Abstract
Periodontal disease is a chronic infectious disease, which is characterized by the damaged dental hard tissue by lactic acid generated by microorganisms after the fermentation of carbohydrates rich diet. The risk of periodontal disease is known to be higher in diabetic patients. We compared the diversity of five commonly occurring dental bacteria including Porphyromonas gingivalis, Tannerella forsythia, Capnocytophaga ochracea, Prevotella intermedia, and Aggregatibacter actinomycetemcomitans in 14 type-2 diabetic patients and equal numbers of healthy controls. The subgingival samples were collected using sterile paper points. We used 16S rRNA sequence specific primers for PCR-based identification of dental bacteria. Our results showed that A. actinomycetemcomitans was completely absent in control subjects but present in 43% of diabetic patients. C. ochracea was highly prevalent in diabetic patients (100%) as compared to controls (28.5%). The frequency of other three bacterial species was also higher in diabetic patients than control subjects. These findings indicate that dental bacteria are highly prevalent in subgingival pockets of diabetic patients. Therefore, proper monitoring of diabetic patients for dental care is important to prevent bacterial growth and its sequela in risky individuals. Further case-control studies using larger sample size would help in validating the association between oral diseases and diabetes.
Keywords: Dental bacteria, Identification, PCR, Diabetes
1. Introduction
In healthy individuals, the internal tissues are devoid of pathogens, whereas the external tissues like oral cavity are exposed to various kinds of microorganisms (Zarco et al., 2012). The oral cavity is the starting point of the gastrointestinal tract. Because of the frequent supply of food through oral cavity, it creates a favourable environment for nurturing and harbouring microorganisms. The surfaces of oral cavity are covered with diverse microorganisms, constructing the proverbial bacterial biofilm and maintain a dynamic ecological balance with the host body. Any disruption in this equilibrium causes oral disease (Patil et al., 2013). In healthy oral cavity, there is a distinctive predominant bacterial flora that are not only highly diverse in nature but also subject and site-specific (Aas et al., 2005). There are numerous microorganisms that come under the broader term oral microflora and include bacteria, fungi, most protozoa but rarely viruses. The diversity and counts of microflora vary according to food habits, salivary features and antibiotic regimens used for treating infections (Javed et al., 2013). Oral bacteria have been associated in the pathogenesis of highly prevalent oral diseases such as dental caries (Martinez-Martinez et al., 2019, Bergamo et al., 2019, Almusawi et al., 2018) and periodontal disease (Dahlen et al., 2019, Genco et al., 2019). A clear understanding about the molecular diversity of microflora is important for pre-emption and redemption of associated health conditions. The phenotypic methods of bacterial identification based on microscopy and culture techniques are less sensitive, subjective and time consuming (Srinivasan et al., 2015). On the other hand, polymerase chain reaction (PCR) provides the highest sensitivity for the detection of microorganisms (Santos et al., 2004).
It is important to note that oral bacteria associated with periodontal disease are mainly anaerobic and therefore they are very difficult to cultivate (Chen and Slots, 2000). Genotyping of microorganisms using 16S rRNA gene sequencing has been recognized as a more sensitive, specific and accurate method for bacterial identification (Clarridge, 2004). By virtue of the well-conserved regions in 16S rRNA gene among different biological species, comparison of these sequences provides a useful tool for studying molecular diversity and evolutionary phylogenetics (Slabbinck et al., 2010). Rampini et al. (2011) have shown that 16S rRNA gene is highly useful for detection of culture-negative bacteria in patients pre-treated with antibiotic drugs. Molecular methods for species specific identification of oral bacteria can be based on sequence-specific primers (Khan, 2012), probes (Salminen et al., 2015) or direct sequencing (Jiang et al., 2016, Riggio et al., 2007).
