Abstract
Background
Late HIV diagnosis is detrimental both to the individual and to society. Strategies to improve early diagnosis of HIV must be a key health care priority. We examined whether nation-wide electronic registry data could be used to predict HIV status using machine learning algorithms.
Methods
We extracted individual level data from Danish registries and used algorithms to predict HIV status. We used various algorithms to train prediction models and validated these models. We calibrated the models to mimic different clinical scenarios and created confusion matrices based on the calibrated models.
Findings
A total 4,384,178 individuals, including 4,350 with incident HIV, were included in the analyses. The full model that included all variables that included demographic variables and information on past medical history had the highest area under the receiver operating characteristics curves of 88·4% (95%CI: 87·5% – 89·4%) in the validation dataset. Performance measures did not differ substantially with regards to which machine learning algorithm was used. When we calibrated the models to a specificity of 99·9% (pre-exposure prophylaxis (PrEP) scenario), we found a positive predictive value (PPV) of 8·3% in the full model. When we calibrated the models to a sensitivity of 90% (screening scenario), 384 individuals would have to be tested to find one undiagnosed person with HIV.
Interpretation
Machine learning algorithms can learn from electronic registry data and help to predict HIV status with a fairly high level of accuracy. Integration of prediction models into clinical software systems may complement existing strategies such as indicator condition-guided HIV testing and prove useful for identifying individuals suitable for PrEP.
Funding
The study was supported by funds from the Preben and Anne Simonsens Foundation, the Novo Nordisk Foundation, Rigshospitalet, Copenhagen University, the Danish AIDS Foundation, the Augustinus Foundation and the Danish Health Foundation.
Keywords: HIV prevention, HIV diagnosis, Machine learning, Registry data, Big data
Introduction
Over half of individuals diagnosed with HIV in the European Region are diagnosed in a late stage of infection (i.e. with a CD4 count below 350 cells/µL at time of diagnosis) [1]. Early diagnosis is important for several reasons; it improves life expectancy among people living with HIV (PLWH) [2], reduces risk of onward HIV transmission [3], and lowers associated health care costs [4]. Moreover, there is now a large body of evidence from randomized controlled trials that early initiation of antiretroviral therapy (ART) lowers risk of acquired immunodeficiency syndrome (AIDS) and non-AIDS events [5,6].
One way to diagnose PLWH at an early stage may be through universal screening in which all individuals, that encounter the health care system, are offered testing. In the Western European Region, where the prevalence of HIV is low, such an approach may be associated with a poor cost-benefit ratio [7]. Thus, alternative strategies have been proposed, such as indicator-condition guided testing. Such guidance, which in part builds on studies on HIV prevalence within specific medical conditions [8], recommends HIV testing in any condition, which may indicate the presence of HIV [9]. Although this approach offers several advantages, it does not consider all the prior health data of a given individual. Moreover, it does not consider age, sex or any temporal patterns of prior medical diseases, and it does not include those medical conditions that would be associated with a lower risk of HIV transmission. Including this kind of information into decision algorithms of HIV prediction may not only improve discriminative abilities but may potentially also help identify individuals at increased risk of acquiring HIV, including individuals who might be pre-exposure prophylaxis (PrEP) candidates.
Machine learning algorithms, a set of mathematical or algorithmic tools which extract generalizable patterns from large data sets in order to make predictions about the outcome in new, unseen cases, are rapidly growing research areas which have also found their way into HIV research. It has so far primarily been used to predict progression of HIV and ART resistance and for ART optimization [10], [11], [12]. In the present study, we applied machine learning algorithms that learn from nation-wide historic electronic registry data to make prediction of HIV status and evaluated the potential of such prediction models as a screening tool and to identify suitable candidates for PrEP. Focusing on assessing the general applicability of these types of methods and thus establishing a reference baseline performance, we focused our investigation on simpler general-purpose modeling frameworks over detailed models of higher performance but reduced ability to generalize.
Methods
Setting
As of December 31, 2016, Denmark had an adult population of approximately 4·6 million people, 6200 (∼0·1%) people diagnosed with HIV, and an estimated 600 (∼0·01%) individuals with undiagnosed HIV infection [13]. PLWH are treated in eight specialized HIV-care centers and are typically seen as outpatients at intended intervals of 12–24 weeks. Combination antiretroviral treatment is provided free of charge at the HIV-centers.
Data sources
The unique 10-digit personal identification number assigned to all Danish residents at birth or immigration was used to avoid multiple registrations and to track individuals in the following registries.
The Danish National Hospital Registry (DNHR) was established in 1977 and stores information on all inpatient and outpatient admission to hospitals in Denmark. Diagnoses are classified according to the ICD 8th revision (ICD8) until December 31, 1993, and 10th revision (ICD10) thereafter. The registry has been expanded gradually so that it now also contains outpatient data, data from emergency wards, and data from psychiatric admissions dating back to 1995, which led us to use data on admission dating back to Jan 1, 1995.
The Danish Civil Registration System was established in 1968 and stores information on all Danish residents. From this registry we extracted data on date of birth, sex, residency, marital status and migration.
The Employment Classification Module provides information on the occupation and employment status of the population chargeable with tax. From this registry we extracted data on main source of income
Educational Classification Module includes data on successfully completed educational attainments collected directly from all Danish educational institutions for all Danish residents. Ninety-seven percent of the Danish-born population and 85–90% of the immigrant population has non-missing data. Educational attainment is classified according to a Danish modified version of the International Standard Classification of Education 2011 [14].
