Abstract

Heparin is a polysaccharide-based anticoagulant agent, which is widely used in surgery and blood transfusion. However, overdosage of heparin may cause severe side effects such as bleeding and low blood platelet count. Currently, there is only one clinically licensed antidote for heparin: protamine sulfate, which is known to provoke adverse effects. In this work, we present a stable and biocompatible alternative for protamine sulfate that is based on serum albumin, which is conjugated with a variable number of heparin-binding peptides. The heparin-binding efficiency of the conjugates was evaluated with methylene blue displacement assay, dynamic light scattering, and anti-Xa assay. We found that multivalency of the peptides played a key role in the observed heparin-binding affinity and complex formation. The conjugates had low cytotoxicity and low hemolytic activity, indicating excellent biocompatibility. Furthermore, a sensitive DNA competition assay for heparin detection was developed. The detection limit of heparin was 0.1 IU/mL, which is well below its therapeutic range (0.2–0.4 IU/mL). Such biomolecule-based systems are urgently needed for next-generation biocompatible materials capable of simultaneous heparin binding and sensing.
Introduction
Heparin is a highly charged glycosaminoglycan (GAG) mainly composed of uronic acid and glucosamine subunits.1 Ever since its first medical use in the 1930s, heparin has found widespread applications as the first naturally occurring polysaccharide-based drug.2 The medical role of heparin is primarily based on its anticoagulant ability. It can complex with thrombin inhibitors such as antithrombin III with high affinity. This further inactivates thrombin, factor Xa, and other coagulation factors leading to an interrupted blood-clotting cascade.3 Commonly, the dosage of heparin needs to be maintained within 2–8 IU/mL during cardiovascular surgery and 0.2–1.2 IU/mL in postoperative and long-term care. Maintaining heparin concentrations at sufficient levels will prevent thrombosis while avoiding the risk of serious side effects, including heparin-induced thrombocytopenia, caused by excessive heparin.4−6 Hence, it is of vital importance to monitor heparin concentrations and also neutralize its anticoagulant effect when blood clotting needs to be recovered. Currently, heparin neutralization is predominantly achieved by an arginine-rich shellfish protein, protamine sulfate (PS), which is also the only clinically licensed antidote. Anti-Xa assay and activated partial thromboplastin time assay (aPTT) are the most desirable methods to monitor blood coagulation.7−9
Protamine sulfate binds to heparin efficiently through the electrostatic interaction between its cationic arginine groups and anionic sulfonate groups of heparin.10 Despite the high binding efficiency using PS involves serious drawbacks that should not be overlooked. For example, it can cause severe adverse effects including anaphylactic reactions, and it is ineffective in the removal of low molecular weight heparin.11−14 Therefore, great effort has been devoted to the development of new heparin antidote candidates.2 Most of these rely on the development of cationic molecules since heparin has the highest negative charge among all known biomacromolecules.15 Methylene blue is one of the earliest and simplest compounds that have been studied.16 However, its binding efficiency is largely hindered in physiological conditions due to the low charge density.17 Other small molecular systems have also been reported, such as surfen, delparantag, and foldamers.18−21 Owing to their multivalency effects, cationic oligopeptides,22,23 synthetic polymers,24−27 and self-assembled systems28−31 have drawn attention.
As mentioned above, another important area in maintaining sufficient heparin levels is the detection technique. Traditional clotting time-based assays have been proven accurate, but they are rather time-consuming. Therefore, real-time heparin detection methods would be highly desirable, and numerous luminescent,32 colorimetric,33,34 and fluorescent35−38 sensors have been developed along these lines. Fluorescence-based methods have been established using both turn-on35,37 and turn-off36 approaches.
Serum albumin (SA) is broadly recognized as a biocompatible vector for the delivery of drugs.39−41 In addition, many SA-based conjugates have been reported to be nontoxic and nonimmunogenic.40,42−45 In 2005, an albumin-based anticancer drug, Abraxane, was first approved by the U.S. Food and Drug Administration (FDA), and continuous effort has been put into the development of albumin-based drugs.46,47 In this work, albumin-based heparin-binding compounds were developed. They are based on SA proteins that are conjugated with a variable number of heparin-binding peptides (HBPs). The heparin-binding peptide is known from the fibroblast growth factor (FGF) and has a dissociation constant of ∼134 pM with heparin.48 Single (1SP) and multiple (3SP and 7SP) peptide-conjugated SAs were synthesized, and the binding ability of the products to heparin was evaluated with methylene blue (MB) displacement assay, dynamic light scattering (DLS), and anti-Xa assay in selected buffers and human blood plasma. It was found that the heparin-binding capacity increased with the amount of peptides attached. Under physiological conditions, 3SP and 7SP maintained moderate heparin-binding ability, which could be attributed to the multivalency effect,49−51 while 1SP showed negligible heparin binding. The complexes were also visualized with atomic force microscopy (AFM). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye assay and hemolysis assay results revealed negligible toxicity to human dermal fibroblasts (HDF) cells and human red blood cells (RBCs) within the tested concentration range. Finally, a fluorescence-based heparin detection method was developed through complexing 7SP and chemically modified DNA (22 nucleotide (nt)-long DNA with a fluorescein (6-FAM) and black hole quencher (BHQ1) at the 5′ and 3′ ends, respectively). A calibration curve for quantitative heparin level detection was constructed, and it was found that the detection limit is <0.1 IU/mL, which is well below the concentration required during cardiovascular surgery (0.2–1.2 IU/mL).
