Abstract
Transfer RNAs (tRNA) are important molecules that involved in protein translation machinery and acts as a bridge between the ribosome and codon of the mRNA. The study of tRNA is evolving considerably in the fields of bacteria, plants, and animals. However, detailed genomic study of the cyanobacterial tRNA is lacking. Therefore, we conducted a study of cyanobacterial tRNA from 61 species. Analysis revealed that; cyanobacteria contain thirty-six to seventy-eight tRNA gens per genome that encodes for 20 tRNA isotypes. The number of iso-acceptors (anti-codons) ranged from thirty-two to forty-three per genome. tRNAIle with anti-codon AAU, GAU, and UAU was reported to be absent from the genome of Gleocapsa PCC 73,106 and Xenococcus sp. PCC 7305. Instead, they were contained anti-codon CAU that is common to tRNAMet and tRNAIle as well. The iso-acceptors ACA (tRNACys), ACC (tRNAGly), AGA, ACU (tRNASer), AAA (tRNAPhe), AGG (tRNAPro), AAC (tRNAVal), GCG (tRNAArg), AUG (tRNAHis), and AUC (tRNAAsp) were absent from the genome of cyanobacterial lineages studied so far. A few of the cyanobacterial species encode suppressor tRNAs, whereas none of the species were found to encode a selenocysteine iso-acceptor. Cyanobacterial species encode a few putative novel tRNAs whose functions are yet to be elucidated.
Keywords: Cyanobacteria, tRNA, Translation, Iso-acceptors, Anti-codon, Codon
Abbreviations: tRNA, transfer RNA; U, uridine; Ψ, pseudouridine; A, adenine; C, cytosine; G, guanine
1. Introduction
The study of tRNA biology has brought an unprecedented level of understanding as well as various novel discoveries in the fields of genetics and molecular biology. The principal function of tRNA is to transfer the amino acids to the ribosome machinery during the process of protein synthesis (Hopper and Phizicky, 2003, Kirchner and Ignatova, 2014). Essentially, tRNA functions as an adaptor molecule and delivers amino acids to the ribosome by reading the genetic information encoded within the mRNA (Mohanta and Bae, 2017, Ribas de Pouplana and Dedon, 2014, XiaoLong and EnDuo, 2013). The tRNAs are universally found in every cell type and are the most abundant small non-coding cellular molecules, constituting approximately 4–10% of all cellular RNA. The importance and evolution of tRNA are based on its comprehensive relationship to the origin and development of the genetic code; the discovery of the universal nature of the genetic code in prokaryotes and eukaryotes has been incredibly important to the field of molecular biology (Koonin and Novozhilov, 2009). In 1966, Robert W. Holley first proposed the clover leaf model of tRNA containing four arms and three loops. These components are the acceptor arm, the D-arm, the D-loop, the anti-codon arm, the variable loop, the Ψ-arm, and the Ψ-loop (Clark, 2006, Kirchner and Ignatova, 2014). Nuclear encoded eukaryotic and prokaryotic tRNAs have a variable length of 73–90 nucleotides (nts) (Kirchner and Ignatova, 2014, Sharp et al., 1985). The acceptor arm contains 7 bp, the D-arm 3–4 bp, the anti-codon (AC) arm 5 bp, the variable loop 4–23 bp, the Ψ-arm 5 bp and the Ψ-loop 7 bp. In some species, tRNA has variable numbers of nucleotide residues in the D-loop and the variable arms (Kirchner and Ignatova, 2014). Regardless, the variable arm starts after nucleotide 44, and the anti-codon is always numbered as nucleotides 34, 35, and 36, while the C-C-A tail is always numbered 74, 75, and 76 (Kirchner and Ignatova, 2014). The acceptor arm provides the point of attachment for the amino acid through the C-C-A tail. The D-arm is named for the presence of an unusual pyrimidine nucleotide called dihydrouracil, and it contains a synthetase site that recognizes the amino acid activating enzyme. The anti-codon loop reads the nucleotide codon of mRNA. The Ψ-arm is named for the presence of the uracil-pseudouridine nucleotide and plays a crucial role in the recognition of the ribosome. The genetic code includes sixty-four codons, of which sixty-one are sense codons, and three are nonsense codons. The sense codons are read by sixty-one tRNA iso-acceptors. To date, all of this information has been obtained from studies confined to the bacterial, plant and animal kingdoms, and only very limited genomic data are available on cyanobacterial tRNAs. The availability of the complete genome sequence of several cyanobacterial species prompted us to examine the cyanobacterial tRNA sequences. To this end, we have identified and analysed the tRNAs from sixty-one cyanobacterial species, and here, we reported the novel genomics details of cyanobacterial tRNAs.
2. Materials and methods
2.1. Retrieval of genomic tRNA sequence data
The annotated genomic tRNA sequences of the cyanobacterial species were downloaded from the Joint Genome Institute (JGI) Genome Portal (Nordberg et al., 2014). Based upon the availability of the genome sequence data at the start of this study, we downloaded the tRNA sequences from sixty-one cyanobacterial species (Supplementary data 1). Further details of the species used in this study are shown in Supplementary Table 1.
2.2. tRNA analysis
The genomic tRNA sequences of all the species were submitted to the tRNAScan-SE servers to determine whether all the downloaded sequences encoded tRNA or not. The analysis revealed the presence of a clover leaf-like structure tRNA in the studied sequences, thus confirming that the sequences encode tRNA. The statistical parameters used for the analysis of tRNAs in tRNAScan-SE were as follows; sequence source: bacterial/general tRNA; search mode: default; query sequence: formatted (FASTA); genetic code for tRNA isotype prediction: universal (Lowe and Eddy, 1997). The tRNAScan-SE server was used for the analysis because tRNAScan-SE produces only one false positive report per 15 gigabases (Lowe and Eddy, 1997). tRNAScan-SE is one of the most successful programs for identifying tRNAs in genome sequences. tRNAScan-SE utilizes EufindtRNA to search for conserved non-coding tRNA sequences and evaluates the co-variance in the tRNA. The algorithm of tRNAScan-SE can detect and identifying tRNAs at 99–100% accuracy with a very low error rate.
2.3. Prediction of tRNA clover leaf-like structure and novel tRNA
The clover leaf-like structures of cyanobacterial tRNAs were predicted using the tRNAScan-SE server, and the resulting novel tRNA structures were retained for further analysis. The numbers of nucleotides in the acceptor arm, the D-arm, the D-loop, the anti-codon arm, the variable loop, the Ψ-arm and the Ψ-loop were recorded by manual counting for individual tRNAs by observing the tRNA structures that resulted from tRNAScan-SE server.