Study population
We included all Danish individuals that: i) had at least one hospital visit between January 1, 1995, until December 31, 2016, ii) had a Danish personal identification number, and iii) were aged ≥ 16 years at the hospital visit. We excluded individuals with i) an HIV diagnoses registered before 16 years of age and ii) an HIV diagnosis before Jan 1, 1995.
Statistics
We defined an index visit which was the last registered hospital visit or the first registered hospital visit with an HIV-diagnosis. For the index visit we computed 167 variables associated with medical history. These variables were a slightly modified version of categories used previously [15]. The categories and the associated ICD8/ICD10 codes are provided as online supplementary material. Some ICD codes were omitted from the chosen disease categories used for the models. All codes have been highlighted in online supplementary material Table S1. For each index visit we identified all hospital visits up until three months before the index visit. Based on these visits we calculated a score for each disease category that was 10 minus the number of years between each hospital visit and the index visit, e.g. if an individual was hospitalized five years prior to the index visit with a diagnosis of pneumococcal pneumonia (PP) the variable PP was assigned a value of 10 - 5 = 5, if the time was equal to or exceeded 10 years the variable was assigned the value 0. We also included a variable that indicated how many hospital visits the individual had the last year prior to the index visit. Additional variables included were age (>16, 16–25, 25–35, 35–45, 45–55 and 55+), sex (male and female), place of birth (Denmark, Scandinavia, other and unknown), highest educational attainment (primary school, high school, vocational internships and main course, short higher education, middle length higher education, bachelor and long length higher education, PhD programs, unknown), marital status (married, divorced, widowhood, registered partnership, cancelled registered partnership, longest living of two partners and unknown), main source of income (self-employed, employed spouse, wage earner with own business, wage earner without business, wage earner with support, senior citizen with own business, senior citizen, other and unknown) and place of residency (capital region, large city region, hinterland region, provincial region, rural region and unknown). For more details on the variables please refer to Table 1 and Table S1.
Table 1.
Clinical characteristics of the training and validation cohort.
| Training cohort | Validation cohort | |
|---|---|---|
| Total number of individuals | 2,972,264 | 1,411,914 |
| HIV+, n (%) | 3,063 (0·1%) | 1,287 (0·1%) |
| Male, n (%) | 1,470,420 (49%) | 652,912 (46%) |
| Age | ||
| 15–25, n (%) | 383,521 (13%) | 149,525 (11%) |
| 25–35, n (%) | 33,662 (11%) | 232,098 (16%) |
| 35–45, n (%) | 439,223 (15%) | 168,687 (12%) |
| 45–55, n (%) | 408,877 (14%) | 203,669 (14%) |
| 55+, n (%) | 1,406,981 (47%) | 657,935 (47%) |
| Country of origin | ||
| Denmark, n (%) | 2,623,960 (88%) | 1,288,389 (91%) |
| Scandinavia, n (%) | 34,075 (1%) | 13,315 (1%) |
| Other Countries, n (%) | 280,223 (9%) | 100,519 (7%) |
| Unknown, n (%) | 34,006 (1%) | 9,691 (1%) |
| Highest Educational Attainment | ||
| Primary school, n (%) | 929,426 (31%) | 403,871 (29%) |
| High school, n (%) | 160,838 (5%) | 92,277 (7%) |
| Vocational internships and main course, n (%) | 829,897 (28%) | 417,074 (30%) |
| Short higher education, n (%) | 89,474 (3%) | 44,397 (3%) |
| Middle length higher education, n (%) | 322,035 (11%) | 165,348 (12%) |
| Bachelor and long higher education, n (%) | 192,771 (6%) | 95,981 (7%) |
| PhD Programs, n (%) | 10,187 (<1%) | 4,256 (<1%) |
| Unknown, n (%) | 437,636 (15%) | 188,710 (13%) |
| Marital status | ||
| Married, n (%) | 1,331,231 (45%) | 622,066 (44%) |
| Divorced, n (%) | 313,083 (11%) | 154,192 (11%) |
| Widowhood, n (%) | 364,476 (12%) | 163,582 (12%) |
| Registered partnership, n (%) | 4,280 (<1%) | 1,863 (<1%) |
| Cancelled registered partnership, n (%) | 1,322 (<1%) | 625 (<1%) |
| Longest living of two partners, n (%) | 330 (<1%) | 125 (<1%) |
| Unknown, n (%) | 85,289 (3%) | 33,911 (2%) |
| Main source of income | ||
| Self-employed, n (%) | 93,678 (3%) | 41,650 (3%) |
| Employed spouse, n (%) | 3,273 (<1%) | 1,654 (<1%) |
| Wage earner with own business, n (%) | 39,663 (1%) | 18,179 (1%) |
| Wage earner without business, n (%) | 1,174,574 (40%) | 610,584 (43%) |
| Wage earner with support, n (%) | 6,074 (<1%) | 2,989 (<1%) |
| Senior citizen with own business, n (%) | 21,963 (1%) | 9,666 (1%) |
| Senior citizen, n (%) | 1,059,706 (36%) | 499,156 (35%) |
| Others, n (%) | 524,385 (18%) | 211,455 (15%) |
| Unknown, n (%) | 48,948 (2%) | 16,581 (1%) |
| Place of residence | ||
| Capital region, n (%) | 761,306 (26%) | 363,318 (26%) |
| Large city region, n (%) | 352,530 (12%) | 174,090 (12%) |
| Hinterland region, n (%) | 467,034 (16%) | 219,237 (16%) |
| Provincial region, n (%) | 657,852 (22%) | 311,164 (22%) |
| Rural region, n (%) | 648,253 (22%) | 311,194 (22%) |
| Unknown, n (%) | 85,289 (3%) | 32,911 (2%) |
The response variable in all models was an HIV diagnosis during the index visit. HIV was defined as one a of the following diagnoses: ICD8: 07983 or ICD10: B20-B24.9. The validity of an HIV diagnosis in LPR has previously been shown to be very high [16].
