Abstract
Fluorescence polarization microscopy, which can visualize both position and orientation of fluorescent molecules, is useful for analyzing architectural dynamics of proteins in vivo, especially that of cytoskeletal proteins such as actin. Fluorescent phalloidin conjugates and SiR-actin can be used as F-actin orientation probes for fluorescence polarization microscopy, but a lack of appropriate methods for their introduction to living specimens especially to tissues, embryos, and whole animals hampers their applications to image the orientation of F-actin. To solve this problem, we have developed genetically encoded F-actin orientation probes for fluorescence polarization microscopy. We rigidly connected circular permutated green fluorescent protein (GFP) to the N-terminal α-helix of actin-binding protein Lifeact or utrophin calponin homology domain (UtrCH), and normal mEGFP to the C-terminal α-helix of UtrCH. After evaluation of ensemble and single particle fluorescence polarization with the instantaneous FluoPolScope, one of the constructs turned out to be suitable for practical usage in live cell imaging. Our new, genetically encoded F-actin orientation probe, which has a similar property of an F-actin probe to conventional GFP-UtrCH, is expected to report the 3D architecture of the actin cytoskeleton with fluorescence polarization microscopy, paving the way for both the single molecular orientation imaging in cultured cells and the sub-optical resolution architectural analysis of F-actin networks analysis of F-actin in various living systems.
Keywords: cytoskeleton, actin, actin-binding protein, fluorescence polarization, live cell imaging, single-molecule orientation
Introduction
Actin, one of the major cytoskeletal proteins, has fundamental roles in cellular processes, i.e. cell division, motility, intracellular transport and the maintenance of cell morphology [1]. Although a variety of probes or optical methods has been developed to observe actin filaments in living cells [2], it is challenging to resolve the spatial organization of actin filaments in their dense networks because of the spatial resolution limit of light microscopy methods.
Fluorescence emitted from fluorescent molecules such as fluorescent dyes/proteins is polarized along with the axis of the fluorophore’s transition dipole moment [3]. Fluorescence polarization microscopy is a method to obtain the orientation information of the fluorophores by analyzing the fluorescence polarization [4,5]. If F-actin is labeled with fluorescent molecules in a rotationally constrained manner (constrained tagging), the orientation of the averaged alignment of actin filaments can be determined as an ensemble orientation of fluorescence polarization observed with fluorescence polarization microscope (FPM). Moreover, even the orientation of a single actin filament with the length shorter than optical resolution (~200 nm) can also be determined when its labeling density is sufficiently sparse. Such fluorescent labeling of actin filaments has been achieved by applying fluorescently labeled phalloidins. Fluorescent phalloidins, however, easily interfere with the dynamics of actin, as they stabilize F-actin by inhibiting its depolymerization [6]. In addition, their membrane impermeability necessitates laborious delivery procedures such as microinjection, precluding their broader applications. SiR-actin [7] is another probe that is membrane permeable and easy to use for staining actin cytoskeleton in cultured cells but does not work well in thick tissues and embryos due to its limited penetration into specimens.
To circumvent these problems, we devised genetically encoded fluorescent probes that specifically bind to F-actin. As the direct fusion of a fluorescent protein to an actin-molecule has been reported to cause problems in actin dynamics [8–11], we utilized two actin-binding proteins, Lifeact [12] and utrophin calponin homology domain (UtrCH) [13], as scaffolds for constrained green fluorescent protein (GFP) labeling for FPM.
Methods
Plasmid construction
cpGFPCT3/10
The cpGFPCT3/10 (hereinafter, cpGFP) sequence was designed from GFP1-10 OPT and GFP11 M3 [14]. An EAAAKA helix motif [15] was added to the C-terminal helix of cpGFP in order to check the effect of helix elongation on the orientation of GFP fluorescence. The sequence of open reading frame (ORF) is shown in supplementary data.
cpGFP-Lifeact
The cpGFP sequence was subcloned into a modified pcDNA3 [16] previously reported. Lifeact cDNA was synthesized (Integrated DNA Technologies, Coralville, IA, USA) and inserted into the C-terminus of cpGFP in the modified pcDNA3. Variations with different linkers were constructed with polymerase chain reaction (PCR) mutagenesis using PrimeSTAR (Takara Bio, Kusatsu, Japan) according to the manufacturer’s instructions. An example sequence (ORF of L18) is shown in supplementary data.
cpGFP-UtrCHΔN
The backbone was amplified from pcDNA3.1(+) (Thermo Fischer Scientific, Waltham, MA, USA) by PCR. The first fragment, cDNA of UtrCHΔN, was synthesized (Integrated DNA Technologies). The second fragment, the sequence coding cpGFP and an amino acid linker was amplified from a vector containing cpGFP using a forward primer that has a homology arm to pcDNA3.1(+) and a reverse primer that has a linker sequence (reverse complement) and UtrCHΔN-homology arm. These two fragments were inserted into the backbone by Gibson Assembly [17]. Variations with different linkers were constructed using different primers for the second fragment. An example sequence (ORF of GU14) is shown in supplementary data.
UtrCHΔC-mEGFPΔN
The backbone was made by digestion of pcDNA3.1(+) with HindIII and NotI. The first fragment, UtrCH cDNA, was amplified from GFP-UtrCH (Addgene Plasmid #26737). The second fragment, mEGFP cDNA, was amplified from a vector containing mEGFP. Those two fragments were inserted into the backbone by Gibson Assembly. Variations with different linkers were constructed with PCR mutagenesis using PrimeSTAR. An example sequence (ORF of UG7) is shown in supplementary data.
Cell culture and sample preparation
HeLa cells were cultured on 96 well glass bottom plates (AGC TECHNO GLASS, Shizuoka, Japan) at 37°C in Medium 199 without phenol red (Thermo Fisher Scientific) containing 5% fetal bovine serum (Biowest, Nuaillé, France). The day before the microscopic observation, cells were transfected with each plasmid using Avalanche-Everyday Transfection Reagent (EZ biosystems, College Park, MD, USA; 0.2 μL reagent and 200 ng plasmid in 20 μL Opti-MEM (Thermo Fisher Scientific) per well). When necessary, we mixed SiR-actin (Spirochrome Ltd, Stein am Rhein, Switzerland) at the same time so that the final concentration would be 200 nM. When Atto565-phalloidin (Merck, Kenilworth, NJ, USA) was used, cells were fixed with 3% PFA in phosphate buffered saline (PBS) for 5 min, washed with PBS and 100 mM glycine, permeabilized with 0.1% TritonX-100 in PBS for 5 min, stained with Atto565-phalloidin for 20 min and then washed and mounted in SlowFade Diamond (Thermo Fisher Scientific).
