Abstract
CD8+ T cells play a central role in antitumour immunity, which often exhibit ‘exhaustion’ in the setting of malignancy and chronic viral infection due to T cell immunoglobulin and mucin domain 3 (TIM3) and myeloid‐derived suppressor cells (MDSCs). Our team previously found that overactive MDSCs and exhausted TIM3+CD8+ T cells were observed in myelodysplastic syndromes (MDS) patients. However, it is not obvious whether MDSCs suppress CD8+ T cells through TIM3/Gal‐9 pathway. Here, Gal‐9, as the ligand of TIM3, was overexpressed in MDSCs. The levels of Gal‐9 in bone marrow supernatants, serum and culture supernatants of MDSCs from MDS patients were elevated. CD8+ T cells from MDS or normal controls produced less perforin and granzyme B and exhibited increased early apoptosis after co‐culture with MDSCs from MDS. Meanwhile, the cytokines produced by CD8+ T cells could be partially restored by TIM3/Gal‐9 pathway inhibitors. Furthermore, CD8+ T cells produced less perforin and granzyme B after co‐culture with excess exogenous Gal‐9, and the function of CD8+ T cells was similarly restored by TIM3/Gal‐9 pathway inhibitors. Expression of Notch1, EOMES (associated with perforin and granzyme B secretion), p‐mTOR and p‐AKT (associated with cell proliferation) was decreased in CD8+ T cells from MDS after co‐culture with excess exogenous Gal‐9. These suggested that MDSCs might be the donor of Gal‐9, and TIM3/Gal‐9 pathway might be involved in CD8+ T cells exhaustion in MDS, and that TIM3/Gal‐9 pathway inhibitor might be the promising candidate for target therapy of MDS in the future.
Keywords: CD8+ T cells, exhaustion, myelodysplastic syndrome
1. INTRODUCTION
Myelodysplastic syndromes (MDS) are malignant bone marrow disorders characterized by ineffective haematopoiesis and high risk to transformation into acute myeloid leukaemia. Increased malignant clones and cellular immunodeficiency might contribute to the pathogenesis of MDS.1, 2, 3, 4, 5 Cytotoxic T lymphocytes (CTLs) play a central role in tumour immunity.6 Adoptive T cell immunotherapy in neoplastic disorders has been restricted by CD8+ T cells ‘exhaustion’ due to some immune checkpoint inhibitors, such as T cell immunoglobulin and mucin domain 3 (TIM3), programmed death‐1 (PD‐1), cytotoxic T lymphocyte antigen 4 (CTLA‐4) and lymphocyte‐activation gene 3 (LAG3).7, 8
Among these immune checkpoints, the interaction of TIM3 with its ligand galectin‐9 (Gal‐9) promotes Tc1 cells9 apoptosis, CD8+T cells exhaustion,10, 11 malignant cell proliferation12 and myeloid‐derived suppressor cells (MDSCs) amplification.13 In humans, MDSCs are usually defined as CD14−CD11b+ cells or the cells which express the myeloid marker (CD33) and are lack of lineage maturation markers and HLA‐DR14 (Lin−HLA‐DR−CD33+ cells). In previous research, increased Lin−HLA‐DR−CD33+ MDSCs in MDS patients were observed, and this cell population was obviously separated by flow cytometry than other labelled cells. We also found that CD8+ T cells could be inhibited by MDSCs, which were ‘crazy’ in MDS patients. CD8+ T cells with less INF‐γ overexpressed TIM3 in MDS patients. Further, TIM3+CD8+ T cells produce less perforin and granzyme B.1, 2 The hypothesis that CD8+ T cells exhaustion might be induced by MDSCs through TIM3/Gal‐9 pathway drove us to detect Gal‐9 expression in MDSCs, and the level of perforin and granzyme B in CD8+ T cells after co‐culture with MDSCs or excess exogenous Gal‐9 or TIM3/Gal‐9 pathway inhibitors. We also explored Notch1, eomesodermin (EOMES), phospho‐mTOR (p‐mTOR) and phospho‐AKT (p‐AKT) proteins, which are related to the function and proliferation of CD8+ T cells,15, 16 to verify that TIM3/Gal‐9 pathway might be involved in CD8+ T cells exhaustion induced by MDSCs in MDS patients.
2. MATERIALS AND METHODS
2.1. Patients
Fifty‐two treatment‐naive Chinese MDS patients (32 males and 20 females, median age 64, Table 1) who were newly diagnosed according to the WHO classification17 in the Hematology Department of Tianjin Medical University General Hospital from January 2017 to August 2019 were enrolled, including patients with MDS with single lineage dysplasia (MDS‐SLD, n = 4), MDS with multilineage dysplasia (MDS‐MLD, n = 4), MDS‐RS (MDS with ring sideroblasts) with single lineage dysplasia (MDS‐RS‐SLD, n = 3), MDS‐RS with multilineage dysplasia (MDS‐RS‐MLD, n = 5), MDS with isolated del (5q) (n = 2), MDS with excess blasts 1 (MDS‐EB‐1, n = 20), MDS with excess blasts 2 (MDS‐EB‐2, n = 13) and MDS unclassifiable (MDS‐U, n = 1). Thirty‐one healthy volunteers (mean age 44.48 ± 14.87) were enrolled. There was no difference in the sex ratio or age between MDS patients and normal healthy volunteers (P > .05).
Table 1.
