task 3.
Task 3 (selection): For this task, each available data item (e.g., glucose levels, insulin dosage regimen) was shown with an accompanying checkbox. The reviewer was directed to “select the information you consider pertinent when preparing to present this case at morning rounds.” The reviewer selected relevant data items by toggling the accompanying checkbox to the checked state. Data on information-seeking behavior were collected during Task 3; data selections made by the reviewers were recorded in the LEMR database and used to create the training data set.
3.4. Derivation of statistical models
Patient data and selections made by reviewers recorded in the LEMR system were processed into a representation that is suitable for machine learning. We applied three machine learning algorithms and computed the performance of the models on the training data.
3.4.1. Data representation and processing
A predictor variable denotes any patient data item and includes observations, measurements, actions, or other information that are available from the EMR system. Examples of predictor variables include diagnosis, demographics, laboratory test results, vital sign measurements, ventilator settings, and medication administrations. A predictor value is the value that a predictor variable takes in a patient. For example, consider a patient with diabetes mellitus in whom glucose levels are recorded in the EMR. Then diagnosis = diabetes mellitus denotes that the predictor variable diagnosis has the value diabetes mellitus and glucose = 85, 100, 90, 105 mg/dL denotes that the predictor variable glucose consists of a series of serum glucose levels over a period of time.
A target variable (or simply target) is any patient data item that a clinician can potentially seek as relevant for a specific task in a specific patient. Any observation, measurement, action, or other information that is available for a patient in the EMR and sought by a clinician is a target. To distinguish a target from a predictor variable with the same name, the word “target” is appended to the name of the target. Thus, glucose refers to a predictor variable that consists of glucose levels in a patient while glucose target refers to a target that encodes clinician information-seeking behavior. A target, in a given context, takes only two values; it is assigned the value yes if it is available for a patient and a clinician sought it for the given task, and it is assigned the value no if it is available for a patient, but a clinician did not seek it. It is not defined if it was not measured for the patient. For example, for a patient with diabetes mellitus, glucose target = yes denotes that the target variable glucose target contained clinical data and was sought by a clinician. Consider a different patient who has kidney failure and glucose levels are recorded but are not sought by a clinician. Then, glucose target = no denotes that the target variable glucose target contained clinical data but was not sought by a clinician. Finally, glucose target = missing denotes that glucose levels were not measured for a patient and, therefore, this target is not available for selection by a reviewer. Since target data are not readily available in currently deployed EMR systems, we obtained them from clinician reviewers in a laboratory setting using the LEMR system, as described in the previous section.
Predictor variables include simple atemporal variables (e.g., diagnosis), as well as more complex variables captured as time series measurements (e.g., glucose). We use the term variables to denote raw patient data items that are extracted from the EMR system (e.g., glucose levels) and the term features to denote functions of those variables (e.g., most recent measurement of a glucose time series). We construct features from predictor variables as described below [23]:
Atemporal variables comprised of diagnosis and demographics that included age, height, weight, body mass index, sex, race and length of ICU stay. For each atemporal variable, we generate a single feature that is assigned a single value for a patient for the duration of ICU stay (e.g., sex = female).
Complex variables consisting of time series data, including laboratory test results, vital signs, ventilator settings and medication administrations. For each medication variable, we generate four features including an indicator of whether the drug is currently prescribed, the time elapsed between first administration and the current time, the time elapsed between the most recent administration and the current time, and the dose at the most recent administration. For each laboratory test result, vital sign and ventilator setting, we generate up to 35 features including an indicator of whether the event or measurement ever occurred, the value of the most recent measurement, the highest value, the lowest value, the slope between the two most recent values, and 30 other features. The complete list of features derived from each type of complex variable is shown in Supplementary Table 1.
Target variables are treated like atemporal predictor variables when they are translated into features. Thus, the glucose target (i.e., whether glucose levels are sought in a given context) is translated into a single target feature (glucose target = yes, no, or undefined) in contrast to the glucose predictor which is expanded into a set of 35 features.
A patient instance (or simply instance or sample) is a vector of (predictor) feature values and corresponding target values derived from data from a subinterval of a patient’s ICU stay defined from the time of admission to the ICU to the current time. The vector of feature values summarizes the evolution of the patient’s clinical status from the time of admission to the current time. A data set (e.g., a training data set) is a collection of patient instances.
3.4.2. Model derivation
In the data representation just described, the temporal aspects of the predictor variables are implicitly summarized in the vector of feature values, and such a representation enables standard machine learning methods to be applied. To train a predictive model for the glucose target, for example, we train on all feature values and the corresponding glucose target values (yes or no) of a set of instances to predict if glucose level is relevant. By changing the target, a predictive model is trained for each laboratory test, vital sign, and ventilator setting, and medication.
Two practical challenges with deriving models in this data set are the large number of missing values and high dimensionality. We imputed missing values of features using two different methods. In the first method, they were imputed with the median. In the second method, continuous predictor variables were imputed with linear regression and discrete predictor variables were imputed with logistic regression. Both regression methods used all available features to predict a feature of interest. Both imputation methods were applied, creating two distinct data sets (a median imputed data set and a regression imputed data set).
