Abstract
Stress granules (SGs) are primarily composed of mRNAs that stall at translation initiation and usually appear in the cytoplasm under unusual physiological or pathological conditions such as hypoxia, oxidative stress, and viral infection. Recent studies have indicated that several components of SGs participate in tumourigenesis and cancer metastasis through tumour‐associated signalling pathways as well as other mechanisms. Furthermore, some chemotherapy drugs have been reported to induce SGs. Thus, the roles of SGs in cancer treatment have attracted considerable interest. Importantly, disturbing the recruitment of SGs components or microtubule polymerization, as well as other strategies that can abolish SGs formation, is reported to inhibit tumour progression, suggesting that targeting SGs could be a promising strategy for cancer treatment. In this review, we summarize the relationship between SGs and cancer, as well as recent advances in targeting SGs, in the interest of providing new opportunities for cancer treatment.
Abbreviations
- 5‐Fu
5‐fluorouracil
- Caprin1
cell cycle associated protein 1
- eIF2A
eukaryotic initiator factor 2A
- ER stress
endoplasmic reticulum stress
- FUS
fused in sarcoma
- G3BP1
Ras‐GTPase‐activating protein SH3 domain‐binding protein 1
- HDAC6
histone deacetylase 6
- HDACs
histone deacetylases
- HRI
haem‐regulated inhibitor
- Hsp40
heat shock protein 40
- Hsp70
heat shock protein 70
- mRNPs
messenger ribonucleoprotein particles
- mTOR
mammalian target of rapamycin
- NDs
neurodegenerative diseases
- PERK
PKR‐like endoplasmic reticulum kinase
- PKR
RNA‐dependent protein kinase
- PRD
prion‐related domain
- PTMs
post‐translation modifications
- RBPs
RNA‐binding proteins
- RRMs
RNA recognition motifs
- SGs
stress granules
- SIRT6
sirtuin 6
- TIA‐1
T‐cell‐restricted intracellular antigen‐1
- UPS
ubiquitin proteasome system
- USP10
ubiquitin specific peptidase 10
1. INTRODUCTION
During the eukaryotic gene expression process, RNA translation is thought to be a key process that regulates RNA modification, stability, location, protein function, and chromatin structure (Rizvi & Smith, 2017). Stresses such as hypoxia, viral infection, heat shock, and oxidative stress can induce multiple tissue and organ damages, as well as disordered protein translation, all of which are called “integrated stress responses (ISRs)” (Anderson, Kedersha, & Ivanov, 2015). Composed of non‐translated mRNAs, cytoplasmic messenger ribonucleoprotein particles (mRNPs) are components of the ISR and emerge when translation initiation is stalled, subsequently forming granules to ensure cell adaptation to stress conditions. These RNA granules can usually be divided into four types: processing bodies, stress granules (SGs), neuronal granules, and germ cell granules (Anderson & Kedersha, 2006). Among these granules, SGs are the most well studied. Given that SGs assemble when translation initiation is stalled, they usually occur in the cytoplasm, containing various RNA‐binding proteins (RBPs), non‐translated RNAs, types of translation initiation factors, poly(A)‐binding protein, and ribosomal subunits.
SGs assemble immediately once stresses are encountered and are cleaned up once the stresses disappear (Reineke & Neilson, 2019). As SGs are a non‐typical type of multifunctional membrane‐less organelles, the formation of SGs is a highly regulated and dynamic process. SGs are usually triggered by the serine 51 phosphorylation of https://www.guidetopharmacology.org/GRAC/DatabaseSearchForward?searchString=eukaryotic+initiator+factor+&searchCategories=all&species=none&type=all&comments=includeComments&order=rank&submit=Search+Database 2A (eIF2A; Buchan & Parker, 2009; Gilks et al., 2004), which represents the initiation of 48‐s ribosomal subunit disassembly and translation arrest. Considered to be one of the most pivotal components of SGs, the phosphorylation of Ras‐GTPase‐activating protein SH3 domain‐binding protein 1 (G3BP1) affects SG formation (Mahboubi & Stochaj, 2017). Moreover, the post‐translation modifications (PTMs) of mRNPs components are also closely associated with SGs assembly (Jayabalan et al., 2016; Kwon, Zhang, & Matthias, 2007). Neddylation enhances SG assembly in response to cellular oxidative stress caused by arsenite and promotes polysomic disassembly to facilitate SG formation after translation arrest (Jayabalan et al., 2016). Furthermore, several reports have shown that oxidative stress may disturb SGs formation (Lian & Gallouzi, 2009). All of the evidence mentioned above has indicated that the regulation of SG assembly is complicated but precise.
It is generally accepted that there are two models and multiple steps involved in SG assembly, all of which have already been well described in several related reviews (Anderson et al., 2015; Panas, Ivanov, & Anderson, 2016; Protter & Parker, 2016). In the traditional “cores first” model, core granule forms ahead of the outer shell. First, untranslated mRNPs form oligomers through mRNP interaction and modification in mammalian cells (Mahboubi & Stochaj, 2017). RBPs, such as G3BP1 and T‐cell‐restricted intracellular antigen‐1 (TIA‐1) bind to nuclear mRNA to form mRNPs, and this complex subsequently moves out of the nucleus, being translated or keeping maintained at specific regions in the cytoplasm (Anderson & Kedersha, 2008; Fasken & Corbett, 2005). Second, the oligomer grows into small granule core of approximately 200 nm by connecting with additional parts, which likely occurs in processing bodies and transforms out soon afterwards. Third, the core particles are formed as advanced structure and become surrounded by a dynamic and liquid‐like shell in a microtubule ‐dependent manner (Chernov et al., 2009).
The other model is called the “liquid–liquid phase separations first” model which means that granules come into being before the core is formed (Nott et al., 2015). Untranslated mRNPs are divided into droplets that are held together by weak interactions (Wheeler, Matheny, Jain, Abrisch, & Parker, 2016). The next step is similar to that of the first model: the droplets grow larger, accompanied by some untranslated mRNPs. For the high concentration of protein droplets in the granules, the cores emerge gradually, driven by protein–protein interactions (Lin, Protter, Rosen, & Parker, 2015).
SGs assembly and RBPs imbalance are well‐documented to have a strong relationship with several physiological pathological conditions such as neurodegenerative diseases (NDs), cancer progression, and viral infections (Chen & Liu, 2017; Li, King, Shorter, & Gitler, 2013; Malinowska, Niedzwiedzka‐Rystwej, Tokarz‐Deptula, & Deptula, 2016). Given that SGs have attracted considerable attentions in recent years, the correlations between NDs and SGs have already been widely reviewed (Chen & Liu, 2017; Li et al., 2013; Mahboubi & Stochaj, 2017; Reineke & Lloyd, 2013), while reviews about the role of SGs in cancer are largely falling behind. Inspired by the review of Anderson et al. (2015) about the explicit relationship between RNA granules and cancers, in this review, we mainly focus on recent advances in targeting SGs for cancer therapy, in the interest of providing new opportunities for cancer treatment.
2. SGS FORMATION AND TUMOURIGENESIS ARE RECIPROCALLY REGULATED PROCESSES
Up‐regulation of several SGs components has been observed in different kinds of tumours, such as pancreatic cancer (Sim, Irollo, & Grabocka, 2019), sarcoma (Somasekharan et al., 2015), hepatocellular carcinoma (Adjibade et al., 2015), and malignant gliomas (Vilas‐Boas Fde et al., 2016; Table 1). Grabocka and Bar‐Sagi (2016) reported that SG formation was markedly promoted in mutant https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2824 tumours, which was regulated by the secretion of the signalling lipid molecule 15‐deoxy‐Δ12,14 https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2692. SG formation enhancement in turn endowed the chemoresistance of Kras‐mutant cells. It has been indicated that SG formation in glioma cells occurred in an eIF2A‐dependent manner, which might be responsible for the resistance of malignant gliomas to chemotherapeutic drugs (Vilas‐Boas Fde et al., 2016). Overexpression of Y‐box‐binding protein 1 could up‐regulate G3BP1 and was related to the poor survival rate in human sarcomas. In addition, G3BP1 facilitated the tumour metastasis in a mouse model (Somasekharan et al., 2015).
Table 1.