Several studies have reported significantly high prevalence of periodontitis (Idrees et al., 2014, Javed et al., 2013) and dental caries (Al-Sadhan, 2006, Zahrani, 2005) in adults, children, and older population of Saudi Arabia. Moreover, putative role of specific oral bacterial species has also been suggested in many systemic diseases, including diabetes (Taylor et al., 2013, Schara et al., 2013) and cardiovascular disease (Serra et al., 2014, Suzuki et al., 2015). Poor oral health may adversely affect diabetic glycemic control and subsequent medical complications (Moore, 2002). The biofilm built in the oral cavity has been associated with health and disease (Zarco et al., 2012). The bacterial diversity of the oral biofilm is variable among individuals and influences by several factors including age, diet, oral hygiene and genetics (Marsh and Devine, 2011). The diversity and pattern of bacterial exposure in early life of an individual play an important role in the gut colonization of bacteria (Adlerberth and Wold, 2009). A reduced bacterial diversity in the gut has been found to correlate with higher risks of chronic diseases like allergy and asthma (Ege et al., 2011).
Although the implication of molecular methods for identification and diversity analysis of oral microflora is of great concern, there are very few studies undertaken in Saudi Arabia in this particular field. The aim of this study was to compare the diversity of common dental bacteria in non-diabetic and diabetic patients, using the sequence-specific primers. This study is focused on 5 species of periodontal pathogenic bacteria including Porphyromonas gingivalis (P. gingivalis), Tannerella forsythia (T. forsythia), Capnocytophaga ochracea (C. ochracea), Prevotella intermedia (P. intermedia) and Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans).
2. Materials and methods
2.1. Subjects and study design
This study was conducted on 28 unrelated Saudi Arabian female subjects. One group included 14 type-2 diabetic patients with periodontal disease recruited consecutively from Clinic of Dentistry, Section of periodontology, King Saud University, Riyadh, Saudi Arabia. The other group composed of 14 healthy non-diabetic subjects, this group served as control group. All subjects gave an informed written consent before the study, answered a set of questionnaire and underwent clinical and physical examinations. Periodontal examinations were carried out by qualified dentists.
The inclusion criteria were female gender, residents of Riyadh, age range 18–70 years, presence of > 20 intact natural teeth, features of periodontitis (loss of attachment ≥ 5 mm), and type-2 diabetes. The exclusion criteria were the use of antibiotic therapy during the last 2 months and presence of any systemic disease that may influence the periodontal condition. The study protocol was approved by the Research Ethics Committee of the Faculty of Science, King Saud University, Riyadh, Saudi Arabia.
2.2. Sample collection and bacterial lysis
The subgingival samples were collected from four sites (about 5 mm depth) from tooth pockets by a professional dentist in the morning time, using separate sterile paper points (size 40, taper 0.04) from META Company, South Korea. A single vertical stroke was used under quality control procedures for each member of case and control group. All the samples were promptly kept in individual sterile Eppendorf tubes containing Tris-EDTA buffer (pH 7.6). For PCR amplification, 10 μl of lysis buffer was mixed with 90 μl of subgingival sample and the solution was boiled for 5 min. An aliquot of the lysate (4 μl) was used for PCR amplification of each sample.
2.3. Bacterial identification by PCR
PCR detection for all target bacteria was performed using species specific primers. The primers for the 16S ribosomal DNA sequences were selected and purchased from Eurofins Genomics, Germany. The primer pairs used for the PCR amplification and the expected product sizes are given in Table 1.
Table 1.
Sequences of PCR primers.