To reduce dimensionality, we applied a two-step feature selection procedure that is similar to the procedure described in Hauskrecht et al. [23] First, for each set of features constructed from a single variable (e.g., blood glucose levels that is translated into a set of features that include the value of the most recent measurement, the slope between the two most recent values, and other features), we evaluate if the set is predictive of the target by itself. Any set of features with an area under the Receiver Operator Characteristic (AUROC) curve of less than 0.6 is removed. The features that remain after the first step are reduced further using recursive feature elimination and cross-validation (RFECV in scikit-learn version 0.19 [27]). The final set of features is used for model construction. Feature selection is target specific, so it was done separately for each target variable.
Three different machine learning algorithms were applied: lasso logistic regression, support vector machines, and random forests. Models were constructed and evaluated using leave-one-out cross-validation. Imputation and feature selection steps were performed within the cross folds. For all methods, we used implementations in scikit-learn Version 0.19 [27]. The best performing combination of imputation method and machine learning algorithm for each target was considered for inclusion in the evaluation study.
3.5. Evaluation
To evaluate the LEMR system, we applied high performing models derived from the training set to an evaluation data set, and the model predictions were assessed by critical care medicine physicians.
We randomly selected 18 ICU patients who were not included in the training data set and who were admitted between May 2012 and December 2012 to an ICU and had a diagnosis of either AKF (9 patients) or ARF (9 patients). Models that obtained a cross-validated precision of at least 0.67 and recall of at least 0.50 on the training data set were applied to the new cases. We chose these thresholds to balance between highlighting accuracy and completeness. Using the LEMR system in evaluation mode, we showed the evaluation patient cases (with predicted-to-be-relevant data items highlighted) to critical care medicine physicians who reviewed the cases and provided feedback using the following procedure:
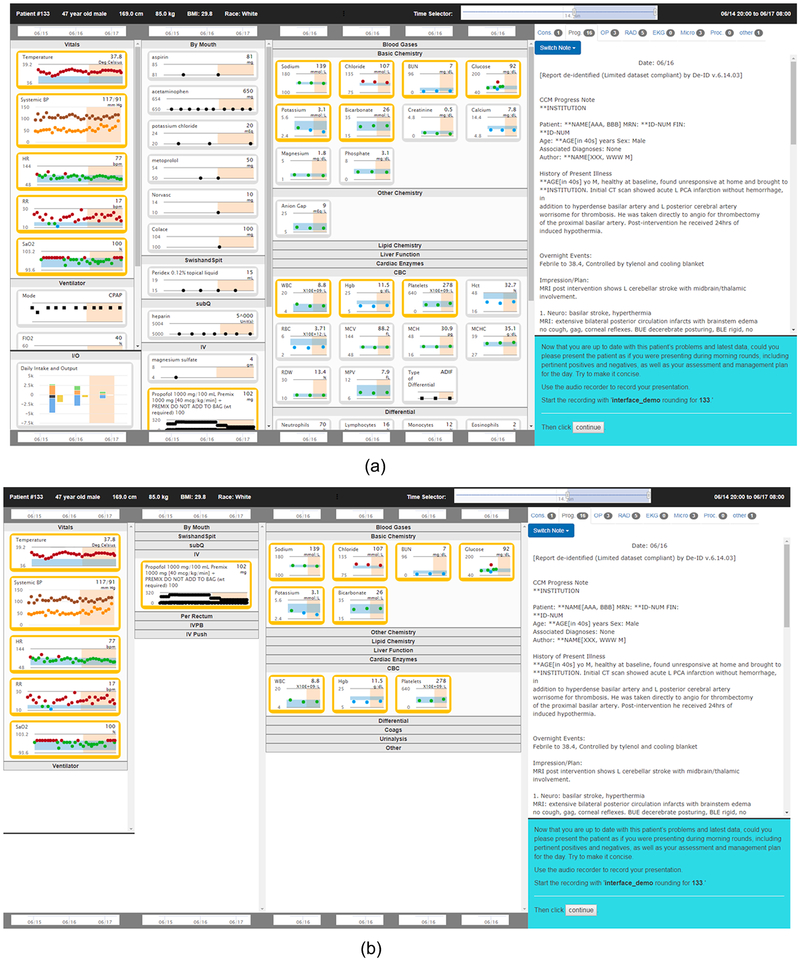
The evaluation consisted of three arms that included a control arm and two intervention arms. In the control arm, the procedure for reviewing the case was similar to the procedure used in the training phase. In the highlights in place intervention arm, the selected models were applied to the case and patient data that were predicted to be relevant were highlighted using a orange-colored box (see Fig. 3, top panel). In the highlights only intervention arm, patient data were highlighted as in the other intervention arm, and, in addition, data that were not predicted to be relevant were removed from the interface (see Fig. 3, bottom panel).
Fig. 3.