Overview of SGs assembly in human cancer cells
| Tumour types (human) | Physiological function | References |
|---|---|---|
| Pancreatic cancer | Regulated by mutant KRAS gene, promoting survival of cancer cells | Grabocka & Bar‐Sagi, 2016 |
| Hepatocellular carcinoma | Resistance to chemotherapeutic drugs like sorafenib | Adjibade et al., 2015 |
| Glioma/glioblastoma | Assembly in the eIF2A‐dependent way, promoting survival of cancer cells | Vilas‐Boas Fde et al., 2016 |
| Sarcoma | YB1 and G3BP1 participating in related biological pathways, associated with the poor prognosis | Somasekharan et al., 2015 |
| Mantle cell myeloma | Assembly in response to bortezomib while resistant to bortezomib‐induced apoptosis | Fournier et al., 2010 |
| Colorectal cancer | Induced by 5‐fluorouracil (5‐Fu) or bortezomib in the eIF2A‐dependent way, promoting survival of cancer cells | Fournier et al., 2010 |
| Mazroui et al., 2007 | ||
| Head and neck cancers | Induced by MG132 or 5‐Fu in the eIF2A‐dependent way, promoting survival of cancer cells | Mazroui et al., 2007 |
| Kaehler et al., 2014 | ||
| Prostate cancer | Induced by the chemotherapeutic drug sodium selenite in the ROS‐dependent way, promoting cell death instead of cell survival | Fujimura, Sasaki, & Anderson, 2012 |
Cancer cells are usually exposed to hypoxia, nutrient starvation, and high osmotic stress conditions for the high metabolic demands of proliferation (Ackerman & Simon, 2014). Thus, the tumour micro‐environment is typically full of a variety of stresses, such as high concentration of ROS and hypoglycaemia, almost all of which could strongly trigger SG formation (Mahboubi & Stochaj, 2017; Figure 1). SGs begin to assemble in response to the stresses mentioned above, which orchestrates cellular adaptation to the hostile environment. In addition, the formation of SGs is reported to regulate several important canonical signalling pathways, such as https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2109 and https://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=288 (Arimoto, Fukuda, Imajoh‐Ohmi, Saito, & Takekawa, 2008; Kedersha et al., 2002; Thedieck et al., 2013; Figure 1). It is well known that mTOR complex 1 plays an essential role in cell metabolism and survival. When amino acids are depleted, mTORC1 is inactivated and released from the lysosomal membrane to the cytoplasm and then accumulates in the SGs in an eIF2A phosphorylation‐dependent manner (Kedersha, Ivanov, & Anderson, 2013). Thus, SGs might inhibit cancer cell growth signalling by changing the location and activity of mTOR complex 1. Furthermore, Arimoto et al. (2008) claimed that hypoxia‐induced SGs inhibited apoptosis in cancer cells by suppressing the stress‐responsive MAPK signalling pathway, which might be related to the chemotherapy resistance (Figure 1).
Figure 1.
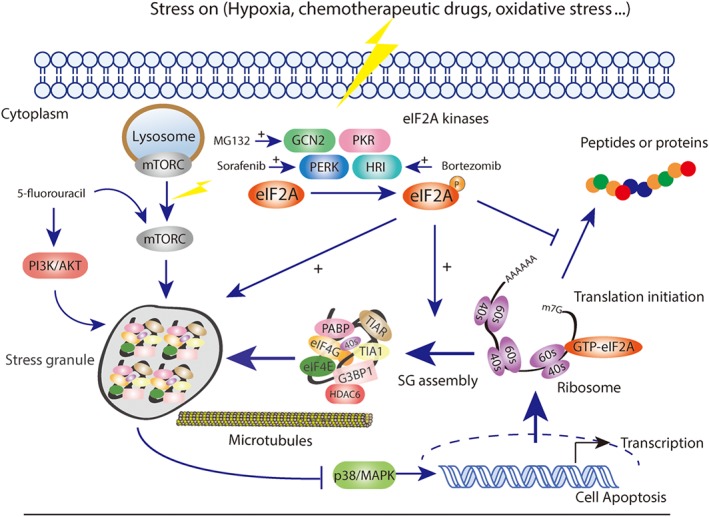
SGs formation and tumourigenesis are reciprocally regulated processes
3. THE MECHANISMS OF CHEMOTHERAPY DRUG‐INDUCED SGS ASSEMBLY
Apart from the pathophysiological conditions, many studies have associated cancer cell survival with the assembly of SGs in response to chemotherapy drugs, which could in turn aggravate cancer. Upon different stress conditions, eIF2 phosphorylation is regulated by different kinds of kinases (Wek, Jiang, & Anthony, 2006) including the double stranded http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2016, PKR‐like endoplasmic reticulum kinase (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2017) Hamanaka, Bennett, Cullinan, & Diehl, 2005), http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2015, and general control non‐derepressible 2 (https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2018) (Hamanaka et al., 2005; Holcik, 2015). Chemotherapeutic drugs are usually considered to induce SG assembly through activating the phosphorylation kinases mentioned above (Figure 1).
3.1. Sorafenib
As a well‐known TK inhibitor, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5711 is widely used for hepatocarcinoma and has already been proposed as a chemotherapeutic stress of SG formation. Rahmani et al. reported that treatment with sorafenib in human leukaemia cells induced severe endoplasmic reticulum stress (ER stress) accompanied by the activation of PERK and the subsequent phosphorylation of eIF2A. Inhibition of PERK activity or blocking the phosphorylation of eIF2A could significantly enhance the lethality of sorafenib treatment (Rahmani et al., 2007). Adjibade et al. (2015) further validated these results by showing that sorafenib‐induced ER stress was the main activator of PERK, which promoted the production of p‐eIF2A and SGs formation as a result. Further experiments showed that treatment with specific inhibitors of PERK or knocking down PERK in MEFs cells could block the formation of SGs, thereby enhancing the therapeutic effect of sorafenib. All of the evidence above has confirmed that in the presence of sorafenib, PERK is the main phosphorylation kinase of eIF2A, which is activated by ER stress and could induce SG formation as a result, thereby contributing to the chemoresistance of cancer cells to sorafenib.
3.2. Bortezomib
https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6391 is a boronate inhibitor of the 26S proteasome that has been approved for a range of haematological tumours but has no effect on solid tumours (McConkey & Zhu, 2008; Richardson, 2004; Richardson, Hideshima, & Anderson, 2003). McEwen et al. (2005) first noted that MEF cells that were deficient in https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2015 had no ability to form SGs after arsenite treatment, resulting in apoptosis and cell death as a result. Subsequently in 2010, Fournier, Gareau, and Mazroui (2010) proposed that bortezomib could strongly induce SG formation through the phosphorylation of eIF2A in cancer cells. Further research showed that this stress response to bortezomib was significantly inhibited by the depletion of HRI, a typical type of eIF2A phosphorylation kinase, which showed that activation of HRI is the prerequisite of bortezomib‐induced SGs. Because the related signalling pathways of the chemoresistance to bortezomib are complex, further studies are surely needed to determine whether SG formation is the most critical one.
3.3. 5‐Fluorouracil
As a long‐established chemotherapy drug, 5‐fluorouracil (5‐FU) strongly inhibits DNA replication and cell proliferation, and is widely used in the treatment of solid tumours, especially in the first‐line therapy of gastric cancer (Longley, Harkin, & Johnston, 2003). The increasing drug resistance problem of 5‐FU has seriously weakened its therapeutic effect, with multiple signalling pathways being involved, such as the PI3K/Akt and mTOR. Kaehler, Isensee, Hucho, Lehrach, and Krobitsch (2014) proposed that 5‐Fu treatment induced SGs assembly in an RNA‐incorporated way; in further experiments, these researchers found an important SG component named RACK1, which was associated with cell survival and apoptosis and was the key regulator of 5‐FU‐induced SG assembly.
Accordingly, based on the close relationship between SGs and cancer, disturbing the formation of SGs can not only influence the tumour progression but also sensitize cancer cells to chemotherapeutic agents, which represents a promising strategy for cancer treatment.
4. INTERFERING WITH SG FORMATION CAN EFFECTIVELY HINDER TUMOUR PROGRESSION
Given that SG assembly is a highly fine regulated process, recent studies have proposed the following methods to target SGs for cancer therapy.
4.1. Regulating the activity of eIF family
4.1.1. eIF2A
Because SGs assembly is primarily triggered by the phosphorylation of the initiation factor eIF2A, it is clearly an effective strategy to inhibit SGs formation by targeting eIF2A phosphorylation. Inhibiting the phosphorylation of eIF2A sensitized glioma cells to chemotherapy (Vilas‐Boas Fde et al., 2016). Brain tumours and malignant gliomas with high lethal rates usually respond poorly to chemotherapeutic drugs. Vilas‐Boas Fde et al. (2016) showed that the dominant negative mutant of phosphorylated eIF2A reduced the assembly of SGs and enhanced the response to chemotherapeutic agents, such as bortezomib, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5343, or https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6815 in the treatment of tumours mentioned above. However, inhibiting the phosphorylation of eIF2A itself has effects on cancer therapy for some additional reasons, such as apoptosis. A selective eIF2A dephosphorylation inhibitor salubrinal (Boyce et al., 2005), promoted apoptosis induced by a proteasome inhibitor in leukaemia and myeloma cells (Drexler, 2009; Teng et al., 2014), which contributed to cancer treatment (Figure 2). Notably, salubrinal inhibited apoptosis in neural cells triggered by ER stress and a recent study about neurodegeneration treatment revealed that overexpressing a specific phosphorylated eIF2A phosphatase, GADD34, or knocking down prion protein expression by lentivirus infection could reduce eIF2A phosphorylation level and thereby rescue cells from the activation of unfolded protein response (Moreno et al., 2012; Figure 2). These discrepancies might be attributable to the different genetic backgrounds between tumour cells and neuronal cells. Thus, whether the phosphorylation of eIF2A is favourable for cancer treatment is dependent on specific cancer types.
