Abstract
Background
As the spread of carbapenemase-producing Enterobacteriaceae poses a critical threat to public health, rapid detection of carbapenemase genes is urgently required for prompt initiation of appropriate antimicrobial therapy and infection control. We evaluated the performance of Xpert Carba-R v.2 (Cepheid, USA) compared with that of culture-based conventional PCR.
Methods
Using the results of 5,479 consecutive clinical rectal swabs, discrepant analysis (enriched culture followed by PCR) was performed for all discordant samples (N=100), which were Carba-R v.2-positive and culture-negative.
Results
Among the samples, 206 carbapenemase genes (3.6%) were detected by Carba-R v.2. The sensitivity and specificity were 95.0% and 98.1%, respectively. The positive predictive value (PPV) and negative predictive value (NPV) were 49.0% and 99.9%, respectively. Following discrepant analysis, the PPV increased to 73.5% and the low PPV (8.1%) of the 86 non-KPC improved to 48.8%. Among the 105 discrepancies, NDM was the most frequently observed (N=56), followed by KPC (N=26), VIM (N=10), IMP (N=8), OXA-48 (N=5). The threshold cycle values between discordant vs. concordant and resolved groups were significantly different (P<0.001).
Conclusions
Carba-R v.2 is a rapid and sensitive method for detecting carbapenemase-encoding genes compared with culture-based conventional PCR. Most of our discrepant results were non-KPC genes. Thus, the clinical significance of the non-KPC positive cases detected by Carba-R v.2 should be investigated. This assay would be useful for deciding whether to isolate pre-exposed patients in hospital settings, based on the high specificity and NPV.
Keywords: Xpert Carba-R v.2, Performance, Carbapenemase-producing Enterobacteriaceae, KPC, NDM, Infection control
INTRODUCTION
The global spread of carbapenem-resistant Enterobacteriaceae (CRE) and infections caused by CRE are urgent health issues causing high mortality and morbidity; hence, the WHO has identified CRE as a cause of global concern [1]. Of the carbapenem resistance mechanisms, carbapenemase genes on transmissible plasmid escalate the probability of dissemination. Patient monitoring and implementation of appropriate isolation measures depending on carbapenemase-producing Enterobacteriaceae (CPE) isolate colonization or infection status are crucial for preventing the spread of CPE, especially on admission and rapid inter-facility or inter-ward transfer.
CPE detection using the culture-based reference method is recommended by the US Centers for Disease Control and Prevention (CDC) and takes at least 72 hours [2]. A longer turn-around time increases medical expenses, as patients with a high risk of CPE, such as those with pre-exposure, need to be separated. Xpert Carba-R assay v. 2 (Cepheid, Sunnyvale, CA, USA) is a multiplex real-time PCR assay that has been upgraded to detect not only imipenem (IMP)-, Klebsiella pneumoniae carbapenemase (KPC)-, New Delhi metallo-β-lactamase (NDM)-, Verona integron-mediated metallo-β-lactamase (VIM)-, and oxacillinase (OXA)-48-type, but also OXA-48 variants (OXA-181 and OXA-232) in less than one hour. Most previous studies have evaluated the Carba-R (both v.1 and v.2) using limited numbers of clinical samples or contrived samples consisting of clearly defined carbapenemase-producing organisms [3,4,5,6]. Therefore, we evaluated the performance of the upgraded version, Carba-R v.2, in a large number of clinical samples from two general hospitals.
MATERIALS AND METHODS
Patients and samples
A total of 5,479 (5,379 from Severance Hospital, Seoul, Korea and 100 from Ilsan Hospital, Goyang, Korea) rectal swab samples were collected from high-risk patients who were admitted to intensive care units, CPE-exposed patients, or patients who were transferred from long-term care facilities to the two general hospitals from August 2016 to December 2017. We performed both the Carba-R v.2 and culture-based conventional PCR to detect five carbapenemase genes (IMP, KPC, NDM, VIM, and OXA-48) in all samples. Requested assays were consecutively included. Multiple genes in a sample were separately counted in the performance comparison of each carbapenemase gene.
This prospective study was approved by the Institutional Review Board of Severance Hospital (4-2016-1124) and Ilsan Hospital (NHIC-2017-03-005). Informed consent was waived as this study did not include any personal information of the patients.
Carbapenemase gene detection
Xpert Carba-R assay v.2
The assay was conducted according to the manufacturer's instructions within one hour of sample reception. The initially suggested threshold cycle (Ct) value was ≤38. Assay information, including Ct value of each sample was collected and reviewed to find an optimal Ct value for detecting carbapenemase genes by using receiver operating characteristic (ROC) curve.