The LEMR user interface showing highlighted data. The top panel shows the highlights in place version of the LEMR interface, which has in-place, orange highlighting of patient data. The bottom panel shows the highlights only version of the interface, in which only the highlighted patient data are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
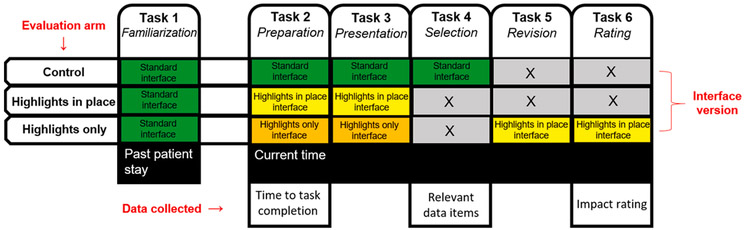
The LEMR system was used in the evaluation mode and reviewers completed the following tasks. An overview of the tasks that constitute the case review procedures for the three arms is shown in Fig. 4, and we give details of the tasks below.
Fig. 4.
Tasks that constitute the case review procedure in the three arms of the evaluation. The standard interface is shown in Fig. 2, and the highlights in place and highlights only versions of the interface are shown in Fig. 3. The last row shows data that were collected during three of the tasks.
Task 1 (familiarization; for all three arms). This task is equivalent to the familiarization task in the training data collection (see Section 3.3).
Task 2 (preparation; for all three arms). An additional day (from 8:00 am on the day selected for the past patient stay to 8:00 am on the next day i.e., “current time”) of the patient’s data were added to the display. The reviewer was prompted with “24-hours have passed” and directed to “use the available information to prepare to present the case during morning rounds.” After preparation was complete, the reviewer clicked on a button to advance to Task 3.
Task 3 (presentation; for all three arms). The reviewer was prompted with “now that you are up to date with this patient’s problems and latest data, please present the patient as if you were presenting during morning rounds, including pertinent positives and negatives, as well as your assessment and management plan for the day. Try to make it concise.” The verbal presentation was recorded with an audio recorder. After completing the presentation, the reviewer clicked a button that either advanced to Task 4 (if in the control arm), advanced to the next patient case (if in the highlights in place arm), or advanced to Task 5 (if in the highlights only arm).
Task 4 (selection; for the control arm). This task is equivalent to the selection task in the training data collection when data on information-seeking behavior were collected (see Section 3.3). In the interface, each available data item (e.g., glucose levels, insulin dosage regimen) was shown with an accompanying checkbox. The reviewer was directed to “select the information you consider pertinent when preparing to present this case at morning rounds.” The reviewer selected relevant data items by toggling the accompanying checkbox to the checked state. The reviewer then clicked a button to advance to the next patient case.
Task 5 (revision; for the highlights only arm). The goal of this task was to identify relevant data that was previously hidden. The reviewer was shown the case using the highlights in place interface version - i.e., the hidden data were revealed - and was prompted with “additional information is now displayed. Considering the additional information, if you would like to revise your presentation, please do so now.” Revisions to the rounding presentation were recorded using an audio recorder. After finishing the revisions (or opting not to revise), the reviewer clicked on a button to advance to Task 6.
Task 6 (rating; for the highlights only arm). The goal of this task was to assess the clinical impact of relevant data that were previously hidden. The reviewer was prompted with “if you revised your presentation, rate the clinical impact those revisions would have on patient care.” Clinical impact was selected on a three-point scale: “no impact”, “minor impact”, and “major impact”, and included a fourth option labeled “no revisions.” The reviewer then clicked a button to advance to the next patient case.
In the control arm, a reviewer completed Tasks 1, 2, 3, and 4 in order; in the highlights in place arm, a reviewer completed Tasks 1, 2, and 3 in order; and in the highlights only arm, a reviewer completed Tasks 1, 2, 3, 5, and 6 in order (see Fig. 4). Data collected during the evaluation included time to task completion during Task 2, a list of relevant data items in Task 4, and, based on data revealed in Task 5, a rating of the LEMR system’s clinical impact in Task 6.
3.6. Data analysis
For evaluation, we used a fractional factorial design to assign 12 reviewers to each complete case review tasks for all of 18 patient cases using, where a given reviewer reviewed a given case using only one of the three arms (see Supplemental Fig. 1). We performed several analyses to evaluate several aspects of the LEMR system including model performance, impact on time to task completion, adequacy of data items that were highlighted, and clinical impact of items that were not highlighted.
We report the performance of models using precision and recall calculated on the evaluation data set. For example, for a glucose model that predicts if glucose level is relevant or not, precision is the number of patient cases for which glucose was predicted to be relevant and was indeed relevant divided by the number of patient cases in which glucose was predicted to be relevant. Similarly, recall is the number of patient cases in which glucose was predicted to be relevant and was indeed relevant divided by the number patient cases in which glucose was indeed relevant. The data items selected in Task 4 served as a gold standard for relevancy in calculating precision and recall.
The impact of the highlighted items on time to task completion was calculated using one-way ANOVA with post hoc analysis. Homogeneity of variance by Bartlett's test was calculated before performing ANOVA to verify that the variance did not differ among groups. Post hoc analysis was performed using Tukey’s Honest Significant Difference test which also assumes homogeneity of variance. For the statistical analyses, we used the following functions implemented in R version 3.3.3: Bartlett.test() from the stats package, aov() from the stats package, and HSD.test() from the agricolae package.