3. Results and discussion
3.1. Cyanobacterial tRNA gene families are diverse among species
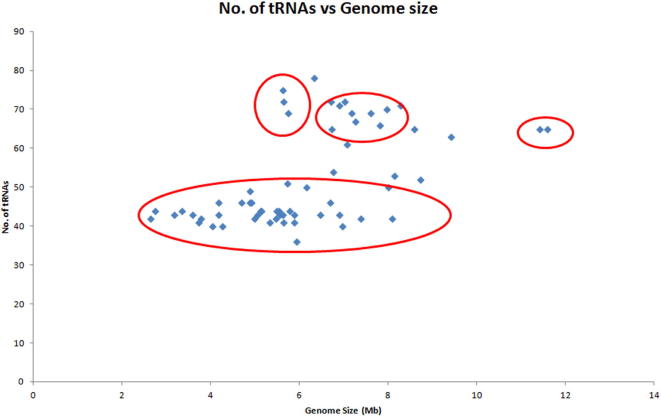
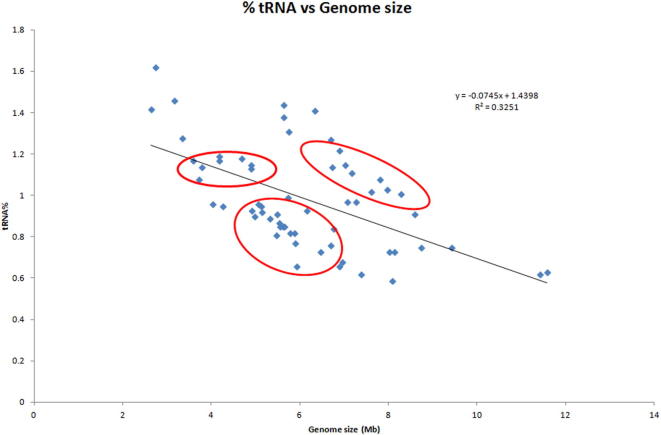
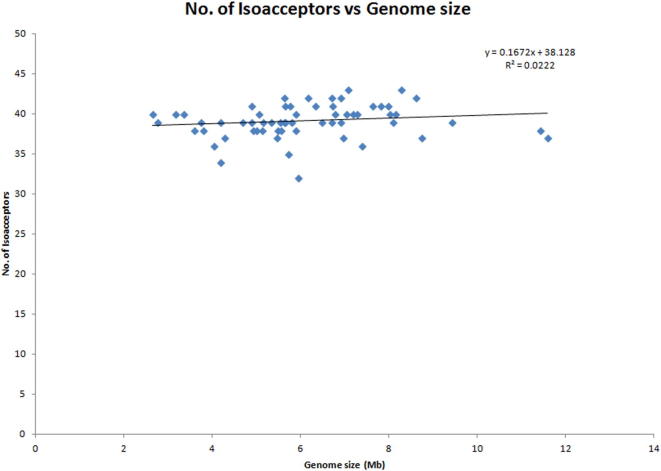
Analysis revealed, the cyanobacterial tRNA gene family ranged from thirty-six to seventy-eight tRNAs per genome (Supplementary Table 1). Xenococcus sp. PCC 7305 encoded the lowest number (36), while Nostoc sp. PCC 7107 encoded the highest number (78) of tRNAs (Mohanta et al., 2017). Cluster analysis of the number of tRNAs per genome showed four clusters (Supplementary Fig. 1). Most of the species were clustered within 40–50 tRNAs per genome, and some were clustered within 60–70 tRNAs, whereas only a few were clustered within 70–80 tRNAs per genome (Supplementary Fig. 1). The abundance of tRNAs in the cyanobacterial genome ranged from 0.59% to 1.62% with Fischerella sp. PCC 9605 having the lowest (0.59%) abundance and Synechococcus elongatus PCC 7942 having the highest (1.62%) (Supplementary Table 1). Although the genome sizes of Calothrix sp. PCC 7103 and Calothrix desertica were 11.584 Mb and 11.416 Mb, respectively, the number of tRNAs accounted for them was only 0.63% and 0.62% of their genomes (Supplementary Table 1). Cluster analysis revealed that the distribution of tRNA percentage per genome is very dynamic (Supplementary Fig. 2). The number of iso-acceptors in the cyanobacterial genome ranged from thirty-two to forty-three (Supplementary Table 1). The lowest number (32) of iso-acceptors was found in Xenococcus sp. PCC 7305, while the highest number (43) of iso-acceptors was found in Oscillatoria nigro-viridis PCC7112. These numbers show that cyanobacteria do not encode all of the iso-acceptors in their genome and that none of the species were found to contain all of sixty-one iso-acceptors. At least twelve species were found to contain pseudo-tRNAs in their genomes (Supplementary Table 1). The Nostoc sp. PCC 7524 and Scytonema hofmannii UTEX 2349 suppressor tRNAs contain CUA anti-codon, whereas the Oscillatoria nigro-viridis PCC 7112 suppressor tRNA contains UUA. Cluster analysis revealed that the number and distribution of tRNA iso-acceptor fall within the range of 32 to 43 per genome, showing a greater correlation of iso-acceptor number than of the genome size (Supplementary Fig. 3).
The number of individual tRNA family members (isotypes) in the genomes of different species ranged from zero to 10 (Supplementary Table 2). The tRNAIle gene with AAU, GAU, and UAU anti-codons were found to be absent from the genome of Gleocapsa sp. PCC 73,106 and Xenococcus sp. PCC 7305 (Supplementary Table 2). Analyses revealed that, cyanobacterial tRNAIle also encode CAU anti-codon that codes for tRNAMet as well. In total, 53 tRNA genes were found to encode CAU anti-codon (Table 1). The absence of tRNA genes from certain genomes might be due to the independent deletion. The sequence constraints of tRNA function suggest that tRNAs are by large intolerant to mutation. Mutation in tRNA can occur at any stem or loop of the tRNA, and it is reported that more than 230 mutations have been detected in human tRNA and that all of them were found to be associated with various diseases (Guy et al., 2014, Yarham et al., 2010). Therefore, there is a possibility of mutation in the anti-codon loop, and hence absence of AAU, GAU, and UAU anti-codons in tRNAIle in the genome of cyanobacteria. The absence of AAU, GAU, and UAU anti-codons in tRNAIle forced the genome to encode the isoleucine codon by CAU anti-codon. In some organisms, the UCU tRNA anticodon translate to AGA and AGG codons. Hence, the UCU tRNA improves the efficiency of the translation machinery for the AGG codon. A few anti-codons were also found very rarely in the cyanobacterial tRNA. For example, the anti-codons AGU (threonine) AAU (isoleucine) and AGC (alanine) were found only once in the sixty-one studied cyanobacterial species, whereas AUA (tyrosine) and UAU (isoleucine) were found only twice, and CUC (glutamate) was found only thrice (Table 1). Each of the AUU (asparagine), AAG (leucine), and UCG (arginine) iso-acceptors were found only four times. Thus, the iso-acceptors AGU, AUA, AAU, UAU, CUC, AUU, AAG, and UCG are the rarest forms of iso-acceptor in the cyanobacteria. Earlier Marck and Grosjean (2002) reported regarding the presence of CAU anti-codons in tRNAIle which is in accordance with our finding (Marck and Grosjean, 2002). Nucleotide C34 at wobble position of the anti-codon modified to lysidine and lysidine modified tRNA CAU gets charged in-vitro by isoleucine. When modified lysidine is replaced by unmodified C nucleotide, the resulting CAU anti-codon aminoacylated by methionine instead of isoleucine (Marck and Grosjean, 2002). Nucleotide at 44th position (1st position of variable arm) of tRNA CAU possibly play important role for lysinylation (Marck and Grosjean, 2002).
Table 1.
Anti-codon table and distribution of iso-acceptors in cyanobacterial genome. From the studied species we found that, from 64 possible iso-acceptors, cyanobacterial kingdom encodes only 54 iso-acceptors. The iso-acceptors ACC of tRNAGly, AGA and ACT of tRNASer, AAA of tRNAPhe, AGG of tRNAPro, AAC of tRNAVal, AUG of tRNAHis, and AUC of tRNAAsp were found to be absent in the cyanobacterial lineage. No tRNA gene was found to contain the above mentioned iso-acceptors. This absence suggests that these iso-acceptors might have been lost from the tRNA lineage of cyanobacteria.