Culture-based conventional PCR
This method has been considered as the gold standard for detecting carbapenemase genes. The culture-based assay recommended by the US CDC includes overnight enrichment in 5 mL of trypticase soy broth (TSB; Becton Dickinson Co., Sparks, MD, USA) with a 10 µg ertapenem disk (BBL, Becton Dickinson Co.), followed by overnight subculture on a MacConkey agar plate (Asan Pharmaceutical, Seoul, Korea) [7]. All colonies within the inhibition zone were picked and boiled for 10 min for DNA extraction followed by PCR to detect five carbapenemase genes (IMP, KPC, NDM, VIM, and OXA-48), as previously described [8].
Discrepant analysis
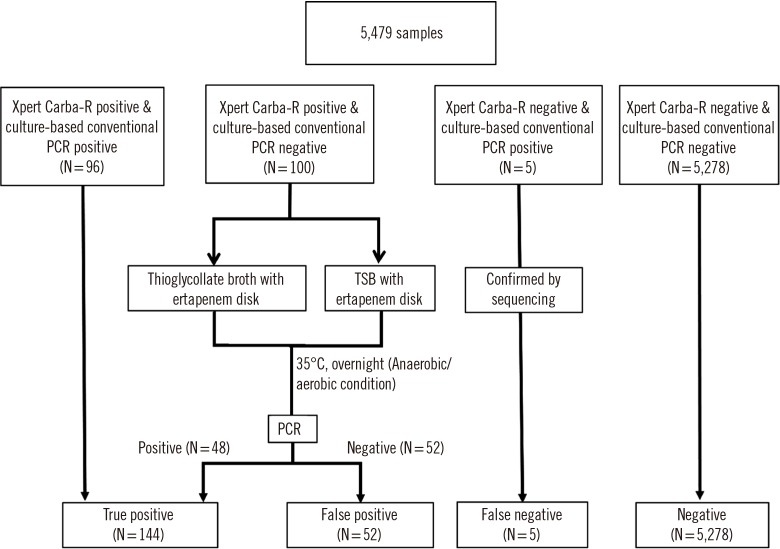
We attempted to resolve the discrepant cases as follows (Fig. 1): For Carba-R v.2 negative but culture-positive cases, the PCR product was confirmed by sequencing using the same primers. The sequencing was performed by a commercial sequencing service (Macrogen Inc, Seoul, Korea). For culture-negative but Carba-R v.2-positive cases, we conducted an additional assay, referred to as discrepant analysis. To identify aerobic or anaerobic organisms carrying carbapenemase, remaining samples stored in a refrigerator (at 4℃ for three to four days) were cultured in TSB and thioglycollate broth. Enriched PCR was performed with 20 µL of incubated broth. In cases of negative results, conventional PCR was conducted again after a five-day incubation to detect fastidious bacterial growth. Based on the discrepant analysis results, all samples were divided into three groups: the concordant group, which showed concordant Carba-R v.2 and culture results; the resolved group that initially showed discrepancy between the two assays, which was resolved following discrepant analysis; and the discordant group, which showed unresolved discrepancies
Fig. 1. Study flow chart and discrepant analysis protocol.
Abbreviation: TSB, tryptic soy broth.
Statistical analysis
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) including 95% confidence interval were calculated. The weighted Kappa value was calculated to compare the two assays. The Ct values of the concordant, resolved, and discordant groups were analyzed using the Kruskal-Wallis test and Mann-Whitney U-test. The median and interquartile range (IQR) were derived from the data summary. P values were calculated using Kruskal-Wallis test and designated threshold of significance was 0.05. The Ct value of each gene group was analyzed using the Mann-Whitney U-test. ROC curve was also analyzed to calculate optimal cut-off Ct value. Statistical analyses were performed using MedCalc Statistical Software version 18 (MedCalc Software Bvba, Ostend, Belgium; http://www.medcalc.org; 2018).