Results and Discussion
Preparation of SA–Peptide Conjugates
For conjugating SA and peptides, an efficient and selective thiol maleimide reaction was used.52−54 Bovine serum albumin used in this work has a free and solvent-exposed cysteine (Cys34), which acts as a suitable and easily accessible conjugation handle.55 Additional advantages including stability, low toxicity, and long circulation time make SA an ideal candidate as a scaffold and shielding group for bioconjugates.45 For the preparation of single peptide-conjugated SA (1SP), an N-terminal maleimide-modified peptide was mixed with SA and left reacting overnight (Figure 1a). According to the analysis of the crude 1SP mixture with semipreparative high-performance liquid chromatography (HPLC) using a heparin affinity column (Figure S1a), the conversion was approximately 50%.56 Peak fractions were collected, desalted, and finally lyophilized. The purity of 1SP was confirmed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF, Figure 1b) and HPLC (Figure 1d).
Figure 1.
(a) Schematic presentation of the preparation of single (1SP) and multiple (3SP and 7SP) peptide-modified SAs. (b) MALDI-TOF spectra of SA and 1SP: MSA = 66,519 g/mol and M1SP = 68,414 g/mol (theoretical molecular weights: MSA = 66,430 g/mol57 and M1SP = 68,636 g/mol); (c) MALDI-TOF spectra of cross-linker-capped SAs: M5× = 67,502 g/mol and M10× = 68,473 g/mol. Therefore, the estimated number of cross-linkers attached to SAs for 5 and 10 equiv syntheses was 3 and 7, respectively; (d) HPLC analysis of purified 1SP, 3SP, and 7SP using a heparin affinity column. SA was analyzed for comparison.
Free lysines on the SA surface were further used for the synthesis of two different types of antidote: SA with approximately three or seven peptides (3SP or 7SP). A cross-linker, di(N-succinimidyl) 3,3′-dithiodipropionate containing a disulfide bond, was first added to SA in 5 or 10 equiv of excess. Successful conjugation was confirmed by HPLC and MALDI-TOF, which further revealed that the estimated number of cross-linkers attached to SA for 5 and 10 equiv syntheses was 3 and 7, respectively (Figure 1c and Figure S1b,c). Surprisingly, no cross-linked SA was observed in either reactions (Figure S1b,d), which is most likely due to the relatively low protein and high cross-linker concentrations. Next, the disulfide bonds were cleaved using a reducing reagent, tris(2-carboxyethyl)phosphine hydrochloride (TCEP), yielding reactive sulfhydryl groups. Excess TCEP and cleaved cross-linkers were removed with HPLC using a desalting column (Figure S2a,b). The reactive elution fractions were subsequently combined with an N-maleimide-modified HBP to yield 3SP and 7SP. From HPLC data, it was found that the conversion in both cases was quantitative and the products were pure (Figure 1d and Figure S2c,d).
Heparin-Binding Study
Heparin-binding ability was first evaluated using MB displacement assay in a 2 mM Tris-HCl buffer. MB is a known heparin-binding dye with an absorbance peak at 664 nm, which shifts to 568 nm when bound to heparin. In the presence of another but stronger heparin-binder, MB can be displaced from heparin and the absorbance peak at 664 nm is restored (Figure S3a). Therefore, the ratio between A (664 nm)/A (568 nm) can be used to evaluate the heparin-binding efficiency of conjugates (Figure 2) as a function of the mass (mB/mH) or molar (nB/nH) ratios between the binder and heparin.34 SA showed minor heparin-binding efficiency within the tested concentration range, while that of a pure peptide was higher but not saturated. The slight binding of SA could be attributed to electrostatic attraction to heparin due to the large pool of lysines on the surface.58,59 When one peptide was attached to SA (1SP), the heparin-binding ability was significantly enhanced and saturated at a mass ratio of ∼100 equiv (∼28 equiv in the molar ratio, Figure 2). This binding efficiency is even higher than that of plain SA or the plain peptide alone, which indicates a synergistic effect between these two components.60 We can also conclude that the number of conjugates needed to fully release MB was significantly reduced with the increasing number of attached peptides. The saturation point shifts from ∼100 equiv of 1SP to ∼32 equiv of 3SP and ∼12 equiv of 7SP (Figure 2). In Table 1, c50, the concentration required for 50% displacement of MB from its complex, and the positive/negative charge ratio of binder/heparin for 50% MB displacement are compared. The data indicates that all binders achieve MB displacement at comparable ratios and 7SP showed similar heparin-binding capacity with PS when comparing the molar ratios.
Figure 2.

MB displacement assay results in (left) mass and (right) molar ratios of the binder to heparin, measured by the A (664 nm)/A (568 nm) ratio. Heparin concentration: 0.017 mg/mL (0.9 μM). Protamine sulfate was plotted for comparison. Measurements were performed using triplicate samples, and the averaged results with standard deviation are presented.
Table 1. Data of 1SP, 3SP, 7SP, and PS Derived from MB Displacement Assay.
| binder | solvent | nominal charge | c50 [μM]a | +/– ratiob |
|---|---|---|---|---|
| 1SP | 2 mM Tris-HCl | 6+ | 111.1 ± 3.1c | 1.01 ± 0.03 |
| 3SP | 2 mM Tris-HCl | 18+d | 26.0 ± 2.1 | 0.71 ± 0.06 |
| 7SP | 2 mM Tris-HCl | 42+d | 9.7 ± 0.6 | 0.61 ± 0.04 |
| PS | 2 mM Tris-HCl | 24+e | 11.1 ± 0.1 | 0.41 ± 0.01 |
c50 represents the concentration required for 50% displacement of MB from its complex in 2 mM Tris-HCl.