| Oral bacteria | Primer sequence | Product size (bp) |
|---|---|---|
| P. gingivalis | TGT AGA TGA CTG ATG GTG AAA ACC | 197 |
| ACG TCA TCC CCA CCT TCC TC | ||
| T. forsythia | GCG TAT GTA ACC TGC CCG CA | 641 |
| TCG TTC AGT GTC AGT TAT ACC T | ||
| C. ochracea | AGA GTT TGA TCC TGG CTC AG | 185 |
| GAT GCC GTC CCT ATA TAC CAT TAG G | ||
| P. intermedia | TTT GTT GGG GAG TAA AGC GGG | 575 |
| TCA ACA TCT CTG TAT CCT GCG T | ||
| A. actinomycetemcomitans | AGA GTT TGA TCC TGG CTC AG CAC TTA AAG GTC CGC CTA CGT GCC |
593 |
The PCR was carried out in a total volume of 30 μl containing 26 μl of reaction mixture and 4 μl of the lysate sample. The reaction mixture contained 1x PCR buffer (10 mM Tris-HCl pH 8.8, 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100), 2 units of Taq DNA polymerase, 0.2 mM dNTPs and 100 pmol of each primer. The cycling conditions were programmed as an initial denaturation step at 95 °C for 5 min followed by the 35 amplification cycles of denaturation at 95 °C for 1 min, primers annealing at 60 °C for 1 min and primer extension at 72 °C for 1.5 min. After completion of all the cycles, a final extension step at 72 °C for 7 min was programmed, followed by storing the tubes at 4 °C. PCR was carried out in a thermal cycler (Applied Biosystems, USA) as reported earlier (Khan, 2012).
2.4. Statistics
The frequency of dental bacteria between the control and diabetic groups was analysed by Fisher’s exact test using the CalcFisher program (Khan, 2003). P values < 0.05 were considered as statistically significant.
3. Results
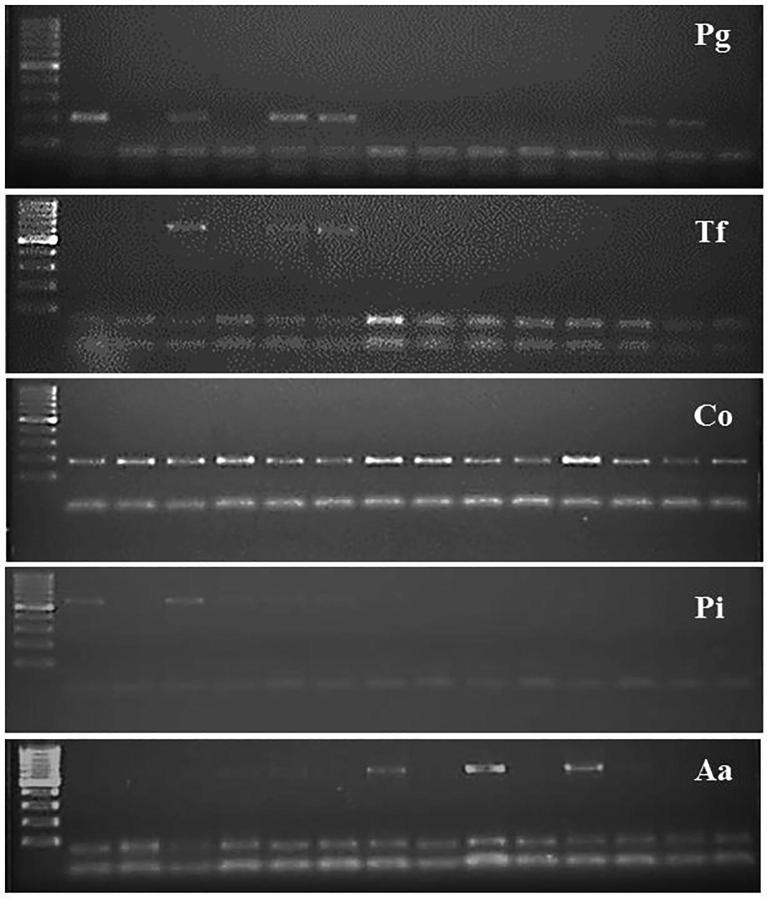
PCR-based identification of dental bacteria using sequences specific primers for each target species resulted intense gel bands of anticipated sizes as follows: P. gingivalis (197 bp), T. forsythia (641 bp), C. ochracea (185 bp), P. intermedia (575 bp), and A. actinomycetemcomitans (593 bp) (Fig. 1). The diversity and frequency of dental bacteria among 14 clinically healthy control subjects and 14 diabetic patients are shown in Table 1. In control subjects, the prevalence of P. gingivalis, T. forsythia, C. ochracea, and P. intermedia was 35.7%, 14%, 28.5% and 14% respectively, whereas A. actinomycetemcomitans was completely absent in this group. In diabetic patients, C. ochracea appeared to be highly prevalent (100%) followed by P. gingivalis (43%), A. actinomycetemcomitans (43%), P. intermedia (35.7%) and T. forsythia (28.5%).
