Abstract
Aim: To investigate the intestinal flora of nonalcoholic fatty liver disease (NAFLD) in Chinese children and adolescents using metagenomic approach.
Methods: All participants underwent magnetic resonance spectroscopy (MRS) to quantify liver fat content. Hepatic steatosis was defined as MRS proton density fat fraction (MRS-PDFF) >5%. A total of 58 children and adolescents were enrolled in this study, including 25 obese NAFLD patients, 18 obese non-NAFLD children, and 15 healthy children. Stool samples were collected and analyzed with metagenomics. We used Shannon index to reflect the alpha diversities of gut microbiota. Wilcoxon rank sum test and Kruskal-Wallis test were performed to evaluate alpha diversities between groups. At last, the differences of gut microbiota composition and functional annotations between obese with and without NAFLD and healthy children were assessed by Kruskal-Wallis test.
Results: Significant differences in gut microbiota composition and functional annotations among three groups of children and adolescents have been observed. Deep sequencing of gut microbiota revealed high abundance of phylum Proteobacteria (Gammaproteobacteria) in obese NAFLD patients, comparing with the control group. Overall, obese children without NAFLD had less abundant Helicobacter and Helicobacter pylori. Compared to the control group, in obese children with NAFLD, the abundance of Bacteroidetes (Alistipes) were significantly reduced. Faecalibacterium prausnitzii was the only species representing a difference between obese children with and without NAFLD. There were not significant differences in terms of alpha diversity among three groups. Functional annotations demonstrated that several pathways were differentially enriched between groups, including metabolism of other amino acids, replication and repair, folding, sorting, degradation, and glycan biosynthesis and metabolism.
Conclusion: Significantly differences are observed in gut microbiota composition and functional annotations between obese children with and without NAFLD in comparison to the healthy children group. The characteristic of gut microbiota in this study may contribute to a further understanding the gut-liver axis of pediatric NAFLD in China.
Keywords: nonalcoholic fatty liver disease, gut microbiota, metagenome, children and adolescents, obese
Introduction
Nonalcoholic fatty liver disease (NAFLD) has emerged as one of the most common chronic liver disorders in children and adolescents, affecting 5–10% of the general child population, and 28–41% of obese children (1). The term NAFLD is also known as a broad spectrum ranging from simple steatosis, nonalcoholic steatohepatitis (NASH), to hepatic cirrhosis. In addition to liver-related disorders, NAFLD is also revealed as a risk factor of chronic kidney disease, cardiovascular diseases, obesity, and diabetes (2–4). What is more, 20% of NASH cases may progress to hepatic cirrhosis, and eventually liver failure (5). Therefore, NAFLD is of great importance on public health, especially in children and adolescents.
The human intestinal tract is the main habitat of microbiota, with ~1014 bacteria existing, which comprises of 100 times more genes than humans. As a group of symbiotes, gut microbiota play an important role in metabolic and immune balance of human beings. First, gut microbiota participate in digestion and absorption of nutrients and production of vitamins and minerals. Second, gut microbiota can affect the production of intestinal hormones such as glucagon-like peptide-1, thereby affecting the metabolism of the host (6). In recent research, we learn that gut microbiota is closely related to the occurrence and development of NAFLD through Gut-liver axis (7, 8). Increasing evidence suggests that gut microbiota connect with hepatic steatosis in several ways (9–11): (1) it affects the appetite signal of the host; (2) it can also increase energy extraction from the intestine; (3) the metabolism of bile acids change, therefore affecting fats and lipid-vitamins obtained in the intestine; (4) the metabolism of choline is affected; (5) it also contributes to increased inflammation in host organisms; (6) intestinal bacterial overgrowth and intestinal permeability increase will lead to bacteria translate into the systemic circulation and endotoxemia.
As early as 2004, Backhed et al. first connected gut microbiota with obesity and NAFLD (12). They found that gut microbiota could not only affect the absorption and storage of energy, but also stimulate the production and infiltration of triglycerides in hepatocytes. Mouzaki et al. (11) diagnosed NAFLD with liver biopsy and further detected the composition of gut microbiota. They demonstrated that gut microbiota in healthy controls were varied from patients with simple fatty liver disease and NASH. The Bacteroides in NASH patients were significantly lower compared to that in simple fatty liver disease patients and healthy controls, and were independent from diet and BMI. While Raman et al. (13) and Li Fan et al. showed that compared with healthy controls, the differences in intestinal flora of NAFLD mainly occurred at family and genus levels, of Firmicutes (14).