| tRNA Isotypes | Iso-acceptors | |||||
|---|---|---|---|---|---|---|
| Polar | ||||||
| Asparagine | AUU (4) | GUU (86) | ||||
| Cysteine | GCA (78) | ACA (0) | ||||
| Glutamine | CUG (11) | UUG (83) | ||||
| Glycine | ACC (0) | GCC (68) | CCC (44) | UCC (76) | ||
| Serine | GGA (63) | AGA (0) | CGA (62) | UGA (66) | ACU (0) | GCU (77) |
| Threonine | AGU (1) | GGU (61) | CGU (61) | UGU (77) | ||
| Tyrosine | AUA (2) | GUA (71) | ||||
| Non-polar | ||||||
| Alanine | AGC (1) | GGC (67) | CGC (50) | UGC (140) | ||
| Isoleucine | AAU (1) | GAU (120) | UAU (2) | CAU (53) | ||
| Leucine | AAG (4) | GAG (68) | CAG (67) | UAG (81) | CAA (77) | UAA (49) |
| Methionine | CAU (146) | |||||
| Phenylalanine | AAA (0) | GAA (92) | ||||
| Proline | AGG (0) | GGG (62) | CGG (58) | UGG (83) | ||
| Tryptophan | CCA (83) | |||||
| Valine | AAC (0) | GAC (59) | CAC (29) | UAC (69) | ||
| Positively charged | ||||||
| Arginine | ACG (71) | GCG (0) | CCG (61) | UCG (4) | CCU (39) | UCU (81) |
| Histidine | AUG (0) | GUG (68) | ||||
| Lysine | CUU (42) | UUU (83) | ||||
| Negatively charged | ||||||
| Aspartic acid | GUC (84) | AUC (0) | ||||
| Glutamic acid | CUC (3) | UUC (84) | ||||
| Others | ||||||
| Suppressor | CUA (2) | UUA (1) | ||||
| Selenocysteine | UCA (0) | |||||
The genomes of Oscillatoria formosa PCC 6407 and Oscillatoria nigro-viridis PCC7112 possess ten tRNALeu genes, which was the highest number of individual tRNA (isotypes) genes found in the cyanobacterial genome (Supplementary Table 2). The cyanobacterial species encoded only one or two tRNAHis genes in their genomes, and the numbers of tRNATyr, tRNACys, and tRNATrp genes varied from one to two per genome (Supplementary Table 2). Only the genomes of Microchaete sp. PCC 7126 and Nostoc sp. PCC 7107 were found to contain three tRNATyr genes. The abundance of tRNAHis and tRNATyr genes was lower than that of tRNACys, tRNATrp, and tRNAAsp genes (Supplementary Table 2). The abundance of tRNALeu genes was found to be much higher than that of tRNASer, tRNAArg and tRNAAla genes (Supplementary Table 2). From a total of sixty-four codons, the cyanobacterial kingdom encoded only fifty-four anti-codons (Table 1). The anti-codons ACA (cysteine), ACC (glycine), AGA and ACU (serine), AAA (phenylalanine), AGG (proline), AAC (valine), CGC (arginine), AUG, (histidine) and AUC (aspartate) were found to be absent. The UCA anti-codon for selenocysteine was also found to be absent in cyanobacterial tRNAs. However, Oscillatoria nigro-viridis PCC 7112 encoded UUA suppressor tRNA, whereas Nostoc sp. PCC 7524 and Scytonema hofmanni UTEX_2349 encoded CUA suppressor tRNA (Table 1). From a total of 146 tRNAMet, 125 were found to encode for tRNAfMet. fMet (N-formylmethionine) is a derivative of methionine where a formyl group is attached to the amino group. tRNAfMet is used for initiation of protein synthesis in prokaryotic organisms which are subsequently removed during post-translational modifications.
3.2. The nucleotide length of tRNASer was greater than that of other tRNAs
The nucleotide composition of the different tRNAs gene varied greatly. We found that the nucleotide length of cyanobacterial tRNAs ranged from fifty-five to ninety-six nucleotides. tRNATyr (gene id: 2508648704) of Xenococcus sp. PCC 7305 was found to be the smallest tRNA gene, containing only fifty-two nucleotides whereas tRNAAsp from Cylindrospermum stagnale (gene id: 2509768958) contained 96 nucleotides. At least 33 tRNASer genes were found to contain 92 nucleotides whereas 12 genes were found to contain 90 nucleotides (Supplementary Table 3). The nucleotide lengths of cyanobacterial tRNASer were higher than those of other tRNAs (Supplementary Table 3). The nucleotide length for tRNALeu ranged from seventy-one to eighty-seven nucleotides. The lengths of tRNACys, tRNAThr, tRNATyr, tRNAPhe, tRNATrp, tRNAArg, tRNAHis, tRNAAsp, and tRNAGlu were strictly confined to seventy-one to seventy-six nucleotides only (Supplementary Table 3). The tRNAAsp from Cylindrospermum stagnale PCC 7417 (gene id: 2509768958) was found to contain ninety-six nucleotides and was the longest tRNA found in cyanobacteria. tRNAGly was found to contain the lowest average number (71.6) of nucleotides (Supplementary Table 3). Nucleotide composition study revealed, adenine, uracil, cytosine, and guanine contained 19.45%, 23.14%, 6.41%, and 31% nucleotides (Supplementary Table 3).
3.3. tRNAs contain a variable number of nucleotides in the arms and loops
Overall, the nucleotide length of the acceptor arm ranged from four to eight nucleotides (Supplementary Table 5). Only two of the 3092 studied tRNAs were found to contain five nucleotides in the acceptor arm, whereas fifteen were found to contain six nucleotides, and two were found to contain eight nucleotides (Supplementary Table 5). In contrast, 99.38% of the tRNAs were found to contain seven nucleotides in the acceptor arm. tRNALys from Fischerella sp. PCC 9431 (gene id: 2512980262) was found to contain four nucleotides each in the acceptor arm, whereas tRNAAsn, tRNALeu, tRNALys, and tRNAMet were found to contain six nucleotides in the acceptor arm (Supplementary Table 5). Interestingly, except for Cylindrospermum stagnale PCC 7417, no other tRNAs from any cyanobacterial species were found to contain eight nucleotides in the acceptor arm (Supplementary Table 5).
Study revealed, cyanobacterial tRNAs were found to contain 3′-CCA tail sequence in the tRNA genes. From 3092 tRNA genes, 916 (29.62%) genes were found to contain 3′-CCA sequence. Previous study led by Ardell and Hou (2016) also reported regarding the presence of 3′-CCA sequence in bacteria (Ardell and Hou, 2016). From the studied 146 tRNAMet genes, 116 (79.45%) genes were found to contain 3′-CCA tail sequence and from 125 tRNAfMet (initiator tRNA) genes, 109 (74.65%) were found to contain 3′-CCA sequence. Ardell and Hou (2016) reported 74.1% of prokaryotic initiator tRNA contain 3′-CCA sequence and the result from cyanobacterial initiator tRNA genes (74.65%) closely match with the report of Adrell and Hou (Ardell and Hou, 2016).
The D-arm of cyanobacterial tRNAs were found to contain two to four nucleotides (Supplementary Table 5). Of 3092 studied tRNAs, only three were found to contain two nucleotides in the D-arm. They were tRNACys from Synechococcus elongatus PCC 7942 (gene id: 640711018) and tRNASer from Cyanothece sp. BH63E ATCC 51,472 (gene id: 2507502103) and from Pleurocapsa sp. PCC7319 (gene id: 2509709995). Overall, 29.04% of cyanobacterial tRNAs were found to contain three nucleotides, whereas 70.86% of tRNAs was found to contain four nucleotides in the D-arm. Although the majority of tRNAAla, tRNAArg, tRNAAsn, tRNACys, tRNAGlu, tRNAGln, tRNAGly, tRNALys, tRNAPhe, tRNAPro, and tRNATyr genes were found to contain four nucleotides, a few were found to contain three nucleotides in the D-arm. This result suggests that these tRNA families inherently possess four nucleotides in the D-arm, and a deletion of one nucleotide most likely caused the others to have only three nucleotides.
The nucleotide length of the D-loop in cyanobacterial tRNAs varied from seven to thirteen (Supplementary Table 5). Of 3092 studied tRNAs, only one tRNAMet from Calothrix sp. PCC 7507 (gene id: 2505802877) was found to contain seven nucleotides in the D-loop. For tRNAAla, the D-loop was found to contain seven to eleven nucleotides (Supplementary Table 5).
The length of the anti-codon arm ranged from two to seven nucleotides. The majority of the tRNA genes contained five nucleotides in the anti-codon arm (Supplementary Table 5). However, some variation was observed in the length of the anti-codon arm in different cyanobacterial tRNAs. All of the cyanobacterial tRNAPhe and tRNAIle genes were found to contain five nucleotides in the anti-codon arm without any variation. Eight of the tRNAAsp genes were found to contain four nucleotides in the anti-codon arm, while the remainder of the genes had only five nucleotides. In the case of tRNALys, one gene was found to contain two nucleotides; ninety-two (72.44%) were found to contain four nucleotides, and thirty-four (26.77%) were found to contain five nucleotides (Supplementary Table 5). tRNAMet genes were found to be the most variable with regard to the nucleotide composition in the anti-codon arm. They contained three to five nucleotides in the anti-codon arm. Among 204 tRNAMet genes, twenty (9.80%) were found to contain three nucleotides, forty-four (21.56%) were found to contain four nucleotides, and the remaining 140 (68.62%) were found to contain five nucleotides.