Fig. 1.
Representative gel images showing the specific bands for different bacteria. Abbreviations are Pg, P. gingivalis; Tf, T. forsythia; Co, C. ochracea; Pi, P. intermedia; Aa, A. actinomycetemcomitans.
There were positive amplifications for P. gingivalis in 5 out of 14 (35%) healthy subjects and in 6 out of 14 (43%) diabetic samples. PCR amplification using sequences specific primers for T. forsythia resulted intense gel bands in only 2 out of 14 (14%) control samples and 4 out of 14 (28.5%) diabetic samples. PCR analysis using 16S rDNA primers confirmed the presence of C. ochracea in all subgingival samples from diabetic patients (100%) while only 4 out of 14 (28.5%) control subjects showed the presence of this bacterial species (Table 1). P. intermedia were detected in 2 of the 14 control subjects (14%) as compared to 5 of 14 (35.7%) in diabetic group. None of the control subjects showed the occurrence of A. actinomycetemcomitans in their subgingival samples whereas 6 of the 14 diabetic patients (42.8%) were found to be positive for A. actinomycetemcomitans (Table 1). The statistical analysis showed significantly higher (P < 0.05) prevalence of C. ochracea and A. actinomycetemcomitans in diabetic patients as compared to control subjects (Fig. 2).
Fig. 2.
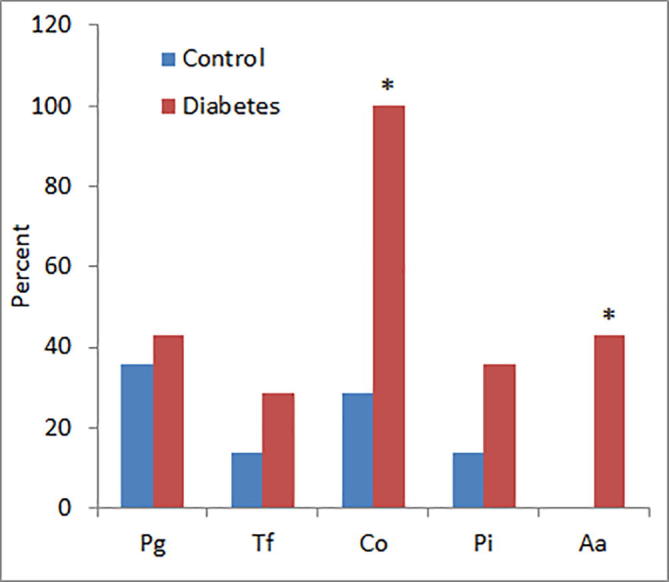
Comparative prevalence of dental bacteria in controls and diabetic patients. *P < 0.05 versus control group using Fisher’s exact test. Abbreviations are Pg, P. gingivalis; Tf, T. forsythia; Co, C. ochracea; Pi, P. intermedia; Aa, A. actinomycetemcomitans.
4. Discussion
The results of this study showed that the occurrence of dental bacteria was more prevalent in subgingival samples from diabetic patients as compared to control subjects (Table 2). Oral bacteria are known to predominantly colonize on different surfaces in oral cavity with the aid of specific adhesins located on the bacterial wall that specifically bind to complementary receptors found on the oral surfaces (Gibbons, 1989). Based on the 16S rDNA sequence data generated from human subgingival plaque samples, the dominant microflora included 347 species that fell into 9 bacterial phyla, while multiple subjects showed the presence of known putative oral bacteria including P. gingivalis, B. forsythus, and T. denticola (Paster et al., 2001). Mager et al. (2003) have reported variable trends of 40 cultivable bacteria on the surfaces of oral tissue, gingival plaques and saliva from healthy individuals. In fact, all the patients with periodontal disease do not necessarily harbour the similar subgingival microorganisms, hence, only a limited number of oral bacteria have been suggested as the predictors for the progression of periodontal damage (van Winkelhoff, 2003).