Figure 2.
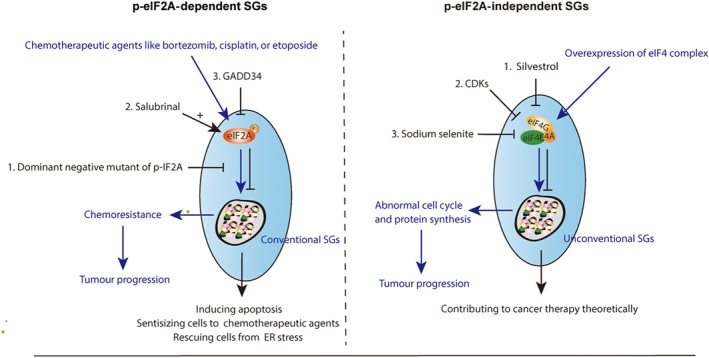
Targeting SGs by regulating the activity of eIF2A or eIF4 complex
4.1.2. eIF4
Although eIF2A is a significant translation initiation factor of SGs formation, not all kinds of SG assembly depend on the serine 51 phosphorylation of eIF2A. Considered as unconventional SGs, eIF2A‐independent SGs assembly is induced by different initiation conditions compared to conventional SGs (Sasaki, Fujimura, & Anderson, 2012). Apart from the eIF2A, eIF4F complex consists of eIF4A, eIF4E, and eIF4G can also regulate SG formation (Pelletier, Graff, Ruggero, & Sonenberg, 2015; Sonenberg & Hinnebusch, 2009), which acts as a core exon junction complex component (Ryu et al., 2019) and controller of translation initiation. Activity or composition alteration of the eIF4F complex can induce non‐canonical SGs, which is not dictated by eIF2A phosphorylation. Thus, targeting the eIF4F complex might be a promising approach to inhibit SG formation. Silvestrol, one of the flavagline derivatives, had a strong effect on the eIF4F complex, inhibiting translation initiation and exhibiting strong anti‐cancer activity in human prostate and breast cancer (Cencic et al., 2009). Given that https://www.guidetopharmacology.org/GRAC/DatabaseSearchForward?searchString=cyclin-dependent+kinases&searchCategories=all&species=none&type=all&comments=includeComments&order=rank&submit=Search+Database could affect on the nonsense‐mediated mRNA decay pathway during the cell cycle by regulating the phosphorylation of eIF4A3, it is possible that cyclin‐dependent kinases modulators could influence eIF4 complex function. In addition, targeting eIF4E by sodium selenite (Sasaki et al., 2012) and loss of eIF4E or eIF4B could also induce unconventional SGs (Ayuso, Martinez‐Alonso, Regidor, & Alcazar, 2016), thereby contributing to the body of knowledge regarding cancer therapy (Figure 2).
Multiple studies have already shown that high expression of eIF4 can lead to abnormal cell cycling as well as accelerate protein synthesis, and thus promote tumourigenesis and malignant progression (Pelletier et al., 2015; Topisirovic, Ruiz‐Gutierrez, & Borden, 2004; Tutar & Tutar, 2008), contrary to the function of eIF2A. Thus, it is likely confirmed that eIF2A‐dependent or eIF4‐dependent types of SGs not only have different components and induction conditions but also exert opposite functions in cellular survival and tumourigenesis (Sasaki et al., 2012).
4.2. Inhibiting the assembly of SGs
Because SGs are formed upon the inhibition of translation initiation, these particles naturally contain multiple types of RBPs, and almost all of these proteins influence SG assembly and disassembly. In addition, some proteins that are not involved in SGs also have effects on the regulation of key steps in SG formation. For example, heat shock protein families in yeasts, such as heat shock protein 40 (Hsp40) and heat shock protein 70 (Hsp70) are both localized in SGs (Cherkasov et al., 2013; Kroschwald et al., 2015) and exert critical roles in SG formation. Overexpression of Hsp70 could inhibit SG assembly while accelerate the SG disassembly (Kroschwald et al., 2015; Mazroui, Di Marco, Kaufman, & Gallouzi, 2007) and different types of Hsp40 regulated SGs through different pathways. In addition, other proteins such as Sis1, an important cytoplasmic Hsp70 co‐chaperone, promoted SG clearance by autophagy; while Ydj1, a type I HSP40 co‐chaperone, promoted this clearance by starting new translation (Tutar & Tutar, 2008; Walters, Muhlrad, Garcia, & Parker, 2015), which was associated with the activity of Hsp70, as well. Overall, proteins associated with SG assembly provide considerable possibilities to target SG components, as well as key steps for cancer treatment.
4.2.1. Targeting the SGs components
TIA‐1
In response to eIF2 phosphorylation, related RBPs including TIA‐1 and TIAR exert self‐aggregation and RNA‐binding functions immediately to promote SG formation (Kedersha et al., 2000). In addition, TIA‐1 could also dynamically transport the stalled initiation complex into SGs foci (Gilks et al., 2004). TIA‐1 involves three N‐terminal RNA recognition motifs (RRMs) and a glutamine‐rich prion‐related domain (PRD) at the C‐terminal (Gilks et al., 2004; Kedersha et al., 2000), which plays an important role in SG condensation. Overexpression of PRD could form small cytoplasmic bodies, and conformational changes in PRD contributed to SG core formation (Han et al., 2012; Figure 3). It has been proposed that Pub1, a yeast homologue of TIA‐1, promoted liquid droplet formation in a temperature‐ and salt‐dependent manner, which were subsequently transformed into rigid structures, similar to RNA granules (Lin et al., 2015). All of the findings described above suggest that targeting TIA‐1 for SG regulation is worth being studied for cancer treatment.
Figure 3.
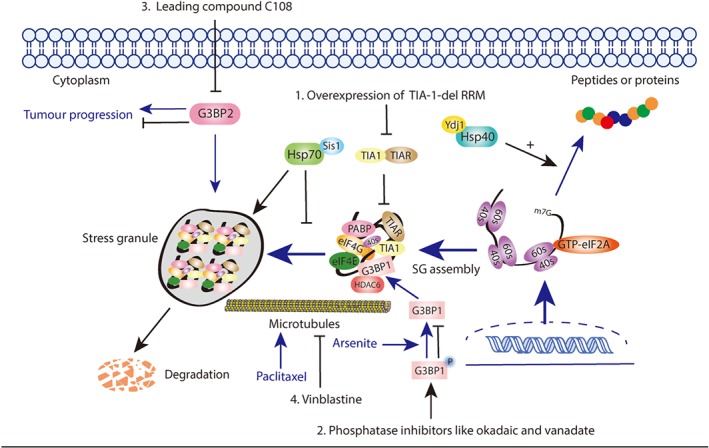
Targeting SGs by regulating SGs components and the key steps of assembly
Overexpression of TIA‐1 truncation protein lacking the RRM domain prevented SG assembly induced by arsenite, as TIA‐1‐del RRM cut off the interaction between TIA‐1 and TIAR (Kedersha, Gupta, Li, Miller, & Anderson, 1999). Cells transfected with the mutant could not form SGs because related mRNAs had no ability to accumulate in the cytoplasm. This result has showed that TIA‐1 inhibited protein expression, which had an inverse correlation with SG assembly, consistent with the finding that RNA triage domains had an influence on protein expression (Kedersha et al., 2000). Thus, the dominant negative mutant of TIA‐1 may be considered a good strategy to target SGs.
G3BP
G3BP1 and G3BP2 are essential for SG formation and both contain RNA‐binding domains, such as RGG or RRM, which means that they might be associated with the RNA interaction (Solomon et al., 2007b). Unlike other SG‐related proteins, G3BP lacks prion‐like domains necessary for SG core formation and the mechanism of G3BP‐regulating SG formation has not been clarified to date. It is well accepted that heat shock or arsenite treatment could induce the dephosphorylation of G3BP at Ser149, which initiateed SG formation (Gallouzi et al., 1998; Tourriere et al., 2003). In contrast, the phosphorylation of G3BP at Ser149 has been shown to inhibit SG formation.