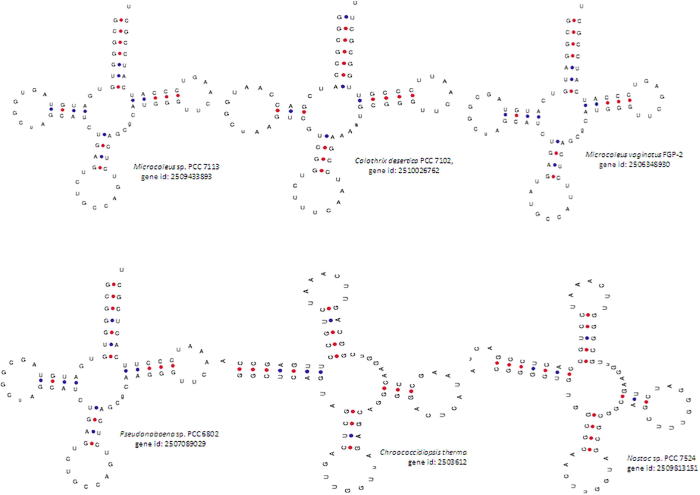
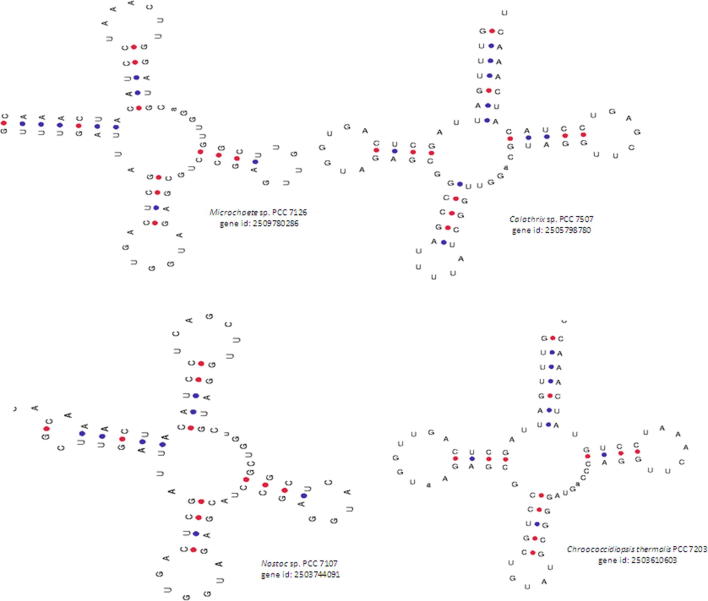
The anti-codon loop was found to contain five to nine nucleotides (Supplementary Table 5). However, more than 95% of the tRNA molecules examined were found to contain seven nucleotides in the anti-codon loop. tRNAGln from Microchaete sp. PCC 7126 (gene id: 2509780286) had five nucleotides in the anti-codon loop. tRNALys from Calothrix sp. PCC 7507 (gene id: 2505798780) and tRNATyr from Chroococcidiopsis thermalis PCC 7203 (gene id: 2503610603) and Nostoc sp. PCC 7107 (gene id: 2503744091) were also found to contain five nucleotides in the anti-codon loop. Among 3092 tRNAs, sixty-nine (2.23%) tRNAs were found to contain nine nucleotides in the anti-codon loop. tRNAHis from Dactylococcopsis salina PCC 8305 (gene id: 2509553070), tRNAPro from Nostoc sp. PCC7524 (gene id: 2509813151), and tRNAThr from Fischerella sp. PCC9431 (gene id: 2512981565) were found to contain at least one tRNA gene with nine nucleotides in the anti-codon loop. tRNAGly from Microcoleus sp. PCC 7113 (gene id: 2509433893), Microcoleus vaginatus FGP-2 (gene id: 2506348930), Oscillatoria acuminate PCC 6304 (gene id: 2509421382), and Pseudanabaena sp. PCC 6802 (gene id: 2507089029) were found to contain nine nucleotides in the anti-codon loop. tRNALeu genes from Leptolyngbya sp. PCC 7375 (gene id: 2509846846), Leptolyngbya sp. PCC 7375 (gene id: 2509846848), and Pseudanabaena sp. PCC 6802 (gene id: 2507089039) were also found to contain nine nucleotides in the anti-codon loop. Of a total of 204 tRNAMet genes, sixty-one tRNAs were found to contain nine nucleotides in the anti-codon loop. Surprisingly, not a single tRNA was found to contain an even number (six, eight or ten) of nucleotides in the anti-codon loop.
The length of the variable loop of the cyanobacterial tRNAs ranged from three to twenty-three nucleotides (Supplementary Table 5). The variable loop of tRNACys of Synechococcus elongatus PCC 7942 (gene id: 640711018) was found to contain only three nucleotides and, interestingly, that was found to contain only two nucleotides in the D-arm. For tRNAAla, all genes were found to contain five nucleotides in the variable loop with a minor variation. In the case of tRNAAsn, most of the sequences were found to contain five nucleotides, while twenty of them were found to contain six nucleotides. tRNAAsp of Microcoleus sp. PCC 7113 gene (gene id: 2509433873) was found to contain only four nucleotides. The length of the variable loop in tRNACys genes ranged from three to seven nucleotides (Supplementary Table 5). tRNAGlu genes were found to contain either four or five nucleotides, and most of them were found to contain four nucleotides. The variable loop of the tRNAGln was found to contain four to seven nucleotides. Among 193 tRNAGly genes, 113 (58.54%) were found to contain five nucleotides, while the remaining eighty (41.45%) were found to contain four nucleotides. Seven of the tRNAHis genes were found to contain six nucleotides, while the remainder of the tRNAHis genes were found to contain five nucleotides in the variable loop. Among tRNAIle genes, eighteen (14.51%) were found to contain five nucleotides, and 106 (85.48%) were found to contain six nucleotides. The variable loops of tRNALeu genes varied from five to eighteen nucleotides: the number of nucleotides in the tRNALeu variable loop could be five, six, seven, nine, eleven, twelve, thirteen, fourteen, fifteen, sixteen and seventeen. These data showed that tRNALeu genes had eleven different nucleotide variants in the variable loop. tRNALys genes contained five (27.55%) or six (71.65%) nucleotide while tRNAMet genes were found to contain five to seventeen nucleotides in the variable loop. Among ninety-one tRNAPhe genes, except for two, all others were found to contain five nucleotides in the variable loop. The variable loops of tRNAPro genes were found to contain five to six nucleotides. The variable loops of tRNASer genes were very diverse and contained four to twenty-three nucleotides (Supplementary Table 5). The variable loops of tRNAThr genes were found to contain four to six nucleotides. The majority of tRNAThr genes were found to contain five nucleotides, while only seven of them were found to contain four nucleotides, and only six were found to contain six nucleotides (Supplementary Table 5). tRNATrp genes contained four to six nucleotides in the variable loops. Among eighty-three tRNATrp genes, fourteen were found to contain four nucleotides; one was found to contain six nucleotides, while the remaining sixty-eight (81.92%) were found to contain five nucleotides. tRNATyr genes were found to contain five to eighteen nucleotides in the variable loops. For tRNAVal, all genes were found to contain five nucleotides in the variable loop. Importantly, it was found that only tRNALeu, tRNASer and tRNATyr genes contained ten or more nucleotides in their variable loops, whereas the remainder of the tRNA genes contained less than ten nucleotides in their variable loops. Previous reports suggest that the variable loop is less critical for the biological function of tRNA (Brennan and Sundaralingam, 1976, Kjems et al., 1989). However, the length of the variable arm is important for the recognition of tRNA by aminoacyl tRNA synthetase, and it also helps to maintain the stability of the tRNA. However, the long variable loop consisting of a helical stem loop emerges from the deep groove side of the dihydrouridine helix (Brennan and Sundaralingam, 1976). The long variable loop most possibly resulted from retaining a splicing-deficient intron (Kjems et al., 1989).