LLC-PK1 cells were cultured on cell culture plates at 37°C in Medium 199 without phenol red containing 10% fetal bovine serum. The day before the microscopic observation, cells were transferred to glass bottom dishes, and at the same time (before cells were attached to the substrate) were transfected with each plasmid using Lipofectamine 3000 (Thermo Fisher Scientific). When Atto565-phalloidin was used, experiments were carried out in the same way as in the case of HeLa cells. For analyzing the fluorescence polarization orientation of Alexa Fluor (AF) 488-phalloidin bound to F-actin, we introduced AF488 phalloidin into LLC-PK1 cells by using a beads-loading method [18]. The final concentration of AF488 phalloidin was estimated to be 50–100 nM.
Fluorescence microscopy
Epi-fluorescence images were acquired using a Nikon Eclipse TiE inverted microscope (Nikon, Tokyo, Japan). Illumination was provided by SOLA light engine (Lumencor, Beaverton, OR, USA). Images were acquired with an iXon 3 897 EMCCD camera (Andor Technology, Belfast, UK) or Zyla 4.2 PLUS sCMOS camera (Andor Technology) using the Nikon imaging software NIS Elements Advanced Research. Objective lenses, PlanApoλ 10× 0.45 N.A. dry, PlanFluor 40× 0.75 N.A. dry, PlanApo VC 60× 1.40 N.A. oil; Nikon, were used.
Fluorescence polarization microscopy
We built a polarization microscope with a polarization beam splitting system at the detection side of a Nikon Eclipse TiE microscope [19]. The splitting system was assembled in G-Ang base 241 (G-Angstrom, discontinued due to manufacturer closedown). We used a 40× objective lens (PlanFluor 40× 0.75 N.A. dry; Nikon) for fluorescence polarization imaging. In order to analyze the fluorescence polarization of actin filaments labeled with our probes, the light from the primary image plane of the microscope (the location of a slit in the supplementary data online) is recollimated by a collimating lens (Thorlabs, NJ, USA). We split the fluorescence polarization images by using a linear polarization beam splitter (PBS; SURUGA SEIKI, Shizuoka, Japan) that was placed at the conjugate plane of the back focal plane of the objective lens. The linear polarization beam splitter splits fluorescence into two orthogonal polarization orientations, 0° (hereinafter we call as horizontal orientation) and 90° (hereinafter we call as vertical orientation). Split fluorescence beams were steered by two mirrors (Thorlabs) and were recombined along the same optical axis at the second PBS. The two images with orthogonal polarization orientations were focused on the same sCMOS camera (Zyla 4.2 PLUS) side by side. The collimating lens and focusing lens added 2× magnification to the final images. The respective images were labeled with pseudo-colors (horizontal polarization, green; vertical polarization, magenta) and merged with the Nikon imaging software NIS Elements Advanced Research. To confirm that our FPM setup works, we observed actin filaments of fixed LLC-PK1 cells stained with Atto565-phalloidin whose fluorescence polarization was reported to be parallel to actin filaments [20] (see supplementary data online).
Instantaneous FluoPolScope
The mechanisms of instantaneous FluoPolScope were reported previously [21]. We used this imaging system for quantitatively analyzing the properties of fluorescence polarization observed in F-actin labeled with different orientation probes. In the instantaneous FluoPolScope, we analyzed fluorescence polarization by splitting the emission according to polarization orientations 0°, 45°, 90° and 135°, and finally focusing onto the four quadrants of an EMCCD camera (iXon plus, Andor). The polarization of fluorescence was characterized by its degree of polarization, called the polarization factor (p) and the orientation (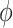 ) of its polarization. The fluorescence intensity recorded in each polarization quadrant (In) is related to the average intensity across the four quadrants (I), polarization factor, orientation and background intensity (Ibg) as follows: In = I[1 + pCos (2n − 2
) of its polarization. The fluorescence intensity recorded in each polarization quadrant (In) is related to the average intensity across the four quadrants (I), polarization factor, orientation and background intensity (Ibg) as follows: In = I[1 + pCos (2n − 2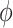 )] + Ibg,() where n = 0°,45°, 90°, 135°.
)] + Ibg,() where n = 0°,45°, 90°, 135°.
Retrieval of the orientation and polarization factor of particles is efficiently expressed in terms of Stokes parameters of the polarization-resolved emission:
S 0 = 0.25 (I0 + I45 + I90 + I135) – Ibg = I, S1 = I0-I90, S2 = I45 – I135. (2)
While the estimates of S1, S2 are independent of the isotropic background intensity Ibg, their uncertainty is impacted by the shot noise of the background.
The above relationship can be written in matrix form as S = M(I – Ibg),() where I = [I0, I45, I90, I135]T is the column vector of input intensities, S = [S0, S1, S2]T is the column vector of Stokes parameters and M, called instrument matrix, is the matrix of coefficients that relate these two vectors according to Eq. (3). The matrix M in above equation is replaced by the instrument matrix that is generated using the tangential polarizer as described in the supplementary information of the previous report by Mehta et al. [21].