Data in a related research study showed that both the wide type and the non‐phosphorylatable mutant S149A G3BP had interactions with histone deacetylase 6 (HDAC6); however, the phosphomimetic mutant S149E G3BP failed to coprecipitate with HDAC6 (Kwon et al., 2007). Thus, enhancing the phosphorylation of G3BP could block the initiation of SG formation, which provided new approaches to the regulation of SGs formation. For example, arsenite treatment caused dephosphorylation of G3BP and subsequently increased its interaction with HDAC. Moreover, the interaction of G3BP and https://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2618 was weaker after treatment with phosphatase inhibitors, such as http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5349 and vanadate (Kedersha et al., 2016; Figure 3). In addition, phosphatase inhibitors could also increase the G3BP phosphorylation level (Tourriere et al., 2001). All of the methods described above could change the phosphorylation states of G3BP and then regulate SG formation, promoting the investigation of homologous approaches for cancer therapy.
Furthermore, it has been proposed that SG formation might be associated with the initiation of breast cancer because the important SG component G3BP2 could stabilize the squamous cell carcinoma antigen recognized by T‐cell 3 mRNA and promote tumourgenesis (Gupta et al., 2017; Figure 3). These data showed that the lead compound, C108, identified by high‐throughput chemical screening could alleviate the function of G3BP2 and inhibit the activity of tumour‐initiating cells (Gupta et al., 2017). Thus, combining compound C108 and its derivatives with standardized treatment might be considered a beneficial method for patients with breast cancer.
4.2.2. Targeting the key steps of SG assembly
SGs usually assemble and disassemble rapidly in response to environmental stress, which is contradictory with the hindered diffusion rules in cytoplasm (Lukacs et al., 2000). Thus, how such rapid aggregation can form attracts considerable interest. As previous studies reported, macromolecule transportation, such as recruitment of mRNPs in the cytoplasm, depended on cytoskeleton‐active transport (Bomsel, Parton, Kuznetsov, Schroer, & Gruenberg, 1990). mRNPs are transported along with microtubule motor proteins, and several studies have found that microtubule integrity was important for SG assembly (Ivanov, Chudinova, & Nadezhdina, 2003). Disruption of microtubules with the microtubule‐depolymerizing drug http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6851 could trigger SG disappearance in CV‐1 cell (Ivanov et al., 2003; Figure 3). In contrast, the microtubule‐stabilizing drug http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2770 had the opposite function.
The microtubule system is necessary for SG formation, and microtubules can drive small particles to form larger granules through coalescence (Arn & Macdonald, 1998). These results have provided helpful strategies for targeting microtubules to regulate SG formation. Stress conditions, such as UV irradiation and hypoxia‐induced SGs might be partly inhibited by combination therapy with microtubule‐inhibiting drugs, such as vinblastine.
4.3. Regulating PTMs related to SGs assembly
Post‐translational modifications with ubiquitin and ubiquitin‐like proteins are essential for eukaryotic cellular processes. One of the most common modifications is ubiquitination, which regulates various fundamental cellular activities (Hershko & Ciechanover, 1998; Komander & Rape, 2012). In addition, proteins can also be modified with ubiquitin‐like proteins, such as NEDD8 and SUMO, which are called neddylation and sumoylation respectively (van der Veen & Ploegh, 2012). Other PTMs, such as acetylation, phosphorylation, O‐GlcNAc modification (Ohn, Kedersha, Hickman, Tisdale, & Anderson, 2008), and arginine methylation of ribosomal subunits, are also involved in SG assembly, SG protein interactions, disassembly, and clearance (Mahboubi & Stochaj, 2017; Ohn & Anderson, 2010). For example, phosphorylation of fused in sarcoma (FUS) and G3BP1 at Ser149 disturbed the formation of SGs (Tourrière et al., 2003), and phosphorylation of FUS prevented its recruitment to SGs (Han et al., 2012), while dephosphorylation functioned in contrast (Reineke et al., 2017; Tourriere et al., 2003). Most SGs contain arginine methylation motifs , such as RGC motifs, which have effects on the recruitment of SG components (Nott et al., 2015). In addition, G3BP1 was observed to bind to HDAC6 and sirtuin 6 (SIRT6), both of which have extensive enzymatic activity and are required for SGs production. Methylation was determined to promote the RBPs interaction among fragile X mental retardation protein, heterogeneous nuclear ribonucleoprotein, and FUS (Dolzhanskaya, Merz, Aletta, & Denman, 2006; Yamaguchi & Kitajo, 2012) and then to stimulate the SG assembly. As it is well‐known to all that the assembly of SGs is highly complex, PTMs of individual components even make this process further complicated, which actually provides great possibilities to regulate SGs in cancer therapy by targeting PTMs.
4.3.1. Ubiquitination
The ubiquitin proteasome system (UPS) is the most important pathway responsible for intracellular protein degradation. The activity of UPS is essential not only for protein homeostasis but also for the translation process through providing reused amino acids that can be produced by SG disassembly after the stress is relieved (Hershko, 2005; Vabulas & Hartl, 2005). Studies have shown that inhibiting the activity of UPS with pharmacological agents, such as proteasome inhibitors https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=8616 or bortezomib influenced translation initiation, which could induce a typical stress response and was followed by the overexpression of the heat shock protein Hsp72 (Fournier et al., 2010; Schewe & Aguirre‐Ghiso, 2009; Figure 4). In addition, UPS has also been shown to be involved in RNA metabolism. All of these observations showed that UPS was associated with the formation and disassembly of SG, suggesting a promising strategy applied for SG regulation and cancer therapy.
Figure 4.
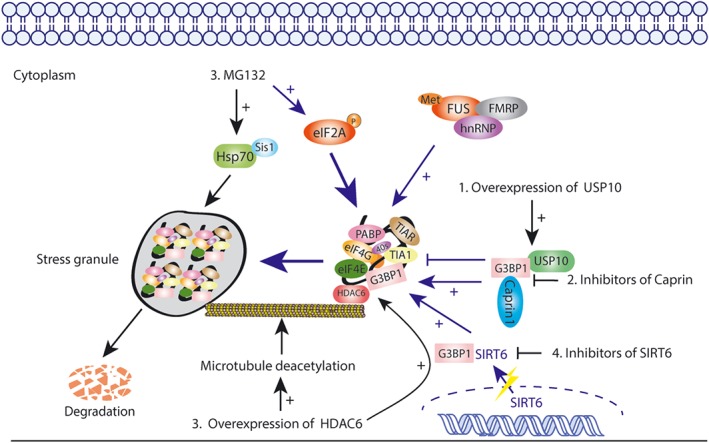
Targeting SGs by regulating related PTMs like ubiquitinations, phosphorylations, and acetylations
Ubiquitin specific peptidase 10
Multiple studies have shown that G3BP‐dependent SG formation is regulated by ubiquitin‐specific peptidase 10 (USP10) and cell cycle‐associated protein 1 (Caprin1), and the former is an important type of deubiquitinase that usually binds with G3BP to inhibit SG formation, while the latter functions oppositely (Solomon et al., 2007a; Figure 4). Overexpression of mCherry‐USP10 inhibited SG formation induced by arsenite, and USP10 competed with Caprin1 for binding with G3BP (Kedersha et al., 2016). Although the binding sites of USP10 and Caprin1 overlapped, USP10 had a Phe‐Gly‐Asp‐Phe region that provided interaction with G3BP and SG inhibition, while Caprin1 did not (Panas et al., 2015). Researchers recombined and purified the His‐tagged G3BP1 and His‐tagged Caprin1 protein through bacteria, aiming to study direct protein–protein interactions in vitro. The results showed that Caprin1 disturbed the interaction between G3BP1 and USP10 in a dose‐dependent manner, and competition existed between Caprin1 and USP10 for binding to G3BP (Kedersha et al., 2016). Furthermore, arsenite could induce the formation of SGs in non‐induced tet‐on GFP‐USP10 cells, but once USP10 was overexpressed by https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6464, SG formation was completely blocked. USP10 also inhibits the cytoplasmic mRNPs condensed into SGs. Thus, targeting USP10 or Caprin to decrease the interaction between G3BP and USP10 may be accepted as a new strategy to regulate SG formation. Small inhibitors of Caprin or the agonists of USP10 warrant further sudy for cancer treatment.
MG132
Although multiple studies have shown that MG132 could strongly induce the formation of SGs in an eIF2‐dependent manner, cells treated with MG132 for 6 hr were found to disassemble SGs and partly restore translation activity, despite of the maintenance of eIF2 phosphorylation (Mazroui et al., 2007). The data showed that sustainable stress of MG132 passed suicide signals to the cell through activating Hsp70, which contributed to the overexpression of proapoptotic proteins and, eventually, apoptosis at last (Mazroui et al., 2007; Figure 4). Therefore, identifying the factors involved in MG132‐induced SG disassembly could provide better approaches to promote cancer cell transformation from survival to apoptosis, which would help to improve cancer treatment.