Table 2.
Distribution of oral bacteria in healthy subjects and diabetic patients.
| SN | Oral bacteria | Control | Diabetes | P-Value |
|---|---|---|---|---|
| 1 | P. gingivalis | 5 (35.7%) | 6 (43.0%) | 0.720 |
| 2 | T. forsythia | 2 (14.0%) | 4 (28.5%) | 0.406 |
| 3 | C. ochracea | 4 (28.5%) | 14 (100%) | 0.00007 |
| 4 | P. intermedia | 2 (14.0%) | 5 (35.7%) | 0.230 |
| 5 | A. actinomycetemcomitans | 0 (0.0%) | 6 (43.0%) | 0.0079 |
The prevalence of two bacterial species, Capnocytophaga ochracea and Aggregatibacter actinomycetemcomitans, was significantly higher in diabetic patients as compared to normal subjects (Fig. 2). Capnocytophaga is a gram-negative bacterium that has been linked in the pathogenesis of periodontal disease (Jolivet-Gougeon et al., 2007). These microorganisms are able to produce various enzymes with the tendency to periodontal breakdown and therefore considered as opportunistic pathogens. Their high frequency in diabetic group may be attributed to the anaerobic conditions of the subgingival pocket due to high glucose levels that would cause excessive periodontal damage in diabetic patients (Lion et al., 1996). An interesting observation is that all the diabetic patients harboured C. ochracea whereas only 4 out of 14 control subjects showed the presence of C. ochracea anaerobes in their sub-gingival pockets (Fig. 2). These results are in agreement with an earlier study reporting that Capnocytophaga species are frequently isolated in adult periodontitis subjects while their frequency and counts are significantly higher in the diseased sites of diabetic as compared to non-diabetic periodontitis patients (Ciantar et al., 2005).
None of the control subjects showed the presence of A. actinomycetemcomitans in their subgingival pockets whereas 43% of diabetic patients harboured this pathogen. A. actinomycetemcomitans has a vital role in the initiation of periodontal disease and if not treated properly, tooth turns to be mobile and severely damaged because of the progressive bone and attachment loss leading to ultimate tooth extraction (Slots, 1976, Henderson et al., 2010). Interestingly, A. actinomycetemcomitans can migrate to and colonize in the heart tissue and has been suggested as a risk factor in the development of cardiac disease (Haraszthy et al., 2000, Taylor-Robinson et al., 2002). Hence, timely detection of A. actinomycetemcomitans in oral samples would have a greater impact on health issues extending beyond the denture. Our results are consistent with previous reports that demonstrated high prevalence of A. actinomycetemcomitans in patients with diabetes mellitus (Murakami et al., 2013). In our study, subgingival samples from 28.5% of diabetic patients were T. forsythia positive as compared to 14% healthy controls. T. forsythia bacterium is an anaerobic, Gram-negative species of Cytophaga Bacteroidetes family, linked with the development of periodontal disease (Brennan et al., 2007). Deng et al. (2011) failed to observe any association between T. forsythia and periodontal status whereas the occurrence of P. gingivalis and A. actinomycetemcomitans was independently and significantly associated with chronic periodontitis.
In conclusion, the findings of this study showed that dental bacteria are more prevalent in diabetic patients than non-diabetic subjects. Particularly, the prevalence of C. ochracea and A. actinomycetemcomitans was significantly higher in diabetic patients as compared to healthy individuals. Further studies with larger sample size should be conducted to confirm a more detailed description of bacterial species associated with oral disease in diabetic patients which can throw light on a variety of etiological microorganisms. Recent studies have advocated that patients with periodontal disease are more susceptible to metabolic syndrome (Kim et al., 2019, Doğan et al., 2019). Severe periodontal disease may also put diabetic patients at increased risk of diabetic complications (Veena et al., 2018). For a long time, it was thought that bacteria were the causative factors for the association between periodontal disease and systemic disease. However, recent researches have demonstrated that inflammation caused by bacterial invasion may be responsible for such association (Hegde and Awan, 2019, Balaji et al., 2018). Efforts should be made to focus on treating the inflammation besides targeting periodontal bacteria for more efficient management of oral disease.