Adequacy of highlighted data was evaluated by comparing and summarizing the number of patient data items displayed, highlighted, and manually selected in each case during Task 2 through Task 4. We compared the performance of model-based highlighting to the performance of random highlighting. For random highlighting, for each case, we randomly selected h data items (where h is the number of items highlighted by the models) from a set of n data items (where n is the total number of displayed items). We also evaluated the clinical impact of items that were not highlighted from the ratings that reviewers provided in Task 6.
4. Results
4.1. Reviewers
All reviewers were ICU physicians trained in critical care medicine, including fellows and attending physicians. Details of the reviewers are shown in Table 1 for both the training and evaluation phases. Five reviewers who participated in the training phase also participated in the evaluation phase.
Table 1.
Characteristics of reviewers.
| Phase of study | Number of reviewers | Average number of years since medical school graduation |
Average number of years spent in ICU |
Average number of weeks per year spent rounding in the ICU |
|---|---|---|---|---|
| Training | 11 | 5.3 (3.0–10.0) | 1.8 (0.3–7.0) | 34 (26–42) |
| Evaluation | 12 | 5.4 (3.0–11.0) | 1.7 (0.6–4.0) | 36 (28–44) |
4.2. Performance of models
Details of the training and evaluation data sets are shown in Table 2. The training data set was assembled from 178 patient cases and 1864 raw predictor variables. After feature construction, there were a total of 30,770 features. A feature was removed if it was missing in every patient case, had no variability, or the values were identical to the values of another feature in every case. Application of these criteria provided a final set of 6935 predictor features.
Table 2.
Characteristics of training and evaluation data sets.
| Number of cases | Number of predictor features | Number of target features | Number of models | |
|---|---|---|---|---|
| Training data set | 178 | 6935 | 865 | 80 |
| Evaluation data set | 72* | 6935 | 25 | 25 |
18 distinct patient cases with each case reviewed independently by four reviewers.
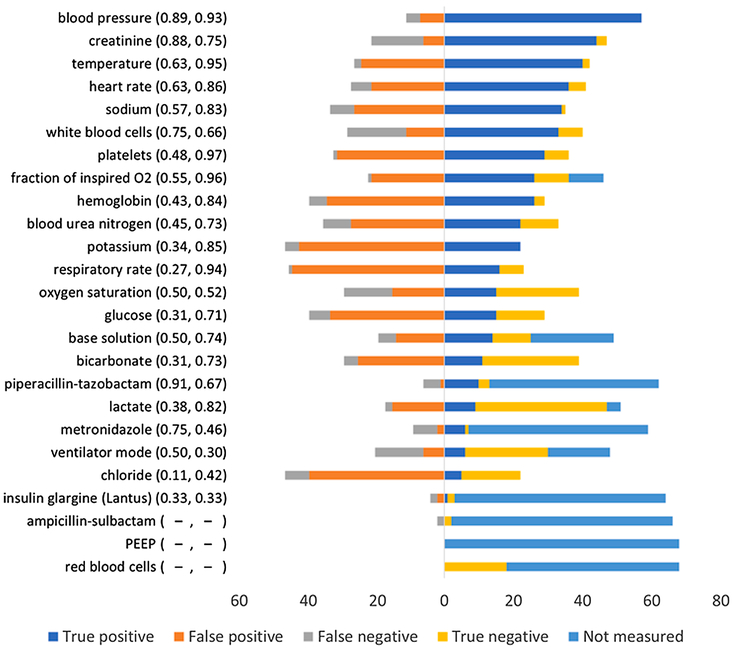
The data set contained 865 target features. A model was constructed for a target feature if it was available in 20 or more cases and if it was selected in 5 or more patient cases. We constructed models for the 80 targets for which sufficient training data were available. The performance of some of these models has been published previously [28]. We selected 25 high performing models that had precision ≥0.67 and recall ≥0.50 and used them in the evaluation study (see Fig. 5). Precision ranged from 0.11 to 0.91 and recall ranged from 0.30 to 0.97. For three of the models (ampicillin-sulbactam, positive end-expiratory pressure (PEEP), and red blood cells) precision and recall are not reported because the target variable of each model was not measured in an adequate number of patient cases in the evaluation data set.
Fig. 5.
Performance of models in the evaluation study. Each row represents a distinct model that was tested in the evaluation study. The name of the target variable with precision and recall (in parenthesis) of the model on the evaluation data set is on the left. The color bar on the right shows the number of true positives, false positives, false negatives, true negatives, and “not measured” for each model. “Not measured” denotes the number of cases where the target variable was either not measured or not available in the evaluation data set. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4.3. Time to task completion
Time to task completion was measured in Task 2 when a reviewer spent time in preparing to present the case at morning rounds. The Bartlett test of homogeneity of variances showed no statistically significant difference in the variance of time to task completion among the three arms (Bartlett's K-squared = 2.17, df = 2, p-value = 0.34); therefore, both ANOVA and Tukey’s tests are appropriate. Summary statistics of the time to task completion in each arm are shown in Table 3.
Table 3.