Clinical features of MDS patients were studied here
| Sample | Sex | Age | WHO subtype | Blasts of BM (%) | Cytogenetics | IPSS |
|---|---|---|---|---|---|---|
| 001 | Female | 61 | MDS‐EB‐1 | 6 | 7p‐,18p‐ | 2 |
| 002 | Male | 65 | MDS‐MLD | 1 | (‐) | 0.5 |
| 003 | Male | 55 | MDS‐EB‐2 | 16.5 | (‐) | 2 |
| 004 | Male | 67 | MDS‐U | 1 | ‐7 | 1.5 |
| 005 | Male | 68 | MDS‐EB‐2 | 14.5 | 8q‐,‐5,‐7 | 3 |
| 006 | Female | 29 | MDS‐MLD | 1 | (‐) | 0 |
| 007 | Female | 52 | MDS‐EB‐1 | 5 | (‐) | 0 |
| 008 | Male | 66 | MDS‐RS‐MLD | 0.5 | (‐) | 0.5 |
| 009 | Female | 74 | MDS‐EB‐2 | 17 | 5q‐,7q‐,+8 | 3 |
| 010 | Male | 63 | MDS‐EB‐1 | 7 | 8q‐ | 1.5 |
| 011 | Female | 65 | MDS‐EB‐1 | 5.5 | 5q‐,7q‐,+8 | 2 |
| 012 | Male | 68 | MDS‐MLD | 4 | 7q‐,‐7 | 1.5 |
| 013 | Male | 35 | MDS‐RS‐SLD | 1 | (‐) | 0 |
| 014 | Female | 63 | MDS‐EB‐1 | 7.5 | (‐) | 1 |
| 015 | Female | 65 | MDS with del (5q) | 0.5 | 5q‐,+8 | 0.5 |
| 016 | Female | 52 | MDS‐EB‐2 | 16 | (‐) | 2.0 |
| 017 | Female | 72 | MDS‐RS‐MLD | 0 | (‐) | 0.5 |
| 018 | Male | 30 | MDS‐EB‐1 | 7.5 | 20q‐ | 2.0 |
| 019 | Male | 64 | MDS‐EB‐2 | 11 | (‐) | 2.0 |
| 020 | Male | 60 | MDS‐EB‐1 | 5.5 | (‐) | 1 |
| 021 | Male | 54 | MDS‐EB‐2 | 19 | +8 | 2.5 |
| 022 | Male | 52 | MDS‐EB‐2 | 12 | (‐) | 2 |
| 023 | Male | 61 | MDS‐EB‐1 | 9 | (‐) | 1.0 |
| 024 | Female | 45 | MDS‐EB‐1 | 6.5 | +8 | 2.0 |
| 025 | Male | 26 | MDS‐SLD | 1 | (‐) | 0 |
| 026 | Female | 66 | MDS‐MLD | 0 | (‐) | 0.5 |
| 027 | Male | 52 | MDS‐EB‐2 | 11 | (‐) | 2 |
| 028 | Female | 48 | MDS‐EB‐2 | 16 | (‐) | 2 |
| 029 | Male | 64 | MDS‐EB‐1 | 5 | (‐) | 1 |
| 030 | Male | 76 | MDS‐EB‐1 | 5 | (‐) | 1.0 |
| 031 | Male | 59 | MDS‐EB‐1 | 9 | (‐) | 1.0 |
| 032 | Male | 55 | MDS‐EB‐1 | 5 | (‐) | 1.0 |
| 033 | Female | 59 | MDS‐EB‐1 | 8 | t(2;12)(p12;p12) | 1.5 |
| 034 | Female | 67 | MDS‐EB‐1 | 6.5 | (‐) | 1.0 |
| 035 | Female | 68 | MDS‐RS‐MLD | 4 | (‐) | 0.5 |
| 036 | Male | 84 | MDS‐RS‐MLD | 0 | (‐) | 0.5 |
| 037 | Male | 65 | MDS‐EB‐1 | 5.5 | (‐) | 1 |
| 038 | Male | 44 | MDS‐SLD | 1 | (‐) | 0 |
| 039 | Male | 64 | MDS‐EB1 | 5.5 | (‐) | 1 |
| 040 | Male | 71 | MDS‐RS‐MLD | 3 | 7q‐ | 1.5 |
| 041 | Male | 39 | MDS‐EB1 | 7 | (‐) | 0.5 |
| 042 | Female | 67 | MDS‐RS‐SLD | 2 | (‐) | 0 |
| 043 | Male | 62 | MDS‐EB2 | 16 | (‐) | 2 |
| 044 | Male | 69 | MDS‐SLD | 3 | 7q‐,20q‐ | 1.5 |
| 045 | Female | 66 | MDS‐EB2 | 18 | 5q‐,20q‐ | 2 |
| 046 | Male | 68 | MDS‐RS‐SLD | 1 | 7q‐ | 1 |
| 047 | Female | 73 | MDS‐EB1 | 7 | (‐) | 1 |
| 048 | Male | 65 | MDS‐EB2 | 15.5 | (‐) | 2 |
| 049 | Male | 74 | MDS‐EB1 | 9 | (‐) | 1 |
| 050 | Male | 62 | MDS‐EB2 | 16 | (‐) | 2 |
| 051 | Female | 81 | MDS with del (5q) | 1 | 5q‐ | 1 |
| 052 | Female | 59 | MDS‐SLD | 0 | ‐7 | 1.5 |
The study was approved by the Ethics Committee of Tianjin Medical University General Hospital. Informed written consents were obtained from all the patients and normal controls following the Declaration of Helsinki.
2.2. Flow cytometry analysis
2.2.1. Membrane markers and intracellular markers detection
One hundred microlitres of peripheral blood and bone marrow samples from MDS patients and healthy volunteers were placed in heparin‐containing anticoagulant tubes. The specimens were incubated with 15 µL of Lin/HLA‐DR/CD33 and their isotype control antibodies at 4°C for 30 minutes. After incubation, erythrocytes were lysed with 2 mL of erythrolysin for 10 minutes, centrifuged at 400 g for 5 minutes and washed twice with phosphate buffered saline (PBS). After permeabilizing the cell membrane using an IntraSure Kit (BD Biosciences), 5 μL galectin‐9 monoclonal antibody was added to the cells, incubated for 20 minutes at 4°C in the dark and washed twice with PBS. Finally, 5 × 105 cells per tube were detected by flow cytometry. After CD8+ T cells and MDSCs were co‐cultured, they were co‐incubated with CD3/CD8 antibodies in the same way. For intracellular staining, the samples were incubated with perforin and granzyme B antibodies after permeabilizing the cell membrane. The phenotype of MDSCs was analysed for the cell surface markers Lin, HLA‐DR and CD33. Intracellular expression of galectin‐9 was determined. Perforin or granzyme B expression in co‐cultured CD8+ T cells was analysed. All data were collected on a flow cytometer (Beckman Coulter), and the results were analysed with Kaluza software (Beckman Coulter).
The labelled antibodies included CD3‐APC (SK7, BD Biosciences), CD3‐PE (SK7, BD Biosciences), CD8‐FITC (SK1, BD Biosciences), TIM3‐APC (7D3, BD Biosciences), Lin‐FITC (BD Biosciences), HLA‐DR‐PerCP (L243, BD Biosciences), CD33‐APC (WM53, BD Biosciences), galectin‐9‐PE (9M1‐3, BD Biosciences), perforin‐PE (δG9, BD Biosciences) and granzyme B‐PE (GB11, BD Biosciences). The above antibodies were added as described by the manufacturer.
2.2.2. Detection of apoptosis
An apoptosis assay (FITC Annexin V Apoptosis Detection Kit I, BD Biosciences) was used to detect apoptosis of CD8+T cells co‐cultured with MDSCs. The cells were washed twice with cold PBS and were resuspended in 1× binding buffer at a concentration of 1 × 106 cells/mL. Then, 100 µL of the solution (1 × 105 cells) was transferred to a 5‐mL culture tube, and 5 µL of FITC Annexin V and 5 µL of PI were added. The cells were gently vortexed and incubated for 15 minutes at room temperature (25°C) in the dark. Finally, 400 µL of 1× binding buffer was added to each tube. Analysis was performed by flow cytometry (Beckman Coulter).
2.3. Sorting CD8+ T cells and MDSCs
Ten millilitres of fresh peripheral blood or bone marrow was obtained from MDS patients or normal controls (NC). Human CD8+ immunomagnetic bead solution (Miltenyi Biotec) (50 µL) was added to mononuclear cells, which were obtained by Ficoll gradient centrifugation. The samples were incubated for 15 minutes at 4°C and washed once with buffer, and the suspension cells were passed through the MS column in the magnetic field. Then, the column was removed from the magnet and 1 mL of wash buffer was added to the top of the column and the plunger (in the same package as the column) was immediately used to force the buffer through the column. The collected cells were used for the subsequent experiments.
Ten millilitres of bone marrow was obtained from MDS patients, and red blood cells were lysed with lysing solution (BD Biosciences) and washed with PBS; and then, 40 µL of Lin\HLA‐DR\CD33 antibodies were added to label the surface markers of the MDSCs. The cells were co‐incubated for 30 minutes at 4°C in the dark and washed with PBS. The cells were collected by the FACS Aria II (BD Biosciences).