The polarization factor (dimensionless between 0 and 1) and ensemble orientation (in radian) are obtained from Stokes parameters as follows: p = (S12 + S22)1/2/2 S0,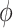 = 0.5tan−1(S2/S1).(4)
= 0.5tan−1(S2/S1).(4)
Results
Circular permutation of superfolder GFP
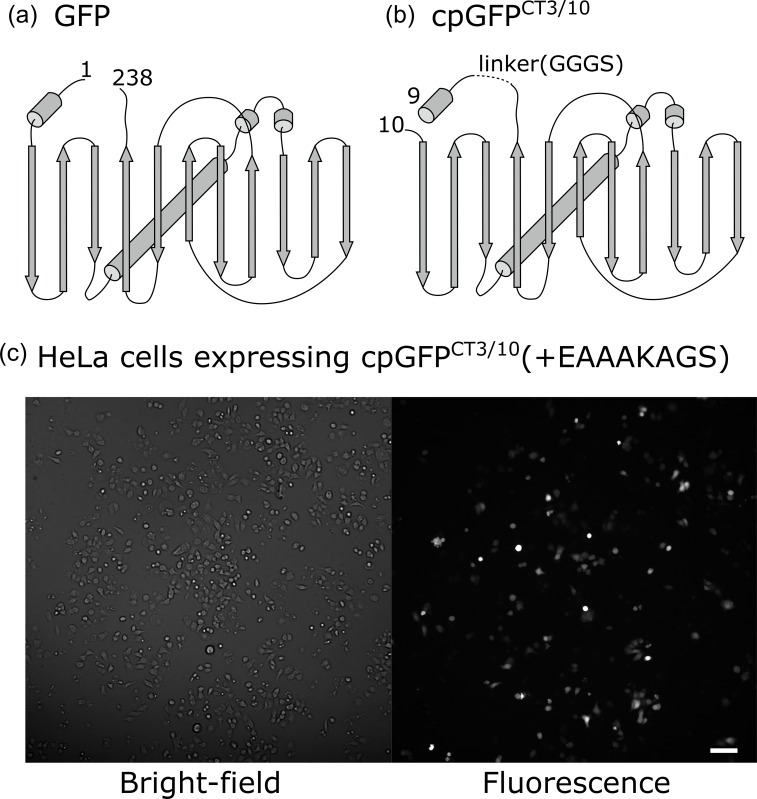
To date, constrained tagging with fluorescent proteins has been achieved mostly by joining the N-terminal 310 helix of GFP with the C-terminal α-helix of a target protein; thereby GFP and the target are linked by a continuous helix [4,22,23]. As far as using this strategy, only proteins with a helix at their C-termini can be the targets for constrained tagging of GFP. As Lifeact forms an α-helix only at the N-terminus [12], we carried out a circular permutation [24] of superfolder GFP [14] at the position just following the original N-terminal helix so that the 310 helix would be located at the C-terminus (Fig. 1a). The resulting circular permutant, cpGFPCT3/10 (hereinafter, cpGFP), emits fluorescence strong enough for imaging (Fig. 1b) when expressed in HeLa cells. Note that in this experiment, an EAAAKAGS amino acid sequence was added to the C-terminal helix of the cpGFP in order to check the influence of the helix elongation on GFP fluorescence. The amino acid sequence EAAAK is reportedly prone to form α-helix [15].
Fig. 1.
Circular permutation of GFP. (a) Schematic of GFP, including superfolder GFP. Rods indicate helices and arrows indicate β-sheets. (b) Schematic of cpGFPCT3/10. In (a) and (b), numbers indicate the first and last amino acids of superfolder GFP and cpGFPCT3/10, respectively. (c) Bright-field (left) and fluorescence (right) images of HeLa cells expressing cpGFPCT3/10(+EAAAKAGS). (Scale bar: 100 μm)
Constrained tagging of Lifeact with cpGFP
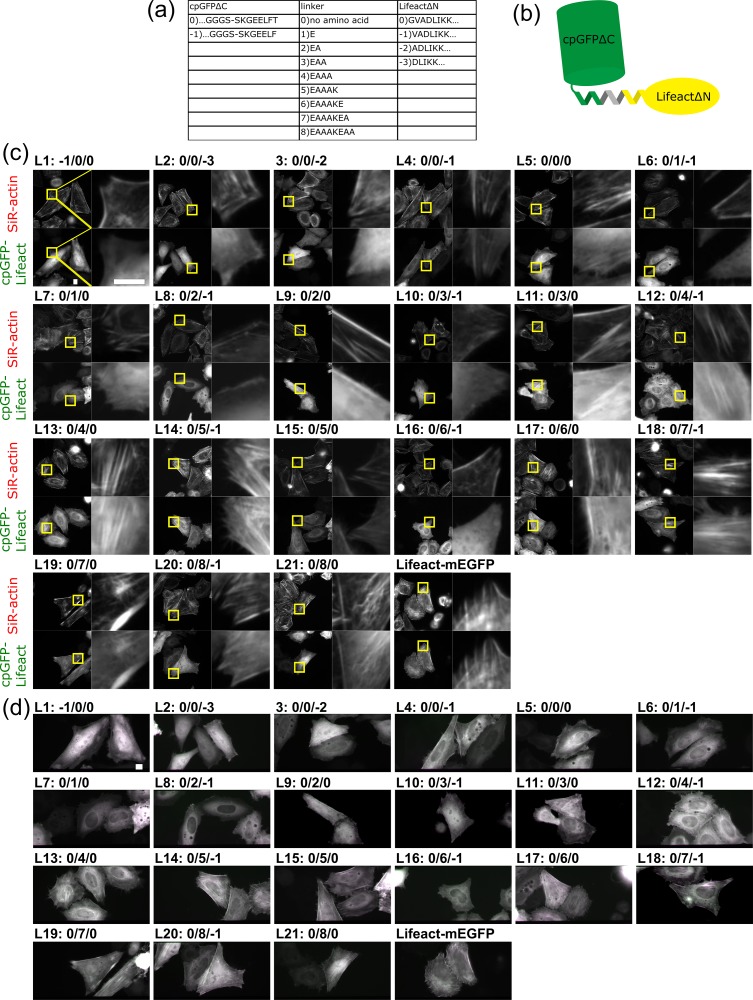
We next constructed expression plasmids in which cpGFP and Lifeact are connected by linkers of various lengths (Fig. 2a and b). We designed nine types of helical linkers based on an EAAAK amino acid sequence [15]. A cpGFP mutant with a single amino acid deletion and Lifeact mutant with an N-terminal amino acid deletion were also used so that in total 21 constructs were tested.
Fig. 2.