The +/– ratio indicates the positive/negative charge ratio of binder–heparin at corresponding c50 values.
The highest concentration of 1SP tested.
We assume that all the attached cross-linkers reacted with HBP and only the charges on HBP were included.
The positive charge of PS was used as reported.29
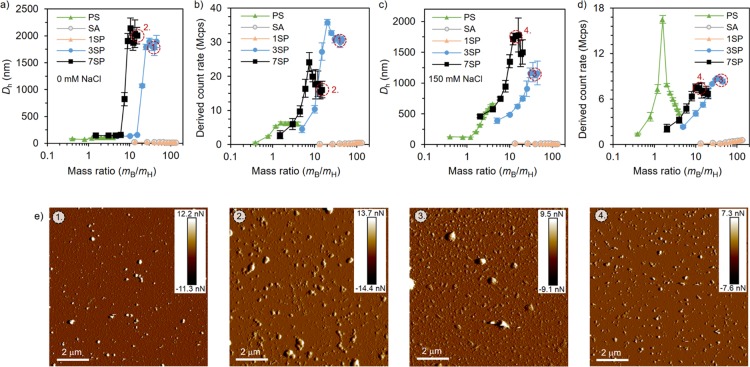
Next, dynamic light scattering (DLS) was employed to confirm the heparin-binding efficiency in the absence of added electrolytes (Figure 3a,b) as well as at a physiological salt concentration (150 mM NaCl, Figure 3c,d). Heparin solutions (0.02 mg/mL) were titrated with the conjugates, and the increased scattering intensity (derived count rate) and hydrodynamic diameter (Dh) were used as indicators for binding and complex formation. In the plain buffer, the hydrodynamic diameters of heparin titrated with SA or 1SP yielded complexes with sizes of ∼20 nm within the whole concentration range (Figure 3a). Much higher values were recorded when heparin was titrated with 3SP or 7SP. This is expected as the one binding site on 1SP results in complex coacervate core micelles, while the multiple binding sites on 3SP and 7SP can cross-link with heparin and therefore form larger complexes.61 As for the derived count rate, solutions titrated with PS, 3SP, and 7SP increased sharply at the low mass ratios and decreased after a certain mass ratio was reached (Figure 3b), which is a typical phenomenon when oppositely charged polyelectrolytes are complexed.62 These peaks indicate a neutral charge balance, and they agree well with the saturation points obtained from the MB displacement assay.
Figure 3.
Titration of heparin (0.02 mg/mL) with binders (a, b) in the 2 mM Tris-HCl buffer and (c, d) at the physiological salt concentration (2 mM Tris-HCl, 150 mM NaCl) was monitored with DLS. The hydrodynamic diameter (a, c) and derived count rate (b, d) were used as indicators for the successful complexation. Titrations were performed using triplicate samples, and the averaged results with standard deviation are presented. (e) Peak force tapping mode AFM images (insets show the peak force error scale) of heparin neutralized with (1, 3) 3SP (mB/mH = 40) and (2, 4) 7SP (mB/mH = 15) in the presence of 0 mM (1, 2) and 150 mM (3, 4) NaCl. The corresponding DLS samples have been marked on the data.
At the physiologically relevant salt concentration, the binding affinities of all compounds were slightly weakened because of the shielding effect of counter ions.51 Nevertheless, both hydrodynamic diameter values and derived count rates remained high for heparin complexes formed with 3SP or 7SP (Figure 3c,d).63 SA and 1SP showed a small increase in the count rate and no change in size, indicating minor complex formation with heparin at these conditions. The free peptide presented heparin-binding ability in the Tris-HCl buffer, as indicated by the increased diameter and count rate, but the binding was not observed in the presence of 150 mM NaCl (Figure S4).
The morphologies of heparin complexed with 3SP and 7SP were directly visualized with AFM (Figure 3e and Figure S5). Both complexes were well dispersed on the silica surface and adopted roughly spherical morphologies with sizes up to ∼1 μm. No obvious size change was observed in buffers with or without NaCl. The size of the complexes observed with AFM is slightly smaller than with DLS, which could be attributed to drying and consequent shrinking of the complexes.64
Finally, in vitro heparin neutralization efficiency was evaluated in plasma using a chromogenic anti-Xa assay (Figure 4). Again, it can be observed that the neutralization efficiency increased with the number of conjugated peptides. SA and 1SP showed negligible neutralization efficiency, which is consistent with the DLS results in 150 mM NaCl. Compared to SA and 1SP, 3SP and 7SP showed much better performance. 3SP reached as high as 60% neutralization at ∼50 equiv, while 7SP showed full heparin neutralization at ∼25 equiv.
Figure 4.

Heparin neutralization measured with the anti-Xa assay in plasma. Experiments were performed using triplicate samples, and the averaged results with standard deviation are presented.