RESULTS
Of 5,479 samples, 206 carbapenemase genes were detected in 196 samples (3.6%) by Carba-R v.2 (Table 1; Fig. 1). Concordant Carba-R v.2 and culture-based conventional PCR results were observed for 96 positive and 5,278 negative samples. In total, 1.8% (100/5,479) of the samples were Carba-R v.2-positive but culture-negative, while <0.1% (5/5,479) of samples were Carba-R v.2 negative but culture-positive. Carba-R v.2-negative, culture-positive cases confirmed by sequencing were considered as false negative. Of the 100 Carba-R v.2-positive but initially culture-negative samples, the presence of carbapenemase genes (KPC [N=18], NDM [N=28], VIM [N=3], IMP [N=2], OXA-48 [N=2]; 53 genes in total) was confirmed in 46 samples following anaerobic culture supplementation and enriched PCR. In contrast, 52 samples remained discordant, as the target carbapenemase genes were not recovered from the broth culture. We observed 10 (0.18%) samples harboring multiple carbapenemase types: NDM, IMP, and VIM (N=1); KPC and NDM (N=6); KPC and IMP (N=1); NDM and OXA (N=1); and KPC and VIM (N=1).
Table 1. Comparison of overall performance of the Xpert Carba-R v.2 and culture-based PCR method.
| Carbapenemase genes detected with Xpert Carba-R v.2 | Culture-based conventional PCR | |||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Results | ||||
| Before discrepant analysis | Positive | 96 | 100 | 196 |
| Negative | 5 | 5,278 | 5,283 | |
| Total | 101 | 5,378 | 5,479 | |
| After discrepant analysis | Positive | 144 | 52 | 196 |
| Negative | 5 | 5,278 | 5,283 | |
| Total | 149 | 5,330 | 5,479 | |
A total of 653 patients underwent multiple serial assays (two [N=501], three [N=123], four [N=16] five [N=9], six [N=3], and seven assays [N=1]), and 21 patients had a positive result more than once. Of these 21 patients, six patients (one KPC, two VIM, and three NDM) were Carba-R v.2-positive but culture-negative, with high Ct values (median Ct, 37.1); their Carba-R v.2 results converted to negative in subsequent assays.
The sensitivity and specificity of the Carba R were 95.0% and 98.1%, respectively (Table 2). The weighted Kappa value was 0.63 (0.57–0.70). Following discrepant analysis, the sensitivity and specificity improved to 96.7% and 99.0%, respectively (Table 2). The weighted Kappa value increased to 0.83 (0.79–0.87). KPC was the most frequently detected gene type (N=120, 58.3%), followed by NDM (N=61, 29.6%), VIM (N=10, 4.9%), IMP (N=9, 4.4%), and OXA-48 (N=6, 2.9%). The performance was fairly variable according to gene type (Table 2). A total of 95 KPC-harboring microorganisms were recovered from culture. Klebsiella pneumoniae was the most common species recovered in culture (85, 89.4%), followed by Escherichia coli (5, 5.3%), Citrobacter freundii (2, 2.3%), Citrobacter koseri (1, 1.0%), and Enterobacter kobei (1, 1.0%). For NDM, two K. pneumoniae, two Citrobacter amalonaticus, and one E. coli strains were recovered. For both IMP and OXA, one K. pneumoniae strain was recovered.
Table 2. Sensitivity, specificity, PPV, and NPV according to carbapenemase type with discrepant analysis.
| Carbapenemase type (N) | % Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | |
|---|---|---|---|---|---|
| Total (206)* | At initial analysis | 95.0 (88.8–98.4) | 98.1 (97.7–98.5) | 49.0 (44.0–53.9) | 99.9 (99.8–100) |
| After discrepant analysis | 96.7 (92.3–99.0) | 99.0 (98.7–99.3) | 73.5 (67.8–78.4) | 99.9 (99.7–100) | |
| KPC (120) | At initial analysis | 94.9 (88.6–98.3) | 99.6 (99.3–99.7) | 79.7 (72.4–85.4) | 99.9 (99.8–100) |
| After discrepant analysis | 95.7 (90.3–-98.6) | 99.9 (99.7–99.9) | 93.3 (87.5–96.6) | 99.9 (99.8–99.9) | |
| NDM (61) | At initial analysis | 100 (47.8–100) | 99.0 (98.7–99.2) | 8.2 (6.4–10.3) | 100 |
| After discrepant analysis | 100 (89.4–100) | 99.5 (99.3–99.7) | 54.1 (44.4–63.5) | 99.9 (99.8–99.6) | |
| VIM (10) | At initial analysis | - | 99.8 (99.6–99.9) | - | 100 |
| After discrepant analysis | 100 (29.2–100) | 99.9 (99.7–99.9) | 30.0 (17.0–47.3) | 100 | |
| IMP (9) | At initial analysis | 100 (2.5–100) | 99.8 (99.7–99.9) | 11.1 (5.9–19.9) | 100 |
| After discrepant analysis | 100 (29.2–100) | 99.9 (99.8–99.9) | 33.3 (18.3–52.6) | 100 | |
| OXA-48 (6) | At initial analysis | 100 (2.5–100) | 99.9 (99.8–100) | 16.7 (7.7–32.4) | 100 |
| After discrepant analysis | 100 (29.2–100) | 99.9 (99.8–100) | 50 (24.4–75.6) | 100 | |
| Non-KPC (86)† | At initial analysis | 100 (59.0–100) | 99.6 (99.5–99.7) | 8.1 (6.6–9.9) | 100 |
| After discrepant analysis | 100 (91.6–100) | 99.8 (99.7–99.8) | 48.8 (41.5–56.2) | 100 | |
*The number of positive samples was counted; samples harboring multiple genes were counted as one sample.