We used logistic regression with an elastic net penalty function (GLMnet) as regularizer to fit models with increasing variable complexity. A regularizer is a tool which automatically penalizes complex models over simpler ones during model fitting, and the particular type used here (elastic net) allows for the automatic exclusion of variables with little impact on model performance (these variables are marked with a “-“ in Table S1); thus, the role of the regularizer is to find the model that best fits the data with the minimal number of variables. The elastic net is a regularized regression method that combines lasso and ridge regression penalties, the hyperparameter α controls which mix between the two penalties is used, ie. if α = 0 pure ridge penalty is used and if α = 1 pure lasso penalty is used [17]. We used 10-fold cross validation to determine the value of λ in the penalty function and used 10-fold cross validation to determine the mix between the penalty functions (the α-value). When a model was fit, we used the model to calculate a risk score of HIV for each individual included. The risk scores were used to create receiver operating characteristics (ROC) curve for all the models. Furthermore, we calculated area under receiver operating characteristics curve (AUROC) in order to compare the models. The ROC curves were used to calibrate the models to certain values of sensitivity and specificity by calculating their corresponding risk scores. We additionally calculated sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV).
To evaluate how much information was gained by including an increasing number of variables, we fitted three different models with the elastic net penalty function. Each model was fitted independently, i.e. regularization was done separately for each model and parameters in the model calculated independently. The variables introduced in the three different models were: i) age, sex and sexually transmitted infections (STIs, see Table 1 for a complete list), ii) age, sex, place of birth, educational attainment, marital status, place of residency and main source of income and iii) all included in ii and medical history, this was the final model used. The total number of index visits was randomly divided into a training set (70%) and a validation set (30%). The validation set was used to make out-of-sample validation on the prediction algorithm.
To evaluate whether different machine learning algorithms performed better with regards to discriminative ability, we also fitted simple logistic regression, random forest with random undersampling (1 HIV-positive to 10 HIV-negative individuals) to balance the dataset, logistic regression with lasso regularizer (GLMLasso) and logistic regression with ridge regularizer (GLMRidge), and logistic regression with elastic net regularizer with synthetic minority oversampling technique (SMOTE) to balance dataset, on all variables and compared these using AUROC. We tested differences in performance of the models on the models that were calibrated to a sensitivity of 90% with McNemars test.
We created calibrated confusion matrices of the final model to mimic three clinical scenarios. The first scenario is when a high certainty of an HIV diagnosis is required (model calibrated to a specificity of 99·9%), e.g. when evaluating potential candidates of PrEP, the specific score of 99·9% was chosen to be high but other than that is arbitrary. The second scenario is the optimal trade-off between sensitivity and specificity (model optimized according to the highest value of the Youden's index (sensitivity + specificity - 1)) [18]. The third scenario is when a large coverage of the diagnosis is required (model calibrated to a sensitivity of 90%), e.g. a screening setting, the 90% sensitivity was chosen to reflect the first ‘90′ in the WHO ’90-90–90′. The calibration was done by calculating the risk score that yields the desired performance characteristic (eg. sensitivity of 90%) in the training data set, and then applying this risk score to the validation data set and then calculate the actual sensitivity, specificity, NPV and PPV. Of note is that the calibration of the model is made on the training set, and the confusion matrix is calculated on the validation dataset. Therefore, the actual performance values may differ slightly from what the model was calibrated to.
Statistical analyses were performed using R version 3·5·0 (R Development Core Team (2018). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria) [19].
Results
We included 4,384,178 individuals from the Danish population with at least one hospital visit. These data were divided into a training set with 2,972,264 hospital visits and a validation set with 1,411,914 hospital visits. We identified 4,350 PLWH that were included in the analyses, in either the training data or the validation data. For exact characteristics of the two datasets please refer to Table 1.
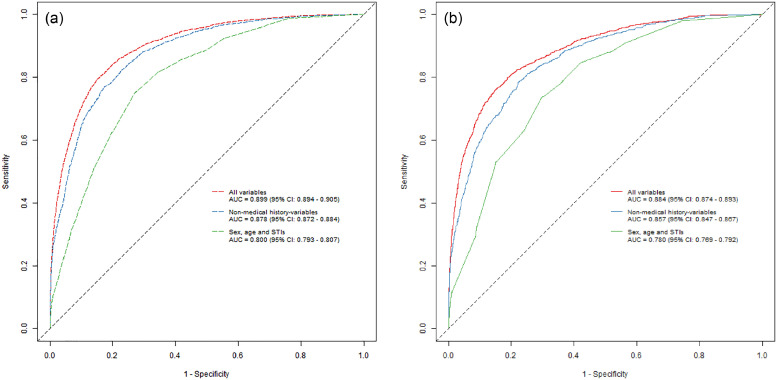
We found that an increasing number of variables included in the model resulted in increasing predictive performance. For the model including age, sex and STIs (model I), we found an AUC of 0·780 (95% CI: 0·769–0·792) in the validation set. For the full model (model III), which included additional information about medical history, we found an AUC of 0·884 (95% CI: 0·874–0·893) in the validation set (Fig. 1a and b).