Combining all the results from the binding assays, we found that the SA conjugated with multiple peptides (3SP and 7SP) showed significant multivalency effects, leading to excellent heparin-binding ability. 7SP showed better performance than that of PS when comparing the molar ratios, which underlines the potential of 7SP to substitute PS. On the other hand, the heparin-binding capacity of unmodified SA and the peptide was only moderate in the Tris-HCl buffer and negligible at the physiological salt concentration. When the two components were conjugated in a 1:1 ratio (1SP), significant enhancement was achieved but only in the absence of added electrolytes and plasma. Considering that there are 30–35 lysines available for modification on the SA surface, it is foreseeable that even more efficient heparin binders could be achieved through further optimization.
Biocompatibility Study
The biocompatibility of conjugates, pure PS, and heparin was first studied with an MTT cell viability assay. Human dermal fibroblasts (HDF) cells were incubated for 4, 14, or 24 h with the free compounds (PS, heparin, HBP, SA, 1SP, 3SP, and 7SP) and the same components complexed with heparin. As can be observed from Figure 5 and Figure S6, SA and heparin did not induce any decrease in cell viability at 100 μg/mL after 24 h of incubation. Increased cytotoxicity was observed with PS as the incubation time and concentration increased: the cell viability dropped to 50% after 24 h of incubation (100 μg/mL PS). The free peptide followed a similar trend, but the drop at 100 μg/mL was not as drastic as in the case of PS. For 1SP, no toxicity was observed. However, when the number of peptides conjugated to SA was increased, cell viability decreased with the increasing concentration and incubation time. In general, 3SP and 7SP showed similar influence on fibroblasts as PS. When 1 μg/mL (∼0.2 IU/mL) heparin was first neutralized by the compounds and subsequently incubated with fibroblasts, only minor toxicity was observed (Figure 5).
Figure 5.

Effect of the heparin binders on cell viability evaluated with MTT assay using HDF cells. (left) Cells were incubated with free binders at four concentrations for 24 h. (right) Heparin of 1 μg/mL was first neutralized by binders and then incubated with the cells for 4, 14, or 24 h. Measurements were performed using triplicate samples, and the averaged results with standard deviation are presented.
Aside from MTT assay, hemolysis assay was utilized to evaluate biocompatibility of the studied compounds under application-relevant conditions. Since heparin has been widely used in both surgery and blood transfusion, it is of vital importance to assess the influence of the heparin-binding conjugates on the stability of RBCs.65,66 Hemolysis data in Figure 6 shows that the conjugates induced negligible hemolysis on RBCs up to 100 μg/mL. When 5 μg/mL (1.1 IU/mL) heparin was first fully neutralized by 3SP or 7SP and thereafter incubated with RCBs, no noticeable hemolysis was observed. Combining the hemolysis and MTT assay results, we can conclude that the biocompatibility of the SA–peptide conjugates is comparable to that of the commercial PS.
Figure 6.

Hemolysis assay with RBCs; results indicate that both (a) binders alone and (b) binder–heparin (5 μg/mL) complexes cause negligible red blood cell hemolysis. Solutions of 1× phosphate-buffered saline (PBS) and 1% Triton X-100 were used as the negative and positive control, respectively. Measurements were performed using triplicate samples, and the averaged results with standard deviation are presented.
Switch-On Heparin Detection
With the help of a dye-modified DNA oligonucleotide and the 7SP, we were able to design a detection system for ultralow amounts of heparin. A non-self-complementary 22 nt-long DNA sequence (0.2 μM), modified with a fluorescent dye (6-FAM) at the 5′ end and a quencher dye (BHQ1) at the 3′ end, was first complexed with 7SP through electrostatic interactions. This led to a significant drop in 6-FAM fluorescence intensity due to formation of the complex where both dyes are at close proximity (blue squares in Figure 7). It was found that the maximum quenching effect was reached at 9 μg/mL 7SP already, resulting in roughly one-sixth of the initial fluorescence intensity (FL, off). When heparin was introduced, DNA in the 7SP–DNA complex was replaced with heparin owing to the high binding efficiency between heparin and 7SP, yielding free unbound DNA strands and 7SP–heparin complexes. As a result, the fluorescence intensity increased as a function of heparin concentration (orange triangles in Figure 7). It can be observed that, within the range of 0–5.5 μg/mL heparin, the fluorescence intensity rises almost linearly with respect to the increasing heparin concentration. The initial fluorescence level is regained at 5.5 μg/mL (FL, on), showing that quantitative detection of heparin is feasible with this system. Moreover, the detection limit reaches 0.5 μg/mL (∼0.1 IU/mL), which is well below the lowest therapeutic level (0.2 IU/mL), thus making the 7SP–DNA complex a highly sensitive heparin detector.4−6 In addition, the selectivity of the detector complex toward heparin was compared to two other GAG analogues, hyaluronic acid and chondroitin sulfate (Figure S7). As expected, in the same concentration range, no clear fluorescence change was observed with chondroitin sulfate, while hyaluronic acid showed only a moderate increase (∼2-fold enhancement). Plausibly, this difference in the obtained fluorescence changes can be attributed to the charge densities as the charge density of the hyaluronic acid is higher than that of chondroitin sulfate. Nevertheless, according to these results, the detector complex is very selective toward heparin (∼6-fold fluorescence enhancement with heparin and only ∼2-fold enhancement with hyaluronic acid).
Figure 7.