In recent years, with increasing prevalence of obesity and NAFLD in children and adolescents, drawing more attention of researchers to these populations. Zhu et al. performed 16S RNA sequencings to analyze the composition of gut microbiota in Caucasian children in a controlled diet with NASH, obesity, and healthy controls in the United States (15). They suggested that alcohol-producing bacteria Escherichia was significantly increased in NASH children. Comparing with healthy controls, children with NASH and obesity had increased Bacteroidetes and decreased Firmicutes. The differences of gut microbiota between NASH patients and obese children were exhibited in Proteobacteria, Enterobacteriaceae, and Escherichia. Michail et al. (16) conducted metagenomics and proteomics to examine the composition, function, and metabolism of intestinal flora in obese children with and without NAFLD against the healthy children controls. Gammaproteobacteria, Prevotella and ethanol were observed to have a significant increase in NAFLD patients. In addition, there was an increase in the pathways involved in energy metabolism and lipid metabolism in children with NAFLD, compared with the healthy controls. Researchers in Italy (5) analyzed the gut microbiota in children with simple fatty liver disease, NASH, obesity, and healthy children but got a different result. Compared with healthy groups, Actinobacteria were increased in NAFLD children, while Bacteroidetes were decreased, which was contrary to the research by Zhu et al. (15). There was no significant difference in gut microbiota composition among the groups of children with simple fatty liver disease, NASH, and obesity, which was consistent with the previous study by Zhu et al.
Researchers have demonstrated that gut microbiota of NAFLD patients could undergo a compositional and functional change, but the differences of gut microbiota in obesity, NAFLD, and healthy children have no consistency. Furthermore, studies are rarely reported in the case of Chinese pediatric NAFLD. Therefore, the present study used metagenome to clarify the composition and function of gut microbiota in obese children with and without NAFLD against healthy children in China in order to unravel the relationship of liver-gut axis and its function in children NAFLD.
Materials and Methods
Subjects
The research was approved by the Human Ethics Committee of Shenzhen Children's Hospital (Supplemental Document Ethic Committee Approval). A total of 58 participants were recruited in the study during May 2017 to July 2018, including 43 obese [body mass index (BMI) above age- and gender-specific 95th percentile] and 15 healthy controls. The average age of participants is 13.7 years, ranging from 9 to 17 years. In view of their diagnosis in our previous study (17), children and adolescents were stratified into obesity with (n = 25) or without (n = 18) NAFLD, and matched healthy controls (n = 15). All subjects with antibiotics or probiotics history in the past 3 months were exclude. All authors of this paper had access to the study data and reviewed and approved the final manuscript.
Magnetic Resonance Spectroscopy (MRS)
Children and adolescents underwent single-voxel MRS scanning using 3.0 T MR unit (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) in accordance with previous research (17). Hepatic steatosis is defined as MRS proton density fat fraction (MRS-PDFF) > 5%.
Fecal Sample Collection, DNA Extraction and Metagenomic Sequencing
Before collecting, methods and notes were explained by researchers. Samples were collected using a sterile kit and frozen at −80°C immediately until detection.
All stool samples were analyzed in Beijing Genomics Institute (BGI, Shenzhen, China). Briefly, DNA was isolated from 58 fecal samples and an average of 5.92 Gb of sequence was obtained from each subject. All DNA fragments were purified by QIA quick PCR Purification Kit (Qiagen) during library construction. Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System were used to qualify and quantify the sample libraries. The qualified libraries were then sequenced using Illumina HiSeq System.
Metagenomic Analysis
In order to acquire accurate sequences, we removed sequences with a 90% similarity to the human genome. Then we assembled all samples to obtain reads which were more than 300 bp for further analysis. With CD-Hit software (18), genes were combined and clustered. Finally, we got 2,053,172 non-redundant genes. All these genes were blasted against with Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using the Diamond software (19) to obtain function annotations. The taxonomic composition was performed using MEGAN software (20) based on NR databases. Alpha diversity was calculated using Shannon index based on species profile.