†Non-KPC includes NDM, VIM, IMP, and OXA-48.
Abbreviations: CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; VIM, Verona integron-mediated metallo-β-lactamase; IMP, imipenem; OXA-48, oxacillinase-48.
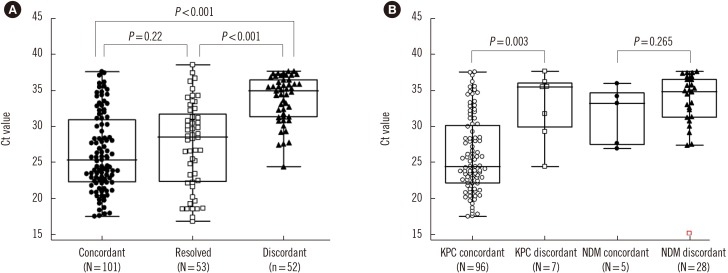
In discrepant cases, NDM was the most frequently observed (Carba-R v.2-positive and culture-negative/total Carba-R v.2-positive, 28/61, 45.9%), followed by KPC (8/120, 1.2%), VIM (7/10, 70%), IMP (6/9, 66.6%), and OXA-48 (3/6, 50%). We categorized the samples into three groups: concordant, resolved, and discordant, as described above, with median Ct values of 25.4, 28.5, and 35.0, respectively. The Ct values of the three groups were statistically different (P<0.001), except for the concordant and resolved following discrepant analysis groups (P=0.22; Fig. 1A). According to carbapenemase type, the difference in Ct value was statistically significant for KPC (P=0.003), but not for NDM (P=0.265; Fig. 2B).
Fig. 2. Comparison of number, median Ct value and interquartile range in (A) the concordant group at initial assay (25.4, 22.2–30.9), resolved discrepancy group after discrepant analysis (28.5, 22.3–31.7), and discordant groups (52, 35.0, 31.4–37.7) and (B) the concordant (24.4, 22.2–30.1) and discordant (35.5, 29.9–36.0) KPC groups and concordant (32.2, 27.5–34.6) and discordant (34.8, 31.3–36.5) NDM groups.
Abbreviations: Ct, threshold cycle; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase.
ROC curve analysis yielded a Ct value of 28.8 as the optimal cut-off for KPC with 72.5% sensitivity and 87.5% specificity, when the concordant and resolved groups were considered as positive. When this cut-off value was applied to our results, the PPV improved to 82.1%. Satisfactory cut-offs were not derived for the other carbapenemase genes, owing to the limited number of assays
DISCUSSION
The spread of carbapenemase genes poses a public health threat, increasing the need for a rapid molecular method for carbapenemase gene detection [9]. We prospectively evaluated the performance of the newly developed Carba-R v.2 for detection of carbapenemase genes compared with culture-based PCR, using a large number of clinical rectal swab samples. The overall performance of Carba-R v.2 was reliable, except for the PPV; this could result in unnecessary isolation of patients, causing extra medical resource consumption and patient discomfort.
To achieve effective infection control, PPV, and NPV are important to assess the positive and negative results of each carbapenemase gene detection assay. The lower PPV of Carba-R v.2-positive, culture-negative cases might be caused by false positive results due to nonspecific binding, lower bacterial load in the samples, reduced bacterial viability owing to inappropriate sample collection/transportation, or uncultivable organisms harboring carbapenemase genes [10]. To resolve these issues, we performed discrepant analysis; 48% (48/100) of the Carba-R v.2-positive, culture-negative cases converted to culture-positive. These cases indicate the existence of carbapenemase genes; however, the carbapenemase gene-harboring organisms were not recovered using the initial culture method. The advantages of the molecular method over culture-based reference methods in detecting carbapenemase genes are that it is rapid and sensitive. Therefore, culture-based conventional PCR should be complemented by a molecular screening assay [11,12].