Constrained tagging of Lifeact with cpGFP. (a) Names and sequences of cpGFPΔC mutants, linkers, and LifeactΔN mutants. (b) Schematic of constrained cpGFP-Lifeact. (c) Binding of cpGFP-Lifeact constructs to F-actin bundles. In the presence of 200 nM SiR-actin, HeLa cells expressing each construct were imaged by epifluorescence microscopy. (Scale bar: 10 μm) (d) Fluorescence anisotropy of cpGFP-Lifeact constructs on F-actin structures. HeLa cells expressing each construct were imaged by FPM in the presence of 200 nM SiR-actin. (Scale bar: 10 μm)
We expressed these fusion constructs in HeLa cells and checked the colocalization of their products with actin filaments by double staining with SiR-actin (Fig. 2c). At the same time, we analyzed the fluorescence anisotropy of fusion proteins on stress fibers with FPM (Fig. 2d). Stress fibers colored in magenta or green mean their fluorescence anisotropy and the successful constrained tagging of Lifeact with cpGFP. Among 21 constructs (L1–21), 9 constructs only weakly colocalized with actin filaments (L1, 2, 3, 6, 7, 8, 9, 10 and 16), while the other 12 constructs clearly colocalized with actin filaments (L4, 5, 11, 12, 13, 14, 15, 17, 18, 19, 20 and 21). L18 showed vertical polarization orientation (colored magenta) on stress fibers that aligned horizontally, which means the polarization orientation is perpendicular to the length of actin filaments. Constructs that have a shorter linker (≤2 amino acids; L1–9 but L4 and 5) showed very weak localization to F-actin with high cytosolic background. Constructs with a longer linker (≥4 amino acids; L12–21 but L16) showed stronger binding to F-actin than those with a shorter linker, but the binding affinity was still low compared with that of original Lifeact-mEGFP. As Lifeact is a short peptide that consists of only 17 amino acids, it is likely that cpGFP closely positioned to Lifeact interfered with the interaction between F-actin and Lifeact, although we cannot exclude the possibility that the binding preference of Lifeact for F/G-actin has been altered.
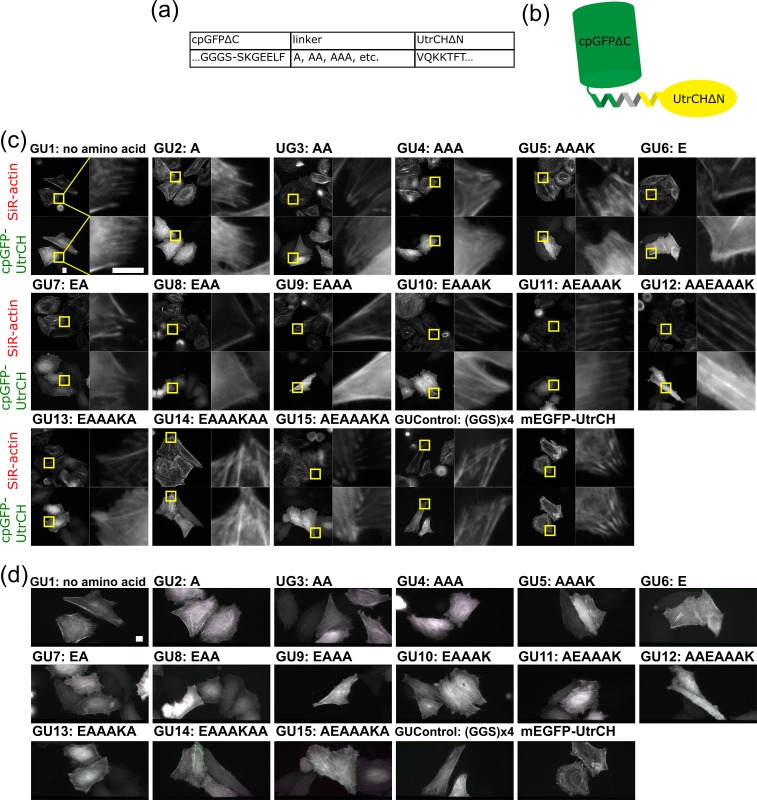
Constrained tagging of UtrCH with cpGFP
F-actin labeled with L18 construct showed the strongest fluorescence anisotropy among the Lifeact-based constructs, but its fluorescence anisotropy was not strong enough to use as a practical probe for monitoring the orientation of individual actin filaments with FPM. We then tried constrained tagging of an actin-binding protein UtrCH, in which the first 261 amino acids specifically bind to F-actin [13], with GFP. UtrCH has α-helices at both the N- and the C-termini of its actin-binding region (residues 32–46 and 240–252, respectively) [25]. We first tried connecting the C-terminal helix of cpGFP with the N-terminal helix of UtrCH. We designed 15 types of helical linkers based on the EAAAK motif and constructed 15 cpGFPΔC-UtrCHΔN fusions (Fig. 3a and b, GU1–15). We checked their colocalization with actin filaments and the fluorescence anisotropy on the stress fibers in the same way as we tested for screening cpGFP-Lifeact constructs (Fig. 3c and d). All constructs showed colocalization with actin filaments, and we found that GU14 showed the highest degree of fluorescence anisotropy on stress fibers by comparing the fluorescence intensity differences between two images of the same stress fibers through vertical and horizontal polarizers as shown in green and magenta pseudo-color images, respectively (Fig. 3d). Similar to the L18 construct, fluorescence polarization orientation of GU14 was perpendicular to the length of the actin filament. In all cpGFPΔC-UtrCHΔN constructs tested, including GUControl that has a flexible linker, we observed that the amounts of diffusive fractions in the cytosol were higher than those observed in the cells expressing the original mEGFP-UtrCH, indicating that the N-terminal 31 amino acids of UtrCH are required for its efficient targeting to F-actin.
Fig. 3.