Statistical Analysis
All data were statistically analyzed using SPSS 22.0 software and R environment. Numerical variables were firstly tested for normality. Age and BMI between three groups were compared by independent-samples T-test. The Chi-square test was performed for gender comparison between groups. The differences between groups in taxonomic composition and function annotations were determined using Kruskal-Wallis test. Alpha diversity was calculated based on Shannon index. Wilcoxon Rank-Sum test or Kruskal-Wallis test were used for diversity differential analysis, depending on group numbers.
Results
Clinical Characteristics
A total of 58 participants were enrolled in the present study with an average age of 13.7 ± 1.8 years (range 9–17 years). According to MRS examination, hepatic steatosis was diagnosed with MRS-PDFF > 5%. We divided all subjects into three groups based on liver fat content and BMI. Among them were obese NAFLD patients (n = 25), obese patients (n = 18), and healthy controls (n = 15). There was no difference in gender and age among these three groups. The basic information of these three groups is summarized in Table 1.
Table 1.
Characteristics of all subjects.
| Obese NAFLD (n = 25) | Obesity (n = 18) | Health controls (n = 15) | |
|---|---|---|---|
| Male | 19 (76.0%) | 12 (66.7%) | 10 (66.7%) |
| Female | 6 (24%) | 6 (33.3%) | 5 (33.3%) |
| Age (years) | 14.1 ± 2.1 | 13.9 ± 1.3 | 13.7 ± 2.0 |
| BMI (kg/m2) | 30.6 ± 3.4* | 28.6 ± 1.8* | 20.2 ± 1.9 |
| MRS-PDFF | 15.5% ± 8.8% | 2.8% ± 1.2% | 1.3% ± 0.26% |
P < 0.05 versus healthy controls.
Taxonomy Comparison of Gut Microbiota at Phylum Level
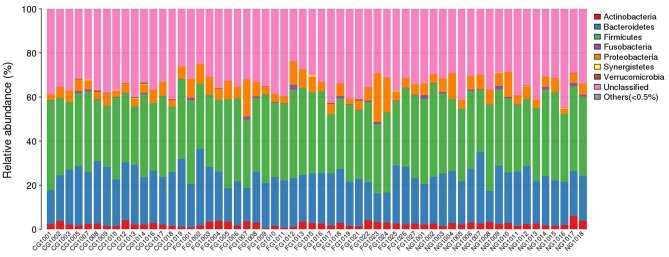
Taxonomic composition of all subjects was analyzed at phylum, class, order, family, genus, and species level. To obtain an accurate taxonomic change in all samples, abundant levels with <0.5% average abundance were classified into others. We observed significant differences between obese patients with and without NAFLD and healthy controls. At phylum level, we found that Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria were the dominant phyla in all samples (Figure 1). Comparing with the healthy controls, the abundance of Proteobacteria was statistically increased and Bacteroidetes was decreased in obese NAFLD children. In contract, Firmicutes and Actinobacteria did not show large variations in three groups of children (Figure 2).
Figure 1.
Phylum distribution of gut microbiota of all subjects. The distribution of bacterial phyla (abundance >0.5%) of each individual is presented as bar chart in terms of percentage weight.
Figure 2.