2.4. Cells culture
2.4.1. Culture MDSCs
MDSCs were sorted by flow cytometry and then cultured with 10% foetal bovine serum (FBS) (containing 60 mg/L penicillin and 100 mg/L streptomycin) (Gibco) in the presence of 50 ng/mL recombinant human (rh) granulocyte‐monocyte colony‐stimulating factor (GM‐CSF) (PeproTech Inc) at 37°C in a 5% CO2 incubator. The culture supernatants were collected after 48 hours for ELISA.
2.4.2. Co‐culture of CD8+ T cells with MDSCs
To investigate whether MDSCs affect the function of CD8+ T cells via the TIM3/Gal‐9 pathway, 105 CD8+ T cells and MDSCs (>95% Lin−HLA‐DR−CD33+ cells) at a ratio of 1:2 were supplemented with 10% foetal bovine serum (FBS) (containing 60 mg/L penicillin and 100 mg/L streptomycin) and co‐cultured at 37°C in a 5% CO2 incubator for 24‐48 hours. The effect of MDSCs on CD8+ T cells was analysed by adding 10 µg/mL TIM3 inhibitor (F38‐2E2, BioLegend) and 10 µg/mL Gal‐9 inhibitor (9M1‐3, BioLegend). The expression of perforin and granzyme B in CD8+ T cells was detected by flow cytometry, and the apoptosis of CD8+ T cells was assessed by using an Apoptosis Kit (BD Biosciences).
2.4.3. Co‐culture of CD8+ T cells with exogenous Gal‐9
CD8+ T cells selected by magnetic beads were co‐cultured with 2 ng/mL exogenous recombinant human Gal‐9 (R&D Systems) with 10% FBS (containing 60 mg/L penicillin and 100 mg/L streptomycin) at 37°C in a 5% CO2 incubator for 24‐48 hours. Then, the expression of perforin, granzyme B and pathway‐related proteins in CD8+ T cells were detected with or without TIM3/Gal‐9 inhibitors.
2.5. Proliferation assay
CD8+ T cells from normal controls were stained with 2.5 μmol/L carboxyfluorescein diacetate succinimidyl ester (CFSE, BD Biosciences) for 10 minutes in a 37°C water bath and washed with 9× the original volume of PBS. The cells were pelleted by centrifugation, the supernatant was decanted, and 10 mL of complete medium with 10% FBS was added to stop the reaction. CFSE‐labelled CD8+ T cells were cultured with 1 µg/mL anti‐CD3 (OKT, eBioscience), 1 µg/mL anti‐CD28 (CD28.2, eBioscience) and 20 U/mL rhIL‐2 (R&D Systems) with or without MDSCs from MDS. The proliferation of CD8+ T cells in NC was determined by measuring CFSE fluorescence using flow cytometry.
2.6. Real‐time PCR
Expression of the Gal‐9 gene in MDSCs was detected by real‐time quantitative PCR. Purified MDSCs from MDS patients and normal controls were lysed using TRIzol reagent (Invitrogen Life Technologies). RNA was reverse‐transcribed using a DNA Synthesis Kit according to the manufacturer's protocol (TAKARA). The primers are listed in Table 2. Reactions were performed using the Bio‐Rad iQ5 Real‐Time System and the respective SYBR Green I kit (TAKARA). The mRNA level of Gal‐9 was normalized to that of β‐actin, and the relative expression of mRNA was calculated by the 2−ΔΔCt method.
Table 2.
Primer sequence of Gal‐9 and β‐actin in PCR
| Target gene | Primer sequences | Annealing temperature (°C) |
|---|---|---|
| Gal‐9 |
F:5′‐CGGTTTGAAGATGGAGGGTA‐3′ R:5′‐CAGGAAGCAGAGGTCAAAGG‐3′ |
59 |
| β‐Actin |
F:5′‐TTGCCGACAGGATGCAGAA‐3′ R:5′‐GCCGATCCACACGGAGTACT‐3′ |
56 |
Abbreviations: F, forward primer; R, reverse primer.
2.7. Enzyme‐linked immunosorbent assay (ELISA)
Bone marrow supernatants, serum and culture supernatants of MDSCs from MDS patients and normal controls were harvested. The levels of Gal‐9 were measured by using a Human Galectin‐9 ELISA Kit (KAMIYA Biomedical Company) according to the manufacturer's instructions.
2.8. Western blot analysis
CD8+ T cells cultured alone and co‐cultured with exogenous Gal‐9 were collected and lysed in RIPA buffer supplemented with complete protease inhibitors and phosphatase inhibitors (Roche). Protein levels were quantified using a BCA Kit. Proteins were separated by electrophoresis on 4%‐12% precast gels (Bio‐Rad Laboratories) and transferred to nitrocellulose membranes (Millipore Corp). The membranes were blocked in 5% skimmed milk (BD Biosciences) or 5% bovine serum albumin (BSA) (Solarbio) and then incubated overnight at 4°C with primary antibodies against Notch1, p‐mTOR, p‐AKT, EOMES and GAPDH. After washing the membranes 3 times in TBST (Tris‐HCl containing NaCl and Tween 20), the membranes were incubated with the relevant horseradish peroxidase–conjugated secondary antibodies (1:5000 dilution, Cell Signaling Technology). A summary of the primary antibodies mentioned above is provided in Table 3. The reaction was detected using Super ECL Plus Detection Reagent. Protein levels were normalized to GAPDH.
Table 3.
Primary antibodies
| Antibodies | Species | Type | Dilution | Source |
|---|---|---|---|---|
| Notch1 | Rabbit | Monoclonal IgG | 1:1000 | CST |
| p‐mTOR | Rabbit | Monoclonal IgG | 1:1000 | CST |
| p‐AKT | Rabbit | Monoclonal IgG | 1:2000 | CST |
| EOMES | Rabbit | Monoclonal IgG | 1:1000 | CST |
| GAPDH | Rabbit | Monoclonal IgG | 1:1000 | CST |
Abbreviations: CST, Cell Signaling Technology (Danvers, MA, USA); EOMES, eomesodermin; p‐AKT, phospho‐AKT; p‐mTOR, phospho‐mTOR.
2.9. Statistical analysis
Prism statistical software (v7.00; GraphPad Software, Inc, La Jolla, CA, USA) was applied for data analysis. Statistical differences between two groups were analysed using Student's t test. One‐way ANOVA was used to compare three groups or more. All data are presented as the mean ± standard error of the mean (normal distribution data) or median (25% percentile‐75% percentile) (non‐normal distribution data).
3. RESULTS
3.1. Galectin‐9 was highly expressed in MDSCs, and Gal‐9 was also elevated in bone marrow supernatants, serum and culture supernatants of MDSCs from MDS patients compared to those from normal controls
Gal‐9 is highly expressed on tumour cells and Th1 cells. The expression of Gal‐9 in MDSCs in MDS is still not obvious. Therefore, we measured the expression of Gal‐9 in MDSCs (Lin−HLA‐DR−CD33+ cells) by using flow cytometry and RT‐PCR. The median percentage of Gal‐9 in MDSCs from MDS patients (n = 29, samples: 001‐031, except 005 and 018) was significantly higher than that in NC (17.21% (8.74%‐35.77%) vs 2.61% (1.19%‐3.39%), P < .0001****), but there was no difference between the low‐risk and high‐risk groups (16.86% (7.15%‐39.19%) vs 20.72% (10.74%‐36.35%), P = .7148) (Figure 1A‐C). Gal‐9 mRNA in MDSCs from MDS patients (n = 12, samples: 001, 004‐007, 016, 018, 024, and 031‐034) was higher than that in NC (3.49 (2.09‐12.57) vs 0.99 (0.50‐2.50), P < .05*) (Figure 1D).