Fig. 1.
Training and validation sample ROC curves. The figures show the training (a) sample and validation sample (b) performance of the three different models based on the best performing algorithm (GLMridge). Each point on the graphs represents a sensitivity and a specificity for a particular cut-off with regards to risk-score calculated by using the parameters generated by fitting the different models.
When we calibrated the models to a sensitivity of 90%, the actual sensitivity of model III was 86·2%. Specificity was 69·9% for model III and only 43·5% for model I. The model, which included information on birth, educational attainment, marital status, place of residency and main source of income, but not medical history (model II), had a sensitivity of 86·1% and a specificity of 65·4% (Table 2a). The PPV of model III was 0·26%, for model II it was 0·23% and for the model I it was 0·15%. Thus, for the full model, 384 individuals would have to be tested to find one undiagnosed PLWH. For model I and model II, 681 and 441 individuals would have to be tested, respectively (Table 2a).
Table 2.
Confusion matrices and performance characteristics (GLMRidge algorithm).
| Table 2a: Algorithms calibrated to a sensitivity of 0·90 (high coverage - screening) | ||||||
| Age, sex and STIs | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 1,172 | 797,138 | 798,310 | Sensitivity | 0·911 (0·894 - 0·926) | |
| Test- | 115 | 613,489 | 613,604 | Specificity | 0·435 (0·434 - 0·436) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0015 (0·0014 - 0·0016) | |
| NPV | 0·9998 (0·9998 - 0·9998) | |||||
| Age, sex, origin of birth, educational attainment, marital status, place of residence and main source of income | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 1,108 | 487,880 | 488,988 | Sensitivity | 0·861 (0·841 - 0·879) | |
| Test- | 179 | 922,747 | 922,926 | Specificity | 0·654 (0·653 - 0·655) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0023 (0·0021 - 0·0024) | |
| NPV | 0·9998 (0·9998 - 0·9998) | |||||
| Age, sex, origin of birth, educational attainment, marital status, place of residence, main source of income and medical history | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 1,110 | 424,654 | 425,764 | Sensitivity | 0·862 (0·842 - 0·881) | |
| Test- | 177 | 985,973 | 986,150 | Specificity | 0·699 (0·698 - 0·700) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0026 (0·0025 - 0·0028) | |
| NPV | 0·9998 (0·9998 - 0·9998) | |||||
| Table 2b: Algorithms calibrated to a specificity of 0·999 (high risk population) | ||||||
| Age, sex and STIs | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 35 | 1,667 | 1,702 | Sensitivity | 0·027 (0·019 - 0·038) | |
| Test- | 1,252 | 1,408,960 | 1,410,212 | Specificity | 0·999 (0·999 - 0·999) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0206 (0·0144 - 0·0285) | |
| NPV | 0·9992 (0·9991 - 0·9992) | |||||
| Age, sex, origin of birth, educational attainment, marital status, place of residence and main source of income | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 77 | 1,042 | 1,119 | Sensitivity | 0·060 (0·048 - 0·074) | |
| Test- | 1,210 | 1,409,585 | 1,410,795 | Specificity | 0·999 (0·999 - 0·999) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0688 (0·0547 - 0·0853) | |
| NPV | 0·9992 (0·9991 - 0·9992) | |||||
| Age, sex, origin of birth, educational attainment, marital status, place of residence, main source of income and medical history | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 104 | 1,156 | 1,260 | Sensitivity | 0·081 (0·067 - 0·097) | |
| Test- | 1,183 | 1,409,471 | 1,410,654 | Specificity | 0·999 (0·999 - 0·999) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0825 (0·0679 - 0·0991) | |
| NPV | 0·9992 (0·9991 - 0·9992) | |||||
| Table 2c: Algorithms calibrated to maximal Youdens Index (Sensitivity + Specificity – 1) | ||||||
| Age, sex and STIs | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 952 | 424,326 | 425,278 | Sensitivity | 0·740 (0·715 - 0·763) | |
| Test- | 335 | 986,301 | 986,636 | Specificity | 0·699 (0·698 - 0·700) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0022 (0·0021 - 0·0024) | |
| NPV | 0·9997 (0·9997 - 0·9998) | |||||
| Age, sex, origin of birth, educational attainment, marital status, place of residence and main source of income | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 919 | 246,857 | 247,776 | Sensitivity | 0·714 (0·689 - 0·739) | |
| Test- | 368 | 1,163,770 | 1,164,138 | Specificity | 0·825 (0·824 - 0·826) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0037 (0·0035 - 0·004) | |
| NVP | 0·9997 (0·9996 - 0·9997) | |||||
| Age, sex, origin of birth, educational attainment, marital status, place of residence, main source of income and medical history | ||||||
| HIV+ | HIV- | Total | ||||
| Test+ | 981 | 211,973 | 212,954 | Sensitivity | 0·762 (0·738 - 0·785) | |
| Test- | 306 | 1,198,654 | 1,198,960 | Specificity | 0·850 (0·849 - 0·850) | |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0046 (0·0043 - 0·0049) | |
| NPV | 0·9997 (0·9997 - 0·9998) |
The tables depict confusion matrices and actual sensitivities, specificities, positive predictive values (PPVs) and negative predictive values (NPVs) of the best performing GLMridge model. The models are calibrated according to the risk score that yields the desired value in the training data, i.e. when sensitivities are calculated on the validation set the actual sensitives and specificities may differ slightly.