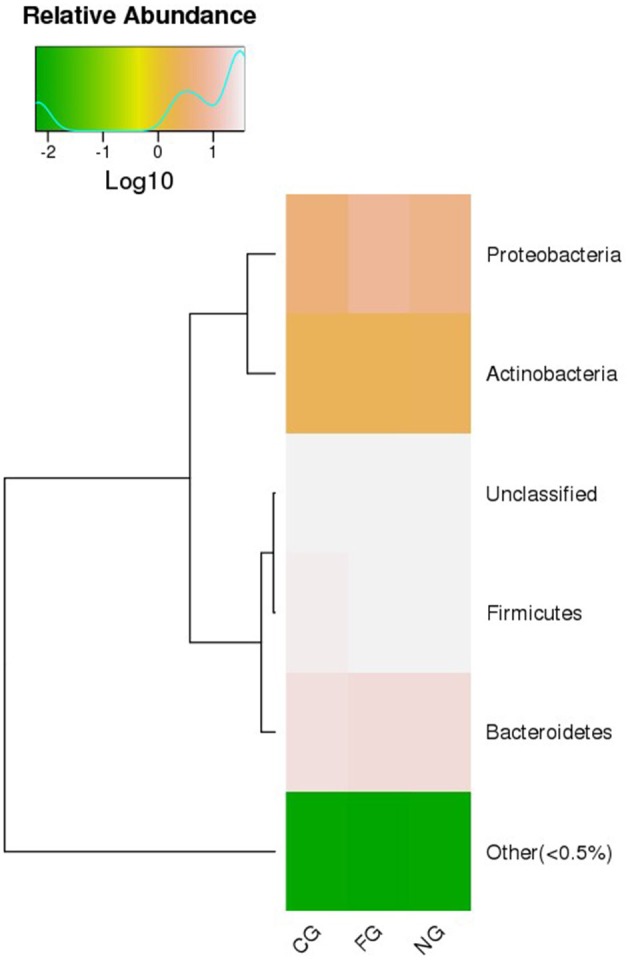
Heat map of gut microbiota between groups at phylum level. Group comparison and clustering of major bacteria phyla and their relative abundance. CG, healthy controls; FG, obese NAFLD group; NG, obese non-NAFLD group.
Taxonomy Comparison of Gut Microbiota at Class, Genus, and Species Level
Within phylum Firmicutes, class Negativicutes, genus Phascolarctobacterium, and species Phascolarctobacterium succinatutens exhibited a significant increase in obese NAFLD children, compared with the healthy controls (Supplemental Table 1). In contrast, genus Lactobacillus, Oscillibacter, and Ruminiclostridium showed a strong decrease in obese NAFLD patients. Additionally, we observed that Faecalibacterium prausnitzii was the only species representing differences between obese children with and without NAFLD. The Pathogenic genus Clostridium showed no statistical difference among these groups.
Decreased abundance of Bacteroidetes was mainly due to the decreased abundance in class Bacteroidia, genus Alistipes, and Paraprevotella in the obese children with or without NAFLD (Supplemental Table 1). Besides, at species level, we found that the abundance of Bacteroides clarus and Odoribacter splanchnicus were reduced in obese children groups with and without NAFLD. Interestingly, Parabacteroides johnsonii only exhibited a decrease in the obese NAFLD group, compared with the healthy group.
The increased representation of Proteobacteria in obese NAFLD group was mostly explained by the increased class Gammaproteobacteria, genus Klebsiella, Kluyvera, and species Klebsiella pneumoniae and Kluyvera ascorbata (Supplemental Table 1). It is worth noting that genus Helicobacter and species Helicobacter pylori were present and decreased in obese non-NAFLD children, compared with the healthy control group.
Furthermore, it is noteworthy that there were not any differences in phylum Actinobacteria in the three groups of children, which was in contrast to previous studies (15).
Ecological Diversities of Gut Microbiota
In the present study, we used Shannon index to assess the Alpha diversity of the community. Three groups of subjects did not demonstrate statistical difference in diversity (Table 2).
Table 2.
Comparison of Shannon-index between groups.
| Group | Statistical method | P-value |
|---|---|---|
| FG vs. CG vs. NG | Kruskal-Wallis | 0.3 |
| FG vs. NG | Wilcoxon Rank-Sum | 0.6 |
| FG vs. CG | Wilcoxon Rank-Sum | 0.1 |
| CG vs. NG | Wilcoxon Rank-Sum | 0.3 |
CG, healthy controls; FG, obese NAFLD group; NG, obese non-NAFLD group.