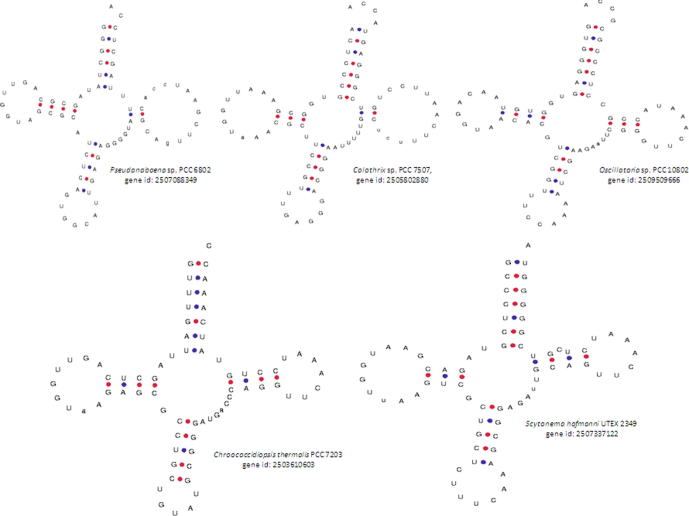
The length of the Ψ-arm ranged from two to six nucleotides. tRNALeu of Calothrix sp. PCC 7507 (gene id: 2505802880) had only two nucleotides in the Ψ-arm. tRNATyr from Chroococcidiopsis thermalis PCC 7203 (gene id: 2503610603) was found to contain only four nucleotides in the Ψ-arm. Apart from these tRNAs, all other tRNA genes were found to contain five nucleotides in the Ψ-arm. The fact that the Ψ-arm is the ribosome recognition site and, apart from a few tRNAs, all Ψ-arms were found to contain five nucleotides suggests that it is a conserved structural feature of the tRNA molecule.
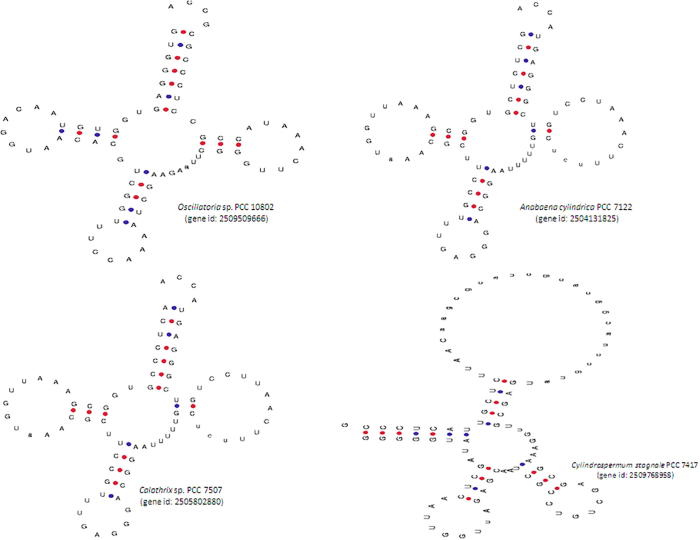
The length of the Ψ-loop ranged from two to thirty nucleotides in the cyanobacterial tRNA. Approximately 3087 tRNA genes (99.83%) were found to contain seven nucleotides in the Ψ-loop. The tRNATrp gene from Oscillatoria sp. PCC 10,802 (gene id: 2509509666) was found to contain nine nucleotides in the Ψ-loop (Fig. 1). Among tRNALeu genes, the Ψ-loops of Anabaena cylindrica PCC 7122 (gene id: 2504131825) and Calothrix sp. PCC 7507 (gene id: 2505802880) were found to contain thirteen nucleotides (Fig. 1). The Ψ-loop, along with the Ψ-arm, recognizes the ribosome during protein synthesis. The presence of five nucleotides in the Ψ-arm and seven nucleotides in the Ψ-loop is a characteristic feature of tRNAs. However, the presence of a lower or a higher number of nucleotides in the Ψ-arm and the Ψ-loop is very intriguing. The existence of non-standard Ψ-arms and Ψ-loops suggests that further novel translational mechanisms that remain to be elucidated might be occurring in these species.
Fig. 1.
The nucleotide variation in the Ψ-loop of cyanobacterial tRNAs. The Ψ-loop of cyanobacterial tRNA possesses seven nucleotides. However, a few of the cyanobacterial tRNAs were found to contain more than seven nucleotides in this loop. The Ψ-loop from Oscillatoria sp. PCC 10,802 (gene id: 2509509666) was found to contain nine nucleotides, whereas Anabaena cylindrica PCC 7122 (gene id: 2504131825) and Calothrix sp. PCC 7507 (gene id: 2505802880) were found to contain thirteen nucleotides. The Ψ-loop of Cylindrospermum stagnale PCC 7417 (gene id: 2509768958) tRNA contained twenty-nine nucleotides.
3.4. Based on the D-loop and the variable loop, cyanobacterial tRNA can be classified into two classes
tRNA research has received considerable attention since the inception of the clover leaf-like model by Robert W. Holley in 1965. Initially, tRNAs were classified based on the amino acid they transferred to the translation machinery. Further classification was then performed based on the number and position of introns present in the tRNA (Tocchini-Valentini et al., 2009). The acceptor arm, the D-arm, the D-loop, the anti-codon arm, the anti-codon loop, the variable loop, the Ψ-arm and the Ψ-loop all contain a specified number of nucleotides. The presence of specific numbers of nucleotides in different parts of the tRNA has its own significance and functional importance. Hence, it was very important for us to classify the tRNAs according to the distribution nucleotides in different parts of the tRNA molecule. As a result, we have classified the cyanobacterial tRNAs based on the nucleotide composition in the D-arm and the variable loop. Based on the number of nucleotides in the D-arm and the variable loop, we were able to classify these tRNAs into two classes, referred to as class I and class II tRNAs (Table 2). The D-arm of class I tRNAs has 2–4 nucleotides, whereas the D-arm of class II tRNAs has only four nucleotides. Similarly, the variable loops in class I tRNA have 0–7 nucleotides, whereas the tRNAs of class II have 8–23 nucleotides. Based on the D-arm nucleotide numbers, the tRNAs in class I were tRNAAla, tRNAArg, tRNAAsn, tRNACys, tRNAGlu, tRNAGln, tRNAGly, tRNAHis, tRNALeu, tRNALys, tRNAMet, tRNAPhe, tRNAPro, tRNASer, and tRNATyr, whereas those in class II were tRNAAsp, tRNAIle, tRNAThr, tRNATrp and tRNAVal. However, based on the variable loop nucleotide numbers, the tRNAs in class I were tRNAAla, tRNAArg, tRNAAsn, tRNAAsp, tRNACys, tRNAGlu, tRNAGln, tRNAGly, tRNAHis, tRNAIle, tRNALys, tRNAPhe, tRNAPro, tRNAThr, tRNATrp, and tRNAVal, and those in class II were tRNALeu, tRNAMet, tRNASer, and tRNATyr (Table 2). Class I tRNAs have a short variable loop with no helical stem and hairpin structure, whereas class II tRNAs might possess an additional stem and hairpin loop.
Table 2.