When we calibrated the models to a specificity of 99·9%, model III had the highest sensitivity (8·1%). The specificity did not differ substantially between the three models (Table 2b).
When we calibrated the models with regards to the highest Youden's Index, model III outperformed the other models in all metrics. Thus, sensitivity was 76·2%, 74% and 71%, specificity was 85·0%, 82·5% and 69·9% and PPV was 0·46%, 0·37% and 0·22% for model III, II and I, respectively. Thus, for the full model, 226 individuals would have to be tested in order to find one undiagnosed PLWH. For model I and model II, 446 and 269 individuals would have to be tested, respectively (Table 2c).
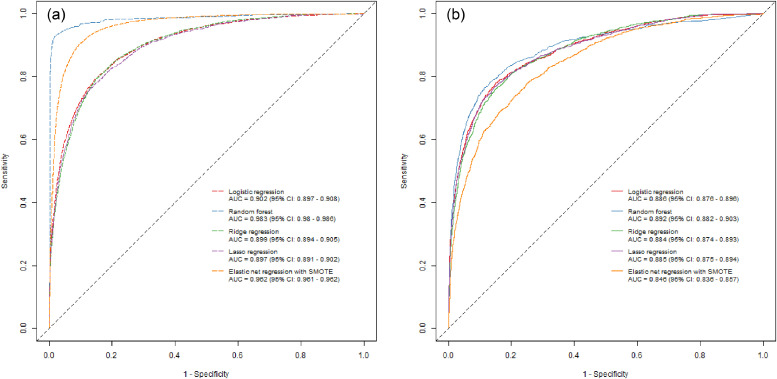
When we compared different machine learning algorithms, we found that the random forest algorithm performed slightly better with an AUC of 0·892 (95% CI: 0·882 − 0·903), however, the CIs where overlapping with the other machine learning algorithms. Regardless, the improved performance of the random forest could indicate that some covariate interactions are present. The logistic regression with elastic net penalty and SMOTE to balance the data set performed worst with an AUC of 0·846 (95% CI: 0·836 − 0·857). When we calibrated the model from the random forest algorithm to different sensitivities and specificities the actual sensitivities and specificities in the validation data was substantially different from what they were calibrated to, i.e. the actual sensitivity in the validation data set was 37·5%, and the specificity was 95·4% (Table 3). This was not the case for the other machine learning algorithms (GLMlasso, GLMridge and simple logistic regression). When we calibrated the final models from the different machine learning algorithms to a sensitivity of 90% we found that the ridge regression performed best, and differed highly from all other algorithms (McNemars test: P-value < 0·001) except the simple logistic regression from which we found no statistically significant difference (McNemars test: P-value 0·494). When we compared the models from the machine learning algorithms (GLMlasso and GLMridge) with a simple logistic regression, we found that the simple logistic regression model had performance characteristics that did not differ substantially from any of the machine learning algorithms (Fig. 2).
Table 3.
Confusion matrices and performance characteristics of different algorithms.
| 3a: simple logistic regression algorithm sensitivity of 0·90 (high coverage - screening) | |||||
| HIV+ | HIV- | Total | |||
| Test+ | 1,107 | 424,879 | 425,986 | Sensitivity | 0·860 (0·840 - 0·879) |
| Test- | 180 | 985,748 | 985,928 | Specificity | 0·699 (0·698 - 0·700) |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·026 (0·0024 - 0·0027) |
| NPV | 0·9998 (0·9998 - 0·9998) | ||||
| 3b: Random forest algorithm sensitivity of 0·90 (high coverage - screening) | |||||
| HIV+ | HIV- | Total | |||
| Test+ | 483 | 68,763 | 69,246 | Sensitivity | 0·375 (0·349 - 0·402) |
| Test- | 804 | 1,408,960 | 1,411,016 | Specificity | 0·954 (0·953 - 0·954) |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0070 (0·0063 - 0·0067) |
| NPV | 0·9994 (0·9994 - 0·9995) | ||||
| 3c: Lasso regression algorithm sensitivity of 0·90 (high coverage - screening) | |||||
| HIV+ | HIV- | Total | |||
| Test+ | 1,120 | 439,787 | 440,907 | Sensitivity | 0·870 (0·851 - 0·888) |
| Test- | 167 | 970,840 | 971,007 | Specificity | 0·688 (0·687 - 0·689) |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0025 (0·0023 - 0·0027) |
| NPV | 0·9998 (0·9998 - 0·9999) | ||||
| 3d: Ridge regression algorithm sensitivity of 0·90 (high coverage - screening) | |||||
| HIV+ | HIV- | Total | |||
| Test+ | 1,110 | 424,654 | 425,764 | Sensitivity | 0·860 (0·840 - 0·879) |
| Test- | 177 | 985,973 | 986,150 | Specificity | 0·699 (0·698 - 0·700) |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0026 (0·0025 - 0·0028) |
| NPV | 0·9998 (0·9998 - 0·9998) | ||||
| 3e: Elastic net penalty regression (α = 0.992) algorithm with Synthetic minority oversampling (SMOTE) and sensitivity of 0·90 (high coverage - screening) | |||||
| HIV+ | HIV- | Total | |||
| Test+ | 723 | 124,770 | 425,764 | Sensitivity | 0·562 (0·534 - 0·589) |
| Test- | 564 | 1,285,857 | 986,150 | Specificity | 0·912 (0·911 - 0·912) |
| Total | 1,287 | 1,410,627 | 1,411,914 | PPV | 0·0058 (0·0054 - 0·0062) |
| NPV | 0·9996 (0·9995 - 0·9996) |
The tables depict confusion matrices and actual sensitivities, specificities, positive predictive values (PPVs) and negative predictive values (NPVs) of four different algorithms. The models are calibrated according to the risk score that yields the desired value in the training data, i.e. when sensitivities are calculated on the validation set the actual sensitives and specificities may differ slightly.