Functional Annotations of Gut Microbiota
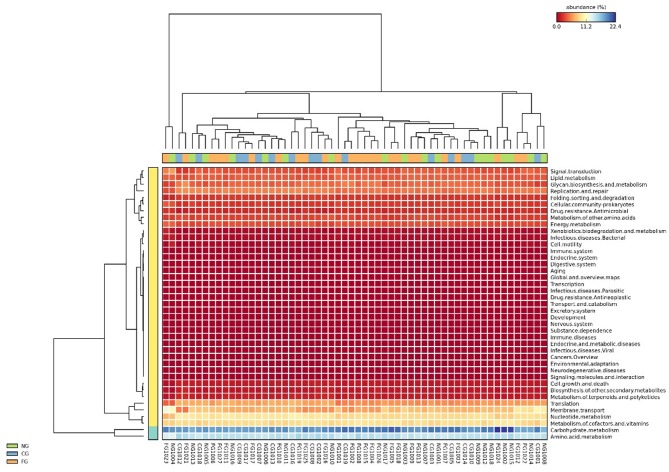
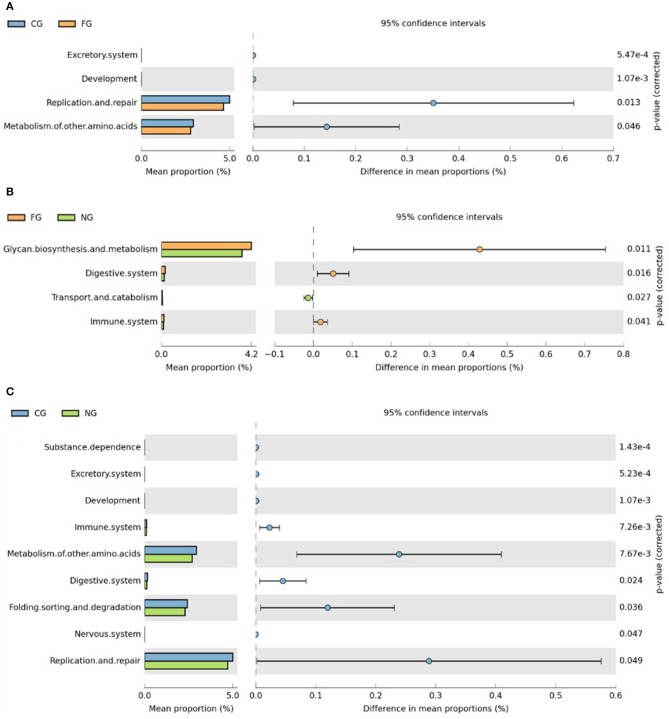
Subsequently, we further assessed functional annotations of gut microbiota via Diamond software against pathway information from the KEGG database. In total, by using Wilcoxon rank-sum test, we observed 923 KEGG orthologies (KOs) in three groups (Figure 3). Compared to healthy controls, pathways such as replication and repair and metabolism of other amino acids in KEGG levels 2 were significantly decreased in obese children with and without NAFLD (Figures 4A,C). The folding, sorting, and degradation pathway decreased in obese patients as well, in comparison to the healthy group. Furthermore, pathways like the digestive system, immune system, glycan biosynthesis, and metabolism showed considerable differences between the two obese groups, with higher abundance in the group of obese NAFLD patients (Figure 4B).
Figure 3.
Heat map of gut microbiota KEGG functional annotations. The KEGG pathway annotation was carried out for each individual sample. The abundance of each KEGG term was then clustered to represent as a heatmap.
Figure 4.
KEGG category comparisons among three groups of children. (A) Differences between obese NAFLD and healthy controls in KEGG functional annotations. (B) Differences between obese patients with and without NAFLD in KEGG functional annotations; (C) Differences between obese non-NAFLD and healthy controls in KEGG functional annotations. CG, healthy controls; FG, obese NAFLD group; NG, obese non-NAFLD group.
Discussion
In the present study, we have assessed gut microbiota of obese children with and without NAFLD in comparison to healthy controls. With metagenomic analysis, we learnt that microbial composition of three groups was significantly different at levels of phylum, class, genus, and species. At phylum level, obese NAFLD children exhibited increased Proteobacteria and decreased Bacteroidetes, compared to the healthy group, while Firmicutes showed no difference between groups. Dating back to 2006, Turnbaugh et al. (21) found that the ratio of Firmicutes to Bacteroidetes increased in obese mice. In their research, they came to the hypothesis that Firmicutes were a group of microbiomes relating to obesity. Genes related to the metabolism of fat and indigestible polysaccharides were detected in Firmicutes. In contrast, the population of Bacteroidetes is strongly increased in lean mice and humans, suggesting its representative role of body composition (22). The ratio of Firmicutes to Bacteroidetes also increased in our study. Thus, we postulated that the ratio of Firmicutes to Bacteroidetes could be a potential biomarker of NAFLD.