Figure 1.
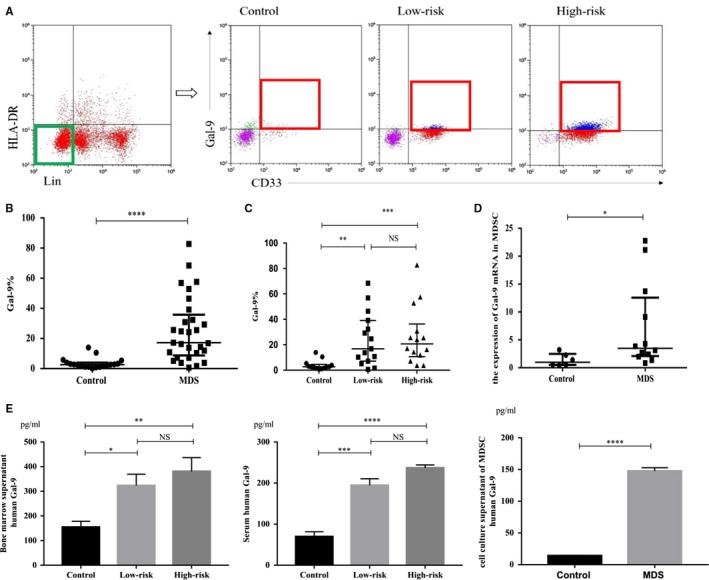
The expression of Gal‐9 in MDSCs. (A) The cells in the green rectangle are Lin−HLA‐DR− cells, and the cells in the red rectangle are Gal‐9+Lin−HLA‐DR−CD33+ cells. Representative flow cytometry scatter diagrams of Gal‐9 expression in Lin−HLA‐DR−CD33+ cells are shown in control, low‐risk or high‐risk MDS patients. UR upper right quadrant, LR lower right quadrant. Gal‐9+Lin−HLA‐DR−CD33+ cells/Lin−HLA‐DR−CD33+ cells = UR/(UR + LR). (B and C) The dot diagrams present the percentage of Gal‐9+Lin−HLA‐DR−CD33+ cells/Lin−HLA‐DR−CD33+ cells in MDS patients (including low‐risk and high‐risk MDS patients) and in controls. (D) Gal‐9 mRNA in MDSCs by quantitative RT‐PCR. (E) Gal‐9 in bone marrow supernatants, serum and MDSC culture supernatants. *P < .05, **P < .01, ***P < .001, ****P < .0001, NS = not significant
The levels of Gal‐9 in bone marrow supernatants, serum (n = 15, samples: 038‐052) and culture supernatants of MDSCs (n = 3, samples: 048, 049, 050) from both low‐risk and high‐risk MDS were significantly higher than those in NC (bone marrow supernatant: 323.6 ± 42.6 pg/mL vs 381 ± 55.98 pg/mL vs 154.9 ± 23.72 pg/mL, P = .0042**; serum: 194.5 ± 16.13 pg/mL vs 237.1 ± 7.18 pg/mL vs 69.81 ± 11.93 pg/mL, P < .0001****; culture supernatants of MDSCs: 147.7 ± 5.24 pg/mL vs 14.2 ± 0.55 pg/mL, P < .0001****) (Figure 1E), but there was no difference between the low‐risk and high‐risk groups. These data suggested that MDSCs might release Gal‐9 into the bone marrow microenvironment and exert immunosuppressive effects.
3.2. Perforin, granzyme B and proliferation of CD8+ T cells in normal controls decreased after co‐cultivation with MDSCs, while the early apoptosis of CD8+ T cells increased
To demonstrate the inhibitory effects of MDSCs on immune cells, we co‐cultured CD8+ T cells from NC with MDSCs from MDS patients and analysed perforin and granzyme B (killer cytokine of CD8+ T cells) by flow cytometry. The expression of perforin and granzyme B in CD8+ T cells from NC co‐cultured with MDSCs from MDS patients (n = 9, samples: 038‐046) was lower than that of CD8+ T cells from NC that were cultured alone (perforin: 16.02 ± 5.86% vs 22.02 ± 5.96%, P = .0271*; granzyme B: 20.1% (14.08%‐37.12%) vs 31.68% (24.17%‐42.32%), P = .0605) (Figure 2A,B). The early apoptosis of CD8+ T cells from NC co‐cultured with MDSCs from MDS patients (n = 8, samples: 039‐046) was significantly higher than that of CD8+ T cells from NC that were cultured alone (38.66 ± 7.91% vs 16.18 ± 3.84%, P = .0075**) (Figure 2C). In order to examine whether the MDSCs in MDS could inhibit the proliferation of CD8+ T cells in NC, CD8+ T cells from NC were labelled with CFSE and subsequently cultured in the presence or absence of MDSCs from MDS patients (n = 5, samples: 045‐049). The mean fluorescence intensity (MFI) of CFSE in the co‐cultured group showed a slight increase compared with that of the CD8+ T cells cultured alone (953075 (533581‐4433061) vs 757364 (330311‐1596830), P > .05) (Figure 2D); there was an upward trend in co‐cultured group as seen in the flow cytogram, although there was no significant difference(Figure 2E).
Figure 2.