Classification of cyanobacterial tRNAs based on the number of nucleotides in the D-arm and variable loop regions. Based on nucleotide variation in the D-arm, the tRNAs that contained two to four nucleotides in the D-arm were classified as class I, whereas those contained four nucleotides in the D-arm were classified as class II tRNAs. Similarly, based on the nucleotide variation in the variable region, the tRNAs that contained zero to seven nucleotides were classified as class I, whereas those that contained eight to twenty-three nucleotides were classified as class II tRNAs.
| tRNA class | No. of nucleotides in D-arm | tRNA |
|---|---|---|
| Class I | 2–4 | Ala, Arg, Asn, Cys, Glu, Gln, Gly, His, Leu, Lys, Met, Phe, Pro, Ser, Tyr |
| Class II | 4 | Asp, Ile, Thr, Trp, Val |
| No. of nucleotides in the variable region | ||
| Class I | 0–7 | Ala, Arg, Asn, Asp, Cys, Glu, Gln, Gly, His, Ile, Lys, Phe, Pro, Thr, Trp, Val |
| Class II | 8–23 | Leu, Met, Ser, Tyr |
3.5. Cyanobacteria encode putative novel tRNAs
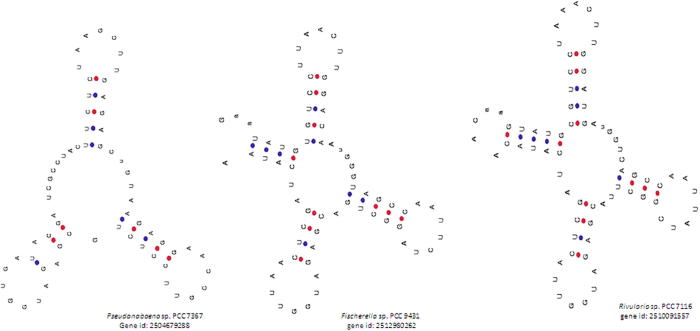
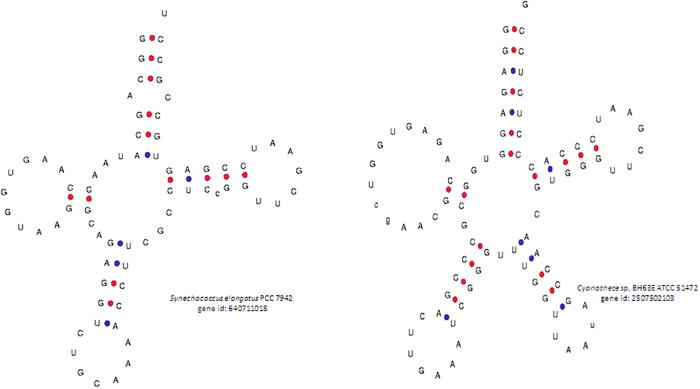
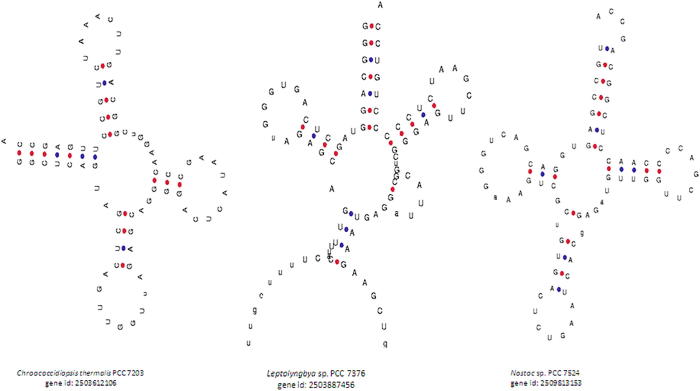
The acceptor arm usually contains seven nucleotides. However, in our study, we found a few tRNAs that contained zero (Pseudanabaena sp. PCC 7367, gene id: 2504679288), four (Fischerella sp. PCC 9431, gene id: 2512980262); six (Rivularia sp. PCC 7116, gene id: 2,510,091,557 and others) or eight (Cylindrospermum stagnale PCC 7417, gene id: 2,509,767,605 and 2509767604) nucleotides (Supplementary Table 5, Fig. 2). At least fifteen tRNAs were found to contain six nucleotides in the acceptor arm (Supplementary Table 5). The amino acids bind to the 3′ end of the acceptor arm during the translation process, and the presence of a smaller number nucleotide in the acceptor arm might impact the binding efficiency of the tRNA to the cognate amino acid. Usually, the D-arm contains three to four nucleotides. However, in our study, we found a few tRNAs that contained only two nucleotides in the D-arm (Synechococcus elongatus PCC 7942, gene id: 640711018; Cyanothece sp. BH63E ATCC 51472, gene id: 2507502103) (Fig. 3). Bovine and human mitochondrial tRNA are reported to lack the D-arm (Arcari and Brownlee, 1980, de Bruijn et al., 1980, Salinas-Giegé et al., 2015). In general, the anti-codon arm contains five nucleotides. However, we observed that cyanobacterial tRNAs contained three (Chroococcidiopsis thermalis PCC 7203, gene id: 2503612106; Cyanobacterium aponinum PCC 10605, gene id: 2503745044, etc.), four or six (Leptolyngbya sp. PCC 7376, gene id: 2503887456; Nostoc sp. PCC 7524, gene id: 2509813153) nucleotides instead of the usual five nucleotides in the anti-codon arm (Fig. 4). Several of the tRNAs were found to have three or six nucleotides in the anti-codon arm. The tRNAs found to contain four nucleotides in the anti-codon arm were tRNAAsp, tRNAGln, tRNAGly, tRNALeu, tRNALys, tRNAMet, tRNAPro, tRNAThr and tRNATyr (Supplementary Table 5). However, except for tRNAMet, no tRNAs were found to possess three nucleotides in the anti-codon arm.
Fig. 2.
Variation in nucleotide composition of the acceptor arms of cyanobacterial tRNAs. Canonical tRNA possesses seven nucleotides in the acceptor arm. However, in a few cyanobacterial species, the acceptor arm was found to be absent, while in others, the acceptor arm contained four, six or eight nucleotides. In Pseudanabaena sp. PCC 7367 (gene id: 2504679288), the acceptor arm was found to be absent, whereas Fischerella sp. PCC 9431 (gene id: 2512980262) contained four and Rivularia sp. PCC 7116 (gene id: 2510091557) contained six nucleotides.
Fig. 3.
Variation in nucleotide composition of the D-arms of cyanobacterial tRNAs. The D-arm of canonical tRNA possesses three to four nucleotides. However, a few cyanobacterial tRNAs were found to possess only two nucleotides in the D-arm. The D-arms of Synechococcus elongatus PCC 7942 (gene id: 640711018) and Cyanothece sp. BH63E ATCC 51,472 (gene id: 2507502103) contained only two nucleotides.
Fig. 4.
The anti-codon loop of cyanobacterial tRNAs with five nucleotides. Several of the cyanobacterial tRNAs were found to possess only five nucleotides instead of the usual seven nucleotides in the anti-codon loop. The cyanobacterial tRNAs that contained five nucleotides in the anti-codon loop possessed either four or five nucleotides in the anti-codon arm.
The anti-codon loop of tRNAs contains seven nucleotides. However, dramatic variation in the nucleotide composition was observed in our study. We discovered several tRNAs that contained more or fewer than seven nucleotides in the anti-codon loop. Some of the tRNAs were found to contain nine nucleotides in the anti-codon loop (Microcoleus sp. PCC 7113, gene id: 2509433893; Calothrix desertica PCC 7102, gene id: 2510026762; Calothrix sp. PCC 7507, gene id: 2505801260; Microcoleus vaginatus FGP-2, gene id: 2506348930; Pseudanabaena sp.PCC6802, gene id: 2507089029; Chroococcidiopsis thermalis PCC 7203, gene id: 2503612106; Nostoc sp. PCC 7524, gene id: 2509813151) (Fig. 5). The tRNAs those were found to contain nine nucleotides in the anti-codon loop had a variable number of nucleotides in the anti-codon arm (Fig. 5). The tRNAs those were found to contain only five nucleotides in the anti-codon loop were from the species Microchaete sp. PCC 7126 (gene id: 2509780286), Calothrix sp. PCC 7507 (gene id: 2505798780), Chroococcidiopsis thermalis PCC 7203 (gene id: 2503610603), and Nostoc sp. PCC 7107 (gene id: 2503744091) (Supplementary Figure 4). Interestingly, none of the tRNAs were found to contain either six or eight nucleotides in the anti-codon loop. Also, of interest, the anti-codon loop was found to be absent in Leptolyngbya sp. PCC 7376 (gene id: 2503887456) (Fig. 4), and Nostoc sp. PCC 7107 (gene id: 2503742551). The presence of such variation in the nucleotide sequences in the anti-codon loop suggests that there might be some novel protein translation machinery in cyanobacteria that remains to be elucidated.