In Bacteroidetes, compared to the healthy group, genus Alistipes (associated with metabolism of plant cell wall polysaccharides and resistant starch) were decreased in obese NAFLD patients. It was in line with research by Zhu et al. (15) and Jiang et al. (23). In compensated and decompensated cirrhosis patients, the abundance of Alistipes decreased as well (24). Thus, Alistipes may be a group of beneficial microbiomes.
Within Proteobacteria, Gammaproteobacteria were observed to be increased in obese NAFLD patients in this study. Gammaproteobacteria are thought to be involved in choline metabolism and might be inhibited in high levels of choline (25). Besides, the relative abundance of Gammaproteobacteria is negatively correlated with liver fat content. However, a contrasting result was present in our study, which may due to the differences in age, religion, and diet of the test subjects. We hypothesized that dietary habits are related to the composition of gut microbiota. Therefore, we can restrict the diet in all subjects for further analysis. Furthermore, genus Helicobacter and species Helicobacter pylori of Proteobacteria were found to be differentially accumulated between obese and healthy children. Researchers in Korea (26) and China (27) found that Helicobacter pylori infection was closely related to the development of NAFLD based on large population research. Eradication of Helicobacter pylori could be a new method to deal with NAFLD. However, many studies do not find any connection between Helicobacter pylori infection and NAFLD (28, 29). Similarly, we did not observe significant changes between NAFLD patients and healthy children, either. Differently, in this study, we directly detected the Helicobacter pylori from stool samples, while most of the above studies detected Helicobacter pylori via serum antibody or urea test. Therefore, further study is needed to assess the relationship of NAFLD and intestinal Helicobacter pylori.
Although there was no significant difference in Firmicutes between three groups in this study. The abundance of probiotic Lactobacillus and anti-inflammatory species Faecalibacterium prausnitzii have been reported to be decreased in obese NAFLD children. Besides, the abundance of Faecalibacterium prausnitzii has been observed to be decreased in NASH patients (15, 30). To our knowledge, Faecalibacterium prausnitzii belong to beneficial microbiomes Clostridium, which can be developed into a potential kind of probiotic. The ecological imbalance of intestinal Faecalibacterium prausnitzii is closely related to inflammatory bowel disease, irritable bowel syndrome and type 2 diabetes (31, 32).
In the current study, ecological diversities of gut microbiota in three groups of subjects did not indicate differences. The same consequence was observed in study by Zhu et al. (15). Gut microbial alpha diversity between NASH patients, obese children and healthy controls were not statistically different. While Del Chierico et al. (5) conducted metagenomic analysis to assess gut microbiota of patients with simple fatty liver disease and NASH, obese, and healthy children. They found that the degree of alpha diversity was highest in healthy children, followed by obese children, NASH children, and simple fatty liver disease patients. Therefore, further study is necessary to analyze the ecological diversities in NAFLD.
Through functional annotations, we observed that several pathways were differentially enriched among these groups, including metabolism of other amino acids, replication and repair, folding, sorting, degradation, and glycan biosynthesis, metabolism and so on, whereas pathways of carbohydrate, lipid, and amino acid metabolism showed no differences. Further investigation of enriched gene clusters may be required to unravel the potential functional module related to obesity or NAFLD. In addition, researchers in Italy have found that urinary metabolomics of obese and NAFLD children are significantly different (33). Therefore, we can also analyze the urinary metabolomics of these children to further explore the metabolism of Chinese children with NAFLD.
The current study has some limitations in experimental design and data analysis. First, the number of children and adolescents studied was relatively small. It was difficult to restrict the same diet in all subjects, which may affect the composition and metabolism of gut microbiota. Second, we assessed gut microbiota via fecal samples instead of intestinal samples. It could not reflect all gut microbiota in the intestine exactly. However, lack of any non-invasive methods to directly obtain samples in the intestine could be the main cause. Therefore, we chose fecal samples to represent gut microbiota. Third, we did not evaluate serum metabolic markers as we described in a previous study (17). Further investigation is needed to assess the connection between gut microbiota and these serum factors.