Number of observations, mean, standard deviation, and range of time to task completion in the three arms of the evaluation study.
| Arm | Number of observations | Mean time to task completion (sec) | Standard deviation | Range |
|---|---|---|---|---|
| Control | 72 | 140.4 | 76.9 | 9.1–513.3 |
| Highlights in place | 72 | 146.4 | 75.3 | 22.9–362.2 |
| Highlights only | 72 | 114.9 | 65.3 | 21.5–334.6 |
The ANOVA test showed a statistically significant difference in time to task completion among the three arms (F2,213 = 3.82; p-value = 0.02). Pairwise results of Tukey’s Honest Significant Difference test are as follows: the times in the control arm and the highlights in place arm were not statistically significantly different (α = 0.1; p-value = 0.87), but the time in the highlights only arm was statistically significantly lesser than the times in the control arm (α = 0.1; p-value = 0.09) and in the highlights in place arm (α = 0.1; p-value = 0.03). Overall, the results provide evidence that clinicians used less time when preparing a summary for presentation at morning rounds when fewer data are available, as was the case in the highlights in place arm.
4.4. Adequacy of highlighted data items
Fig. 6 shows the number of data items available for each of the 18 patient cases, the number of data items highlighted for each case, and the minimum, maximum, and average number of data items that were selected as relevant. We expected the number of items selected to be substantially smaller than the number of items available, and the LEMR system is based on the premise that only a subset of all available patient data is relevant and will be sought by clinicians in the context of a task. Supporting this premise, we found that the cases had, on average, 108.9 data items available, and reviewers selected 22.6 of those items.
Fig. 6.
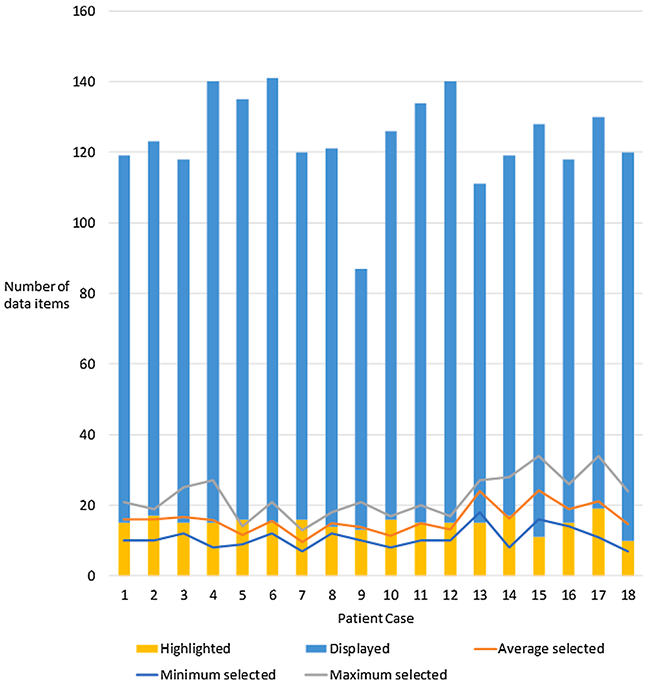
Number of data items that were highlighted, displayed, and manually selected (average, minimum, and maximum) for each case in the evaluation study. Reviewers selected data items manually that they considered are relevant for the task of presenting at morning rounds.
We examined the correspondence between the number of highlighted data items and the number of items sought as relevant for each case. We found the number of highlighted items to be within the range of the number of items selected for 14 of the cases (average highlighted = 15.1 and average selected = 15.7). The remaining four cases were within two and three items of the maximum, and two and five items of the minimum number of selected items (average highlighted = 14.5 and average selected = 17.4).
The precision and recall of the LEMR system based on all 25 models were computed under three scenarios and are shown in Table 4. In the first scenario, ‘Model active patient data,’ precision and recall were computed using only 25 types of data items for which a predictive model was available in the evaluation study (i.e., the 25 models shown in Fig. 5). The performance in this scenario provides an estimate of the performance of the LEMR system if the system had usable predictive models for every data item and can be considered as an upper bound for the performance.
Table 4.
Summary of performance of models in the evaluation study under three scenarios.
| Scenario | Precision [95% CI] | Recall [95% CI] |
|---|---|---|
| Model active patient data | 0.52 [0.49, 0.54] | 0.77 [0.75, 0.80] |
| All patient data | 0.52 [0.49, 0.54] | 0.43 [0.41, 0.45] |
| Randomly selected highlights | 0.15 [0.00, 0.33] | 0.14 [0.00, 0.29] |
In the second scenario, ‘All patient data,’ precision and recall were computed using all data items (i.e., including targets for which models were not available). Note that in this scenario precision is the same as before at 0.52 because the number of highlighted items is unchanged. However, recall decreases from 0.77 to 0.43 because the number of relevant data items that are considered has increased. In the third scenario, ‘Randomly selected highlights,’ precision and recall were computed using data that were randomly selected from the available data. The precision and recall in this scenario are statistically significantly lower than in the previous two scenarios and can be considered as a lower bound for the performance of the LEMR system.
4.5. Clinical impact
In the highlights only arm, reviewers rated the clinical impact of data items that were not highlighted by the LEMR system. After completion of the rounding presentation using the highlights only version of the interface (highlights only arm, Task 3), reviewers were shown ah patient data and provided the opportunity to revise the rounding presentation (highlights only arm, Task 5), and were then asked to rate the impact of those revisions (highlights only arm, Task 6). The ratings are shown in Table 5. In 54.1% of the cases, the unseen data did not lead to a revision of the presentation or the revision was considered to have had no clinical impact (see Table 5). However, in 18.1% of the cases, the reviewers made a revision to the rounding presentation that would have had a major impact on clinical care of the patient.