Acknowledgements
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding the Research Group No. RGP-009.
Footnotes
Peer review under responsibility of King Saud University.
References
- Aas J.A., Paster B.J., Stokes L.N., Olsen I., Dewhirst F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlerberth I., Wold A.E. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Almusawi M.A., Gosadi I., Abidia R., Almasawi M., Khan H.A. Potential risk factors for dental caries in type-2 diabetes patients. Int. J. Dent. Hyg. 2018;16:467–475. doi: 10.1111/idh.12346. [DOI] [PubMed] [Google Scholar]
- Al-Sadhan S.A. Dental caries prevalence among 12–14year-old School children in Riyadh: A 14 year follow-up study of the Oral Health Survey of Saudi Arabia Phase I. Saudi Dent J. 2006;18:2–7. [Google Scholar]
- Balaji S.K., Lavu V., Rao S. Chronic periodontitis prevalence and the inflammatory burden in a sample population from South India. Indian J Dent Res. 2018;29:254–259. doi: 10.4103/ijdr.IJDR_335_17. [DOI] [PubMed] [Google Scholar]
- Bergamo A.Z.N., Matsumoto M.A.N., Nascimento C.D. Microbial species associated with dental caries found in saliva and in situ after use of self-ligating and conventional brackets. J. Appl. Oral Sci. 2019;27 doi: 10.1590/1678-7757-2018-0426. e20180426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R.M., Genco R.J., Wilding G.E., Hovey K.M., Trevisan M., Wactawski-Wende J. Bacterial species in subgingival plaque and oral bone loss in postmenopausal women. J. Periodontol. 2007;78:1051–1061. doi: 10.1902/jop.2007.060436. [DOI] [PubMed] [Google Scholar]
- Chen C., Slots J. Microbiological tests for Actinobacillus actinomycetemcomitans and Porphyrornonas gingivalis. Periodontol. 2000;20:53–64. doi: 10.1111/j.1600-0757.1999.tb00157.x. [DOI] [PubMed] [Google Scholar]
- Ciantar M., Gilthorpe M.S., Hurel S.J., Newman H.N., Wilson M., Spratt D.A. Capnocytophaga spp. in periodontitis patients manifesting diabetes mellitus. J. Periodontol. 2005;76:194–203. doi: 10.1902/jop.2005.76.2.194. [DOI] [PubMed] [Google Scholar]
- Clarridge J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen G., Basic A., Bylund J. Importance of virulence factors for the persistence of oral bacteria in the inflamed gingival crevice and in the pathogenesis of periodontal disease. J. Clin. Med. 2019;8(9) doi: 10.3390/jcm8091339. pii: E1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T., Wang L., Lv J., Pang J., Liu B., Du Y., Ke J. Association of three bacterial species and periodontal status in Chinese adults: an epidemiological approach. J. Clin. Microbiol. 2011;49:184–188. doi: 10.1128/JCM.01819-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Doğan E.S.K., Doğan B., Fentoğlu Ö., Kırzıoğlu F.Y. The role of serum lipoxin A4 levels in the association between periodontal disease and metabolic syndrome. J. Periodontal Implant Sci. 2019;49:105–113. doi: 10.5051/jpis.2019.49.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ege M.J., Mayer M., Normand A.C., Genuneit J., Cookson W.O., Braun-Fahrländer C. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 2011;24:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- Genco R.J., LaMonte M.J., McSkimming D.I. The subgingival microbiome relationship to periodontal disease in older women. J. Dent. Res. 2019;98:975–984. doi: 10.1177/0022034519860449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons R.J. Bacterial adhesion to oral tissues: a model for infectious diseases. J. Dent. Res. 1989;68:750–760. doi: 10.1177/00220345890680050101. [DOI] [PubMed] [Google Scholar]
- Haraszthy V.I., Zambon J.J., Trevisan M., Zeid M., Genco R.J. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 2000;71:1554–1560. doi: 10.1902/jop.2000.71.10.1554. [DOI] [PubMed] [Google Scholar]
