Abstract
Clinical trials testing Janus kinase-1 (JAK1) inhibitors in cancers are under way. Whether the JAK1 mRNA levels in breast tumors correlates with outcome has not been evaluated. JAK1 expression was analyzed via the Oncomine database and Tumor IMmune Estimation Resource site. Tumor tissues from 57 breast cancer patients were used for qRT-PCR and immune infiltration assessment. JAK1 expression was significantly lower in breast invasive carcinoma compared with adjacent normal tissues. Public databases (Kaplan-Meier plotter and PrognoScan) showed that low JAK1 expression was associated with poorer survival. Data from The Cancer Genome Atlas (TCGA) showed that high JAK1 expression was associated with increased survival in both TNM I-II and TNM III-IV patients. JAK1 was inversely correlated with tumor size status, lymph node status, and TNM of breast cancer patients. JAK1 levels were correlated with the T cell transcript-enriched LYM metagene signature and was significantly lower in the low tumor infiltrating lymphocytes (TILs) group. JAK1 expression levels had significant positive correlations with infiltrating levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in breast cancer and not with other B cells. In conclusion, JAK1 mRNA levels were correlated with prognosis and immune infiltrating levels in breast cancer.
Keywords: JAK1, breast cancer, prognosis, immune infiltrates, TIL
INTRODUCTION
Janus kinases are a family of non-receptor tyrosine kinases which are involved in autoimmune diseases and malignancies [1, 2]. Janus kinase-1 (JAK1) is one of the Janus kinase family members. JAK1 is essential for IL-6-class inflammatory cytokine signaling, plays a critical role in metastatic cancer progression, and mediates the persistent oncogenic activation of STAT3 in mammary cancer cells that are driven by ERBB2 receptor tyrosine kinase signaling [3]. JAK1-deficient cell lines were found to be more tumorigenic than wild-type cells [4]. The evidence indicates that JAK1 works as either an oncogene or a tumor suppressor under certain conditions or cell contents [5]. Clinical trials testing of JAK1 inhibitors in advanced solid tumors, including breast cancer, are under way [6]. JAK1 is also expressed in diverse cell types, including immune cells. A recent study has shown that JAK/STAT inhibition acts on the tumor microenvironment to increase production of protumorigenic inflammatory factors in breast cancer patients, which promotes therapeutic resistance [7]. Whether JAK1 levels in breast cancer tissues are associated with tumor immune infiltrates and clinical outcomes has not been evaluated.
Breast cancer mortality remains the second leading cause of female cancer-related deaths worldwide [8]. Extensive efforts are underway to develop molecular signatures and targeted therapies for specific subsets of breast cancer patients [9]. In recent decades, the prognostic and predictive value of mRNA expression has become more attractive. Studies of the transcriptome, including mRNA levels, in primary breast tumors have been useful for predicting intrinsic subtypes, tumor grade, drug responsiveness, and risk of recurrence, each of which can be used as a prognostic tool [10–12].
Here, we evaluated the association between tumor JAK1 mRNA levels and breast cancer patients’ prognosis in public databases such as the Kaplan-Meier plotter, PrognoScan database, and the Human Protein Atlas database. Moreover, we also investigated the correlation of JAK1 mRNA levels with clinicopathological characteristics and tumor-infiltrating immune cells of breast cancer patients. Our findings shed light on the important role of JAK1 in breast cancer as well as providing a potential relationship and an underlying mechanism between JAK1 and tumor-immune interactions.
RESULTS
The mRNA expression levels of JAK1
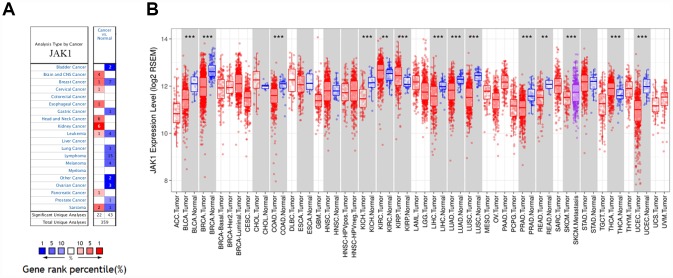
The Oncomine database analysis revealed that JAK1 mRNA expression of breast cancer increased in 1 data set and decreased in 7 data sets compared to the normal tissues (Figure 1A). In addition, JAK1 mRNA expression was lower in bladder cancer, gastric cancer, lung cancer, ovarian cancer, prostate cancer, melanoma, and lymphoma tumors. Higher expression was observed in brain and CNS, cervical, esophageal, head and neck, kidney, and pancreatic cancers in some data sets. To further evaluate JAK1 expression of breast cancer, we examined JAK1 expression using TCGA RNA-sequencing data (Figure 1B). JAK1 expression was significantly lower in breast invasive carcinoma (BRCA) compared with adjacent normal tissues. The results were similar in basal, HER2+, and luminal breast cancer subtypes.