Fluorescence switch-on detection of heparin in the 2 mM Tris-HCl buffer with the 7SP–DNA complex. Blue squares (on the left-hand side) show the titration of 0.2 μM DNA with 7SP. Orange triangles (on the right-hand side) show the fluorescence recovered upon adding heparin to the solution of the 7SP–DNA complex. The insets show schematic steps of the process. Titrations were performed using triplicate samples, and the averaged results with standard deviation are presented.
Conclusions
In this study, we expanded the application space of SA to heparin binding. Assays showed that the heparin-binding ability of the three heparin neutralizers kept mounting as the number of conjugated peptides increased (7SP > 3SP > 1SP). 3SP and 7SP exhibited excellent binding affinity in both the buffer and blood plasma. All SA–peptide conjugates as well as conjugate–heparin complexes showed low or negligible cytotoxicity to HDF cells in application-relevant concentrations. Additionally, hemolysis assay results indicated that no hemolysis was induced on RBCs even with high conjugate concentrations (0.1 mg/mL). Finally, with the help of a dye complex containing 7SP and FAM- and BHQ-modified DNA, we were able to quantitatively detect heparin down to 0.1 IU/mL, which is well below the lower limit of clinically relevant dosage. In summary, all obtained results indicate that these multifunctional (neutralization/detection) biomolecule-based heparin binders are promising alternatives to the commercially available PS, which is arguably efficient but rather contentious due to its adverse effects.
Experimental Section
SA–Peptide Conjugate Synthesis
For the syntheses of 1SP, 1.6 mg (0.72 μmol) of an N-terminal maleimide-modified peptide (sequence: KME KKL HAV PAA KTV KFK, GenScript) was mixed with 40 mg (0.6 μmol) of SA (1.2:1 in a molar ratio) in 1 mL of a 1× phosphate-buffered saline (PBS) buffer and let to react overnight. The crude product was purified with HPLC using a HiTrap Heparin HP 5 mL column (GE Healthcare Life Sciences, elution buffers: buffer A: 10 mM PB buffer, pH 6.0 and buffer B: 10 mM PB buffer, 300 mM NaCl, pH 6.0). The collected fraction was concentrated and dialyzed against Milli-Q water for three days (dialysis tubing cellulose membrane, molecular weight cut-off (MWCO) of 14 kDa, Sigma-Aldrich). The final product was lyophilized, weighed, and dissolved in Milli-Q water for further use.
For the synthesis of 3SP and 7SP, 5 μmol (5×) and 10 μmol (10×) of di(N-succinimidyl) 3,3′-dithiodipropionate in 100 μL of dimethyl sulfoxide (DMSO) were added to 900 μL of PBS buffer solutions containing 1 μmol (66 mg) of SA. These solutions were kept at room temperature overnight. The successful attachment of cross-linkers was determined by HPLC and MALDI-TOF. Before peptide conjugation, 0.3 μmol of each cross-linker-capped SA was first washed three times with PBS using centrifugal filters with an MWCO of 10 kDa (Amicon Ultra). Thirty and 60 μmol of tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (20×× cross-linker, respectively) in 500 μL PBS were added to reduce dithiols. Afterward, the solutions were desalted with a HiTrap desalting column (GE Healthcare Life Sciences, elution buffer: Milli-Q water). Collected fractions were immediately mixed with 1.5 μmol (5×) and 3 μmol (10×) of the N-maleimide-modified peptide and let to react overnight. After the reaction, crude products were purified with HPLC using a HiTrap Heparin HP 5 mL column (GE Healthcare Life Sciences, elution buffers: buffer A: 10 mM PB buffer, pH 6.0; buffer B: 10 mM PB buffer, 1 M NaCl, pH 6.0.). Collected fractions were concentrated and dialyzed against Milli-Q water for three days (dialysis tubing cellulose membrane, MWCO of 14 kDa, Sigma-Aldrich). Final products were lyophilized, weighed, and dissolved in Milli-Q water for further use.
Methylene Blue (MB) Displacement Assay
Eighty microliters of 0.013 mg/mL MB was first mixed with 20 μL of 0.1 mg/mL heparin in a 2 mM Tris-HCl buffer, pH 7.3. Different concentrations of compounds in 20 μL in volume were added to the MB/heparin solution and allowed to equilibrate for 5 min on a shaker. Next, absorption spectra of the samples (400–750 nm) were measured with a BioTek Cytation 3-microplate reader in a 96-well plate at room temperature. The absorbance intensity ratio A (664 nm)/A (568 nm) was used to determine the heparin-binding ability. Measurements were performed using triplicate samples.
Dynamic Light Scattering (DLS) Measurement
DLS measurements were carried out with a Zetasizer Nano ZS device (Malvern Instruments) with a 4 mW He–Ne ion laser at the wavelength of 633 nm and an Avalanche photodiode detector at an angle of 173°. Zetasizer software (Malvern Instruments) was used to attain the data. Cumulant analysis was used to obtain the intensity mean value of the complex size, that is, the hydrodynamic diameter. Experiments were carried out at 25 °C. Heparin solutions were prepared by diluting 10 mg/mL heparin stock solution to 0.02 mg/mL in 0.3 mL of the buffer. The heparin solutions were titrated with 2 μL of sample solutions (different compound concentrations) resulting in total sample volumes of 20 μL. Measurements were carried out in 2 mM Tris-HCl buffer at pH 7.3 with 0 and 150 mM NaCl. After every addition, the samples were allowed to equilibrate for 1 min. Each titration series was carried out three times, and all titration points were measured three times.