More recently, Markmiller et al. (2019) claimed that although SGs formation is associated with UPS or ubiquitinated proteins accumulation, SGs dynamics are not affected by the ubiquitin‐activating enzyme. This article implied that polyubiquitination was dispensable for SG assembly or disassembly, but significant for cellular response signals regulating upon stress. Thus, further studies are needed to provide insight into the relationship between UPS, SGs, and various diseases.
4.3.2. Acetylation
HDAC6
In recent years, reversible protein acetylation has gradually appeared to be a major form of protein modification (Caron, Col, & Khochbin, 2003; Glozak, Sengupta, Zhang, & Seto, 2005). The acetylation of proteins is controlled by histone acetylases as well as histone deacetylases (HDACs). Multiple articles have proposed that HDAC6 strongly associated with tubulin and microtubule network (Boyault, Sadoul, Pabion, & Khochbin, 2007; Hubbert et al., 2002). HDAC6 could interact with G3BP1 both in vitro and in vivo, while other HDACs, such as HDAC1 and HDAC4, failed to be G3BP‐interacting partners (Kwon et al., 2007), implying that HDAC6 might be an essential component of SGs. Data have shown that inactivating mutations of the HDAC6 catalytic domain impaired SG formation, and MEFs lacking HDAC6 also exhibited impaired SG assembly ability (Figure 4). Furthermore, in the absence of intact HDAC6 function, cells could not overcome the fate of apoptosis in oxidative conditions induced by arsenite treatment (Hubbert et al., 2002). Boyault et al. (2006) noted that HDAC6 could deacetylate tubulin and microtubule (Zhang et al., 2003), both of which are important SG components (Figure 4). In addition, HDAC6 not only accelerated the clearance of misfolded proteins and then protected cells from stress‐induced apoptosis but also interacted with AAA ATPase p87/VCP, which was critical for misfolded protein degradation in the proteasome (Boyault et al., 2006). Thus, it is convincing that HDAC6 is necessary in the formation of SGs and cell stress response, and inhibitors targeting HDAC6 deacetylase activity or its interaction with G3BP are worth considering for SG regulation for cancer therapy.
4.3.3. Phosphorylation
SIRT6
As a NAD+‐dependent deacetylase, SIRT6 is found in the nucleus, regulating the structure of chromatin and genomic stability (Michishita et al., 2008). Jedrusik‐Bode et al. (2013) proposed that in response to stress, SIRT6 was localized into the cytoplasm, which was partly dependent on interactions with G3BP and then with the SG components to promote SG assembly. Inhibiting the catalytic activity of SIRT6 or knocking down SIRT6 could strongly impair SG formation and the survival of Caenorhabditis elegans. Further data obtained by these researchers showed that SIRT6 did not affect the acetylation of G3BP. In contrast, the absence of SIRT6 was shown to increase the phosphorylation of G3BP at Ser149, which had already been shown to inhibit SG formation under stress conditions (Tourriere et al., 2003; Figure 4). Thus, targeting SIRT6 could provide a clear strategy to regulate SG formation by changing the phosphorylation status of G3BP.
4.4. Preventing the oxidative stress
Oxidative stress, caused by glucose deprivation (Simons, Mattson, Dornfeld, & Spitz, 2009), https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=244 (Emara et al., 2012; Thedieck et al., 2013), and sodium arsenite, has been proposed to be a significant inducer of SG formation. SGs that respond to different oxidative stress conditions have different components. For example, arsenite‐induced SGs mainly contain the eIF4 complex and require the phosphorylation of eIF2, while the concentrations of eIF3, eIF4E, and eIF4G are relatively low in hydrogen peroxide‐induced SGs (Emara et al., 2012). Emerging evidence has shown that SG formation is helpful for antioxidant activity in mammalian cells. Upon sodium arsenite treatment, cancer cells were more likely to undergo apoptosis without SG formation (Takahashi et al., 2013) . However, once SGs formed, G3BP1 was subsequently inactivated which activated USP10 as a result (Kedersha et al., 2016; Sowa, Bennett, Gygi, & Harper, 2009). Because USP10 usually interacts with multiple polysomic proteins, such as poly(A)‐binding protein (Sowa et al., 2009), its activation could regulate SG formation and balance the ROS level, thereby facilitating tumour survival.
Considering the close relationship of SG formation and oxidative stress, seeking possibilities that target oxidative signal pathways or antioxidant enzymes is a promising method for inhibiting SG formation (Cacciatore, Baldassarre, Fornasari, Mollica, & Pinnen, 2012). For example, https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6737is an antioxidant in plants and animals and can prevent cellular components from damage caused by ROS. Chemical strategies applying analogues or prodrugs to increase the GSH concentration were previously evaluated in vivo and in vitro (Cacciatore et al., 2012; Fujita, Yamafuji, Nakabeppu, & Noda, 2012). Other approaches, such as utilizing medical gases like hydrogen sulfide, hydrogen, and carbon monoxide (Fujita et al., 2012), to rescue ROS levels or directly stimulating related antioxidant enzymes and transcription factors, such as haem oxygenase‐1, SOD, and nuclear factor erythroid related factor 2 have only been approved in neurodegradation disease therapy, all of which need to be confirmed in cancer therapy.
5. CONCLUSION AND FUTURE PERSPECTIVES
Over the past several years, a number of studies have shown that the function of SGs is not simply the temporary storage or degradation controller of mRNA. Indeed, SGs are versatile and important regulators of multiple physiological functions including cancer development (Anderson & Kedersha, 2008). The formation of SGs in eukaryotic cells represents a conserved response, that protects cells from harmful stress conditions and possesses apparent physiological advantages. These protective granules promote cell survival during stress conditions, minimize the expenditure of energy, and simultaneously regulate RNA and protein homeostasis (Arimoto et al., 2008; Panas et al., 2016).
As we reviewed above, it is likely that SG assembly has a strong connection with cancer cell proliferation and metastasis. Targeting SGs through disturbing their formation or assembly, as well as changing the stress condition, can not only influence tumour progression but also sensitize cancer cells to chemotherapeutic agents, which represents a promising strategy for cancer treatment. In addition, hotspot research on human immune checkpoints has shown that microtubule‐targeting drug treatment inhibits programmed https://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=9606 expression in cancer therapy, independent of their antimitotic activity but associated with SG assembly (Franchini et al., 2019). Given that overexpression of immune checkpoint mRNA by stimulated T‐cells leads to SG assembly and chemotherapy resistance, inhibiting the expression of several checkpoint mRNAs by programmed cell death protein 1 and programmed cell death‐ligand 1 inhibitors might be considered a new strategy for cancer treatment in the future.
Last, further research may investigate why SGs exert different functions among cancer cells and neuron cells and whether strategies targeting SGs could be usefully applied in both cancers and NDs. It is clear that SGs share many components with neuronal granules and are associated with neuronal RNA transport, as well as translation, which contributes to ND progression (Batish, van den Bogaard, Kramer, & Tyagi, 2012). Thus, more attention has been paid to the relationship between SGs and NDs, while less attention to SGs and cancer. Recent studies have proposed that neurons exist in the tumour micro‐environment, promoting cancer cell proliferation and metastasis. The secretion of growth factors by cancer cells, in turn, accelerated neuron outgrowth in solid tumours (Jobling et al., 2015). The reciprocal relationship between neurons and cancer cells provides new insights for the potential strategies targeting SGs for cancer and ND treatment. Thus, in the future, understanding the crosstalk between neurons and cancer cells is necessary for clarifying the different functions of SGs among neurons and cancer cells, which might be helpful in developing SGs as a new therapeutic target for cancer treatment.
5.1. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org/, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18(Alexander et al., 2017).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (Grant 81872885 to J.C.), Zhejiang Provincial Natural Science Foundation (Grant Y18H310005 to J.C.), and the Talent Project of Zhejiang Association for Science and Technology (Grant 2018YCGC002 to J.C.).