Constrained tagging of UtrCH with cpGFP. (a) Names and sequences of a cpGFPΔC mutant, linkers, and a UtrCHΔN mutant. (b) Schematic of constrained cpGFP-UtrCH. (c) Binding of cpGFP-UtrCH constructs to F-actin bundles. For experimental details see the legends for Fig. 2c. (Scale bar: 10 μm) (d) Fluorescence anisotropy of cpGFP-UtrCH constructs on F-actin structures. For experimental details see legends for Fig. 2d. (Scale bar: 10 μm)
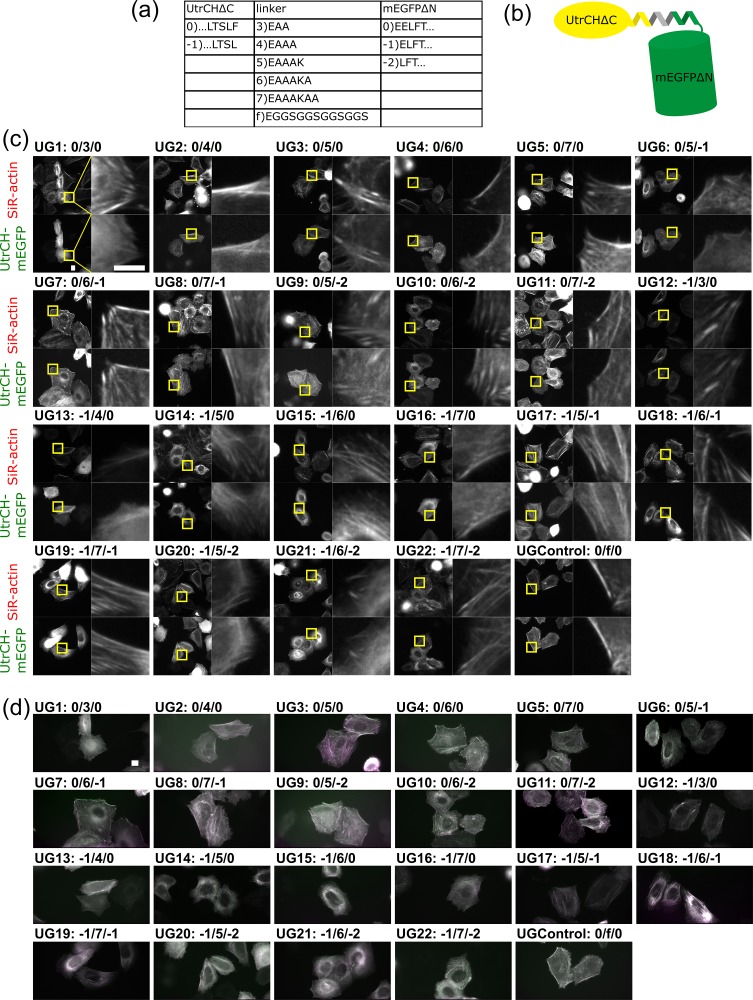
Constrained tagging of UtrCH with mEGFP
The results above lead us to construct the probes by joining the C-terminal helix of UtrCH with the N-terminal helix of mEGFP. We constructed a total of 22 UtrCHΔC-mEGFPΔN fusions (UG1–22, Fig. 4a and b) and tested their performances on F-actin labeling (Fig. 4c and d). All UtrCHΔC-mEGFPΔN constructs well colocalized with F-actin, similar to mEGFP-UtrCH with no amino acid deletion (Fig. 3c). Nine constructs showed a high degree of fluorescence anisotropy on stress fibers (UG3, 5, 7, 9, 10, 11, 17, 18 and 22), among which UG3 and UG7 showed the highest fluorescence anisotropy. Interestingly, the fluorescence polarization orientations of UG3, 5, 7, 9, 11 and 17 were parallel to actin filaments, while those of UG10, 18 and 22 were perpendicular to the filaments. Three constructs that showed the fluorescence polarization perpendicular to actin filaments had the same number of linking amino acids (sum of amino acid deletions and linker length), which suggests that linkers between UtrCHΔC and mEGFPΔN formed helices, as reported by Vrabioiu and Mitchison [22].
Fig. 4.
Constrained tagging of UtrCH with mEGFP. (a) Names and sequences of UtrCHΔC mutants, linkers, and mEGFPΔN mutants. (b) Schematic of constrained UtrCH-mEGFP. (c) Binding of mEGFP-UtrCH constructs to F-actin bundles. For experimental details see the legends for Fig. 2c. (Scale bar: 10 μm) (d) Fluorescence anisotropy of mEGFP-UtrCH constructs on F-actin structures. For experimental details see legends for Fig. 2d. (Scale bar: 10 μm)
Comparison of the fluorescence polarization of fusion constructs on F-actin bundles
We selected four constructs (L18, GU14, UG3, and UG7) that showed obvious fluorescence anisotropy on stress fibers and compared their binding to F-actin and fluorescence anisotropy when bound to F-actin. To this end, we compared their colocalization with actin filaments by staining fixed cells expressing the constructs with Atto565-conjugated phalloidin instead of using SiR-actin in live cells, to exclude any artifacts derived from F-actin stabilizing effect of SiR-actin (Fig. 5a). We expressed each construct in HeLa cells, fixed them with 3% PFA and stained them with Atto565-phalloidin. Unexpectedly, the result we obtained for GU14 turned out to be an artifact of excessively developed actin bundles induced by 200 nM SiR-actin. Co-staining with Atto565-phalloidin revealed that GU14 existed as a diffusive fraction in the cytosol, lacking obvious colocalization with F-actin structures. Other constructs still showed specific binding to F-actin as we had seen in the observations with SiR-actin (Figs 2–4).
Fig. 5.