Atomic Force Microscopy (AFM) Imaging/Measurement
AFM imaging was carried out with Dimension Icon (Bruker) in the ScanAsyst Fluid mode. To prepare the samples for imaging, 15–20 μL of the sample solution (heparin at 0.1 mg/mL) was incubated on a freshly cleaved mica surface for 5 min and allowed to dry. Then, ∼250 μL of the corresponding buffer (2 mM Tris–HCl with or without 150 mM NaCl) was carefully pipetted on the mica to form a meniscus between the AFM scanner and sample surface. During the imaging, the tip scanning velocity was limited to 2–5 μm/s to minimize the drifting of the particles on surface caused by the probe. The images were taken in 256 × 256 resolution, processed, and exported with NanoScope Analysis software ver. 1.9 (Bruker).
Anti-Xa Assay
Heparin neutralization with compounds in plasma was evaluated using a commercial two-stage kit, BiophenTM Anti-Xa (221005). Conjugates of different concentrations were first lyophilized and redissolved in human plasma. Dissolved compounds were then added to the heparin solution in 150 mM NaCl (0.1 mg/mL), giving a final heparin concentration of 0.075 IU/mL and compound/heparin ratios similar to those of the MB replacement assay and DLS measurement. Kit reagents were utilized according to the manufacturer’s instructions. To run the calorimetric assay, 40 μL of the sample solution was added to a 96-well microplate followed by the addition of 40 μL of antithrombin and incubation for 2 min. Then, 40 μL of factor Xa was added and incubated for another 2 min. Afterward, 40 μL of the factor Xa-specific chromogenic substrate was added to the solution and let to react for 2 min. Finally, the reaction was quenched by introducing 80 μL of 2% citric acid. The absorbance at 405 nm was recorded immediately using a BioTek Cytation 3-microplate reader. The anticoagulant activity is inversely proportional to the measured absorption intensity, and the percentage of neutralization was determined using a calibration curve constructed according to the manufacturer’s instructions (Figure S8). Measurements were performed using triplicate samples.
Cell Culture and MTT Assay
Human dermal fibroblasts (HDF) cells (Gibco) were purchased from Fisher Scientific. The cells were then expanded in Dulbecco’s Modified Eagle Medium (DMEM) substituted with 10% fetal bovine serum (FBS) and 100 U/mL penicillin and 100 μg/mL streptomycin. The cells were kept in humidified conditions with 5% CO2 at 37 °C. Once reached the 90% confluency the cells were split using 0.25% ethylenediaminetetraacetic acid (EDTA)-trypsin. Cell passages between 3 and 5 were used for the cell culture studies. Before MTT assay, cells were split into 96-well culture plates (approximately 10,000 cells/well) and incubated for 24 h. After the incubation, the culture media was replaced with 100 μL of compound solutions (0.1–100 μg/mL) or compound/heparin complex solutions in DMEM supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. Based on the DLS data, the amount of PS, 3SP, and 7SP was chosen to fully neutralize heparin, while the highest mass ratios were used for HBP, SA, and 1SP (PS = 2 μg/mL, HBP = 5 μg/mL, SA = 140 μg/mL, 1SP = 140 μg/mL, 3SP = 40 μg/mL, 7SP = 15 μg/mL, and heparin = 1 μg/mL). The cells were then kept in humidified conditions with 5% CO2 at 37 °C for 4, 14, or 24 h. After that, the sample solutions in each well were replaced with 100 uL of complete media and 10 uL of MTT solution (5 mg/mL in PBS). After 4 h of incubation at 37 °C with 5% CO2, the MTT solution was replaced with 100 μL DMSO in each well to dissolve formazan crystals before reading. The absorbance was measured with a microplate reader (Cytation 3, BioTek) at the wavelength of 570 nm. Measurements were carried out using triplicate samples.
Hemolysis Assay
The detailed procedure for the hemolysis assay has been previously reported.67 Generally, freshly donated human red blood cells were purchased (Cambridge Bioscience Ltd., the United Kingdom) and stored at 4 °C. Before samples were added, 1 mL of blood was centrifuged at 500 × g for 5 min and the plasma was removed gently. The remaining red blood cells were washed with 1× PBS three times and redispersed to the initial volume in 1× PBS. The red blood cells were diluted 50× and split into 96-well culture plates (190 μL/well). The concentrated sample (10 μL) or compound/heparin solutions in 1× PBS were added to each well, resulting in the desired final compound concentrations (25–100 μg/mL) or compound/heparin complex concentrations (PS = 10 μg/mL, HBP = 25 μg/mL, SA = 700 μg/mL, 1SP = 700 μg/mL, 3SP = 200 μg/mL, 7SP = 75 μg/mL, and heparin = 5 μg/mL; based on the DLS data, the amount of PS, 3SP, and 7SP was chosen to fully neutralize heparin, while the highest mass ratios were used for HBP, SA, and 1SP). Ten microliters of 20% Triton X-100 in 1× PBS and 10 μL of 1× PBS were added as positive and negative controls, respectively. After incubation at 37 °C for 1 h, the plates were centrifuged for 5 min at 500 × g to pellet intact erythrocytes, and 100 μL of the supernatant from each well was delicately transferred into a clear 96-well plate. The resulting hemoglobin in the supernatant was measured at 540 nm with a microplate reader (Cytation 3, Biotek). The percentage of hemolysis was calculated as follows:
%Hemolysis = [(Asample – Anegative control)/(Apositive control – Anegative control)] × 100
The measurements were performed using triplicate samples.