Fig. 5.
Variation in nucleotide composition of the anti-codon loops of cyanobacterial tRNAs. The anti-codon loops of canonical tRNA possess seven nucleotides. However, some of the cyanobacterial tRNAs were found to possess nine nucleotides in the anti-codon loop. The tRNAs containing nine nucleotides in the anti-codon loop contained either three or four nucleotides in the anti-codon arms. However, none of the cyanobacterial tRNAs were found to possess either six or eight nucleotides in the anti-codon loop.
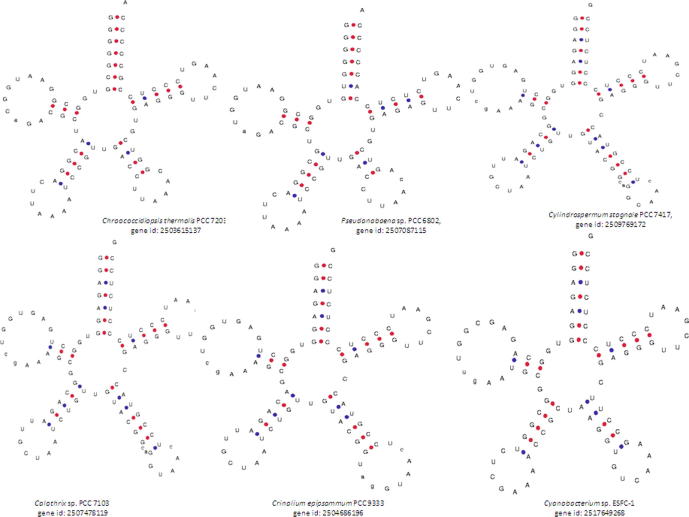
The region of the tRNA between the anti-codon loop and the Ψ-arm, which has a bulge-like structure, is commonly known as the variable loop. In our study, we found that unlike the D-arm, the anti-codon arm and the Ψ-arm, which each contained a loop, the variable region also contained an arm and a loop structure. We referred to this arm as the variable arm and to the loop as the variable loop (Supplementary Fig. 5). Unlike the D-arm, the anti-codon arm and the Ψ-arm, the variable arm has base pairing, and the number of nucleotides in the variable arm ranged from four to seven, whereas the number of nucleotides in the variable loop ranged from five to nine, in contrast to the anti-codon loop. Some of the tRNAs that were found to contain the variable loop were Chroococcidiopsis thermalis PCC7203 (gene id: 2503615137), Pseudanabaena sp.PCC6802 (gene id: 2507087115), Calothrix desertica PCC7102 (gene id: 2510025188), Cylindrospermum stagnale PCC7417 (gene id: 2509769172), Calothrix sp. PCC6303 (gene id:2504098647), Calothrix sp.PCC7507 (gene id: 2505801912), Calothrix sp. PCC7103 (gene id: 2507478119), Crinalium epipsammum PCC9333 (gene id: 2504686196), Cyanobacterium sp. ESFC-1 (gene id: 2517649268), Cyanobacterium stanieri PCC7202 (gene id: 2,503,365,868 and 2503366170), Cylindrospermum stagnale PCC7417 (gene id: 2509768953), Dactylococcopsis salina PCC8305 (gene id: 2509555825), Gloeocapsa sp. PCC7428 (gene id: 2503794465), Leptolyngbya boryana PCC6306 (gene id: 2509803183), Microchaete sp. PCC7126 (gene id: 2509781573), and Prochlorothrix hollandica PCC9006 (gene id: 2509499121) (Supplementary Fig. 5).
The canonical Ψ-arm and the Ψ-loop possess five and seven nucleotides, respectively. However, a few tRNAs were found to contain a diverse number of nucleotides in the Ψ-arm and Ψ-loop as well. The tRNAAla gene of Pseudanabaena sp. PCC 6802 (gene id: 2507088349) and the tRNALeu gene of Calothrix sp. PCC 7507 (gene id: 2505802880) were found to contain only two nucleotides in the Ψ-arm, whereas the Ψ-loop was found to contain thirteen nucleotides. tRNATrp of Oscillatoria sp. PCC 10,802 (gene id: 2509509666) was found to contain three nucleotides in the Ψ-arm and nine nucleotides in the Ψ-loop. The tRNATyr gene from Chroococcidiopsis thermalis PCC 7203 (gene id: 2503610603) and the tRNAGlu gene from Scytonema hofmanni UTEX 2349 (gene id: 2507337122) were found to contain four nucleotides in the Ψ-arm (Supplementary Fig. 6). The tRNACys gene from Synechococcus elongatus PCC 7942 (gene id: 640711018) was found to contain six nucleotides in the Ψ-arm. Interestingly, the tRNAAsp gene from Cylindrospermum stagnale PCC 7417 (gene id: 2509768958) was found to contain twenty-nine nucleotides in the Ψ-loop (Fig. 1). These dynamic variations in the nucleotide length in different parts of the tRNA molecule & novel structure clearly reflect the presence of possible alternate translation machinery in the cyanobacteria. They might also be playing competitive role and performing extra-translational function as well by bypassing ribosomal machinery. Additionally, these could also bring strong tRNA structure and can interfere with the translation process. Francklyn and Minajigi (2010) reported that tRNA is a multi-functional molecule that participates in several processes of the cellular metabolism (Francklyn and Minajigi, 2010). Therefore, the presence of several putative novel forms of cyanobacterial tRNA indicates that cyanobacterial tRNA might be involved in diverse cellular metabolism in addition to the protein translation. Nucleotide sequence change in tRNA can bring changes in the efficiency of aminoacylation and modification of ribosome’s (Dale and Uhlenbeck, 2005, Ledoux et al., 2009, Siegfried et al., 2010). Suppressing the stop codon of isodecoders sequence from the iso-acceptor family of tRNA shows diverse ranges of suppression efficiencies in HeLa cell line (Geslain and Pan, 2010). The isodecoders sequence that do not occur naturally resulted best suppression efficiency, suggesting not all of the naturally occurring iso-decoder are ideal for best translational efficiency (Geslain and Pan, 2010). Charged tRNA can be used as a carrier of activated amino acids whereas uncharged tRNAs that activates protein kinase GCN2 that regulates translation during cellular nutrient availability (Dong et al., 2000). tRNA prevents premature apoptosis by binding to cytochrome C and it has potential to cleave to produce small fragments that can be used as small interfering RNA to regulate translation factors (Ivanov et al., 2011, Mei et al., 2010, Yamasaki et al., 2009). Non-canonical tRNAAsp isodecoders associate with mRNA and regulate alternative 3′ end formation. tRNAs are also interacts with diverse arrays of cellular protein involved in cellular metabolism where tRNA interacts with histone H3K9 or farnesyl-transferase mediated protein modification (Parisien et al., 2013b, Parisien et al., 2013a). To exactly decipher the function of these putative novel tRNAs, first of all it needs to identify their cognate interacting partners that may be protein or RNA.
4. Conclusion
An analysis of cyanobacterial tRNA sequences has revealed the presence of a diverse and dynamic tRNA structure and genomic architecture of cyanobacterial tRNA. The analyses revealed the presence of putative novel tRNA structures in cyanobacteria. The presence of putative novel tRNAs in cyanobacteria is very interesting, and laboratory-based experimental analysis to confirm their functional role can be very promising. However, it is difficult to accurately determine the function of individual tRNA. Although we have gained considerable success in RNA sequencing approach, the presence of post-transcriptional modifications in tRNA makes it difficult to accurately sequence them. The presence of putative novel tRNA in cyanobacteria raises speculation about the presence of diverse evolutionary function that might not be directly linked to translation. Further, the presence of such putative novel tRNA gives a brief idea about their potential to change themselves due to evolutionary pressure.