Fig. 2.
Training and validation sample ROC curves using different machine learning algorithms. The figure shows ROC curves for the training (2a) and validation data (2b) of simple logistic regression, best performing random forest algorithm, the best performing GLMridge algorithm, the best performing GLMLasso algorithm and the best performing logistic regression with elastic net penalty and synthetic minority oversampling technique (SMOTE) prior to analyses.
We assessed whether there was a change in performance of the model over the years (1995–2016) which did not seem to be the case. However, we did see minor fluctuations in sensitivity and specificity as depicted in supplementary Figure S1.
Discussion
The first of the United Nations’ (UN) 90–90–90 targets to end the HIV epidemic is for 90% of PLWH to know their HIV status. Early diagnosis is a prerequisite for achieving these targets. We examined whether machine learning algorithms could learn from existing electronic registry data and help to predict HIV status. Our study shows that such an approach is feasible and that the algorithms had a fairly high accuracy for the prediction of HIV status.
Routine HIV testing of individuals presenting with certain conditions, which may indicate the presence of HIV, has been implemented in European countries guidance on HIV testing [20]. These indicator conditions are defined as conditions in which the prevalence of HIV is >0·1%, i.e. the level at which testing is assumed to be cost-effective [7]. Although this strategy may have facilitated early diagnosis of PLWH across Europe[21], testing rates of individuals presenting with indicator conditions remain low in many settings [22,23], and the number of diagnosed PLWH in Europe is ranging from 28%−98% with only three countries meeting the first “90″ of the UN's targets [24]. Moreover, a significant number of PLWH do not experience an HIV indicator condition in the years prior to diagnosis of HIV [25,26]. Research aimed at developing complementary risk stratification tools are therefore needed.
To our knowledge, this is the first study to apply machine learning methods on nation-wide electronic registry data for the prediction of HIV status. We have shown that this method may help identify undiagnosed PLWH and potentially individuals at increased risk of acquiring HIV. Moreover, the properties of the models seemed to be favorable, and although PPVs was generally low (e.g. 0·26% in the full model where algorithms were calibrated to a high sensitivity and 8·3% in the full model where algorithms were calibrated to a high specificity), this PPV was comparable to currently available manual HIV risk prediction tools which also included very detailed information about sexual preferences and illicit drug use [27]. The difference between model 2 and model 3 were smaller than expected, ie. A fairly high discriminatory value can be achieved by including demographics only, indicating that these variables are very important to include in models that guide clinicians with regards to HIV-testing. However, we did see a substantial difference in that a total number of 441 and 384, respectively, would have to be tested in order to find one PLWH when models were calibrated to a sensitivity of 90%.
There are several barriers to HIV testing including professional barriers (e.g. lack of knowledge of underlying risk factors), and personal barriers (e.g. not disclosing underlying risks due to fear of stigma, discrimination or prosecution) [28]. The present algorithms do not rely on specific information about sexual orientation, whether the individual has a partner with HIV, from a country with high HIV prevalence, or is an intravenous drug user, in which cases a testing may be recommended per se[1]. On the other hand, incorporating such information into the algorithms may potentially improve the discriminative properties of the algorithm.
The use of machine learning algorithms is a growing trend in HIV research. Thus, it has been used to predict progression of HIV and ART resistance and for ART optimization [10], [11], [12]. Recent studies have also shown that machine learning algorithms may be used for surveillance, including prediction of new HIV diagnoses and AIDS incidence based on internet search data [29,30], and there is early evidence that social media data may be used to study parameters such as HIV prevention, testing, and treatment efforts [31]. Our study adds to this growing field. However, whether such algorithms have any potential when applied in a real world setting needs to be further studied. We envision that algorithms could be integrated into clinical systems at hospitals, clinics or in primary care, e.g. by triggering a prompt to offer an HIV test to a given individual. One study evaluated a prototype application which prompted clinicians to add an HIV test when other laboratory tests selected suggested that the patient was at higher risk of HIV infection [32]. This application was found to be both feasible and acceptable among the clinicians. Future work should work toward improving these algorithms, and do external validation of currently available algorithms. This may include exploring other machine learning techniques, redefining or including new predictors and exploring interactions in the present models. Previous studies have suggested that behavioral information such as sexual preference and illicit drug use is useful for HIV prediction[30] and may improve performance of these models further. Future studies should try and make such extensions. However, in our nation-wide setting some of these extensions will prove difficult, e.g. we do not have access to behavioral data or clinical measures of disease for all people living in Denmark.