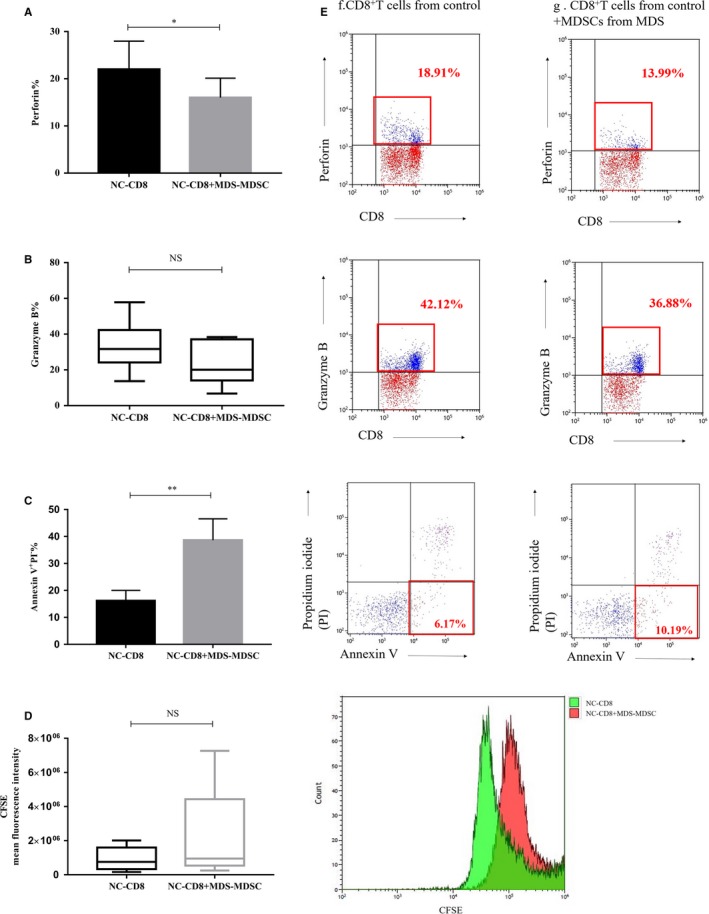
(A‐D) The histogram represents perforin, granzyme B, early apoptosis and the MFI of CFSE in CD8+ T cells from normal controls (NC‐CD8) co‐cultured with MDSCs from MDS patients (MDS‐MDSC) or cultured alone. (E) The cells in the red rectangle represent the expression of perforin, granzyme B and early apoptosis of CD8+ T cells from NC that were cultures alone or co‐cultures with MDSCs from MDS patients. The red flow histogram represents proliferation of CFSE‐labelled NC‐CD8 co‐cultured with MDS‐MDSC, and the green histogram represents NC‐CD8 cultured alone. *P < .05, **P < .01, NS = not significant
3.3. Perforin, granzyme B and early apoptosis of CD8+ T cells from MDS after co‐cultivation with MDSCs
3.3.1. Perforin and granzyme B decreased in CD8+ T cells from MDS patients after co‐cultivation with MDSCs, which was partially restored by TIM3/Gal‐9 pathway inhibitors
We hypothesized that MDSCs might suppress CD8+ T cells through TIM3/Gal‐9 signalling pathway. Therefore, we purified CD8+ T cells from MDS patients and co‐cultured them with MDSCs, F38‐2E2 (TIM3 inhibitor) and 9M1‐3 (Gal‐9 inhibitor) respectively. When the concentration of inhibitor was 10 µg/mL, an obvious effect was observed (Figure 3D). The median perforin and granzyme B in group b were significantly lower compared to that of groups a, c, d and e in 14 samples (numbers: 003‐005, 024‐028 and 035‐040) (perforin%: b: 10.76 ± 1.22% vs a: 19.18 ± 2.37% vs c: 16.68 ± 1.33% vs d: 16.35 ± 1.49% vs e: 16.23 ± 1.61%, P < .0001****; granzyme B%: b: 9.35% (4.02%‐24.21%) vs a: 21.43% (14.59%‐46.90%) vs c: 12.94% (6.52%‐29.45%) vs d: 12.11% (5.20%‐40.99%) vs e: 13.66% (7.18%‐36.79%), P < .0001****) (Figure 3A‐C). The inhibitors had no effect on CD8+ T cells cultured alone (Figure 3E).
Figure 3.
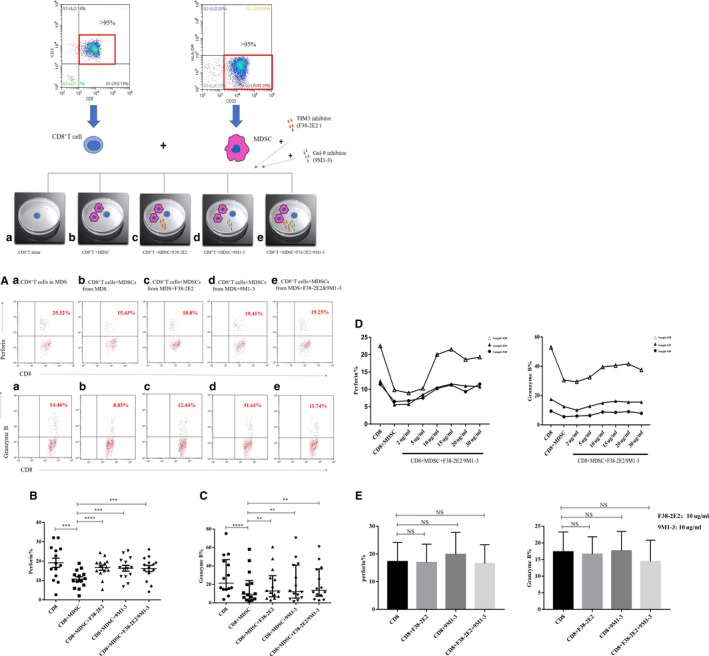
Perforin and granzyme B in CD8+ T cells from MDS patients after co‐culture with MDSCs or TIM3/Gal‐9 inhibitors. a: CD8+ T cells alone; b: CD8+ T cells co‐cultured with MDSCs; c: CD8+ T cells+MDSCs+TIM3 inhibitor (F38‐2E2); d: CD8+ T cells+MDSCs+Gal‐9 inhibitor (9M1‐3); e: CD8+ T cells+MDSCs+F38‐2E2+9M1‐3. (A) The upper right quadrant represents the expression of perforin and granzyme B in CD8+ T cells from MDS patients in the five groups. (B) The expression of perforin in the five groups. (C) The expression of granzyme B in the five groups. (D) Effects of different concentrations (2 µg/mL, 5 µg/mL, 10 µg/mL, 15 µg/mL, 20 µg/mL and 30 µg/mL) of inhibitors on perforin and granzyme B expression in CD8+ T cells in three patients (sample 038, 039, and 040). (E) Perforin and granzyme B expression in CD8+ T cells that were cultured without MDSCs in the presence of inhibitors. *P < .05, **P < .01, ***P < .001, ****P < .0001, NS = not significant
3.3.2. The early apoptosis of CD8+ T cells increased after co‐cultivation with MDSCs and was partially restored by blocking the TIM3/Gal‐9 pathway
We detected early apoptosis of CD8+ T cells from 11 MDS patients (numbers: 003‐005, 027‐031 and 038‐040) after co‐culture with MDSCs and found that early (Annexin V+ PI−) apoptosis was higher than that of CD8+ T cells alone. Furthermore, early apoptosis was reduced after co‐culture with the TIM3 inhibitor (F38‐2E2) and Gal‐9 inhibitor (9M1‐3) (early apoptotic cells%: b: 22.04 ± 5.72% vs a: 16.7 ± 4.64% vs c: 17.63 ± 4.67% vs d: 17.43 ± 4.85% vs e: 17.15 ± 4.66%, P = .0002***) (Figure 4A,B).
Figure 4.