Figure 1.
JAK1 expression levels in human cancers. (A) JAK1 in data sets of different cancers in the Oncomine database. (B) JAK1 expression levels in different tumor types from TCGA database were determined by TIMER (*P < 0.05, **P <0.01, ***P < 0.001).
JAK1 mRNA levels predicts prognosis in breast cancer
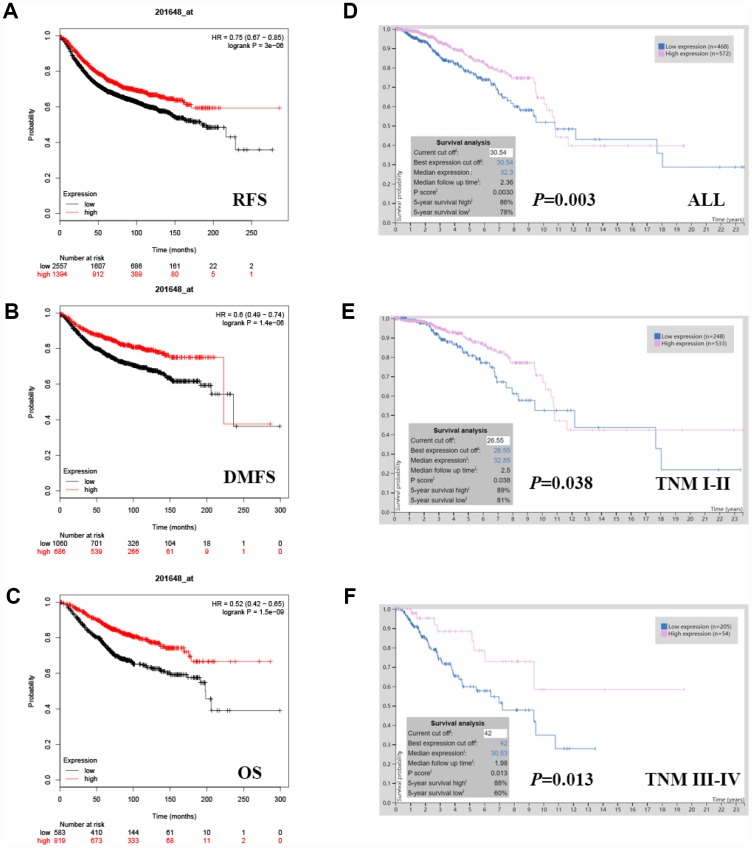
JAK1 expression was evaluated using the PrognoScan (Supplementary Table 1) and was notably found to significantly impact prognosis in breast cancer. Eight cohorts (GSE6532-GPL570, GSE9195, GSE12093, GSE11121, GSE9893, GSE1456-GPL96, GSE3494-GPL96, GSE7390) including different stages of breast cancer showed that high JAK1 expression was associated with a favorable prognosis (Table 1). Similarly, we also found that JAK1 expression was associated with a favorable prognosis in breast cancer patients in the Kaplan-Meier plotter database, which is based on Affymetrix microarrays (Figure 2A–2C; RFS HR[95% CI] = 0.75[0.67-0.85], P = 0.0074; DMFS HR[95% CI] = 0.6[0.49-0.74], P = 0.0035; OS HR[95% CI] = 0.52[0.42-0.65], P = 0.0002). In addition, the RNA sequencing data from TCGA were also used to confirm the JAK1 prognostic value via the Human Protein Atlas database (5-year survival high 86%, 5-year survival low 78%, P = 0.0030, Figure 2D). High JAK1 expression was associated with increased survival in both TNM I-II (P = 0.038, Figure 2E) and TNM III-IV (P = 0.013, Figure 2F) breast cancer patients. Therefore, it is conceivable that low JAK1 expression could be a risk factor for a poor prognosis in breast cancer patients.