Certain ethical issues may arise when using individual characteristics for clinical prediction including the ability of an individual to consent for the use of their information [33]. Implementation of algorithms into clinical practice should take such issues into account and any benefits should outweigh the risks of harm to patients. As previously discussed, the present algorithms do not include information about substance abuse, sexual orientation or gender minority status which may be more acceptable to patients than if such information was to be included
There are several limitations to this study. First, the algorithms assume that PLWH and uninfected individuals are classified correctly. This is not the case as an estimated number of 600/5,800,000 (∼0·01%) individuals in Denmark are undiagnosed PLWH[13], under the assumption that undiagnosed individuals with HIV resemble the PLWH in the current study, the sensitivity and specificity of the models should not change, however PPV would decrease and NPV would increase. For obvious reason, we do not know the medical history of those PLWH that are currently undiagnosed. Second, our algorithm, in its current form, does not have universal applicability. The type and extent of electronic registry data will inevitably vary by country, the regions with the highest number of undiagnosed PLWH may not have access to this type of data. Moreover, there may be some issues in relation to the use of ICD codes due to potential systematic biases in their use, and the distribution of risk factors may vary from country to country. Also we only included data from the secondary health care sector, the majority of health care is provided in the primary sector, so exploring possibilities with data from primary health care would be valuable as and some variables look promising e.g. frequency of visits [34]. Furthermore, although Danish registry data provides a unique data capture of all individuals living in Denmark, the quality and completeness of the data may be impeded by a few factors. We only included PLWH diagnosed after 1995 which represents the date the Danish HIV Cohort Study was initiated. Moreover, there may be a slight loss to follow up (<1%?) and we may not have complete information about past medical history from immigrants. Moreover, we have not yet studied implementation and we are unaware of the potential epidemiological impact or the cost-effectiveness of introducing such an approach. Thus, the aim of this work is not to convey that these algorithms is a standalone strategy, or should replace existing HIV identification strategies, such as indicator-guided HIV testing, but rather to present a novel and complementary method for early HIV identification. Future studies may try to compare the combined effect of algorithmic prediction and indicator condition guided testing. Even in the case that algorithmic prediction would only provide a little additional benefit, future studies in the field may reveal novel insights in the risk characterization of PLWH. The strength of the study includes a nation-wide population-based design including PLWH and uninfected individuals with identical electronic registry data from well-validated and comprehensive registries.
In conclusion, machine learning algorithms can learn from nation-wide electronic registry data and help to identify undiagnosed PLWH with a fairly high level of accuracy. Moreover, these algorithms may help identify individuals who may be suitable for PrEP. Future studies are needed to further evaluate the usefulness and effects of these algorithms and should be aimed toward improving the algorithms. This may include exploring other machine learning techniques to improve model performance (e.g. models intrinsically capable of hand When we calibrated the final models ling non-linearities of which neural nets (plain or more advanced) or gradient boosting could be starting points; models catering to the time-series nature of the problem (e.g. survival analysis); or models based on deeper analysis of the covariance structure), redefining or including new predictors and exploring interactions in the present models.
Declaration of Competing Interest
MGA: No conflicts of interest. AR: No conflicts of interest. LHO: No conflicts of interest. SV: This co-author has taken up full-time employment with the pharmaceutical company Novo Nordisk A/S during the final stages of manuscript preparation. NO: No conflicts of interest.
Acknowledgments
This work was supported by the Preben and Anne Simonsens Foundation, the Novo Nordisk Foundation, Rigshospitalet, Copenhagen University, the Danish AIDS Foundation, the Augustinus Foundation, and the Danish Health Foundation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eclinm.2019.10.016.
Appendix. Supplementary materials
References
- 1.https://ecdc.europa.eu/sites/portal/files/documents/20171127-Annual_HIV_Report_Cover%2BInner.pdf. Accessed January 10, 2019.
- 2.Nakagawa F., Lodwick R.K., Smith C.J. Projected life expectancy of people with HIV according to timing of diagnosis. AIDS. 2012;26:335–343. doi: 10.1097/QAD.0b013e32834dcec9. [DOI] [PubMed] [Google Scholar]
- 3.Hall H.I., Holtgrave D.R., Maulsby C. HIV transmission rates from persons living with HIV who are aware and unaware of their infection. AIDS. 2012;26:893–896. doi: 10.1097/QAD.0b013e328351f73f. [DOI] [PubMed] [Google Scholar]
- 4.Sanders G.D., Bayoumi A.M., Sundaram V. Cost-effectiveness of screening for HIV in the era of highly active antiretroviral therapy. N Engl J Med. 2005;352:570–585. doi: 10.1056/NEJMsa042657. [DOI] [PubMed] [Google Scholar]
- 5.Lundgren J.D., Babiker A.G., Gordin F. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med. 2015;373:795–807. doi: 10.1056/NEJMoa1506816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Group T.A.S., Danel C., Moh R. A trial of early antiretrovirals and isoniazid preventive therapy in Africa. N Engl J Med. 2015;373:808–822. doi: 10.1056/NEJMoa1507198. [DOI] [PubMed] [Google Scholar]
- 7.Yazdanpanah Y., Sloan C.E., Charlois-Ou C. Routine HIV screening in France: clinical impact and cost-effectiveness. PLoS ONE. 2010;5:e13132. doi: 10.1371/journal.pone.0013132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan A.K., Raben D., Reekie J. Feasibility and effectiveness of indicator condition-guided testing for HIV: results from HIDES I (HIV indicator diseases across Europe study) PLoS ONE. 2013;8:e52845. doi: 10.1371/journal.pone.0052845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.http://www.hiveurope.eu/Portals/0/Documents/Guidance.pdf.pdf?ver=2014-01-29-113626-000. Accessed January 10, 2019.