Gao X, Jiang L, Gong Y, et al. Stress granule: A promising target for cancer treatment. Br J Pharmacol. 2019;176:4421–4433. 10.1111/bph.14790
REFERENCES
- Ackerman, D. , & Simon, M. C. (2014). Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends in Cell Biology, 24, 472–478. 10.1016/j.tcb.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjibade, P. , St‐Sauveur, V. G. , Quevillon Huberdeau, M. , Fournier, M. J. , Savard, A. , Coudert, L. , … Mazroui, R. (2015). Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget, 6, 43927–43943. 10.18632/oncotarget.5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , … CGTP Collaborators (2017). The concise guide to PHARMACOLOGY 2017/18: Enzymes. British Journal of Pharmacology, 174(Suppl 1), S272–s359. 10.1111/bph.13877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. , & Kedersha, N. (2006). RNA granules. The Journal of Cell Biology, 172, 803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. , & Kedersha, N. (2008). Stress granules: The Tao of RNA triage. Trends in Biochemical Sciences, 33, 141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson, P. , Kedersha, N. , & Ivanov, P. (2015). Stress granules, P‐bodies and cancer. Biochimica et Biophysica Acta, 1849, 861–870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto, K. , Fukuda, H. , Imajoh‐Ohmi, S. , Saito, H. , & Takekawa, M. (2008). Formation of stress granules inhibits apoptosis by suppressing stress‐responsive MAPK pathways. Nature Cell Biology, 10, 1324–1332. 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- Arn, E. A. , & Macdonald, P. M. (1998). Motors driving mRNA localization: New insights from in vivo imaging. Cell, 95, 151–154. 10.1016/S0092-8674(00)81745-6 [DOI] [PubMed] [Google Scholar]
- Ayuso, M. I. , Martinez‐Alonso, E. , Regidor, I. , & Alcazar, A. (2016). Stress granule induction after brain ischemia is independent of eukaryotic translation initiation factor (eIF) 2α phosphorylation and is correlated with a decrease in eIF4B and eIF4E proteins. The Journal of Biological Chemistry, 291, 27252–27264. 10.1074/jbc.M116.738989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish, M. , van den Bogaard, P. , Kramer, F. R. , & Tyagi, S. (2012). Neuronal mRNAs travel singly into dendrites. Proceedings of the National Academy of Sciences of the United States of America, 109, 4645–4650. 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsel, M. , Parton, R. , Kuznetsov, S. A. , Schroer, T. A. , & Gruenberg, J. (1990). Microtubule‐ and motor‐dependent fusion in vitro between apical and basolateral endocytic vesicles from MDCK cells. Cell, 62, 719–731. 10.1016/0092-8674(90)90117-W [DOI] [PubMed] [Google Scholar]
- Boyault, C. , Gilquin, B. , Zhang, Y. , Rybin, V. , Garman, E. , Meyer‐Klaucke, W. , … Khochbin, S. (2006). HDAC6‐p97/VCP controlled polyubiquitin chain turnover. The EMBO Journal, 25, 3357–3366. 10.1038/sj.emboj.7601210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault, C. , Sadoul, K. , Pabion, M. , & Khochbin, S. (2007). HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene, 26, 5468–5476. 10.1038/sj.onc.1210614 [DOI] [PubMed] [Google Scholar]
- Boyce, M. , Bryant, K. F. , Jousse, C. , Long, K. , Harding, H. P. , Scheuner, D. , … Yuan, J. (2005). A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science, 307, 935–939. 10.1126/science.1101902 [DOI] [PubMed] [Google Scholar]
- Buchan, J. R. , & Parker, R. (2009). Eukaryotic stress granules: The ins and outs of translation. Molecular Cell, 36, 932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciatore, I. , Baldassarre, L. , Fornasari, E. , Mollica, A. , & Pinnen, F. (2012). Recent advances in the treatment of neurodegenerative diseases based on GSH delivery systems. Oxidative Medicine and Cellular Longevity, 2012, 240146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, C. , Col, E. , & Khochbin, S. (2003). The viral control of cellular acetylation signaling. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 25, 58–65. 10.1002/bies.10202 [DOI] [PubMed] [Google Scholar]
- Cencic, R. , Carrier, M. , Galicia‐Vazquez, G. , Bordeleau, M. E. , Sukarieh, R. , Bourdeau, A. , … Pelletier, J. (2009). Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS ONE, 4, e5223 10.1371/journal.pone.0005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , & Liu, B. (2017). Relationships between stress granules, oxidative stress, and neurodegenerative diseases. Oxidative Medicine and Cellular Longevity, 2017, 1809592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov, V. , Hofmann, S. , Druffel‐Augustin, S. , Mogk, A. , Tyedmers, J. , Stoecklin, G. , & Bukau, B. (2013). Coordination of translational control and protein homeostasis during severe heat stress. Current Biology: CB, 23, 2452–2462. 10.1016/j.cub.2013.09.058 [DOI] [PubMed] [Google Scholar]
- Chernov, K. G. , Barbet, A. , Hamon, L. , Ovchinnikov, L. P. , Curmi, P. A. , & Pastre, D. (2009). Role of microtubules in stress granule assembly: Microtubule dynamical instability favors the formation of micrometric stress granules in cells. The Journal of Biological Chemistry, 284, 36569–36580. 10.1074/jbc.M109.042879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhanskaya, N. , Merz, G. , Aletta, J. M. , & Denman, R. B. (2006). Methylation regulates the intracellular protein‐protein and protein‐RNA interactions of FMRP. Journal of Cell Science, 119, 1933–1946. 10.1242/jcs.02882 [DOI] [PubMed] [Google Scholar]
- Drexler, H. C. (2009). Synergistic apoptosis induction in leukemic cells by the phosphatase inhibitor salubrinal and proteasome inhibitors. PLoS ONE, 4, e4161 10.1371/journal.pone.0004161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara, M. M. , Fujimura, K. , Sciaranghella, D. , Ivanova, V. , Ivanov, P. , & Anderson, P. (2012). Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochemical and Biophysical Research Communications, 423, 763–769. 10.1016/j.bbrc.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken, M. B. , & Corbett, A. H. (2005). Process or perish: Quality control in mRNA biogenesis. Nature Structural & Molecular Biology, 12, 482–488. 10.1038/nsmb945 [DOI] [PubMed] [Google Scholar]
- Fournier, M.‐J. , Gareau, C. , & Mazroui, R. (2010). The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell International, 10, 12 10.1186/1475-2867-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini, D. M. , Lanvin, O. , Tosolini, M. , Patras de Campaigno, E. , Cammas, A. , Pericart, S. , … Fournié, J. J. (2019). Microtubule‐driven stress granule dynamics regulate inhibitory immune checkpoint expression in T cells. Cell Reports, 26, 94–107 e107. [DOI] [PubMed] [Google Scholar]
- Fujimura, K. , Sasaki, A. T. , & Anderson, P. (2012). Selenite targets eIF4E‐binding protein‐1 to inhibit translation initiation and induce the assembly of non‐canonical stress granules. Nucleic Acids Research, 40(16), 8099–8110. 10.1093/nar/gks566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, K. , Yamafuji, M. , Nakabeppu, Y. , & Noda, M. (2012). Therapeutic approach to neurodegenerative diseases by medical gases: Focusing on redox signaling and related antioxidant enzymes. Oxidative Medicine and Cellular Longevity, 2012, 324256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi, I. E. , Parker, F. , Chebli, K. , Maurier, F. , Labourier, E. , Barlat, I. , … Tazi, J. (1998). A novel phosphorylation‐dependent RNase activity of GAP‐SH3 binding protein: A potential link between signal transduction and RNA stability. Molecular and Cellular Biology, 18, 3956–3965. 10.1128/MCB.18.7.3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks, N. , Kedersha, N. , Ayodele, M. , Shen, L. , Stoecklin, G. , Dember, L. M. , & Anderson, P. (2004). Stress granule assembly is mediated by prion‐like aggregation of TIA‐1. Molecular Biology of the Cell, 15, 5383–5398. 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak, M. A. , Sengupta, N. , Zhang, X. , & Seto, E. (2005). Acetylation and deacetylation of non‐histone proteins. Gene, 363, 15–23. 10.1016/j.gene.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Grabocka, E. , & Bar‐Sagi, D. (2016). Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell, 167, 1803–1813.e1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N. , Badeaux, M. , Liu, Y. , Naxerova, K. , Sgroi, D. , Munn, L. L. , … Garkavtsev, I. (2017). Stress granule‐associated protein G3BP2 regulates breast tumor initiation. Proceedings of the National Academy of Sciences of the United States of America, 114, 1033–1038. 10.1073/pnas.1525387114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka, R. B. , Bennett, B. S. , Cullinan, S. B. , & Diehl, J. A. (2005). PERK and GCN2 contribute to eIF2α phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Molecular Biology of the Cell, 16, 5493–5501. 10.1091/mbc.e05-03-0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, T. W. , Kato, M. , Xie, S. , Wu, L. C. , Mirzaei, H. , Pei, J. , … McKnight, S. L. (2012). Cell‐free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell, 149, 768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR (2018). The IUPHAR/BPS guide to PHARMACOLOGY in 2018: Updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Research, 46, D1091–d1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A. (2005). The ubiquitin system for protein degradation and some of its roles in the control of the cell‐division cycle (Nobel lecture). Angewandte Chemie (International Ed in English), 44, 5932–5943. 10.1002/anie.200501724 [DOI] [PubMed] [Google Scholar]