Table 1. Survival analysis of JAK1 mRNA in breast cancer patients (the PrognoScan).
| Dataset | Endpoint | Number | ln(HR-high / HR-low) | COX P-value | ln(HR) | HR [95% CI-low CI-up] |
| GSE6532-GPL570 | Distant Metastasis Free Survival | 87 | -1.4754 | 0.00994975 | -1.12962 | 0.32 [0.14 - 0.76] |
| Relapse Free Survival | 87 | -1.4754 | 0.00994975 | -1.12962 | 0.32 [0.14 - 0.76] | |
| GSE9195 | Distant Metastasis Free Survival | 77 | -1.47946 | 0.0298085 | -1.45055 | 0.23 [0.06 - 0.87] |
| Relapse Free Survival | 77 | -1.9106 | 0.00153696 | -1.99732 | 0.14 [0.04 - 0.47] | |
| GSE12093 | Distant Metastasis Free Survival | 136 | -1.43922 | 0.00834641 | -1.32906 | 0.26 [0.10 - 0.71] |
| GSE11121 | Distant Metastasis Free Survival | 200 | -1.44394 | 7.21E-05 | -1.88215 | 0.15 [0.06 - 0.39] |
| GSE9893 | Overall Survival | 155 | -1.34982 | 2.38E-05 | -0.85606 | 0.42 [0.29 - 0.63] |
| GSE1456-GPL96 | Relapse Free Survival | 159 | -1.57388 | 0.000196418 | -1.8311 | 0.16 [0.06 - 0.42] |
| Disease Specific Survival | 159 | -1.71397 | 0.000386357 | -2.05196 | 0.13 [0.04 - 0.40] | |
| Overall Survival | 159 | -1.42147 | 0.00026008 | -1.8023 | 0.16 [0.06 - 0.43] | |
| GSE3494-GPL96 | Disease Specific Survival | 236 | -0.669629 | 0.0294883 | -0.992049 | 0.37 [0.15 - 0.91] |
| GSE7390 | Distant Metastasis Free Survival | 198 | -1.99773 | 0.0381827 | -0.565258 | 0.57 [0.33 - 0.97] |
| Relapse Free Survival | 198 | -0.994522 | 0.150523 | -0.335991 | 0.71 [0.45 - 1.13] | |
| Overall Survival | 198 | -2.58196 | 0.0381021 | -0.585365 | 0.56 [0.32 - 0.97] |
Figure 2.
Kaplan-Meier survival curves comparing the high and low expression of JAK1 in breast cancer. In the Kaplan-Meier plotter database, high JAK1 expression was correlated with good. (A) RFS (HR[95% CI] = 0.75[0.67-0.85], P = 3e-06) (B) DMFS (HR[95% CI] = 0.6[0.49-0.74], P = 1.4e-06) and (C) OS (HR[95% CI] = 0.52[0.42-0.65], P = 1.5e-09). In TCGA data, high JAK1 expression was also correlated with good OS. (D) All breast cancers (P = 0.0030), (E) TNM I-II (P = 0.038) (F) TNM III-IV (P = 0.013).
Correlation of JAK1 expression with the clinicopathological characteristics of breast cancer
The JAK1 mRNA levels in 57 breast cancer tissues were further correlated with the clinicopathological characteristics of breast cancer. Based on the mean JAK1 mRNA level, there were 29 patients with high JAK1 expression and 28 patients with low JAK1 expression. As shown in Table 2, the expression of JAK1 was inversely correlated with tumor size status (P = 0.010), lymph node status (P = 0.001), and TNM staging (P = 0.001) of breast cancer patients. No significant correlation was found between JAK1 expression and other clinicopathological factors, including age (P = 0.357), menopausal status (P = 0.514), histological grade (P = 0.662), ER status (P = 0.516), PR status (P = 0.708), HER2 status (P = 0.248), and breast cancer subtype (P = 0.567).
Table 2. Association between JAK1 mRNA and clinicopathological characteristics in breast cancer patients.
| Characteristics | JAK1 mRNA | P-value | |||
| High | Low | ||||
| Age | 0.357 | ||||
| Median (range) | 50 (32-74) | 51 (31-75) | |||
| ≤50 years | 17 | 56.7% | 13 | 43.3% | |
| >50 years | 12 | 44.4% | 15 | 55.6% | |
| Menopausal status | 0.514 | ||||
| Pre | 17 | 54.8% | 14 | 45.2% | |
| Post | 12 | 46.2% | 14 | 53.8% | |
| Tumor size status | 0.010a | ||||
| T1 | 12 | 75.0% | 4 | 25.0% | |
| T2 | 14 | 46.7% | 16 | 53.3% | |
| T3 | 3 | 60.0% | 2 | 40.0% | |
| T4 | 0 | 0.0% | 6 | 100.0% | |
| Lymph nodes status | 0.001a | ||||
| N0 | 21 | 75.0% | 7 | 25.0% | |
| N1 | 5 | 50.0% | 5 | 50.0% | |
| N2 | 2 | 15.4% | 11 | 84.6% | |
| N3 | 1 | 16.7% | 5 | 83.3% | |
| TNM staging | 0.001 | ||||
| I-II | 25 | 67.6% | 12 | 32.4% | |
| III | 4 | 20.0% | 16 | 80.0% | |
| Histological grade | 0.662 | ||||
| I-II | 18 | 54.5% | 15 | 45.5% | |
| III | 11 | 45.8% | 13 | 54.2% | |
| ER status | 0.516 | ||||
| Negative | 10 | 45.5% | 12 | 54.5% | |
| Positive | 19 | 54.3% | 16 | 45.7% | |
| PR status | 0.708 | ||||
| Negative | 9 | 47.4% | 10 | 52.6% | |
| Positive | 20 | 52.6% | 18 | 47.4% | |
| HER2 status | 0.248 | ||||
| Negative | 23 | 56.1% | 18 | 43.9% | |
| Positive | 6 | 37.5% | 10 | 62.5% | |
| Subtype | 0.567a | ||||
| HR-/HER2- | 6 | 66.7% | 3 | 33.3% | |
| HR-/HER2+ | 3 | 33.3% | 6 | 66.7% | |
| HR+/HER2- | 16 | 50.0% | 16 | 50.0% | |
| HR+/HER2+ | 4 | 57.1% | 3 | 42.9% | |
a. Using Fisher’s exact test; P < 0.05, statistically significant.