- 10.Singh Y. Machine learning to improve the effectiveness of ANRS in predicting HIV drug resistance. Healthc Inform Res. 2017;23:271–276. doi: 10.4258/hir.2017.23.4.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riemenschneider M., Heider D. Current approaches in in computational drug resistance prediction in HIV. Curr HIV Res. 2016;14:307–315. doi: 10.2174/1570162x14666160321120232. [DOI] [PubMed] [Google Scholar]
- 12.Zazzi M., Cozzi-Lepri A., Prosperi M.C. Computer-aided optimization of combined anti-retroviral therapy for HIV: new drugs, new drug targets and drug resistance. Curr HIV Res. 2016;14:101–109. doi: 10.2174/1570162x13666151029102254. [DOI] [PubMed] [Google Scholar]
- 13.https://www.ssi.dk/Aktuelt/Nyhedsbreve/EPI-NYT/2017/Uge%2036%20-%202017.aspx. Accessed January 10, 2019.
- 14.http://www.dst.dk/extranet/uddannelsesklassifikation/DISCED-15.pdf. Accessed January 10, 2019.
- 15.Omland L.H., Legarth R., Ahlström M.G., Sørensen H.T., Obel N. Five-year risk of HIV diagnosis subsequent to 147 hospital-based indicator diseases: a Danish nationwide population-based cohort study. Clin Epidemiol. 2016;8:333–340. doi: 10.2147/CLEP.S101288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obel N., Reinholdt H., Omland L.H. Retrivability in the Danish national hospital registry of HIV and hepatitis B and C coinfection diagnoses of patients managed in HIV centers 1995-2004. BMC Med Res Methodol. 2008;8:25. doi: 10.1186/1471-2288-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou H.H. Journal of the Royal Statistical Society, series B (statistical methodology) Wiley; Chichester: 2008. Trevor. regularization and variable selection via the elastic net; pp. 301–320. [Google Scholar]
- 18.Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 19.R Development Core Team . R Foundation for Statistical Computing. Vienna, Austria; 2008. R: a language and environment for statistical computing. [Google Scholar]
- 20.Lazarus J.V., Hoekstra M., Raben D. The case for indicator condition-guided HIV screening. HIV Med. 2013;14:445–448. doi: 10.1111/hiv.12022. [DOI] [PubMed] [Google Scholar]
- 21.Scognamiglio P., Chiaradia G., De Carli G. The potential impact of routine testing of individuals with HIV indicator diseases in order to prevent late HIV diagnosis. BMC Infect Dis. 2013;13:473. doi: 10.1186/1471-2334-13-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elmahdi R., Gerver S.M., Gomez Guillen G., Fidler S., Cooke G., Ward H. Low levels of HIV test coverage in clinical settings in the U.K.: a systematic review of adherence to 2008 guidelines. Sex Transm Infect. 2014;90:119–124. doi: 10.1136/sextrans-2013-051312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruutel K., Lemsalu L., Latt S., TbH Opt. Monitoring HIV-indicator condition guided HIV testing in Estonia. HIV Med. 2018;19:47–51. doi: 10.1111/hiv.12586. [DOI] [PubMed] [Google Scholar]
- 24.Porter K., Gourlay A., Attawell K. Substantial heterogeneity in progress toward reaching the 90-90-90 HIV target in the who European region. J Acquir Immune Defic Syndr. 2018;79:28–37. doi: 10.1097/QAI.0000000000001761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joore I.K., Twisk D.E., Vanrolleghem A.M. The need to scale up HIV indicator condition-guided testing for early case-finding: a case-control study in primary care. BMC Fam Pract. 2016;17:161. doi: 10.1186/s12875-016-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joore I.K., Arts D.L., Kruijer M.J. HIV indicator condition-guided testing to reduce the number of undiagnosed patients and prevent late presentation in a high-prevalence area: a case-control study in primary care. Sex Transm Infect. 2015;91:467–472. doi: 10.1136/sextrans-2015-052073. [DOI] [PubMed] [Google Scholar]
- 27.Smith D.K., Pals S.L., Herbst J.H., Shinde S., Carey J.W. Development of a clinical screening index predictive of incident HIV infection among men who have sex with men in the United States. J Acquir Immune Defic Syndr. 2012;60:421–427. doi: 10.1097/QAI.0b013e318256b2f6. [DOI] [PubMed] [Google Scholar]
- 28.Bond K.T., Frye V., Taylor R. Knowing is not enough: a qualitative report on HIV testing among heterosexual African-American men. AIDS Care. 2015;27:182–188. doi: 10.1080/09540121.2014.963009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q., Chai Y., Li X., Young S.D., Zhou J. Using internet search data to predict new HIV diagnoses in China: a modelling study. BMJ Open. 2018;8 doi: 10.1136/bmjopen-2017-018335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nan Y., Gao Y. A machine learning method to monitor China's AIDS epidemics with data from Baidu trends. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0199697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young S.D., Yu W., Wang W. Toward automating HIV identification: machine learning for rapid identification of HIV-related social media data. J Acquir Immune Defic Syndr. 2017;74:128–131. doi: 10.1097/QAI.0000000000001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chadwick D.R., Hall C., Rae C. A feasibility study for a clinical decision support system prompting HIV testing. HIV Med. 2017;18:435–439. doi: 10.1111/hiv.12472. [DOI] [PubMed] [Google Scholar]
- 33.Cato K.D., Bockting W., Larson E. Did I tell you that? ethical issues related to using computational methods to discover non-disclosed patient characteristics. J Empir Res Hum Res Ethics. 2016;11(3):214–219. doi: 10.1177/1556264616661611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Iguacel R., Pedersen C., Llibre J.M. Primary health care: an opportunity for early identification of people living with undiagnosed HIV infection. HIV Med. 2019;20(6):404–417. doi: 10.1111/hiv.12735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.