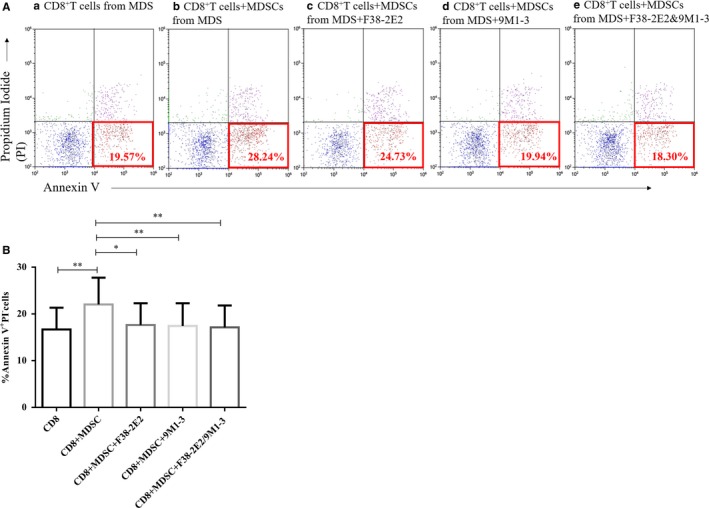
Early apoptosis of CD8+ T cells from MDS patients after co‐culture with MDSCs or TIM3/Gal‐9 inhibitors. (A) Scattered dots in the red rectangle represent early apoptosis of CD8+ T cells from MDS patients in the five co‐culture systems. The a. b. c. d. and e. experimental groups represent CD8+ T cells from MDS, CD8+ T cells+MDSCs from MDS, CD8+ T cells+MDSCs+F38‐2E2, CD8+ T cells+MDSCs+9M1‐3 and CD8+ T cells+MDSCs+F38‐2E2+9M1‐3, respectively. (B) The histogram represents early apoptotic cells. *P < .05, **P < .01, NS = not significant
3.4. Decreased perforin and granzyme B in CD8+ T cells after co‐cultivation with excess exogenous Gal‐9
We speculated that MDSCs might be a source of Gal‐9 due to its overexpression; therefore, we took excess exogenous Gal‐9 in the place of MDSCs and co‐cultured CD8+ T cells with excess exogenous Gal‐9 (recombinant human Gal‐9 [R&D Systems]) and TIM3/Gal‐9 pathway inhibitors.
The perforin and granzyme B in CD8+ T cells purified from MDS patients (n = 9, samples: 026‐028, 034, 036, 037 and 045‐047) were significantly lower after co‐cultivation with excess exogenous Gal‐9 than those in CD8+ T cells alone and were partially restored by TIM3 inhibitor or Gal‐9 inhibitor (perforin%: i: 15.33 ± 2.83% vs a: 21.79 ± 3.35% vs j: 20.63 ± 3.37% vs k: 19.01 ± 3.49%, P < .0001****; granzyme B%: i: 15.95 ± 5.47% vs a: 21.32 ± 6.56% vs j: 20.18 ± 6.44% vs k: 18.38 ± 6.07%, P = .0019**) (Figure 5A‐C).
Figure 5.

Perforin and granzyme B in CD8+ T cells from MDS patients after co‐culture with excess exogenous Gal‐9 or TIM3/Gal‐9 inhibitors. (A) Flow cytometry scatter diagrams of perforin and granzyme B expression in CD8+ T cells from MDS. a: CD8+ T cells alone; i: CD8+ T cells with excess exogenous Gal‐9; j: CD8+ T cells from MDS with excess exogenous Gal‐9 and F38‐2E2; and k: CD8+ T cells from MDS with excess exogenous Gal‐9 and 9M1‐3. (B and C) The histogram represents the expression of perforin and granzyme B in the four groups. *P < .05, **P < .01, ***P < .001, NS = not significant
3.5. Notch1, EOMES, p‐mTOR and p‐AKT decreased in CD8+ T cells after co‐cultivation with excess exogenous Gal‐9
We further explored the mechanism of CD8+T cell suppression by excess exogenous Gal‐9. The transcriptional regulator EOMES plays a critical role in the development and maturation of CTL. Notch1 regulates EOMES by directly binding to the promoter of key effector molecules and then regulates perforin and granzyme B via EOMES. The expression of Notch1 and EOMES in CD8+ T cells (n = 9, samples: 027‐037, except 035,036) treated with excess exogenous Gal‐9 was significantly reduced compared to those of CD8+ T cells alone (Notch1: 5.01 ± 0.98% vs 1.7 ± 0.24%, P < .01**; EOMES: 34.81 ± 7.96% vs 26.37 ± 6.65%, P < .05*) (Figure 6A).
Figure 6.
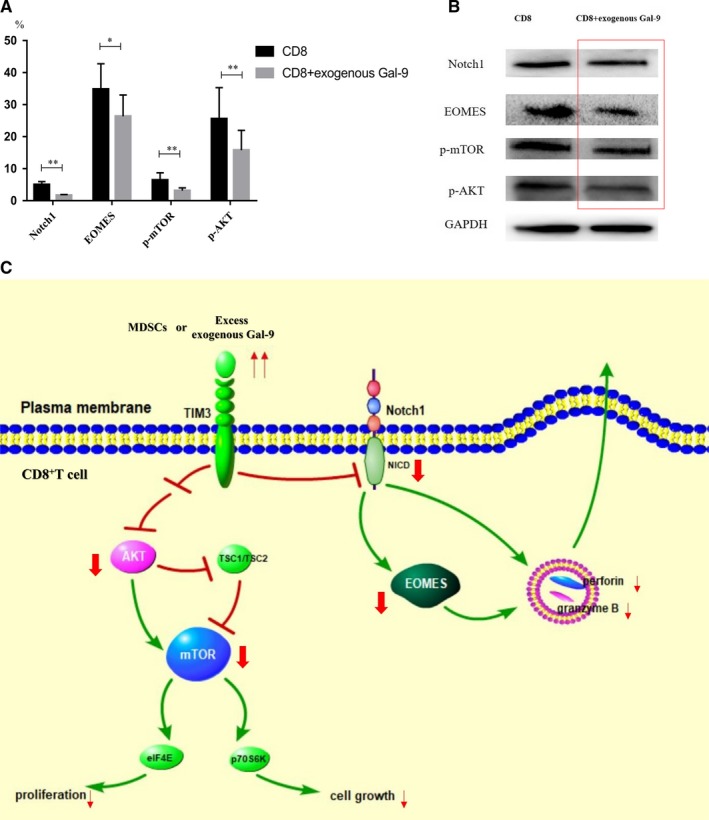
The expression levels of Notch1, EOMES, phospho‐mTOR and phospho‐AKT proteins were reduced in CD8+ T cells after co‐culture with exogenous Gal‐9. (A) The histogram representing the expression of Notch1, EOMES, p‐mTOR and p‐AKT by flow cytometry. (B) The expression of Notch1, EOMES, p‐mTOR and p‐AKT by Western blotting. (C) The possible pathogenesis of CD8+ T cells from MDS patients' suppression by MDSCs or excess exogenous Gal‐9. *P < .05; **P < .01
The mammalian target of rapamycin (mTOR) is a serine/threonine (Thr)‐protein kinase that plays a key role in regulating cell growth and proliferation. The expression of p‐mTOR and p‐AKT in CD8+ T cells (n = 9, samples: 027‐037, except 035,036) treated with excess exogenous Gal‐9 was lower compared to those of CD8+ T cells alone (p‐mTOR: 6.44 ± 2.30% vs 3.15 ± 0.88%, P < .01**; p‐AKT: 25.57 ± 9.73% vs 15.78 ± 6.21%, P < .01**, Figure 6A). A downward trend in Notch1, EOMES, p‐mTOR and p‐AKT was observed in CD8+ T cells after co‐cultivation with excess exogenous Gal‐9 by Western blotting (Figure 6B).