JAK1 mRNA levels are associated with tumor infiltrating lymphocytes
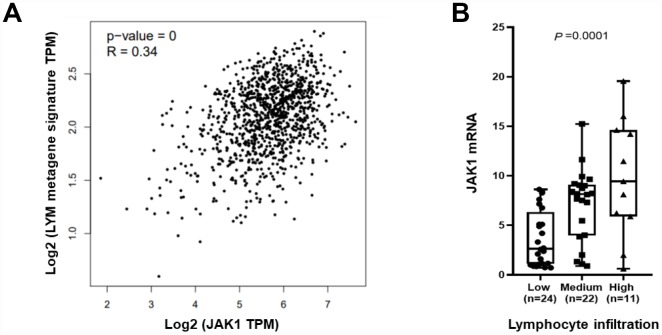
JAK1 is expressed in immune cells and TILs which have been associated with favorable breast cancer prognosis. A lymphocyte-specific immune recruitment (LYM) metagene signature is related to tumor infiltration by lymphocytes and is associated with favorable prognosis in breast cancer. Therefore, we tested whether breast tumor JAK1 mRNA levels correlated with the T cell transcript-enriched LYM metagene signature. The results showed that there was a significant correlation between JAK1 mRNA levels and the LYM metagene in tumor samples from TCGA (Figure 3A). To confirm the correlation, we assessed the presence of TILs in the surrounding stroma of 57 breast cancer cases in which we had already tested the JAK1 mRNA. The presence of TILs ranged from a score of 0 – 90%. According to the presence of TILs, we divided patients into 3 groups: low TILs (less than 1%, 1% to 9%, and 10% to 19%), medium TILs (20% to 49% and 50% to 74%), and high TILs (75% or greater). There were 24 cases with low TILs (42.1%), 22 cases with medium TILs (38.6%), and 11 cases with high TILs (19.3%). We found that JAK1 mRNA levels were statistically significantly lower in the low TILs group compared with the high TILs group (Figure 3B, P = 0.0001).
Figure 3.
JAK1 mRNA levels associated with tumor infiltrating lymphocytes. (A) The average expression of the LYM metagene signature (SASH3, CD53, NCKAP1L, LCP2, IL10RA, PTPRC, EVI2B, BIN2, WAS, and HAVCR2) in each breast cancer sample from TCGA is shown relative to JAK1 mRNA. (B) JAK1 mRNA levels are shown relative to levels of tumor infiltrating lymphocytes in 57 breast cancer samples.