Conclusion
In conclusion, large variations of the composition and functional annotations of gut microbiota of NAFLD patients, obese, and healthy children exist in Chinese children and adolescents. The findings in our study and other studies are noteworthy in the understanding of gut microbiota in pediatric NAFLD. Further analysis is necessary to reveal the consistent relationship and molecular mechanism of gut microbiota in NAFLD pathogenesis.
Data Availability Statement
The datasets generated for this study can be found in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under Bioproject PRJNA578215 and will be released upon the publication of this article.
Ethics Statement
The studies involving human participants were reviewed and approved by Human Ethics Committee of Shenzhen Children's Hospital. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
SZ designed experiments. YZ, JZ, JL, MC, and SZ collected the data and performed the research. YZ, JZ, ZW, and SZ analyzed the data. YZ and MC wrote the manuscript. SZ critically reviewed and revised the manuscript for final submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the participants who recruited patients in this study.
Glossary
Abbreviations
- BMI
body mass index
- KEGG
kyoto encyclopedia of genes and genomes
- KOs
KEGG orthologies
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- MRS
magnetic resonance spectroscopy
- PDFF
proton density fat fraction.
Footnotes
Funding. This work was supported by Shenzhen Science Technology Research and Development Fund from Shenzhen Science Technology and Innovation Commission, Shenzhen, Guangdong, China, Nos. CXZZ20150529144041624 and JCYJ20160429174706491.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2019.00518/full#supplementary-material
References
- 1.Anderson EL, Howe LD, Jones HE, Higgins J, Lawlor D, Fraser A. The prevalence of non-alcoholic fatty liver disease in children and adolescents: a systematic review and meta-analysis. PLoS ONE. (2015) 10:e0140908. 10.1371/journal.pone.0140908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Italian Association for the Study of the Liver AISF position paper on nonalcoholic fatty liver disease (NAFLD): updates and future directions. Dig Liver Dis. (2017) 49:471–83. 10.1016/j.dld.2017.01.147 [DOI] [PubMed] [Google Scholar]
- 3.Targher G, Byrne CD. Non-alcoholic fatty liver disease: an emerging driving force in chronic kidney disease. Nat Rev Nephrol. (2017) 13:297–310. 10.1038/nrneph.2017.16 [DOI] [PubMed] [Google Scholar]
- 4.Targher G, Byrne CD, Lonardo A, Zoppini G, Barbui C. Non-alcoholic fatty liver disease and risk of incident cardiovascular disease: a meta-analysis. J Hepatol. (2016) 65:589–600. 10.1016/j.jhep.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 5.Del Chierico F, Nobili V, Vernocchi P, Russo A, Stefanis C, Gnani D, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. (2017) 65:451–64. 10.1002/hep.28572 [DOI] [PubMed] [Google Scholar]
- 6.Vrieze A, Holleman F, Zoetendal EG, de Vos WM, Hoekstra JB, Nieuwdorp M. The environment within: how gut microbiota may influence metabolism and body composition. Diabetologia. (2010) 53:606–13. 10.1007/s00125-010-1662-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guercio Nuzio S, Di Stasi M, Pierri L, Troisi J, Poeta M, Bisogno A, et al. Multiple gut-liver axis abnormalities in children with obesity with and without hepatic involvement. Pediatr Obes. (2017) 12:446–52. 10.1111/ijpo.12164 [DOI] [PubMed] [Google Scholar]
- 8.Poeta M, Pierri L, Vajro P. Gut-liver axis derangement in non-alcoholic fatty liver disease. Children. (2017) 4:E66. 10.3390/children4080066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paolella G, Mandato C, Pierri L, Poeta M, Di Stasi M, Vajro P. Gut-liver axis and probiotics: their role in non-alcoholic fatty liver disease. World J Gastroenterol. (2014) 20:15518–31. 10.3748/wjg.v20.i42.15518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machado MV, Cortez-Pinto H. Diet, microbiota, obesity, and NAFLD: a dangerous quartet. Int J Mol Sci. (2016) 17:481. 10.3390/ijms17040481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. (2013) 58:120–7. 10.1002/hep.26319 [DOI] [PubMed] [Google Scholar]