- Hegde R., Awan K.H. Effects of periodontal disease on systemic health. Dis. Mon. 2019;65:185–192. doi: 10.1016/j.disamonth.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Henderson B., Ward J.M., Ready D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen? Periodontol 2000. 2010;54:78–105. doi: 10.1111/j.1600-0757.2009.00331.x. [DOI] [PubMed] [Google Scholar]
- Idrees M.M., Azzeghaiby S.N., Hammad M.M., Kujan O.B. Prevalence and severity of plaque-induced gingivitis in a Saudi adult population. Saudi Med. J. 2014;35:1373–1377. [PMC free article] [PubMed] [Google Scholar]
- Javed F., Al-Askar M., Samaranayake L.P., Al-Hezaimi K. Periodontal disease in habitual cigarette smokers and nonsmokers with and without prediabetes. Am. J. Med. Sci. 2013;345:94–98. doi: 10.1097/MAJ.0b013e31824d5337. [DOI] [PubMed] [Google Scholar]
- Jiang S., Gao X., Jin L., Lo E.C. Salivary microbiome diversity in caries-free and caries-affected children. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17121978. pii: E1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet-Gougeon A., Sixou J.L., Tamanai-Shacoori Z., Bonnaure-Mallet M. Antimicrobial treatment of Capnocytophaga infections. Int. J. Antimicrob. Agents. 2007;29:367–373. doi: 10.1016/j.ijantimicag.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Khan H.A. A Visual Basic software for Fisher’s exact test. J. Stat. Soft. 2003;8:1–7. [Google Scholar]
- Khan H.A. Molecular identification and phylogeny of commonly occurring periodontal bacteria using 16S rRNA gene sequences. J. Pure Appl. Microbiol. 2012;6:517–523. [Google Scholar]
- Kim J.S., Kim S.Y., Byon M.J., Lee J.H., Jeong S.H., Kim J.B. Association between periodontitis and metabolic syndrome in a Korean nationally representative sample of adults aged 35-79 years. Int. J. Environ. Res. Public Health. 2019;16(16) doi: 10.3390/ijerph16162930. pii: E2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion C., Escande F., Burdin J.C. Capnocytophaga canimorsus infections in human: review of the literature and cases report. Eur. J. Epidemiol. 1996;12:521–533. doi: 10.1007/BF00144007. [DOI] [PubMed] [Google Scholar]
- Mager D.L., Ximenez-Fyvie L.A., Haffajee A.D., Socransky S.S. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 2003;30:644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- Marsh P.D., Devine D.A. How is the development of dental biofilms influenced by the host? J. Clin. Periodontol. 2011;38:28–35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Martinez R.E., Domínguez-Pérez R.A., Sancho-Mata J., Abud-Mendoza C., Ayala-Herrera J.L., Popoca-Hernandez E.A. The frequency and severity of dental caries, and counts of cariogenic bacteria in rheumatoid arthritis patients. Dent Med. Probl. 2019;56:137–142. doi: 10.17219/dmp/105340. [DOI] [PubMed] [Google Scholar]
- Moore P. The diabetes-oral health connection. Compendium. 2002;23:14–20. [PubMed] [Google Scholar]
- Murakami M., Suzuki J., Yamazaki S. High incidence of Aggregatibacter actinomycetemcomitans infection in patients with cerebral infarction and diabetic renal failure: a cross-sectional study. BMC Infect. Dis. 2013;13:557–566. doi: 10.1186/1471-2334-13-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster B.J., Boches S.K., Galvin J.L., Ericson R.E., Lau C.N., Levanos V.A., Sahasrabudhe A., Dewhirst F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001;18:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S., Rao R.S., Sanketh D.S., Amrutha N. Microbial flora in oral diseases. J. Contemp. Dent. Pract. 2013;14:1202–1208. doi: 10.5005/jp-journals-10024-1477. [DOI] [PubMed] [Google Scholar]