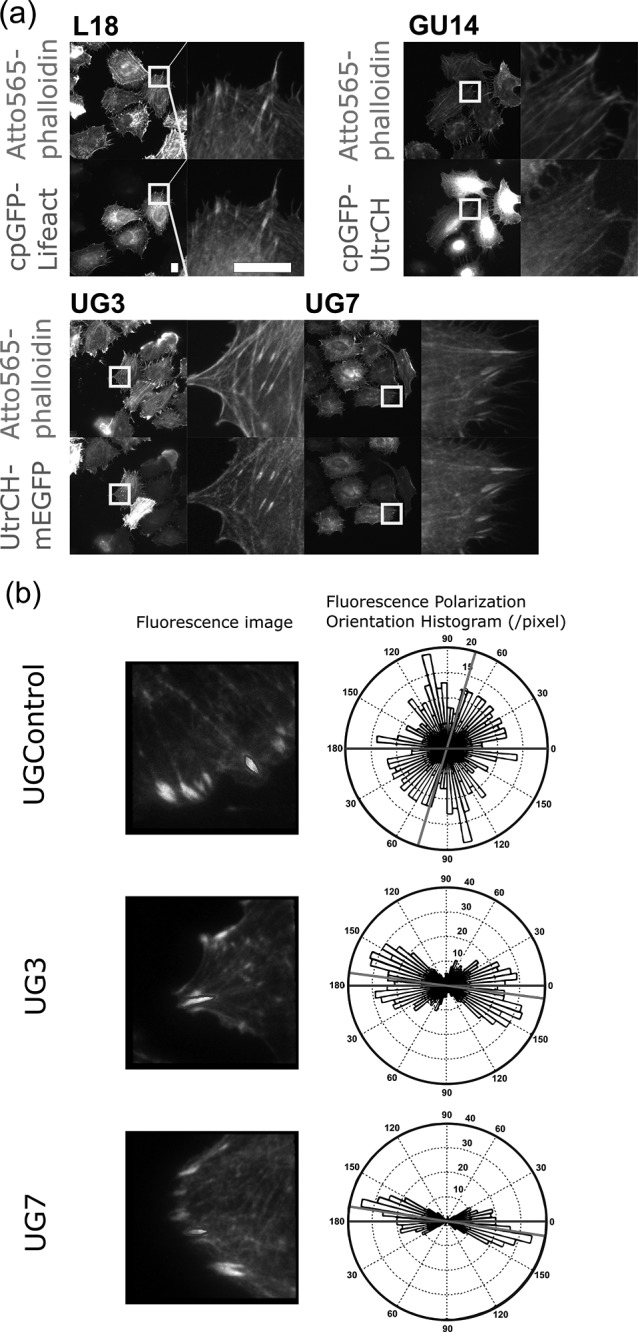
Comparison of the performance of the fusion constructs as F-actin orientation probes. (a) Binding of the constructs to F-actin in the absence of SiR-actin. HeLa cells expressing each construct were fixed with 3% PFA, stained with Atto565-phalloidin and imaged with an epi-fluorescence microscope. (Scale bar: 10 μm) (b) Ensemble fluorescence polarization orientation analysis of genetically encoded F-actin orientation probes. LLC-PK1 cells expressing UGControl, UG3 and UG7 were imaged with the instantaneous FluoPolScope. Focal adhesions (FAs) were selected as the locations for measuring the fluorescence polarization orientations of the constructs. The margins of FAs were traced (ROIs shown by black lines) to select the regions for analysis. For each construct, representative images of FAs in LLC-PK1 cells and a rose plot histogram of ensemble fluorescence polarization orientation measured at the selected FA (enclosed by black lines) are shown. The horizontal black line represents the long axis of the ROI and the gray line represents the mean orientation of ensemble fluorescence polarization in the ROI.
We found that UG3 and UG7 showed higher binding affinity to F-actin structures than those of L18 and GU14 (Fig. 5a) and showed higher fluorescence anisotropy on actin filaments than those observed in cells expressing L18 (Figs 2d and 4d). For quantitative analysis of UG3 and UG7, we further tested the properties of fluorescence anisotropy of these two probes on F-actin by using the instantaneous FluoPolScope, an FPM that analyzes the fluorescence polarization by splitting the emitted fluorescence along four polarization orientations namely 0°, 45°, 90° and 135° [21]. We used two image analyses: (i) pixel-based analysis and (ii) particle-based analysis for evaluating the fluorescence polarization of these probes. As the particle-based analysis requires fine-tuning of observation conditions, we first carried out the pixel-based analysis. We acquired images of LLC-PK1 cells expressing UGControl, UG3 or UG7 by the instantaneous FluoPolScope, and then computed the fluorescence polarization orientation at focal adhesions based on the fluorescence intensities in each pixel of focal adhesions’ images (Fig. 5b). As expected, the rose plots of the fluorescence polarization orientation histogram of UG3 and UG7 show their highest peaks at the orientation parallel to the length of focal adhesions. The fluorescence polarization of UGControl at focal adhesions did not show a particular orientation.
We have noticed that in the pixel-based analysis, Point Spread Function (PSF) of 100 nm beads images were slightly extended along the transmission axis of polarized fluorescence. Polarization-dependent deformation of PSF might cause an artifact in the pixel-based analysis, since the quadrant polarized light images become blurred only to the orientation parallel to the polarization axis [26]. Because of this artifact, we tend to overestimate fluorescence polarization along the length of a fluorescent filament.