Correlation analysis between JAK1 expression and 6 types of infiltrating immune cells
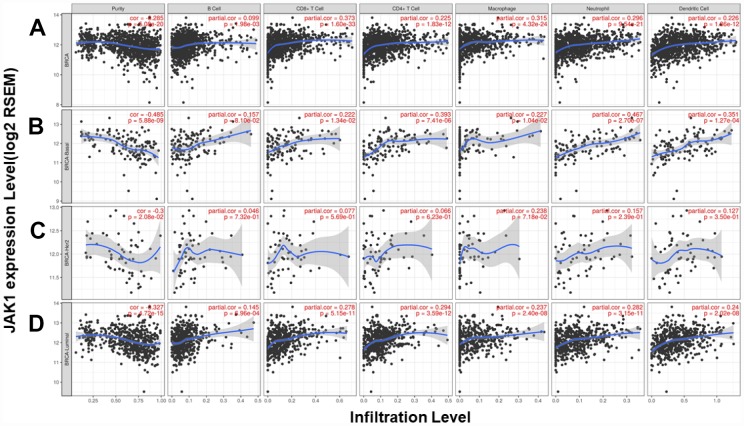
We analyzed the correlation between JAK1 expression and 6 types of infiltrating immune cells (B cells, CD4 T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells). The results showed that JAK1 expression levels had a significantly positive correlation with infiltrating levels of CD8+ T cells (r = 0.373, P = 1.60e-33), CD4+ T cells (r = 0.225, P = 1.83e-12), macrophages (r = 0.315, P = 4.32e-24), neutrophils (r = 0.296, P = 9.54e-21), and dendritic cells (r = 0.226, P = 1.86e-12) in breast cancer and no significant correlations with B cells (r = 0.099, P = 1.98e-03) (Figure 4A). In different breast cancer subtypes, the correlations were not all the same (Figure 4B–4D). In basal-like breast cancer, JAK1 expression levels were positively correlated with infiltrating levels of CD4+ T cells, neutrophils, and dendritic cells. In luminal breast cancer, JAK1 expression levels were positively correlated with infiltrating levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. Since the tumor purity in HER2+ breast cancer was not significant (P = 2.08e-02), the correlations between JAK1 expression and the 6 types of infiltrating immune cells needs further study to confirm these results.
Figure 4.
Correlation of JAK1 expression with immune infiltration level in the TIMER database. (A) Other than B cells, JAK1 expression has a significant positive correlation with infiltrating levels of CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in breast cancer. TIMER database analysis in (B) basal, (C) HER2 and (D) luminal subtypes.
DISCUSSION
Here, we report a study supporting the role of JAK1 in breast cancer. JAK1 mRNA expression was significantly lower in breast invasive carcinoma compared with adjacent normal tissues. JAK1 mRNA levels were inversely correlated with the tumor size status, lymph node status, and TNM staging of breast cancer patients. To our knowledge, this is the first study to report a consistent association between increasing JAK1 mRNA levels and favorable prognosis in breast cancer patients.
JAK1 is required in the JAK/signal transducer and activator of transcription (STAT) pathway for the activation of STAT1 and STAT2 in response to interferon [13] Previous studies have reported that the JAK/STAT pathway had a central role in driving normal and cancer stem cell growth, and deregulation of the pathway was implicated in the promotion of oncogenic phenotypes, including tumorigenesis, proliferation, anti-apoptosis, invasion, angiogenesis, metastasis, and immune evasion [14–16]. In breast cancer, this pathway was altered by the following mechanisms: [17] (1) down-regulation of phosphotyrosine specific phosphatases; [18] (2) down-regulation of negative regulators of STAT; [19] (3) an increase in the amount of IL-6; [20] and (4) activation of other upstream oncogenic pathways, such as c-Src, ERBB1, or PI3K/ mTOR. [21–23]. It is worth noting that JAK kinases mediate signaling for over 20 cytokines, and genomic changes that alter their activity can have diverse effects. For example, loss-of-function JAK1 mutations are suggestive of immune evasion in multiple cancers [24] and have been reported in patients who are unresponsive to immunotherapies [25]. JAK1-deficient mice exhibited perinatal lethality and phenotypes as diverse as defective lymphopoiesis and lack of IFN response [26]. Therefore, high JAK1 mRNA being a good prognostic marker in breast cancer patients may be due to the importance of JAK1 in immune system function.
Another important aspect of this study is that JAK1 was correlated with diverse immune infiltration levels. Through public database analyses, we observed correlations between JAK1 mRNA and the LYM metagene signature (moderate correlation) and levels of infiltrating immune cells (weak and moderate correlation). In previous studies, when RNA seq data were used to analyze the relationship between gene mRNA levels and LYM metagene signature or infiltrating immune cells, the correlation coefficients were mostly uncorrelated or a weak and moderate correlation. Strong correlations were rare [27, 28] To verify the results of the database analyses, we further explored the correlation between JAK1 mRNA levels and TILs using our own breast cancer specimens. We found that JAK1 mRNA levels were statistically significantly lower in the low TILs group compared with high TILs group in our breast cancer patient cohort. This result proved that, to a certain degree, JAK1 mRNA levels could reflect lymphocyte infiltration in breast cancer, although we did not identify the cell type of infiltrating lymphocytes. In this era of stratified medicine, the development of immunological biomarkers is of increasing importance [29]. Recent studies support the integration of TILs in a clinicopathologic prognostic model for breast cancer patients and confirm the excellent survival of patients with high TILs after adjuvant chemotherapy [30]. While clinical trials testing JAK inhibitors are under way, determining how specific inhibition of the individual JAK family members influences the repertoire and antitumor activities of tumor-infiltrating T cells represents an important area for future investigation.