- Rampini S.K., Bloemberg G.V., Keller P.M., Büchler A.C., Dollenmaier G., Speck R.F., Böttger E.C. Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin. Infect. Dis. 2011;53:1245–1251. doi: 10.1093/cid/cir692. [DOI] [PubMed] [Google Scholar]
- Riggio M.P., Aga H., Murray C.A., Jackson M.S., Lennon A., Hammersley N., Bagg J. Identification of bacteria associated with spreading odontogenic infections by 16S rRNA gene sequencing. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2007;103:610–617. doi: 10.1016/j.tripleo.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Salminen A., Kopra K.A., Hyvärinen K., Paju S., Mäntylä P., Buhlin K., Nieminen M.S., Sinisalo J., Pussinen P.J. Quantitative PCR analysis of salivary pathogen burden in periodontitis. Front. Cell. Infect. Microbiol. 2015;5:69. doi: 10.3389/fcimb.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C.F., Sakai V.T., Machado M.A.A.M., Schippers D.N., Greene A.S. Reverse transcription and polymerase chain reaction: principles and applications in dentistry. J. Appl. Oral Sci. 2004;12:1–11. doi: 10.1590/s1678-77572004000100002. [DOI] [PubMed] [Google Scholar]
- Schara R., Skaleric E., Seme K., Skaleric U. Prevalence of periodontal pathogens and metabolic control of type 1 diabetes patients. J. Int. Acad. Periodontol. 2013;15:29–34. [PubMed] [Google Scholar]
- Serra e Silva Filho W., Casarin R.C., Nicolela Jr E.L., Passos H.M., Sallum A.W., Gonçalves R.B. Microbial diversity similarities in periodontal pockets and atheromatous plaques of cardiovascular disease patients. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109761. e109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabbinck B., Waegeman W., Dawyndt P., De Vos P., De Baets B. From learning taxonomies to phylogenetic learning: integration of 16S rRNA gene data into FAME-based bacterial classification. BMC Bioinf. 2010;11:69. doi: 10.1186/1471-2105-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J. The predominant cultivable organisms in juvenile periodontitis. Scand. J. Dent. Res. 1976;84:1–10. doi: 10.1111/j.1600-0722.1976.tb00454.x. [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Karaoz U., Volegova M. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0117617. e0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J., Aoyama N., Aoki M. Incidence of periodontitis in Japanese patients with cardiovascular diseases: a comparison between abdominal aortic aneurysm and arrhythmia. Heart Vessels. 2015;30:498–502. doi: 10.1007/s00380-014-0507-6. [DOI] [PubMed] [Google Scholar]
- Taylor J.J., Preshaw P.M., Lalla E. A review of the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J. Periodontol. 2013;84:113–134. doi: 10.1902/jop.2013.134005. [DOI] [PubMed] [Google Scholar]
- Taylor-Robinson D., Aduse-Opoku J., Sayed P., Slaney J.M., Thomas B.J., Curtis M.A. Oro-bacteria in various atherosclerotic arteries. Eur. J. Clin. Microbiol. Infect. Dis. 2002;21:755–757. doi: 10.1007/s10096-002-0810-5. [DOI] [PubMed] [Google Scholar]
- van Winkelhoff A.J. Microbiology in diagnosis and treatment planning in periodontics. Int. J. Dent Hyg. 2003;1:131–137. doi: 10.1034/j.1601-5037.2003.00031.x. [DOI] [PubMed] [Google Scholar]
- Veena H.R., Natesh S., Patil S.R. Association between diabetic retinopathy and chronic periodontitis-A cross-sectional study. Med. Sci. (Basel) 2018;6(4) doi: 10.3390/medsci6040104. pii: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrani A.A. Dental health status among a sample of elderly dental patients in Riyadh, Saudi Arabia. Saudi Dent J. 2005;17:74–82. [Google Scholar]
- Zarco M.F., Vess T.J., Ginsburg G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]