- Hershko, A. , & Ciechanover, A. (1998). The ubiquitin system. Annual Review of Biochemistry, 67, 425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Holcik, M. (2015). Could the eIF2α‐independent translation be the achilles heel of cancer? Frontiers in Oncology, 5, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert, C. , Guardiola, A. , Shao, R. , Kawaguchi, Y. , Ito, A. , Nixon, A. , … Yao, T. P. (2002). HDAC6 is a microtubule‐associated deacetylase. Nature, 417, 455–458. 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Ivanov, P. A. , Chudinova, E. M. , & Nadezhdina, E. S. (2003). Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Experimental Cell Research, 290, 227–233. 10.1016/S0014-4827(03)00290-8 [DOI] [PubMed] [Google Scholar]
- Jayabalan, A. K. , Sanchez, A. , Park, R. Y. , Yoon, S. P. , Kang, G. Y. , Baek, J. H. , … Ohn, T. (2016). NEDDylation promotes stress granule assembly. Nature Communications, 7, 12125 10.1038/ncomms12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik‐Bode, M. , Studencka, M. , Smolka, C. , Baumann, T. , Schmidt, H. , Kampf, J. , … Bober, E. (2013). The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. Journal of Cell Science, 126, 5166–5177. 10.1242/jcs.130708 [DOI] [PubMed] [Google Scholar]
- Jobling, P. , Pundavela, J. , Oliveira, S. M. , Roselli, S. , Walker, M. M. , & Hondermarck, H. (2015). Nerve‐cancer cell cross‐talk: A novel promoter of tumor progression. Cancer Research, 75, 1777–1781. 10.1158/0008-5472.CAN-14-3180 [DOI] [PubMed] [Google Scholar]
- Kaehler, C. , Isensee, J. , Hucho, T. , Lehrach, H. , & Krobitsch, S. (2014). 5‐Fluorouracil affects assembly of stress granules based on RNA incorporation. Nucleic Acids Research, 42, 6436–6447. 10.1093/nar/gku264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. , Chen, S. , Gilks, N. , Li, W. , Miller, I. J. , Stahl, J. , & Anderson, P. (2002). Evidence that ternary complex (eIF2‐GTP‐tRNA(i)(Met))‐deficient preinitiation complexes are core constituents of mammalian stress granules. Molecular Biology of the Cell, 13, 195–210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. , Cho, M. R. , Li, W. , Yacono, P. W. , Chen, S. , Gilks, N. , … Anderson, P. (2000). Dynamic shuttling of TIA‐1 accompanies the recruitment of mRNA to mammalian stress granules. The Journal of Cell Biology, 151, 1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. , Ivanov, P. , & Anderson, P. (2013). Stress granules and cell signaling: More than just a passing phase? Trends in Biochemical Sciences, 38, 494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. , Panas, M. D. , Achorn, C. A. , Lyons, S. , Tisdale, S. , Hickman, T. , … Anderson, P. (2016). G3BP‐Caprin1‐USP10 complexes mediate stress granule condensation and associate with 40S subunits. The Journal of Cell Biology, 212, 845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. L. , Gupta, M. , Li, W. , Miller, I. , & Anderson, P. (1999). RNA‐binding proteins TIA‐1 and TIAR link the phosphorylation of eIF‐2 α to the assembly of mammalian stress granules. The Journal of Cell Biology, 147, 1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander, D. , & Rape, M. (2012). The ubiquitin code. Annual Review of Biochemistry, 81, 203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Kroschwald, S. , Maharana, S. , Mateju, D. , Malinovska, L. , Nuske, E. , Poser, I. , … Alberti, S. (2015). Promiscuous interactions and protein disaggregases determine the material state of stress‐inducible RNP granules. eLife, 4, e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. , Zhang, Y. , & Matthias, P. (2007). The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes & Development, 21, 3381–3394. 10.1101/gad.461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. R. , King, O. D. , Shorter, J. , & Gitler, A. D. (2013). Stress granules as crucibles of ALS pathogenesis. The Journal of Cell Biology, 201, 361–372. 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, X. J. , & Gallouzi, I. E. (2009). Oxidative stress increases the number of stress granules in senescent cells and triggers a rapid decrease in p21waf1/cip1 translation. The Journal of Biological Chemistry, 284, 8877–8887. 10.1074/jbc.M806372200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Protter, D. S. , Rosen, M. K. , & Parker, R. (2015). Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Molecular Cell, 60, 208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley, D. B. , Harkin, D. P. , & Johnston, P. G. (2003). 5‐Fluorouracil: Mechanisms of action and clinical strategies. Nature Reviews Cancer, 3, 330–338. 10.1038/nrc1074 [DOI] [PubMed] [Google Scholar]
- Lukacs, G. L. , Haggie, P. , Seksek, O. , Lechardeur, D. , Freedman, N. , & Verkman, A. S. (2000). Size‐dependent DNA mobility in cytoplasm and nucleus. The Journal of Biological Chemistry, 275, 1625–1629. 10.1074/jbc.275.3.1625 [DOI] [PubMed] [Google Scholar]
- Mahboubi, H. , & Stochaj, U. (2017). Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochimica et Biophysica Acta ‐ Molecular Basis of Disease, 1863, 884–895. 10.1016/j.bbadis.2016.12.022 [DOI] [PubMed] [Google Scholar]
- Malinowska, M. , Niedzwiedzka‐Rystwej, P. , Tokarz‐Deptula, B. , & Deptula, W. (2016). Stress granules (SG) and processing bodies (PB) in viral infections. Acta Biochimica Polonica, 63, 183–188. [DOI] [PubMed] [Google Scholar]
- Markmiller, S. , Fulzele, A. , Higgins, R. , Leonard, M. , Yeo, G. W. , & Bennett, E. J. (2019). Active protein neddylation or ubiquitylation is dispensable for stress granule dynamics. Cell Reports, 27, 1356–1363.e1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui, R. , Di Marco, S. , Kaufman, R. J. , & Gallouzi, I. E. (2007). Inhibition of the ubiquitin‐proteasome system induces stress granule formation. Molecular Biology of the Cell, 18, 2603–2618. 10.1091/mbc.e06-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey, D. J. , & Zhu, K. (2008). Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resistance Updates: Reviews and Commentaries in Antimicrobial and Anticancer Chemotherapy, 11, 164–179. 10.1016/j.drup.2008.08.002 [DOI] [PubMed] [Google Scholar]
- McEwen, E. , Kedersha, N. , Song, B. , Scheuner, D. , Gilks, N. , Han, A. , … Kaufman, R. J. (2005). Heme‐regulated inhibitor kinase‐mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. The Journal of Biological Chemistry, 280, 16925–16933. 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- Michishita, E. , McCord, R. A. , Berber, E. , Kioi, M. , Padilla‐Nash, H. , Damian, M. , … Chua, K. F. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature, 452, 492–496. 10.1038/nature06736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, J. A. , Radford, H. , Peretti, D. , Steinert, J. R. , Verity, N. , Martin, M. G. , … Mallucci, G. R. (2012). Sustained translational repression by eIF2α‐P mediates prion neurodegeneration. Nature, 485, 507–511. 10.1038/nature11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott, T. J. , Petsalaki, E. , Farber, P. , Jervis, D. , Fussner, E. , Plochowietz, A. , … Baldwin, A. J. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Molecular Cell, 57, 936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn, T. , & Anderson, P. (2010). The role of posttranslational modifications in the assembly of stress granules. Wiley Interdisciplinary Reviews RNA, 1, 486–493. 10.1002/wrna.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn, T. , Kedersha, N. , Hickman, T. , Tisdale, S. , & Anderson, P. (2008). A functional RNAi screen links O‐GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nature Cell Biology, 10, 1224–1231. 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas, M. D. , Ivanov, P. , & Anderson, P. (2016). Mechanistic insights into mammalian stress granule dynamics. The Journal of Cell Biology, 215, 313–323. 10.1083/jcb.201609081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas, M. D. , Schulte, T. , Thaa, B. , Sandalova, T. , Kedersha, N. , Achour, A. , & McInerney, G. M. (2015). Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathogens, 11, e1004659 10.1371/journal.ppat.1004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J. , Graff, J. , Ruggero, D. , & Sonenberg, N. (2015). Targeting the eIF4F translation initiation complex: A critical nexus for cancer development. Cancer Research, 75, 250–263. 10.1158/0008-5472.CAN-14-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter, D. S. W. , & Parker, R. (2016). Principles and properties of stress granules. Trends in Cell Biology, 26, 668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani, M. , Davis, E. M. , Crabtree, T. R. , Habibi, J. R. , Nguyen, T. K. , Dent, P. , & Grant, S. (2007). The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulum stress. Molecular and Cellular Biology, 27, 5499–5513. 10.1128/MCB.01080-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke, L. C. , & Lloyd, R. E. (2013). Diversion of stress granules and P‐bodies during viral infection. Virology, 436, 255–267. 10.1016/j.virol.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke, L. C. , & Neilson, J. R. (2019). Differences between acute and chronic stress granules, and how these differences may impact function in human disease. Biochemical Pharmacology, 162, 123–131. 10.1016/j.bcp.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke, L. C. , Tsai, W.‐C. , Jain, A. , Kaelber, J. T. , Jung, S. Y. , & Lloyd, R. E. (2017). Casein kinase 2 is linked to stress granule dynamics through phosphorylation of the stress granule nucleating protein G3BP1. Molecular and Cellular Biology, 37, e00596–e00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, P. G. (2004). A review of the proteasome inhibitor bortezomib in multiple myeloma. Expert Opinion on Pharmacotherapy, 5, 1321–1331. 10.1517/14656566.5.6.1321 [DOI] [PubMed] [Google Scholar]
- Richardson, P. G. , Hideshima, T. , & Anderson, K. C. (2003). Bortezomib (PS‐341): A novel, first‐in‐class proteasome inhibitor for the treatment of multiple myeloma and other cancers. Cancer Control: Journal of the Moffitt Cancer Center, 10, 361–369. 10.1177/107327480301000502 [DOI] [PubMed] [Google Scholar]
- Rizvi, N. F. , & Smith, G. F. (2017). RNA as a small molecule druggable target. Bioorganic & Medicinal Chemistry Letters, 27, 5083–5088. 10.1016/j.bmcl.2017.10.052 [DOI] [PubMed] [Google Scholar]
- Ryu, I. , Won, Y. S. , Ha, H. , Kim, E. , Park, Y. , Kim, M. K. , … Kim, Y. K. (2019). eIF4A3 phosphorylation by CDKs affects NMD during the cell cycle. Cell Reports, 26, 2126–2139.e2129. [DOI] [PubMed] [Google Scholar]
- Sasaki, A. T. , Fujimura, K. , & Anderson, P. (2012). Selenite targets eIF4E‐binding protein‐1 to inhibit translation initiation and induce the assembly of non‐canonical stress granules. Nucleic Acids Research, 40, 8099–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schewe, D. M. , & Aguirre‐Ghiso, J. A. (2009). Inhibition of eIF2α dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Research, 69, 1545–1552. 10.1158/0008-5472.CAN-08-3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, E. , Irollo, E. , & Grabocka, E. (2019). Evaluating stress granules in pancreatic cancer in vitro and in vivo. Methods in Molecular Biology, 1882, 183–195. 10.1007/978-1-4939-8879-2_17 [DOI] [PubMed] [Google Scholar]
- Simons, A. L. , Mattson, D. M. , Dornfeld, K. , & Spitz, D. R. (2009). Glucose deprivation‐induced metabolic oxidative stress and cancer therapy. Journal of Cancer Research and Therapeutics, 5(Suppl 1), S2–S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, S. , Xu, Y. , Wang, B. , David, M. D. , Schubert, P. , Kennedy, D. , & Schrader, J. W. (2007a). Distinct structural features of caprin‐1 mediate its interaction with G3BP‐1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Molecular and Cellular Biology, 27, 2324–2342. 10.1128/MCB.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, S. , Xu, Y. , Wang, B. , David, M. D. , Schubert, P. , Kennedy, D. , & Schrader, J. W. (2007b). Distinct structural features of Caprin‐1 mediate its interaction with G3BP‐1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Molecular and Cellular Biology, 27, 2324–2342. 10.1128/MCB.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan, S. P. , El‐Naggar, A. , Leprivier, G. , Cheng, H. , Hajee, S. , Grunewald, T. G. , … Sorensen, P. H. (2015). YB‐1 regulates stress granule formation and tumor progression by translationally activating G3BP1. The Journal of Cell Biology, 208, 913–929. 10.1083/jcb.201411047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg, N. , & Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell, 136, 731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa, M. E. , Bennett, E. J. , Gygi, S. P. , & Harper, J. W. (2009). Defining the human deubiquitinating enzyme interaction landscape. Cell, 138, 389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M. , Higuchi, M. , Matsuki, H. , Yoshita, M. , Ohsawa, T. , Oie, M. , & Fujii, M. (2013). Stress granules inhibit apoptosis by reducing reactive oxygen species production. Molecular and Cellular Biology, 33, 815–829. 10.1128/MCB.00763-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Y. , Gao, M. , Wang, J. , Kong, Q. , Hua, H. , Luo, T. , & Jiang, Y. (2014). Inhibition of eIF2α dephosphorylation enhances TRAIL‐induced apoptosis in hepatoma cells. Cell Death & Disease, 5, e1060 10.1038/cddis.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck, K. , Holzwarth, B. , Prentzell, M. T. , Boehlke, C. , Klasener, K. , Ruf, S. , … Baumeister, R. (2013). Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell, 154, 859–874. 10.1016/j.cell.2013.07.031 [DOI] [PubMed] [Google Scholar]
- Topisirovic, I. , Ruiz‐Gutierrez, M. , & Borden, K. L. (2004). Phosphorylation of the eukaryotic translation initiation factor eIF4E contributes to its transformation and mRNA transport activities. Cancer Research, 64, 8639–8642. 10.1158/0008-5472.CAN-04-2677 [DOI] [PubMed] [Google Scholar]
- Tourriere, H. , Chebli, K. , Zekri, L. , Courselaud, B. , Blanchard, J. M. , Bertrand, E. , & Tazi, J. (2003). The RasGAP‐associated endoribonuclease G3BP assembles stress granules. The Journal of Cell Biology, 160, 823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tourrière, H. , Chebli, K. , Zekri, L. , Courselaud, B. , Blanchard, J. M. , Bertrand, E. , & Tazi, J. (2003). The RasGAP‐associated endoribonuclease G3BP assembles stress granules. The Journal of Cell Biology, 160, 823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tourriere, H. , Gallouzi, I. E. , Chebli, K. , Capony, J. P. , Mouaikel, J. , van der Geer, P. , & Tazi, J. (2001). RasGAP‐associated endoribonuclease G3Bp: Selective RNA degradation and phosphorylation‐dependent localization. Molecular and Cellular Biology, 21, 7747–7760. 10.1128/MCB.21.22.7747-7760.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar, L. , & Tutar, Y. (2008). Ydj1 but not Sis1 stabilizes Hsp70 protein under prolonged stress in vitro. Biopolymers, 89, 171–174. 10.1002/bip.20881 [DOI] [PubMed] [Google Scholar]
- Vabulas, R. M. , & Hartl, F. U. (2005). Protein synthesis upon acute nutrient restriction relies on proteasome function. Science, 310, 1960–1963. 10.1126/science.1121925 [DOI] [PubMed] [Google Scholar]
- van der Veen, A. G. , & Ploegh, H. L. (2012). Ubiquitin‐like proteins. Annual Review of Biochemistry, 81, 323–357. 10.1146/annurev-biochem-093010-153308 [DOI] [PubMed] [Google Scholar]
- Vilas‐Boas Fde, A. , da Silva, A. M. , de Sousa, L. P. , Lima, K. M. , Vago, J. P. , Bittencourt, L. F. , … Barcelos, L. S. (2016). Impairment of stress granule assembly via inhibition of the eIF2α phosphorylation sensitizes glioma cells to chemotherapeutic agents. Journal of Neuro‐Oncology, 127, 253–260. 10.1007/s11060-015-2043-3 [DOI] [PubMed] [Google Scholar]
- Walters, R. W. , Muhlrad, D. , Garcia, J. , & Parker, R. (2015). Differential effects of Ydj1 and Sis1 on Hsp70‐mediated clearance of stress granules in Saccharomyces cerevisiae. RNA (New York, NY), 21, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek, R. C. , Jiang, H. Y. , & Anthony, T. G. (2006). Coping with stress: eIF2 kinases and translational control. Biochemical Society Transactions, 34, 7–11. 10.1042/BST0340007 [DOI] [PubMed] [Google Scholar]
- Wheeler, J. R. , Matheny, T. , Jain, S. , Abrisch, R. , & Parker, R. (2016). Distinct stages in stress granule assembly and disassembly. eLife, 5 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, A. , & Kitajo, K. (2012). The effect of PRMT1‐mediated arginine methylation on the subcellular localization, stress granules, and detergent‐insoluble aggregates of FUS/TLS. PLoS ONE, 7, e49267 10.1371/journal.pone.0049267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Li, N. , Caron, C. , Matthias, G. , Hess, D. , Khochbin, S. , & Matthias, P. (2003). HDAC‐6 interacts with and deacetylates tubulin and microtubules in vivo. The EMBO Journal, 22, 1168–1179. 10.1093/emboj/cdg115 [DOI] [PMC free article] [PubMed] [Google Scholar]