5. Data availability
All the data used during this study was taken from publicly available genomic database and details are mentioned in the materials and methods section
Declaration of Competing Interest
Authors don’t have any competing of interest to declare
Acknowledgments
Acknowledgement
The authors would like to extend their sincere appreciation to the Chair, Natural and Medical Sciences Research Center, University of Nizwa for providing necessary support. Authors also like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding the research group NO (RGP-271).
Author’s contribution
TKM: Conceived the idea, performed the experiment and analysis, drafted and revised the manuscript, DY: revised the manuscript, AK: analysed & revised the manuscript, AH: drafted and revised the manuscript, EFA: drafted and revised the manuscript, AAH: Revised the manuscript.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2019.06.004.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary figure 1.
Cluster of number of tRNAs versus cyanobacterial genome size. The Fig. shows that the majority of cyanobacterial species possess 40 to 50 tRNAs per genome, and their genome sizes fall within the range of 3 to 7 Mb. Few species show the presence of 60 to 70 tRNAs per genome. However, the genome sizes of all them are greater than 5 Mb.
Supplementary figure 2.
Percentage distribution of cyanobacterial tRNAs versus genome size. This analysis shows no correlation between percentages (%) of cyanobacterial tRNA abundance versus genome size. However, 0.6 to 1% tRNAs occur within genomes of 4 to 6 Mb.
Supplementary figure 3.
Correlations between the number of tRNA iso-acceptors and genome size in cyanobacteria. The Fig. shows that the majority of the genomes encode 37 to 41 iso-acceptors within a genome size range of 4 to 8 Mb, showing a greater consistency in the iso-acceptor number than in the genome size
Supplementary figure 4.
Variation in nucleotide composition of the anti-codon arms of cyanobacterial tRNAs. The anti-codon arm of canonical tRNA possesses five nucleotides. However, some of the cyanobacterial tRNAs were found to possess three, four or six nucleotides in the anti-codon arm. The anti-codon arm of Chroococcidiopsis thermalis PCC 7203 (gene id: 2503612106) contained three nucleotides, and that of Nostoc sp. PCC 7524 (gene id: 2509813153) contained six nucleotides
Supplementary figure 5.
The variable arm and variable loop in the cyanobacterial tRNAs. The variable region of cyanobacterial tRNA contained zero to twenty-three nucleotides. A few of the class II tRNAs were found to contain a long arm and a bulged-loop-like structure. We named the long arm the variable arm and the bulged loop the variable loop, in contrast to other arms and loops. The bulged-loop-like structure of the variable region resembled the loop-like structures of the D-loop, the anti-codon loop and the Ψ-loops.
Supplementary figure 6.
Nucleotide variation in the Ψ-arms of cyanobacterial tRNAs. In canonical tRNA, the Ψ-arm possesses five and the Ψ-loop possesses seven nucleotides. However, a few of the cyanobacterial tRNAs contained two to four nucleotides in the Ψ-arm.
Genomic tRNA sequences of cyanobacterial species used during the study.
References
- Arcari P., Brownlee G.G. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980;8:5207–5212. doi: 10.1093/nar/8.22.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardell D.H., Hou Y.M. Initiator tRNA genes template the 3’ CCA end at high frequencies in bacteria. BMC Genomics. 2016;17:1–12. doi: 10.1186/s12864-016-3314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T., Sundaralingam M. Structure of transfer RNA molecules containing the long variable loop. Nucleic Acids Res. 1976;3:3235–3251. doi: 10.1093/nar/3.11.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B.F.C. The crystal structure of tRNA. J. Biosci. 2006;31:453–457. doi: 10.1007/BF02705184. [DOI] [PubMed] [Google Scholar]
- Dale T., Uhlenbeck O.C. Amino acid specificity in translation. Trends Biochem. Sci. 2005;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- de Bruijn M.H., Schreier P.H., Eperon I.C., Barrell B.G., Chen E.Y., Armstrong P.W., Wong J.F., Roe B.A. A mammalian mitochondrial serine transfer RNA lacking the “dihydrouridine” loop and stem. Nucleic Acids Res. 1980;8:5213–5222. doi: 10.1093/nar/8.22.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A.G. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- Francklyn C.S., Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010;584:366–375. doi: 10.1016/j.febslet.2009.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geslain R., Pan T. Functional analysis of human tRNA isodecoders. J. Mol. Biol. 2010;396:821. doi: 10.1016/j.jmb.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy M.P., Young D.L., Payea M.J., Zhang X., Kon Y., Dean K.M., Grayhack E.J., Mathews D.H., Fields S., Phizicky E.M. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev. 2014;28:1721–1732. doi: 10.1101/gad.245936.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A.K., Phizicky E.M. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2014;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- Kjems J., Leffers H., Olesen T., Garrett R.A. A unique tRNA intron in the variable loop of the extreme thermophile Thermofilum pendens and its possible evolutionary implications. J. Biol. Chem. 1989;264:17834–17837. [PubMed] [Google Scholar]
- Koonin E.V., Novozhilov A.S. Origin and evolution of the genetic code: the universal enigma. IUBMB Life. 2009;61:99–111. doi: 10.1002/iub.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S., Olejniczak M., Uhlenbeck O.C. A sequence element that tunes E. coli tRNA(GGC)(Ala) to ensure accurate decoding. Nat. Struct. Mol. Biol. 2009;16:359–364. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C., Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta T., Syed A., Ameen F., Bae H. Novel Genomic and Evolutionary perspective of cyanobacterial tRNAs. Front. Genet. 2017;8:200. doi: 10.3389/fgene.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta T.K., Bae H. Analyses of genomic tRNA reveal presence of novel tRNAs in Oryza sativa. Front. Genet. 2017;8:90. doi: 10.3389/fgene.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg H., Cantor M., Dusheyko S., Hua S., Poliakov A., Shabalov I., Smirnova T., Grigoriev I.V., Dubchak I. The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 2014;42:D26–D31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M., Wang X., Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013;10:1853–1867. doi: 10.4161/rna.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M., Wang X., Perdrizet G., Lamphear C., Fierke C.A., Maheshwari K.C., Wilde M.J., Sosnick T.R., Pan T. Discovering RNA-protein interactome by using chemical context profiling of the RNA-protein interface. Cell Rep. 2013;3:1703–1713. doi: 10.1016/j.celrep.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas de Pouplana L., Dedon P. More than an adaptor molecule: the emerging role of tRNA in cell signaling and disease. FEBS Lett. 2014;588:4267. doi: 10.1016/j.febslet.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas-Giegé T., Giegé R., Giegé P. tRNA biology in mitochondria. Int. J. Mol. Sci. 2015;16:4518–4559. doi: 10.3390/ijms16034518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S.J., Schaack J., Cooley L., Burke D.J., Soil D. Structure and transcription of Eukaryotic tRNA gene. Crit. Rev. Biochem. 1985;19:107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Siegfried N.A., O’Hare B., Bevilacqua P.C. Driving forces for nucleic acid pKa shifting in an A+·C wobble: effects of helix position, temperature, and ionic strength. Biochemistry. 2010;49:3225–3236. doi: 10.1021/bi901920g. [DOI] [PubMed] [Google Scholar]
- Tocchini-Valentini G.D., Fruscoloni P., Tocchini-Valentini G.P. Processing of multiple-intron-containing pretRNA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20246–20251. doi: 10.1073/pnas.0911658106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XiaoLong Z., EnDuo W. Transfer RNA: a dancer between charging and mis-charging for protein biosynthesis. Sci. China Life Sci. 2013;56:921–932. doi: 10.1007/s11427-013-4542-9. [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Ivanov P., Hu G., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarham J.W., Elson J.L., Blakely E.L., McFarland R., Taylor R.W. Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. - RNA. 2010;1:304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic tRNA sequences of cyanobacterial species used during the study.
Data Availability Statement
All the data used during this study was taken from publicly available genomic database and details are mentioned in the materials and methods section