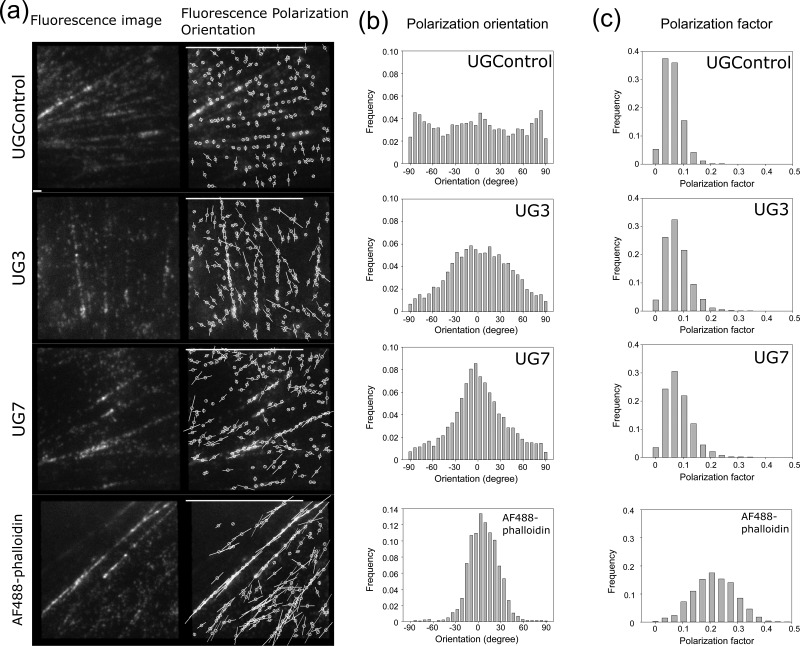
To avoid this artifact, we used a particle-based method that we have established for the analysis of fluorescence polarization of single molecules [21]. We did photobleaching until particles were sparsely distributed so that each measuring area covers the extended Airy disc only from a single particle PSF; thus, the polarization-dependent blur effect was minimized. By fine-tuning of appropriate laser power (2–6 μW/μm2) and exposure time (200 ms), we could observe the single molecules of F-actin binding constructs on actin filaments in live LLC-PK1 cells. We show representative fluorescence particle images (left) and fluorescence polarization maps (right) of LLC-PK1 cells expressing UGControl, UG3,and UG7 from top to bottom, respectively (Fig. 6a). The orientation of a yellow bar on each particle represents the polarization orientation, and the length represents the polarization factor. We created histograms of the orientation of fluorescence polarization (Fig. 6b) and the polarization factor (Fig. 6c) of the fluorescent particles observed on the stress fibers. As a reference, we analyzed the fluorescence polarization and the polarization factors of AF 488 phalloidin (AF488-phalloidin) bound to the stress fibers (Fig. 6a–c, bottom panels), which was introduced into the cells using beads-loading method [18]. There was no significant peak in the orientation histogram of UGControl, although we frequently observed a small peak at the orientation perpendicular to the length of F-actin. The orientation histograms of UG3 and UG7 both had single peaks at around 0o. The peak of the UG7 histogram was sharper than that of UG3 histogram, while it was not as sharp as that of AF488-phalloidin histogram at around 0o. The results we obtained for AF488-phalloidin were consistent with the previous results [21]. The value of polarization factor is high when the 3D angler wobble of the probe with respect to the axis of an actin filament is small, while it is also affected by the inclination angle of the dipole with respect to the focus plane (see Methods for details). The peak of the histogram of the polarization factor for UGControl was lower than those for UG3 and UG7. This is reasonable as the linker for UGControl was designed to be flexible. The polarization factor of AF488-phalloidin was higher than those for UG3 and UG7. Based on these fluorescence polarization analyses, we concluded that, to our knowledge, UG7 is the first genetically encoded orientation probe for F-actin, which works well in live cells.
Fig. 6.
Single particle analysis of fluorescence polarization in F-actin orientation probes. (a) Single particle analysis of the fluorescence polarization orientation and the polarization factor. LLC-PK1 cells expressing UGControl, UG3 and UG7 and LLC-PK1 cells loaded with AF488-phalloidin were imaged with the instantaneous FluoPolScope. Fluorescence images of stress fibers in LLC-PK1 cells (left) and the orientation maps of the polarized fluorescence of F-actin orientation probes bound to stress fibers (right) are shown. The orientation of white bars represents the fluorescence polarization orientation, and the length of the bar represents the polarization factor. Bars on the bottom: 1 μm; bars on top, polarization factor = 1. (b) Histograms of the fluorescence polarization orientations of F-actin orientation probes tested. Fluorescent particles in a series of ~100 frames (total number of particles, >2000) were analyzed for each construct and AF488-phalloidin. Central axes of actin bundles were manually determined on MATLAB. (c) Histograms of the polarization factors of the F-actin orientation probes tested. Datasets are the same as we used for the fluorescence polarization orientation analysis in (b). In (b) and (c), representative histograms are shown.
Discussion
In this study, we rigidly connected circular permutated GFP to the N-terminal α-helix of actin-binding protein Lifeact or UtrCH, and normal mEGFP to the C-terminal α-helix of UtrCH, using α-helical linkers of various lengths, to create a genetically encoded fluorescent probe for F-actin. To our best knowledge, UG7, the final product of our screening, is the first genetically encoded orientation probe for F-actin that is suitable for practical usage in intracellular conditions.
SiR-actin is a membrane-permeable derivative of an F-actin stabilizing drug jasplakinolide, and its fluorescence is polarized with the orientation perpendicular to F-actin; therefore it is applicable to FPM live imaging [27]. While SiR-actin seems to work well in many types of cultured cells [28,29], it is not useful for some species such as yeasts (our unpublished data) and for thick tissues and embryos. In contrast, our new probe can be applied to any living systems with which genetic methods are available. By combining with fluorescence polarization imaging, our genetically encoded F-actin orientation probe will be most useful for analyzing the 3D architecture of F-actin networks in tissue and in vivo.
Although UG7 is already able to report F-actin orientation both in the ensemble and single molecule analyses, further screening of amino acids that compose the linking helix between UtrCH and GFP might achieve higher polarization factor by making the linker more rigid. The fact that UG7 was superior to UG3 while their linkers have the same number of amino acids indicates that the performance of the probe can be drastically improved by optimizing amino acids used for the linker. In fact, it is reported that side chain interactions of amino acids both within the helix and between inside and around the helix can stabilize α-helix structure [30,31]. In addition, changing binding kinetics of UtrCH to F-actin by mutating amino acids in the actin-binding region might facilitate the stable binding to F-actin for longer tracking. The genetic manipulation potential of UG7 and its improvements in future will help us to make this probe greatly useful for analyzing the 3D architecture of F-actin especially in live tissues, and in whole animals/plants in vivo.
Concluding remarks
We developed a novel genetically encoded orientation probe of F-actin, UG7. We demonstrate that UG7 works as an F-actin probe in live cells and reports the molecular orientation of F-actin by its fluorescence polarization. The methods and results described here will be useful for the development of other genetically encoded probes that report molecular orientations and orders in protein assemblies in general.
It is noteworthy that this is the first successful orientationally constrained tagging of GFP to the protein of interest via a binding protein, but not the direct tagging of GFP to the protein of interest. This approach will be useful for developing molecular orientation probes of any proteins of interest and will have the potential to apply any biological molecules other than proteins.
Supplementary Material
Acknowledgements
We are grateful to Dr M. Kawagishi for helpful discussion and to Mrs. M. Taguchi for experiment assistance. We deeply thank Dr Shalin Mehta for his generous permission of using his MATLAB codes that analyze the polarization factor and fluorescence polarization orientation of F-actin probes we have developed.