Table 5.
Clinical impact of not seeing the data that were not highlighted by the LEMR system.
| Did not revise |
No impact | Minor impact |
Major impact |
|
|---|---|---|---|---|
| Number of patient cases (percent) | 33 (45.8%) | 6 (8.3%) | 20 (27.8%) | 13 (18.1%) |
5. Discussion
We developed a learning EMR system that captures clinician information-seeking behavior and uses it to display relevant and context-sensitive data to the user. In a laboratory setting, we collected training data for a set of patient cases, constructed statistical models of clinician information-seeking behavior, and prospectively applied the models to a new set of cases to evaluate the performance of the LEMR system. Our results showed that the precision of the LEMR system was 52%, which is significantly better than random of 15% (see Section 3.4), and recall was as high as 77% when considering only those data items for which a model was available. With more training data we expect to train more usable models, which would likely increase the recall of the system.
The time to task completion decreased significantly when fewer data items were shown (highlights only arm), but did not decrease when all data were shown with highlighting (highlighting in place arm). This result is consistent with other research that has evaluated emphasizing certain aspects of a patient’s record [29]. Although the reduced time in the highlights only arm is encouraging, reviewer comments regarding the potential impact of data not highlighted suggest that this improvement comes at a cost. As reviewers identified 18% of the data items not shown as having a major clinical impact (see Table 5), the highlights only situation has the risk of hiding potentially important data. The highlighting approach is also limited by the accuracy of the models: the LEMR system did not highlight all of the data that clinicians considered to be relevant, but, when compared to random highlighting, the system performed substantially better (see Table 4).
In light of these results, it is important to emphasize that we do not expect a deployed LEMR system to ever completely ‘hide’ information as in the highlights only arm, which was introduced for experimentation purposes. In a deployed system, we expect that clinicians will always retain access to all of the data on a patient. However, highlighting in some situations might be done in-place, as it was in the highlights in place arm, and, in other situations, the highlighted data may be presented separately in an independent display area [30] or even on a separate screen.
Even when used with displays containing all available patient data, the LEMR approach may present risks of unanticipated consequences. Highlights suggesting that certain items are more relevant than others may introduce automation bias (32). Specifically, the clinician might not notice omissions (when relevant data are not highlighted) or commissions (when irrelevant data are incorrectly highlighted). Studies to investigate the possible impacts of this automation bias are currently underway.
Overall, the results support the feasibility of developing a LEMR system that adaptively identifies and highlights patient data that a clinician is likely to seek for a specific task like preparing for morning rounds.
5.1. Limitations
The LEMR system that we evaluated has several limitations. One limitation is the moderate levels of precision and recall of the models that were derived from the training data set. In a previously published analysis, we showed that model performance improved as the number of training cases increased, and it is likely that additional training samples will improve the performance [28]. Larger amounts of training data are needed to not only improve the performance of models but to also improve coverage of the range of clinical conditions. In our experimental setting, we collected training data by having clinicians review patient cases and select relevant data items. This method has the advantage that the selections are highly specific but has the disadvantage of cost (i.e., the clinicians must be compensated for their time). High throughput methods for collecting training data are possible. For example, commercial EMR systems already capture metadata, such as page-visit, mouse, and keyboard logs [31,32]. These logs may be leveraged to automatically infer what data clinicians seek and, therefore, may be useful for building LEMR system models. Eye-tracking is another conceivable method for automatically capturing data items viewed by the user, which we have begun to explore [33]. Assuming that clinicians look at the data they seek, eye-tracking may be used to infer clinician information-seeking behavior. Because EMR systems are used in a wide variety of contexts, a fully trained LEMR system will likely require data from a large number of interactions to develop accurate models.
The LEMR system was evaluated in the context of only two clinical conditions, namely, AKF and ARF; further research is needed to evaluate the generalizability of the models to additional clinical conditions and the applicability of this approach to uncertain and multiple concurrent disease states. In the current study, the predictor and target variables were limited to structured EMR data. Broadening the scope of the models to include clinical text using natural language processing has the potential to improve their performance and to extend the highlighting to clinical text. Another limitation is that the LEMR system was assessed on a single clinical task of developing a clinical summary for morning rounds, and in future work the system will be evaluated on a range of additional tasks.
It is likely that clinicians do not always agree on which data are relevant for a task in the context of a patient. Hence, models that are constructed using data from many clinicians may not perform well because they may not capture the preferences of individual clinicians. If there is considerable variability among information-seeking behavior among clinicians, then models that are clinician-specific will be needed. Further research is required to characterize variability among information-seeking behavior among clinicians. The design of the LEMR user interface itself could have affected the information-seeking behavior and time to task completion; however, it is likely that the differences between highlighting and not highlighting will generalize to displays that are used in deployed EMR systems. Additional research is needed to evaluate our findings in EMR systems that are in actual use.
Another limitation is that the adequacy of the LEMR system for the task of preparing for morning rounds was self-reported by the reviewers in the evaluation study and was not objectively rated by other clinicians. Thus, the results may be systematically biased if the majority of the reviewers were for or against the idea of a computer trying to guide them to relevant clinical data.