We must acknowledge that there are potential limitations in our analysis. In our own breast cancer samples, we did not include patients who were diagnosed in the past 10 years, which could have been used to estimate survival. We only did analysis on hematoxylin and eosin–stained slides for TIL assessment and did not determine the type of infiltrating cells by immunohistochemistry. The relationship between JAK1 mRNA and different immune cell types was based on analysis of sequencing data from public databases. Therefore, subsequent experimental verification is required.
CONCLUSIONS
In summary, our study provides insights into understanding the potential role of JAK1 mRNA in tumor immunology and its prognostic value. JAK1 mRNA levels correlated with prognosis and immune infiltrating levels in breast cancer, indicating that it can be used as a prognostic biomarker. The potential for JAK1 inhibitors to interfere with immune cells should be evaluated.
MATERIALS AND METHODS
Oncomine database analysis
JAK1 gene expression levels in various types of cancers were identified in the Oncomine database (https://www.oncomine.org/resource/login.html) [31]. The threshold was a P-value of 0.01, a fold change of 1.5, a top 10% gene ranking, and the data had to be from mRNA.
JAK1 mRNA levels and survival outcomes in public databases
To investigate the prognostic role of JAK1 mRNA in breast cancer, the Kaplan-Meier plotter [32] (http://www.kmplot.com; P-value < 0.05, FDR< 0.05), PrognoScan database [33] (http://dna00.bio.kyutech.ac.jp/PrognoScan/; the threshold was adjusted to a Cox P-value < 0.05), and the Human Protein Atlas database [34] (http://www.proteinatlas.org; P-value < 0.05) were used to determine the prognostic significance.
Patients and tissue specimens
Tumor tissues from 57 breast cancer patients were collected between September 2017 and March 2018 at Sun Yat-Sen University Cancer Centre and used for quantitative reverse transcriptase PCR (qRT-PCR) and immune infiltration assessment. Our experiments were in accordance with the ethical standards formulated in the Helsinki Declaration. The Ethics Committee of Sun Yat-Sen University Cancer Centre Health Authority approved this study.
Quantitative reverse transcriptase PCR
The resected tissues were immediately cut and stored in RNAlater (Ambion, Austin, Texas, USA). Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). cDNA was synthesized from 2.0 μg of total RNA using random hexamers and SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. qRT-PCR was performed using SYBR Premix Ex Taq II (Takara Bio Inc, Otsu, Japan). The housekeeping gene HMBS was used as an endogenous control. Primer information: JAK1: 5′-CCACTACCGG ATGAGGTTCTA-3' (forward) and 5'-GGGTCTCGA ATAGGAGCCAG-3' (reverse); HMBS: 5′-GGCAATG CGGCTGCAA-3' (forward) and 5'- GGGTACCCAC GCGAATCAC-3' (reverse). Relative quantification was calculated as 2-ΔCt. ΔCt values = target gene mean Ct value - control gene mean Ct value [27].
Assessment of immune infiltration
Stromal lymphocytic infiltration was defined as the percentage of tumor stroma containing infiltrating lymphocytes, hereinafter referred to as “TIL,” which was similar to a previous publication [35]. Intra-tumoral TILs were not included in this study. According to the guidelines for TIL assessment in breast carcinoma [36, 37], analysis on hematoxylin and eosin–stained slides were independently evaluated by 2 board certified breast pathologists. We used semi continuous variables (deciles): less than 1%, 1% to 9%, 10% to 19%, 20% to 49%, 50% to 74%, and 75% or greater [38]. All cases for which the discordance between the 2 pathologists reached greater than 15% were reviewed together until a consensus was reached.
TIMER database analysis
We analyzed JAK1 expression and the correlation of JAK1 expression with the abundance of 6 types of infiltrating immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) in breast cancer patients via The Tumor IMmune Estimation Resource (TIMER) algorithm database (https://cistrome.shinyapps.io/timer/). [39] Tumor purity is a vital factor that influences the analysis of immune infiltration in tumor samples by genomic approaches. [40]
Gene correlation analysis in GEPIA
The online database Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) was used to find the significant correlation of JAK1 expression with a lymphocyte-specific immune recruitment (LYM) metagene signature. Gene expression correlation analysis was performed on The Cancer Genome Atlas (TCGA) expression data. The LYM metagene was part of the winning prognostic model in the Sage Bionetworks DREAM breast cancer prognosis challenge [41, 42].