4. DISCUSSION
T cell exhaustion, a state of T cell dysfunction characterized by diminished cytokine production and elevated apoptosis, was first characterized in the settings of chronic lymphocytic choriomeningitis virus (LCMV) infection.18 T cell exhaustion could also occur in viral infections such as hepatitis C virus and HIV19, 20 as well as in tumour models11, 21 and haematological malignant diseases.22 The exhausted T cells highly express immune checkpoints, such as TIM3, PD‐1 and CTLA‐4.6, 23 Blocking TIM3 and PD‐1 can reverse T cell exhaustion and dysfunction in malignant tumour patients21, 24, 25 or neoplastic mice.26 These data suggested that CD8+ T cells exhaustion in human or mice could be induced by immune checkpoints in the setting of chronic viral infection or malignant diseases.
Increasing malignant clones and cellular immunity defects are involved in the pathogenesis of MDS,1, 2, 3, 4, 5 which is a malignant haematological disease. Daver et al27 found that the efficacy of hypomethylating agent combination with immune checkpoint inhibitors (PD‐1 inhibitor) in MDS was better than hypomethylating agents alone. TIM3, as one of the important immunity checkpoints, interacts with its ligand Gal‐924 could promote Tc1 cell9 apoptosis, CD8+T cell exhaustion,10, 11 malignant cell proliferation12 and MDSC proliferation.13 MDSCs, which are powerful cellular immunity suppressor, could badly inhibit the antitumour immune response mediated by the effector T cells and lead to tumour cells immunological surveillance escape.28, 29
Here, we found that Gal‐9 was also highly expressed in MDSCs from MDS patients and was increasing in the bone marrow supernatant, serum and MDSC culture supernatants. These data suggested that MDSCs could secrete Gal‐9 into the microenvironment and exert an immunosuppressive effect. Hayashi et al30 found that inflammatory pathway proteins that regulate immunity were moderately elevated in both low‐risk and high‐risk MDS patients but were much lower than that in real infectious diseases. These suggested that cellular immunity was insufficient for malignant clonal cells due to cellular immunity tolerance, both in low‐risk and high‐risk MDS groups. We found that there was no difference of Gal‐9 in MDSCs of high‐risk and low‐risk MDS patients, which was consistent with our hypothesis. Therefore, further exploration of the molecular mechanism of Gal‐9 up‐regulation is needed.
Perforin and granzyme B are the most important cytokines in CD8+ T cells against infectious and malignant cells. MDSCs can inhibit CD8+ T cells in esophageal squamous cell carcinoma.8 Here, perforin, granzyme B and proliferation decreased and the early apoptosis of CD8+ T cells increased after CD8+ T cells from controls were co‐cultured with MDSCs. These data implied that MDSCs with excessive Gal‐9 might induce TIM3 overexpression in CD8+ T cells, leading to a downward trend of perforin and granzyme B after CD8+ T cells from MDS were co‐cultured with MDSCs. We hypothesized that MDSCs might be a source of Gal‐9. Therefore, we replaced MDSCs with excessive exogenous Gal‐9 to repeat the above procedure and found that CD8+ T cells indeed produced less perforin and granzyme B after co‐cultivation with excessive exogenous Gal‐9, similar to that after co‐cultivation with MDSCs from MDS patients. The secretion of perforin and granzyme B by CD8+ T cells could be partially restored by TIM3/Gal‐9 pathway inhibitors.
Meanwhile, the early apoptosis of CD8+ T cells after co‐culture with MDSCs was higher than that of CD8+ T cells alone and could be partially restored by TIM3/Gal‐9 pathway inhibitors. It suggested that CD8+ T cells exhaustion might be induced by MDSC through TIM3/Gal‐9 pathway.
The T‐box transcription factor eomesodermin (EOMES) plays an important role in the development and maturation of CTL.31 Notch1 could regulate perforin and granzyme B by affecting EOMES.15, 32 Here, we found that Notch1 and EOMES decreased after co‐cultivation with excessive exogenous Gal‐9. It indicated that the TIM3/Gal‐9 pathway might induce CD8+T cells ‘exhaustion’ by down‐regulating Notch1 and EOMES.
Mammalian target of rapamycin (mTOR) is a serine/threonine (Thr)‐protein kinase that plays a central role in regulating cell growth and proliferation.33 Overactivation of AKT/mTOR after phosphorylation is associated with over‐proliferation of leukaemia cells,16, 34 as well as tumour cells such as liver cancers and pancreatic neuroendocrine tumours.35, 36 Our study found that p‐mTOR and p‐AKT decreased in CD8+ T cells after co‐culture with exogenous Gal‐9, indicating that TIM3/Gal‐9 pathway might suppress CD8+ T cells through down‐regulating AKT/mTOR (Figure 6C).
In conclusion, CD8+ T cells ‘exhaustion’ could be induced by up‐regulating Gal‐9 in MDSCs and TIM3 in CD8+ T cells of MDS patients, which was partially restored by TIM3/Gal‐9 pathway inhibitors. TIM3/Gal‐9 pathway may be involved in CD8+ T cells exhaustion induced by MDSCs in MDS, leading to malignant MDS clone immunological surveillance escape and over‐proliferation. Therefore, TIM3/Gal‐9 pathway inhibitors might be the promising candidate for target therapy of MDS in the future.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHORS' CONTRIBUTIONS
Tao Jinglian and Han Dong contributed equally to performing research, analysing the data and writing the manuscript; Shao Zonghong, Li Lijuan and Fu Rong designed research, and ensured correct analysis of the data, and contributed to the writing of the manuscript; and Gao Shan, Zhang Wei, Yu Hong and Liu Pei assisted in the designing of research, overseeing the collection of the data and contributed to the writing of the manuscript.
ACKNOWLEDGEMENTS
This study was sponsored by Young National Natural Science Fund of China (Grant Number: 81500101), National Natural Science Fund of China (Grant Number: 81570111), Young National Natural Science Fund of China (Grant Number: 81800123) and Tianjin Applied Basic Research Project Fund (Grant Number: 18JCYBJC91700).
Tao J, Han D, Gao S, et al. CD8+ T cells exhaustion induced by myeloid‐derived suppressor cells in myelodysplastic syndromes patients might be through TIM3/Gal‐9 pathway. J Cell Mol Med. 2020;24:1046–1058. 10.1111/jcmm.14825
Jinglian Tao and Dong Han are co‐first authors. Fu Rong, Lijuan LI and Zonghong Shao are co‐corresponding authors.
DATA AVAILABILITY STATEMENT
More detailed data of this study are available from the corresponding author upon request.