5.2. Future work
While the modeling methods used produced positive results, the study was limited to patients with either AKF or ARF and a single clinical task was considered (critical care medicine physicians preparing to present at morning rounds). Future work will investigate a wider range of contexts that include different types of clinicians, performing different clinical tasks, for a range of clinical diagnoses.
The current study examined a single mode of highlighting. Future work will explore other highlighting approaches such as presenting relevant data in a separate area or a combination of in-place highlighting with the use of a separate area. Furthermore, in future work we plan to design and evaluate better ways to present patient data in the user interface.
In this study, we did not measure the LEMR system’s impact on clinician cognitive load. Future studies will evaluate if the LEMR system highlights affect clinician cognitive load, if clinicians using the LEMR system succumb to automation bias and become over-reliant on highlighting [34], and if the LEMR system highlights improve medical decision making.
Implementation of LEMR functionality in a deployed EMR system will need to be evaluated in a live clinical environment. While the development of models can be done off-line using data that are either obtained directly from an EMR or indirectly from a clinical data warehouse, it will be more challenging to apply the models in real-time in the EMR. Models may either be implemented directly in the native EMR environment with custom display modifications or may reside in an independent environment and be applied to data that are acquired through a Fast Healthcare Interoperability Resources (FHIR) interface.
6. Conclusion
To enable improved perception of relevant patient data, we designed a LEMR system that employs a data-driven approach to identify and highlight relevant patient data in a context-specific manner. The LEMR system had moderate precision and recall in highlighting relevant data given a limited number of training cases. With more training data and other developments outlined above, the precision and recall are expected to increase. Clinicians deemed that the data missed by the system would have no or minor impact on patient care in about 82% of the cases. The system has the potential to improve EMR usability and reduce clinician time by intelligently drawing the clinician’s attention to the right data, at the right time. Future work will explore the extensions needed to more fully realize this potential.
Supplementary Material
Acknowledgments
The research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under award numbers T15 LM007059 and R01 LM012095, and by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01 GM088224. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2019.103327.
References
- [1].Engelman D, Higgins TL, Talati R, Grimsman J, Maintaining situational awareness in a cardiac intensive care unit, J. Thoracic Cardiovasc. Surg 147 (2014) 1105–1106. [DOI] [PubMed] [Google Scholar]
- [2].Manor-Shulman O, Beyene J, Frndova H, Parshuram CS, Quantifying the volume of documented clinical information in critical illness, J. Crit. Care 23 (2008) 245–250. [DOI] [PubMed] [Google Scholar]
- [3].Artis KA, Bordley J, Mohan V, Gold JA, Data omission by physician trainees on ICU rounds, Crit. Care Med 47 (2019) 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Koch SH, Weir C, Haar M, Staggers N, Agutter J, Görges M, et al. , Intensive care unit nurses' information needs and recommendations for integrated displays to improve nurses' situation awareness, J. Am. Med. Inform. Assoc 19 (2012) 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fischer G, Context-aware systems: the right information, at the right time, in the right place, in the right way, to the right person, in: Proceedings of the International Working Conference on Advanced Visual Interfaces: ACM, 2012, pp. 287–294. [Google Scholar]
- [6].Effken JA, Loeb RG, Kang Y, Lin Z-C, Clinical information displays to improve ICU outcomes, Int. J. Med. Inf 77 (2008) 765–777. [DOI] [PubMed] [Google Scholar]
- [7].Law AS, Freer Y, Hunter J, Logie RH, McIntosh N, Quinn J, A comparison of graphical and textual presentations of time series data to support medical decision making in the neonatal intensive care unit, J. Clin. Monit. Comput 19 (2005) 183–194. [DOI] [PubMed] [Google Scholar]
- [8].Monroe M, Lan R, Lee H, Plaisant C, Shneiderman B, Temporal event sequence simplification, IEEE Trans. Visual Comput. Graphics 19 (2013) 2227–2236. [DOI] [PubMed] [Google Scholar]
- [9].Pamplin J, Nemeth CP, Serio-Melvin ML, Murray SJ, Rule GT, Veinott ES, et al. , Improving clinician decisions and communication in critical care using novel information technology, Milit. Med (2019). [DOI] [PubMed] [Google Scholar]
- [10].Suermondt H, Tang P, Strong P, Young C, Annevelink J, Automated identification of relevant patient information in a physician’s workstation, Comput. Appl. Med. Care (1993) 229–232. [PMC free article] [PubMed] [Google Scholar]