Statistical analysis
The Chi-squared test and Fisher’s exact test were used to investigate the significance of the correlation of JAK1 expression with clinicopathological features in breast cancer via SPSS for Windows version 23.0 (Chicago, IL, USA). ANOVA was used to identify the JAK1 mRNA levels in different TIL groups. The correlation of gene expression was evaluated using the Spearman's correlation coefficient. The strength of the correlation was determined using the following guide for the absolute value: 0.00–0.29 (weak), 0.30–0.59 (moderate), 0.60–0.79 (strong), and 0.80–1.0 (very strong). P-values < 0.05 were considered statistically significant. [28]
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from High-level Hospital Construction Project (DFJH201921, Bo Chen); Natural Science Foundation of Guangdong Province (2016A030313768, Ning Liao) and Research Funds from Guangzhou Municipal Science and Technology Project (201707010418, Ning Liao); the National Natural Science Foundation of China (81902828, Bo Chen); the Fundamental Research Funds for the Central Universities (y2syD2192230, Bo Chen); and Medical Scientific Research Foundation of Guangdong Province (B2019039, Bo Chen).
Footnotes
CONFLICTS OF INTEREST: No potential conflicts of interest were disclosed.
REFERENCES
- 1.Schwartz DM, Bonelli M, Gadina M, O’Shea JJ. Type I/II cytokines, JAKs, and new strategies for treating autoimmune diseases. Nat Rev Rheumatol. 2016; 12:25–36. 10.1038/nrrheum.2015.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, Hricik T, Bhagwat N, McKenney AS, Papalexi E, Abdel-Wahab O, Rampal R, Marubayashi S, et al. JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov. 2015; 5:316–31. 10.1158/2159-8290.CD-14-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wehde BL, Rädler PD, Shrestha H, Johnson SJ, Triplett AA, Wagner KU. Janus Kinase 1 Plays a Critical Role in Mammary Cancer Progression. Cell Rep. 2018; 25:2192–2207.e5. 10.1016/j.celrep.2018.10.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sexl V, Kovacic B, Piekorz R, Moriggl R, Stoiber D, Hoffmeyer A, Liebminger R, Kudlacek O, Weisz E, Rothammer K, Ihle JN. Jak1 deficiency leads to enhanced Abelson-induced B-cell tumor formation. Blood. 2003; 101:4937–43. 10.1182/blood-2001-11-0142 [DOI] [PubMed] [Google Scholar]
- 5.Yeh YT, Ou-Yang F, Chen IF, Yang SF, Su JH, Hou MF, Yuan SS. Altered p-JAK1 expression is associated with estrogen receptor status in breast infiltrating ductal carcinoma. Oncol Rep. 2007; 17:35–39. 10.3892/or.17.1.35 [DOI] [PubMed] [Google Scholar]
- 6.Beatty GL, Shahda S, Beck T, Uppal N, Cohen SJ, Donehower R, Gabayan AE, Assad A, Switzky J, Zhen H, Von Hoff DD. A Phase Ib/II Study of the JAK1 Inhibitor, Itacitinib, plus nab-Paclitaxel and Gemcitabine in Advanced Solid Tumors. Oncologist. 2019; 24:14–e10. 10.1634/theoncologist.2017-0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irey EA, Lassiter CM, Brady NJ, Chuntova P, Wang Y, Knutson TP, Henzler C, Chaffee TS, Vogel RI, Nelson AC, Farrar MA, Schwertfeger KL. JAK/STAT inhibition in macrophages promotes therapeutic resistance by inducing expression of protumorigenic factors. Proc Natl Acad Sci USA. 2019; 116:12442–51. 10.1073/pnas.1816410116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019; 69:7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 9.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, Yang L, Wang J, Tang H, Xie X. circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics. 2018; 8:4003–15. 10.7150/thno.24106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002; 415:530–36. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- 11.Chen B, Tang H, Chen X, Zhang G, Wang Y, Xie X, Liao N. Transcriptomic analyses identify key differentially expressed genes and clinical outcomes between triple-negative and non-triple-negative breast cancer. Cancer Manag Res. 2018; 11:179–90. 10.2147/CMAR.S187151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G, Zhang L, Chen ZS, Xie X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019; 18:23. 10.1186/s12943-019-0946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell TJ, John S. Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas. Immunology. 2005; 114:301–12. 10.1111/j.1365-2567.2005.02091.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014; 14:736–46. 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- 15.O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015; 66:311–28. 10.1146/annurev-med-051113-024537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchert M, Burns CJ, Ernst M. Targeting JAK kinase in solid tumors: emerging opportunities and challenges. Oncogene. 2016; 35:939–51. 10.1038/onc.2015.150 [DOI] [PubMed] [Google Scholar]