Switch-On Heparin Detection
The quenching effect of 7SP on FAM- and BHQ- modified 22 nt DNA was evaluated by titrating 0.2 μM DNA with 0–9 μg/mL 7SP in 2 mM Tris-HCl buffer. The fluorescence intensity stabilized after 9 μg/mL 7SP. For the heparin replacement assay, 0.2 μM DNA was first quenched with 9 μg/mL 7SP, which was followed by the titration of heparin.
Acknowledgments
We gratefully thank the Academy of Finland (project nos. 308578, 303804, and 267497), Jane and Aatos Erkko Foundation, Sigrid Jusélius Foundation, and Emil Aaltonen Foundation for financial support. This work was carried out under the Academy of Finland’s Centers of Excellence Programme (2014–2019).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b02883.
Supplementary details on reagents and materials, procedures for HPLC and MALDI-TOF, characterization of cross-linker capped SA, deprotection of cross-linker-capped SA, purification of 1SP, 3SP, and 7SP, absorbance spectra of SA, HBP, 1SP, 3SP, and 7SP titrated with heparin in MB displacement assay, DLS results of heparin titrated with HBP, AFM characterization in Tris-HCl buffer, MTT assay of compounds (incubation time of 4 and 14 h), control experiments of titration with hyaluronic acid and chondroitin 4-sulfate in Tris-HCl buffer, and a calibration curve for anti-Xa assay (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Casu B.; Naggi A.; Torri G. Re-Visiting the Structure of Heparin. Carbohydr. Res. 2015, 403, 60–68. 10.1016/j.carres.2014.06.023. [DOI] [PubMed] [Google Scholar]
- Bromfield S. M.; Wilde E.; Smith D. K. Heparin Sensing and Binding-Taking Supramolecular Chemistry Towards Clinical Applications. Chem. Soc. Rev. 2013, 42, 9184–9195. 10.1039/c3cs60278h. [DOI] [PubMed] [Google Scholar]
- Esko J. D.; Selleck S. B. Order Out of Chaos: Assembly of Ligand Binding Sites in Heparin Sulfate. Annu. Rev. Biochem. 2002, 71, 435–471. 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Hirsh J.; Raschke R. Heparin and Low-Molecular-Weight Heparin: The Seventh ACCP Conference on Antithrombotic and Thrombolytic Therapy. Chest 2004, 126, 188S–203S. 10.1378/chest.126.3_suppl.188S. [DOI] [PubMed] [Google Scholar]
- Warkentin T. E.; Levine M. N.; Hirsh J.; Horsewood P.; Roberts R. S.; Gent M.; Kelton J. G. Heparin-Induced Thrombocytopenia in Patients Treated with Low-Molecular-Weight Heparin or Unfractionated Heparin. N. Engl. J. Med. 1995, 332, 1330–1336. 10.1056/NEJM199505183322003. [DOI] [PubMed] [Google Scholar]
- Girolami B.; Girolami A. Heparin-Induced Thrombocytopenia: A Review. Semin. Thromb. Hemostasis 2006, 32, 803–809. 10.1055/s-2006-955463. [DOI] [PubMed] [Google Scholar]
- Balhorn R. The Protamine Family of Sperm Nuclear Proteins. Genome Biol. 2007, 8, 227–234. 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despotis G. J.; Gravlee G.; Filos K.; Levy J. Anticoagulation Monitoring During Cardiac Surgery: A Review of Current and Emerging Techniques. Anesthesiology 1999, 91, 1122–1151. 10.1097/00000542-199910000-00031. [DOI] [PubMed] [Google Scholar]
- Vandiver J. W.; Vondracek T. G. Antifactor Xa Levels Versus Activated Partial Thromboplastin Time for Monitoring Unfractionated Heparin. Pharmacotherapy 2012, 32, 546–558. 10.1002/j.1875-9114.2011.01049.x. [DOI] [PubMed] [Google Scholar]
- Bromfield S. M.; Posocco P.; Fermeglia M.; Pricl S.; Rodríguez-López J.; Smith D. K. A Simple New Competition Assay for Heparin Binding in Serum Applied to Multivalent PAMAM Dendrimers. Chem. Commun. 2013, 49, 4830–4832. 10.1039/c3cc41251b. [DOI] [PubMed] [Google Scholar]
- Horrow J. C. Protamine: A Review of its Toxicity. Anesth. Analg. 1985, 64, 348–361. [PubMed] [Google Scholar]
- Wan S.; LeClerc J.-L.; Vincent J.-L. Inflammatory Response to Cardiopulmonary Bypass: Mechanisms Involved and Possible Therapeutic Strategies. Chest 1997, 112, 676–692. 10.1378/chest.112.3.676. [DOI] [PubMed] [Google Scholar]
- Nybo M.; Madsen J. S. Serious Anaphylactic Reactions due to Protamine Sulfate: A Systematic Literature Review. Basic Clin. Pharmacol. Toxicol. 2008, 103, 192–196. 10.1111/j.1742-7843.2008.00274.x. [DOI] [PubMed] [Google Scholar]