- [11].Zeng Q, Cimino JJ, Zou KH, Providing concept-oriented views for clinical data using a knowledge-based system: an evaluation, J. Am. Med. Inform. Assoc 9 (2002) 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Anders S, Albert R, Miller A, Weinger MB, Doig AK, Behrens M, et al. , Evaluation of an integrated graphical display to promote acute change detection in ICU patients, Int. J. Med. Inf 81 (2012) 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Klimov D, Shahar Y, Taieb-Maimon M, Intelligent visualization and exploration of time-oriented data of multiple patients, Artif. Intell. Med 49 (2010) 11–31. [DOI] [PubMed] [Google Scholar]
- [14].Post AR, Harrison JH Jr, Protempa: a method for specifying and identifying temporal sequences in retrospective data for patient selection, J. Am. Med. Inform. Assoc 14 (2007) 674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rind A, Wang TD, Aigner W, Miksch S, Wongsuphasawat K, Plaisant C, et al. , Interactive information visualization to explore and query electronic health records, Foundat. Trends Human-Computer Interact 5 (2013) 207–298. [Google Scholar]
- [16].Wright MC, Borbolla D, Waller RG, Del Fiol G, Reese T, Nesbitt P, et al. , Critical care information display approaches and design frameworks: a systematic review and meta-analysis, J. Biomed. Informat.: X (2019) 100041. [DOI] [PubMed] [Google Scholar]
- [17].Grosan C, Abraham A, Rule-based Expert Systems, Springer, Intelligent Systems, 2011, pp. 149–185. [Google Scholar]
- [18].Pickering BW, Herasevich V, Ahmed A, Gajic O, Novel representation of clinical information in the ICU: developing user interfaces which reduce information overload, Appl. Clin. Informat 1 (2010) 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nolan ME, Cartin-Ceba R, Moreno-Franco P, Pickering B, Herasevich V, A multisite survey study of EMR review habits, information needs, and display preferences among medical ICU clinicians evaluating new patients, Appl. Clin. Informat 8 (2017) 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nolan ME, Siwani R, Helmi H, Pickering BW, Moreno-Franco P, Herasevich V, Health IT usability focus section: data use and navigation patterns among medical ICU clinicians during electronic chart review, Appl. Clin. Informat 8 (2017) 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Klann JG, Szolovits P, Downs SM, Schadow G, Decision support from local data: creating adaptive order menus from past clinician behavior, J. Biomed. Inform 48 (2014) 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gambino O, Rundo L, Cannella V, Vitabile S, Pirrone R, A framework for data-driven adaptive GUI generation based on DICOM, J. Biomed. Inform 88 (2018) 37–52. [DOI] [PubMed] [Google Scholar]
- [23].Hauskrecht M, Batal I, Hong C, Nguyen Q, Cooper GF, Visweswaran S, et al. , Outlier-based detection of unusual patient-management actions: an ICU study, J. Biomed. Inform 64 (2016) 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hauskrecht M, Valko M, Batal I, Clermont G, Visweswaran S, Cooper GF, Conditional outlier detection for clinical alerting, in: AMIA Annu. Symp. Proc (2010) pp. 286–290. [PMC free article] [PubMed] [Google Scholar]
- [25].Visweswaran S, Mezger J, Clermont G, Hauskrecht M, Cooper GF, Identifying deviations from usual medical care using a statistical approach, in: AMIA Annu. Symp. Proc (2010) pp. 827–831. [PMC free article] [PubMed] [Google Scholar]
- [26].Yount RJ, Vries JK, Councill CD, The Medical Archival System: an information retrieval system based on distributed parallel processing, Inf. Process. Manage 27 (1991) 379–389. [Google Scholar]
- [27].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. , Scikit-learn: Machine learning in Python, J. Mach. Learn. Res 12 (2011) 2825–2830. [Google Scholar]
- [28].King AJ, Cooper GF, Hochheiser H, Clermont G, Hauskrecht M, Visweswaran S, Using machine learning to predict the information seeking behavior of clinicians using an electronic medical record system, in: AMIA Annu. Symp. Proc (2018) pp. 673–682. [PMC free article] [PubMed] [Google Scholar]
- [29].Laker LF, Froehle CM, Windeler JB, Lindsell CJ, Quality and efficiency of the clinical decision-making process: information overload and emphasis framing, Product. Operat. Manage 27 (2018) 2213–2225. [Google Scholar]
- [30].King AJ, Cooper GF, Hochheiser H, Clermont G, Visweswaran S, Development and preliminary evaluation of a prototype of a learning electronic medical record system, in: AMIA Annu. Symp. Proc (2015) pp. 1967–1975. [PMC free article] [PubMed] [Google Scholar]
- [31].Calvitti A, Hochheiser H, Ashfaq S, Bell K, Chen Y, El Kareh R, et al. , Physician activity during outpatient visits and subjective workload, J. Biomed. Inform 69 (2017) 135–149. [DOI] [PubMed] [Google Scholar]
- [32].Wu DT, Smart N, Ciemins EL, Lanham HJ, Lindberg C, Zheng K, Using EHR audit trail logs to analyze clinical workflow: a case study from community-based ambulatory clinics, in: AMIA Annu. Symp. Proc (2017) pp. 1820–1827. [PMC free article] [PubMed] [Google Scholar]
- [33].King AJ, Hochheiser H, Visweswaran S, Clermont G, Cooper GF, Eye-tracking for clinical decision support: A method to capture automatically what physicians are viewing in the EMR, in: AMIA Jt. Summits Transl. Sci. Proc (2017) pp. 512–521. [PMC free article] [PubMed] [Google Scholar]
- [34].Goddard K, Roudsari A, Wyatt JC, Automation bias: empirical results assessing influencing factors, Int. J. Med. Inf 83 (2014) 368–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.