Interestingly, the Ct values of the concordant vs. discordant groups and the resolved vs. discordant groups were statistically different (P<0.001), while those of the concordant and resolved groups were not statistically different (P=0.22). The concordant group had the lowest Ct values, and the discordant group had the highest values. This indicates that, although the Carba-R v.2 is not quantitative, the Ct values can be affected by target gene load, bacterial load, variable copy number of the carbapenemase-carrying plasmid, and the sample amount inoculated into the cartridge [13]. Hence, a low Ct value would be helpful for estimating high carbapenemase gene load; caution is warranted when ruling out carbapenemase genes in cases of discrepant Carba-R v.2 results with a high Ct value, so as to not miss low-level carbapenemase genes, as reported for another real-time PCR evaluation method, Check-Direct CPE (Check-Points, Wageningen, the Netherlands) [14]. That study suggested that a modified cut-off Ct value<35 reduced false positives. Based on our KPC data, 28.8 was identified as the optimum cut-off for reducing false positives from the culture method following ROC curve analysis, with a sensitivity and specificity of 72.5% and 87.5%, respectively. Satisfactory cut-offs were not derived for the other carbapenemase genes, owing to the limited number of assays. To overcome low PPV, the modified cut-off was applied for the simulation with our results. This could reduce false positives and unnecessary isolation of patients, but nullify higher sensitivity, the most important advantage of the molecular method.
Lau, et al. [15] demonstrated a 10–100-fold higher limit of detection (LOD) for NDM-harboring strains in culture and PCR and for VIM in PCR. We observed different performances according to carbapenemase gene. The Ct values of the concordant and discordant groups for KPC genes (not including resolved) were statistically significantly different (P=0.003); however, the NDM values were not significant (P=0.265; Fig. 1B). Based on the association between Ct value and bacterial loading, other factors, such as gene copy number, should also be considered.
Previous studies have reported higher performance than our data [4,9,10,16]. Some studies included contrived samples spiked with higher inoculums of previously well-characterized carbapenemase genes [4,9,16]. Our study represents clinical practice more accurately, as it was performed with a large number of clinical samples.
Negative conversion of carbapenemase genes in multiple serial assays could be due to bacterial eradication, technical borderline or low bacterial loading around the lower LOD, or non-specific false positives. As colonization of carbapenemase gene-harboring organisms has been reported [10], bacterial eradication during the short period of this study is not feasible. Further studies are required to determine whether these discordant results represent a false positive result or a pitfall of the methodology, which is more sensitive than the culture method. Currently, Carba-R v.2-positive and culture-negative patients are considered negative for carbapenemase genes. In areas of lower prevalence, the PPV could be lower. Unfortunately, no guideline has been released regarding the use of molecular methods as a reference, even in low prevalence settings [10]. The clinical impact of Carba-R v.2-positive and culture-negative samples should be further clarified. In such cases, we suggest a conservative approach; the patients should be excluded from the carbapenemase gene cohort, but kept in isolation.
The limitation of this study was that only four carbapenemase genes other than KPC were examined. In addition, since OXA-48 and its variants are not prevalent at our institutes, the improvement of Carba-R v.2 could not be assayed. Further studies with genetically diverse strains are warranted.
As most of our discrepant results were non-KPC genes, more caution is needed for patients who are positive for non-KPC genes. However, Carba-R v.2 would be useful for deciding whether to quarantine pre-exposed patients in hospital settings, owing to the high specificity and NPV. Considering the global increase in CPE, Carba-R v.2 would be helpful for rapid detection of carbapenemase genes and would also enable more efficient infection control.
ACKNOWLEDGEMENTS
None.
Footnotes
AUTHOR CONTRIBUTIONS: JHB and DY wrote the manuscript. JYC and YSP aided sample collection and interpreting the results. JH B, DY, and YAK planned and carried out the simulations. MK and BK conceived and carried out the experiments. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
CONFLICTS OF INTEREST: None.
FUNDING: This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries through the Agricultural Microbiome R&D Program, funded by the Ministry of Agriculture, Food and Rural Affairs (918003-4); by the Korea Health Technology R&D Project through the KHIDI funded by the Ministry of Health & Welfare, Korea (grant number HI17C180,7); and by the BioNano Health Guard Research Center funded by the MSIP of Korea as a Global Frontier Project (H-GUARD_2014M3A6B2060509). This study was also partly supported by Cepheid. The funders had no role in study design, data collection, and interpretation, or in the decision to submit the work for publication.