- 17.Balko JM, Schwarz LJ, Luo N, Estrada MV, Giltnane JM, Dávila-González D, Wang K, Sánchez V, Dean PT, Combs SE, Hicks D, Pinto JA, Landis MD, et al. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med. 2016; 8:334ra53. 10.1126/scitranslmed.aad3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Liu S, Liu G, Dombkowski A, Abrams J, Martin-Trevino R, Wicha MS, Ethier SP, Yang ZQ. Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene. 2012; 31:333–41. 10.1038/onc.2011.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutherland KD, Lindeman GJ, Choong DY, Wittlin S, Brentzell L, Phillips W, Campbell IG, Visvader JE. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004; 23:7726–33. 10.1038/sj.onc.1207787 [DOI] [PubMed] [Google Scholar]
- 20.Bachelot T, Ray-Coquard I, Menetrier-Caux C, Rastkha M, Duc A, Blay JY. Prognostic value of serum levels of interleukin 6 and of serum and plasma levels of vascular endothelial growth factor in hormone-refractory metastatic breast cancer patients. Br J Cancer. 2003; 88:1721–26. 10.1038/sj.bjc.6600956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, Laudano A, Gazit A, Levitzki A, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001; 20:2499–513. 10.1038/sj.onc.1204349 [DOI] [PubMed] [Google Scholar]
- 22.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000; 10:47–50. 10.1016/S0960-9822(99)00268-7 [DOI] [PubMed] [Google Scholar]
- 23.Tavallai M, Booth L, Roberts JL, Poklepovic A, Dent P. Rationally Repurposing Ruxolitinib (Jakafi (®)) as a Solid Tumor Therapeutic. Front Oncol. 2016; 6:142. 10.3389/fonc.2016.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albacker LA, Wu J, Smith P, Warmuth M, Stephens PJ, Zhu P, Yu L, Chmielecki J. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS One. 2017; 12:e0176181. 10.1371/journal.pone.0176181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016; 375:819–29. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998; 93:373–83. 10.1016/S0092-8674(00)81166-6 [DOI] [PubMed] [Google Scholar]
- 27.Miller CP, Thorpe JD, Kortum AN, Coy CM, Cheng WY, Ou Yang TH, Anastassiou D, Beatty JD, Urban ND, Blau CA. JAK2 expression is associated with tumor-infiltrating lymphocytes and improved breast cancer outcomes: implications for evaluating JAK2 inhibitors. Cancer Immunol Res. 2014; 2:301–06. 10.1158/2326-6066.CIR-13-0189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX, Ding H, Pan YL, Rong L. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front Immunol. 2019; 10:6. 10.3389/fimmu.2019.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen XY, Yeong J, Thike AA, Bay BH, Tan PH. Prognostic role of immune infiltrates in breast ductal carcinoma in situ. Breast Cancer Res Treat. 2019; 177:17–27. 10.1007/s10549-019-05272-2 [DOI] [PubMed] [Google Scholar]
- 30.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, Budczies J, Huober J, Klauschen F, Furlanetto J, Schmitt WD, Blohmer JU, Karn T, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018; 19:40–50. 10.1016/S1470-2045(17)30904-X [DOI] [PubMed] [Google Scholar]
- 31.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007; 9:166–80. 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010; 123:725–31. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- 33.Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009; 2:18. 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 35.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, de Azambuja E, Quinaux E, Di Leo A, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013; 31:860–67. 10.1200/JCO.2011.41.0902 [DOI] [PubMed] [Google Scholar]
- 36.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, Perez EA, Thompson EA, Symmans WF, et al. , and International TILs Working Group 2014. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015; 26:259–71. 10.1093/annonc/mdu450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV, Gonzalez-Ericsson PI, Sanders M, Solomon B, Solinas C, et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol. 2017; 24:311–35. 10.1097/PAP.0000000000000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovács A, Stenmark Tullberg A, Werner Rönnerman E, Holmberg E, Hartman L, Sjöström M, Lundstedt D, Malmström P, Fernö M, Karlsson P. Effect of Radiotherapy After Breast-Conserving Surgery Depending on the Presence of Tumor-Infiltrating Lymphocytes: A Long-Term Follow-Up of the SweBCG91RT Randomized Trial. J Clin Oncol. 2019; 37:1179–87. 10.1200/JCO.18.02157 [DOI] [PubMed] [Google Scholar]
- 39.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017; 77:e108–10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013; 4:2612. 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng WY, Ou Yang TH, Anastassiou D. Development of a prognostic model for breast cancer survival in an open challenge environment. Sci Transl Med. 2013; 5:181ra50. 10.1126/scitranslmed.3005974 [DOI] [PubMed] [Google Scholar]
- 42.McCarthy N. Prognostic models: rising to the challenge. Nat Rev Cancer. 2013; 13:378. 10.1038/nrc3530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.