Funding
Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (KAKENHI) (C) and (B) (18K06819 to K.S., 17H04013, 26293038 to S.T.); Fostering Joint International Research (B) (18KK0222 to S.T.); JSPS Bilateral Joint Research Projects (Open Partnership Joint Research Projects) (1006285 to S.T.); National Institutes of Health (R01 GM100160 to T.T.).
References
- 1. Pollard T D, and Cooper J A (2009) Actin, a central player in cell shape and movement. Science 326: 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Melak M, Plessner M, and Grosse R (2017) Actin visualization at a glance. J. Cell Sci. 130: 525–530. [DOI] [PubMed] [Google Scholar]
- 3. Lakowicz J R. (2006) Principles of Fluorescence Spectroscopy (Springer, USA: https://www.springer.com/us/book/9780387312781). [Google Scholar]
- 4. Vrabioiu A M, and Mitchison T J (2006) Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature 443: 466–469. [DOI] [PubMed] [Google Scholar]
- 5. Koike-Tani M, Tani T, Mehta S B, Verma A, and Oldenbourg R (2015) Polarized light microscopy in reproductive and developmental biology. Mol. Reprod. Dev. 82: 548–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper J A. (1987) Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105: 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lukinavičius G, Reymond L, D’Este E, Masharina A, Göttfert F, Ta H, Güther A, Fournier M, Rizzo S, Waldmann H, Blaukopf C, Sommer C, Gerlich D W, Arndt H D, Hell S W, and Johnsson K (2014) Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods 11: 731–733. [DOI] [PubMed] [Google Scholar]
- 8. Doyle T, and Botstein D (1996) Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. 93: 3886–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu J-Q, and Pollard T D (2005) Counting cytokinesis proteins globally and locally in fission yeast. Science 310: 310–314. [DOI] [PubMed] [Google Scholar]
- 10. Vavylonis D, Kovar D R, O’Shaughnessy B, and Pollard T D (2006) Model of formin-associated actin filament elongation. Mol. Cell 21: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carvalho A, Desai A, and Oegema K (2009) Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell 137: 926–937. [DOI] [PubMed] [Google Scholar]
- 12. Riedl J, Crevenna A H, Kessenbrock K, Yu J H, Neukirchen D, Bista M, Bradke F, Jenne D, Holak T A, Werb Z, Sixt M, and Wedlich-Soldner R (2008) Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burkel B M, von Dassow G, and Bement W M (2007) Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskeleton 64: 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cabantous S, Terwilliger T C, and Waldo G S (2005) Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23: 102–107. [DOI] [PubMed] [Google Scholar]
- 15. Arai R, Ueda H, Kitayama A, Kamiya N, and Nagamune T (2001) Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 14: 529–532. [DOI] [PubMed] [Google Scholar]
- 16. Zhao Y, Araki S, Wu J, Teramoto T, Chang Y-F, Nakano M, Abdelfattah A S, Fujiwara M, Ishihara T, Nagai T, and Campbell R E (2011) An expanded palette of genetically encoded Ca2+ indicators. Science 333: 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibson D G, Young L, Chuang R-Y, Venter J C, Hutchison Iii C A, and Smith H O (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- 18. McNeil P L, and Warder E (1987) Glass beads load macromolecules into living cells. J. Cell Sci. 88: 669–678. [DOI] [PubMed] [Google Scholar]
- 19. Sase I, Miyata H, Ishiwata S, and Kinosita K (1997) Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc. Natl. Acad. Sci. U. S. A. 94: 5646–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Valades Cruz C A, Shaban H A, Kress A, Bertaux N, Monneret S, Mavrakis M, Savatier J, and Brasselet S (2016) Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. Proc. Natl. Acad. Sci. 113: E820–E828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mehta S B, McQuilken M, Riviere P J L, Occhipinti P, Verma A, Oldenbourg R, Gladfelter A S, and Tani T (2016) Dissection of molecular assembly dynamics by tracking orientation and position of single molecules in live cells. Proc. Natl. Acad. Sci. U. S. A. 113: E6352–E6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vrabioiu A M, and Mitchison T J (2007) Symmetry of septin hourglass and ring structures. J. Mol. Biol. 372: 37–49. [DOI] [PubMed] [Google Scholar]
- 23. Kampmann M, Atkinson C E, Mattheyses A L, and Simon S M (2011) Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat. Struct. Mol. Biol. 18: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baird G S, Zacharias D A, and Tsien R Y (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 96: 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keep N H, Winder S J, Moores C A, Walke S, Norwood F L, and Kendrick-Jones J (1999) Crystal structure of the actin-binding region of utrophin reveals a head-to-tail dimer. Structure 7: 1539–1546. [DOI] [PubMed] [Google Scholar]
- 26. Quabis S, Dorn R, Eberler M, Glöckl O, and Leuchs G (2000) Focusing light to a tighter spot. Opt. Commun. 179: 1–7. [Google Scholar]
- 27. Spira F, Cuylen-Haering S, Mehta S, Samwer M, Reversat A, Verma A, Oldenbourg R, Sixt M, and Gerlich D W (2017) Cytokinesis in vertebrate cells initiates by contraction of an equatorial actomyosin network composed of randomly oriented filaments. Elife 6: e30867: Balasubramanian, MK (ed.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swaminathan V, Kalappurakkal J M, Mehta S B, Nordenfelt P, Moore T I, Koga N, Baker D A, Oldenbourg R, Tani T, Mayor S, Springer T A, and Waterman C M (2017) Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc. Natl. Acad. Sci. USA. 114: 10648–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nordenfelt P, Moore T I, Mehta S B, Kalappurakkal J M, Swaminathan V, Koga N, Lambert T J, Baker D, Waters J C, Oldenbourg R, Tani T, Mayor S, Waterman C M, and Springer T A (2017) Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 8: 2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dill K A, Ozkan S B, Shell M S, and Weikl T R (2008) The protein folding problem. Annu. Rev. Biophys. 37: 289–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sivaramakrishnan S, Spink B J, Sim A Y L, Doniach S, and Spudich J A (2008) Dynamic charge interactions create surprising rigidity in the ER/K α-helical protein motif. Proc. Natl. Acad. Sci. USA. 105: 13356–13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.