References
- 1.WHO. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: World Health Organization; 2017. [Google Scholar]
- 2.Landman D, Salvani JK, Bratu S, Quale J. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J Clin Microbiol. 2005;43:5639–5641. doi: 10.1128/JCM.43.11.5639-5641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DK, Kim HS, Pinto N, Jeon J, D'Souza R, Kim MS, et al. Xpert CARBA-R assay for the detection of carbapenemase-producing organisms in intensive care unit patients of a Korean tertiary care hospital. Ann Lab Med. 2016;36:162–165. doi: 10.3343/alm.2016.36.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tato M, Ruiz-Garbajosa P, Traczewski M, Dodgson A, McEwan A, Humphries R, et al. Multisite evaluation of Cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol. 2016;54:1814–1819. doi: 10.1128/JCM.00341-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dortet L, Fusaro M, Naas T. Improvement of the Xpert Carba-R Kit for the detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2016;60:3832–3837. doi: 10.1128/AAC.00517-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uwamino Y, Sugita K, Hasegawa N, Nishimura T, Fujiwara H, Iwata S. Rapid detection and typing of carbapenemase genes from carbapenem-resistant Enterobacteriaceae isolates collected in a Japanese hospital using the Xpert Carba-R assay. Jpn J Infect Dis. 2017;70:124–125. doi: 10.7883/yoken.JJID.2015.660. [DOI] [PubMed] [Google Scholar]
- 7.CDC. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing, Klebsiella spp. and E. coli from rectal swabs. Atlanta: Centers for Disease Control and Prevention; 2009. [Google Scholar]
- 8.Ahn S, Sung JY, Kim H, Kim MS, Hwang Y, Jong S, et al. Molecular epidemiology and characterization of carbapenemase-producing Enterobacteriaceae isolated at a university hospital in Korea during 4-year period. Ann Clin Microbiol. 2016;19:39–47. [Google Scholar]
- 9.Moore NM, Cantón R, Carretto E, Peterson LR, Sautter RL, Traczewski MM, et al. Rapid identification of five classes of carbapenem resistance genes directly from rectal swabs by use of the Xpert Carba-R assay. J Clin Microbiol. 2017;55:2268–2275. doi: 10.1128/JCM.00137-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoyos-Mallecot Y, Ouzani S, Dortet L, Fortineau N, Naas T. Performance of the Xpert® Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int J Antimicrob Agents. 2017;49:774–777. doi: 10.1016/j.ijantimicag.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Schechner V, Straus-Robinson K, Schwartz D, Pfeffer I, Tarabeia J, Moskovich R, et al. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J Clin Microbiol. 2009;47:3261–3265. doi: 10.1128/JCM.02368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwaber MJ, Carmeli Y. An ongoing national intervention to contain the spread of carbapenem-resistant Enterobacteriaceae. Clin Infect Dis. 2014;58:697–703. doi: 10.1093/cid/cit795. [DOI] [PubMed] [Google Scholar]
- 13.Burillo A, Marín M, Cercenado E, Ruiz-Carrascoso G, Pérez-Granda MJ, Oteo J, et al. Evaluation of the Xpert Carba-R (Cepheid) assay using contrived bronchial specimens from patients with suspicion of ventilator-associated pneumonia for the detection of prevalent carbapenemases. PLoS One. 2016;11:e0168473. doi: 10.1371/journal.pone.0168473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otter JA, Dyakova E, Bisnauthsing KN, Querol-Rubiera A, Patel A, Ahanonu C, et al. Universal hospital admission screening for carbapenemase-producing organisms in a low-prevalence setting. J Antimicrob Chemother. 2016;71:3556–3561. doi: 10.1093/jac/dkw309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau AF, Fahle GA, Kemp MA, Jassem AN, Dekker JP, Frank KM. Clinical Performance of Check-Direct CPE, a Multiplex PCR for direct detection of bla(KPC), bla(NDM) and/or bla(VIM), and bla(OXA)-48 from perirectal swabs. J Clin Microbiol. 2015;53:3729–3737. doi: 10.1128/JCM.01921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover FC, Canton R, Kop J, Chan R, Ryan J, Weir F, et al. Detection of colonization by carbapenemase-producing Gram-negative bacilli in patients by use of the Xpert MDRO assay. J Clin Microbiol. 2013;51:3780–3787. doi: 10.1128/JCM.01092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]