REFERENCES
- 1. Tao J, Li L, Wang Y, Fu R, Wang H, Shao Z. Increased TIM3+CD8+T cells in Myelodysplastic Syndrome patients displayed less perforin and granzyme B secretion and higher CD95 expression. Leuk Res. 2016;51:49‐55. [DOI] [PubMed] [Google Scholar]
- 2. Tao JL, Li LJ, Fu R, et al. Elevated TIM3+ hematopoietic stem cells in untreated myelodysplastic syndrome displayed aberrant differentiation, overproliferation and decreased apoptosis. Leuk Res. 2014;38:714‐721. [DOI] [PubMed] [Google Scholar]
- 3. Jiang H, Fu R, Wang H, Li L, Liu H, Shao Z. CD47 is expressed abnormally on hematopoietic cells in myelodysplastic syndrome. Leuk Res. 2013;37:907‐910. [DOI] [PubMed] [Google Scholar]
- 4. Jan M, Chao MP, Cha AC, et al. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc Natl Acad Sci USA. 2011;108:5009‐5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Asayama T, Tamura H, Ishibashi M, et al. Functional expression of Tim‐3 on blasts and clinical impact of its ligand galectin‐9 in myelodysplastic syndromes. Oncotarget. 2017;8:88904‐88917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Severson JJ, Serracino HS, Mateescu V, et al. PD‐1+Tim‐3+ CD8+ T lymphocytes display varied degrees of functional exhaustion in patients with regionally metastatic differentiated thyroid cancer. Cancer Immunol Res. 2015;3:620‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Durgeau A, Virk Y, Corgnac S, Mami‐Chouaib F. Recent advances in targeting CD8 T‐cell immunity for more effective cancer immunotherapy. Front Immunol. 2018;9:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen X, Wang L, Li P, et al. Dual TGF‐beta and PD‐1 blockade synergistically enhances MAGE‐A3‐specific CD8(+) T cell response in esophageal squamous cell carcinoma. Int J Cancer. 2018;143(10):2561‐2574. [DOI] [PubMed] [Google Scholar]
- 9. Monney L, Sabatos CA, Gaglia JL, et al. Th1‐specific cell surface protein Tim‐3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415:536‐541. [DOI] [PubMed] [Google Scholar]
- 10. Sakuishi K, Jayaraman P, Behar SM, Anderson AC, Kuchroo VK. Emerging Tim‐3 functions in anti‐microbial and tumor immunity. Trends Immunol. 2011;32:345‐349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim HD, Song GW, Park S, et al. Association between expression level of PD1 by tumor‐infiltrating CD8(+) T cells and features of hepatocellular carcinoma. Gastroenterology. 2018;155:1936‐1950.e17. [DOI] [PubMed] [Google Scholar]
- 12. Kikushige Y, Miyamoto T, Yuda J, et al. A TIM‐3/Gal‐9 autocrine stimulatory loop drives self‐renewal of human myeloid leukemia stem cells and leukemic progression. Cell Stem Cell. 2015;17:341‐352. [DOI] [PubMed] [Google Scholar]
- 13. Sakuishi K, Jayaraman P, Behar SM, Anderson AC, Kuchroo VK. Emerging Tim‐3 functions in antimicrobial and tumor immunity. Trends Immunol. 2011;32:345‐349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng PH, Lee KY, Chang YL, et al. CD14(+)S100A9(+) monocytic myeloid‐derived suppressor cells and their clinical relevance in non‐small cell lung cancer. Am J Respir Crit Care Med. 2012;186:1025‐1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsukumo SI, Yasutomo K. Regulation of CD8(+) T cells and antitumor immunity by notch signaling. Front Immunol. 2018;9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalaitzidis D, Sykes SM, Wang Z, et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten‐loss‐evoked leukemogenesis. Cell Stem Cell. 2012;11:429‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391‐2405. [DOI] [PubMed] [Google Scholar]
- 18. Barber DL, Wherry EJ, Masopust D, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682‐687. [DOI] [PubMed] [Google Scholar]
- 19. Li H, Wu K, Tao K, et al. Tim‐3/galectin‐9 signaling pathway mediates T‐cell dysfunction and predicts poor prognosis in patients with hepatitis B virus‐associated hepatocellular carcinoma. Hepatology (Baltimore, MD). 2012;56:1342‐1351. [DOI] [PubMed] [Google Scholar]
- 20. Rallon N, Garcia M, Garcia‐Samaniego J, et al. HCV coinfection contributes to HIV pathogenesis by increasing immune exhaustion in CD8 T‐cells. PLoS ONE. 2017;12:e0173943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fourcade J, Sun Z, Benallaoua M, et al. Upregulation of Tim‐3 and PD‐1 expression is associated with tumor antigen‐specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. 2010;207:2175‐2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taghiloo S, Allahmoradi E, Tehrani M, et al. Frequency and functional characterization of exhausted CD8(+) T cells in chronic lymphocytic leukemia. Eur J Haematol. 2017;98:622‐631. [DOI] [PubMed] [Google Scholar]
- 23. Zhou G, Sprengers D, Boor PPC, et al. Antibodies against immune checkpoint molecules restore functions of tumor‐infiltrating T cells in hepatocellular carcinomas. Gastroenterology. 2017;153:1107‐1119.e10. [DOI] [PubMed] [Google Scholar]
- 24. Zhou Q, Munger ME, Veenstra RG, et al. Coexpression of Tim‐3 and PD‐1 identifies a CD8+ T‐cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117:4501‐4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu X, Liu J, Cui P, et al. Co‐inhibition of TIGIT, PD1, and Tim3 reverses dysfunction of Wilms tumor protein‐1 (WT1)‐specific CD8+ T lymphocytes after dendritic cell vaccination in gastric cancer. Am J Cancer Res. 2018;8:1564‐1575. [PMC free article] [PubMed] [Google Scholar]
- 26. Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim‐3 and PD‐1 pathways to reverse T cell exhaustion and restore anti‐tumor immunity. J Exp Med. 2010;207:2187‐2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daver N, Boddu P, Garcia‐Manero G, et al. Hypomethylating agents in combination with immune checkpoint inhibitors in acute myeloid leukemia and myelodysplastic syndromes. Leukemia. 2018;32:1094‐1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gorgun GT, Whitehill G, Anderson JL, et al. Tumor‐promoting immune‐suppressive myeloid‐derived suppressor cells in the multiple myeloma microenvironment in humans. Blood. 2013;121:2975‐2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu JF, Wu L, Yang LL, et al. Blockade of TIM3 relieves immunosuppression through reducing regulatory T cells in head and neck cancer. J Exp Clin Cancer Res: CR. 2018;37:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayashi Y, Zhang Y, Yokota A, et al. Pathobiological pseudohypoxia as a putative mechanism underlying myelodysplastic syndromes. Cancer Discov. 2018;8:1438‐1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu Y, Ju S, Chen E, et al. T‐bet and eomesodermin are required for T cell‐mediated antitumor immune responses. J Immunol. 2010;185:3174‐3183. [DOI] [PubMed] [Google Scholar]
- 32. Cho OH, Shin HM, Miele L, et al. Notch regulates cytolytic effector function in CD8+ T cells. J Immunol. 2009;182:3380‐3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu Y, Yoon SO, Poulogiannis G, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science (New York, NY). 2011;332:1322‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guezguez B, Almakadi M, Benoit YD, et al. GSK3 deficiencies in hematopoietic stem cells initiate pre‐neoplastic state that is predictive of clinical outcomes of human acute leukemia. Cancer Cell. 2016;29:61‐74. [DOI] [PubMed] [Google Scholar]
- 35. Adebayo Michael AO, Ko S, Tao J, et al. Inhibiting glutamine‐dependent mTORC1 activation ameliorates liver cancers driven by beta‐catenin mutations. Cell Metab. 2019;29(5):1135‐1150.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Umesalma S, Kaemmer CA, Kohlmeyer JL, et al. RABL6A inhibits tumor‐suppressive PP2A/AKT signaling to drive pancreatic neuroendocrine tumor growth. J Clin Investig. 2019;129(4):1641‐1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
More detailed data of this study are available from the corresponding author upon request.