- 12.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. (2004) 101:15718–23. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. (2013) 11:868–75; e861–3. 10.1016/j.cgh.2013.02.015 [DOI] [PubMed] [Google Scholar]
- 14.Li F, Sun G, Wang Z, Wu W, Guo H, Peng L, et al. Characteristics of fecal microbiota in non-alcoholic fatty liver disease patients. Sci China Life Sci. (2018) 61:770–8. 10.1007/s11427-017-9303-9 [DOI] [PubMed] [Google Scholar]
- 15.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. (2013) 57:601–9. 10.1002/hep.26093 [DOI] [PubMed] [Google Scholar]
- 16.Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol. (2015) 91:1–9. 10.1093/femsec/fiu002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao YZ, Gan YG, Zhou JL, Liu JQ, Cao WG, Cheng SM, et al. Accuracy of multi-echo Dixon sequence in quantification of hepatic steatosis in Chinese children and adolescents. World J Gastroenterol. (2019) 25:1513–23. 10.3748/wjg.v25.i12.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. (2012) 28:3150–2. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. (2015) 12:59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 20.Huson DH, Weber N. Microbial community analysis using MEGAN. Meth Enzymol. (2013) 531:465–85. 10.1016/B978-0-12-407863-5.00021-6 [DOI] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 22.Hartstra AV, Bouter KE, Backhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. (2015) 38:159–65. 10.2337/dc14-0769 [DOI] [PubMed] [Google Scholar]
- 23.Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. (2015) 5:8096. 10.1038/srep08096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao L, Ling Z, Chen D, Liu Y, Yang F, Li L. Disorganized gut microbiome contributed to liver cirrhosis progression: a meta-omics-based study. Front Microbiol. (2018) 9:3166. 10.3389/fmicb.2018.03166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. (2011) 140:976–86. 10.1053/j.gastro.2010.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TJ, Sinn DH, Min YW, Son HJ, Kim JJ, Chang Y, et al. A cohort study on Helicobacter pylori infection associated with nonalcoholic fatty liver disease. J Gastroenterol. (2017) 52:1201–10. 10.1007/s00535-017-1337-y [DOI] [PubMed] [Google Scholar]
- 27.Yu YY, Cai JT, Song ZY, Tong YL, Wang JH. The associations among Helicobacter pylori infection, white blood cell count and nonalcoholic fatty l iver disease in a large Chinese population. Medicine. (2018) 97:e13271 10.1097/MD.0000000000013271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai O, Huang Z, Li M, Zhang C, Xi F, Tan S. Association between Helicobacter pylori infection and nonalcoholic fatty liver disease: a single-center clinical study. Gastroenterol Res Pract. (2018) 2018:8040262. 10.1155/2018/8040262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okushin K, Takahashi Y, Yamamichi N, Shimamoto T, Enooku K, Fujinaga H, et al. Helicobacter pylori infection is not associated with fatty liver disease including non-alcoholic fatt y liver disease: a large-scale cross-sectional study in Japan. BMC Gastroenterol. (2015) 15:25. 10.1186/s12876-015-0247-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis–a longitudinal study. PLoS ONE. (2013) 8:e62885. 10.1371/journal.pone.0062885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira-Halder CV, Faria AVS, Andrade SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol. (2017) 31:643–8. 10.1016/j.bpg.2017.09.011 [DOI] [PubMed] [Google Scholar]
- 32.Martín R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, et al. Functional characterization of novel faecalibacterium prausnitzii strains isolated from healthy volun teers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front Microbiol. (2017) 8:1226. 10.3389/fmicb.2017.01226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troisi J, Pierri L, Landolfi A, Marciano F, Bisogno A, Belmonte F, et al. Urinary metabolomics in pediatric obesity and NAFLD identifies metabolic pathways/metabolites related to dietary habits and gut-liver axis perturbations. Nutrients. (2017) 9:E485. 10.3390/nu9050485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under Bioproject PRJNA578215 and will be released upon the publication of this article.