- Hirsh J.; Warkentin T. E.; Shaughnessy S. G.; Anand S. S.; Halperin J. L.; Raschke R.; Granger C.; Ohman E. M.; Dalen J. E. Heparin and Low-Molecular-Weight Heparin: Mechanisms of Action, Pharmacokinetics, Dosing, Monitoring, Efficacy, and Safety. Chest 2001, 119, 64S–94S. 10.1378/chest.119.1_suppl.64S. [DOI] [PubMed] [Google Scholar]
- Capila I.; Linhardt R. J. Heparin-Protein Interactions. Angew. Chem., Int. Ed. 2002, 41, 391–412. . [DOI] [PubMed] [Google Scholar]
- Sloand E. M.; Kessler C. M.; McIntosh C. L.; Klein H. G. Methylene Blue for Neutralization of Heparin. Thromb. Res. 1989, 54, 677–686. 10.1016/0049-3848(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Kikura M.; Lee M. K.; Levy J. H. Heparin Neutralization with Methylene Blue, Hexadimethrine, or Vancomycin after Cardiopulmonary Bypass. Anesth. Analg. 1996, 83, 223–227. 10.1213/00000539-199608000-00004. [DOI] [PubMed] [Google Scholar]
- Schuksz M.; Fuster M. M.; Brown J. R.; Crawford B. E.; Ditto D. P.; Lawrence R.; Glass C. A.; Wang L.; Tor Y.; Esko J. D. Surfen, A Small Molecule Antagonist of Heparan Sulfate. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 13075–13080. 10.1073/pnas.0805862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuziej J.; Litinas E.; Hoppensteadt D. A.; Liu D.; Walenga J. M.; Fareed J.; Jeske W. In Vivo Neutralization of Unfractionated Heparin and Low-Molecular-Weight Heparin by a Novel Salicylamide Derivative. Clin. Appl. Thromb./Hemostasis 2010, 16, 377–386. 10.1177/1076029610366439. [DOI] [PubMed] [Google Scholar]
- Choi S.; Clements D. J.; Pophristic V.; Ivanov I.; Vemparala S.; Bennett J. S.; Klein M. L.; Winkler J. D.; DeGrado W. F. The Design and Evaluation of Heparin-Binding Foldamers. Angew. Chem. Int. Ed. 2005, 44, 6685–6689. 10.1002/anie.200501279. [DOI] [PubMed] [Google Scholar]
- Kelly C.; Khaja S.; Vena A.; Yu A.; Esson J. M. Polyion-Sensitive Membrane-Based Electrodes for Heparin-Binding Foldamer Analysis. Anal. Chim. Acta 2010, 681, 1–7. 10.1016/j.aca.2010.09.012. [DOI] [PubMed] [Google Scholar]
- Wakefield T. W.; Andrews P. C.; Wrobleski S. K.; Kadell A. M.; Fazzalari A.; Nichol B. J.; Vanderkooi T.; Stanley J. C. Reversal of Low-Molecular-Weight Heparin Anticoagulation by Synthetic Protamine Analogues. J. Surg. Res. 1994, 56, 586–593. 10.1006/jsre.1994.1093. [DOI] [PubMed] [Google Scholar]
- Hernaiz M. J.; LeBrun L. A.; Wu Y.; Sen J. W.; Linhardt R. J.; Heegaard N. H. Characterization of Heparin Binding by a Peptide from Amyloid P Component Using Capillary Electrophoresis, Surface Plasmon Resonance and Isothermal Titration Calorimetry. Eur. J. Biochem. 2002, 269, 2860–2867. 10.1046/j.1432-1033.2002.02964.x. [DOI] [PubMed] [Google Scholar]
- Kamiński K.; Płonka M.; Ciejka J.; Szczubiałka K.; Nowakowska M.; Lorkowska B.; Korbut R.; Lach R. Cationic Derivatives of Dextran and Hydroxypropylcellulose as Novel Potential Heparin Antagonists. J. Med. Chem. 2011, 54, 6586–6596. 10.1021/jm200380w. [DOI] [PubMed] [Google Scholar]
- Välimäki S.; Khakalo A.; Ora A.; Johansson L.-S.; Rojas O. J.; Kostiainen M. A. Effect of PEG-PDMAEMA Block Copolymer Architecture on Polyelectrolyte Complex Formation with Heparin. Biomacromolecules 2016, 17, 2891–2900. 10.1021/acs.biomac.6b00699. [DOI] [PubMed] [Google Scholar]
- Reyes-Ortega F.; Rodríguez G.; Aguilar M. R.; Lord M.; Whitelock J.; Stenzel M. H.; Román J. S. Encapsulation of Low Molecular Weight Heparin (Bemiparin) into Polymeric Nanoparticles Obtained from Cationic Block Copolymers: Properties and Cell Activity. J. Mater. Chem. B 2013, 1, 850–860. 10.1039/C2TB00194B. [DOI] [PubMed] [Google Scholar]
- Kalaska B.; Miklosz J.; Kamiński K.; Musielak B.; Yusa S.-I.; Pawlak D.; Nowakowska M.; Szczubiałka K.; Mogielnicki A. The Neutralization of Heparan Sulfate by Heparin-Binding Copolymer as a Potential Therapeutic Target. RSC Adv. 2019, 9, 3020–3029. 10.1039/C8RA09724K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Lord M. S.; Stenzel M. H. A Polyion Complex Micelle with Heparin for Growth Factor Delivery and Uptake into Cells. J. Mater. Chem. B 2013, 1, 1635–1643. 10.1039/c3tb00360d. [DOI] [PubMed] [Google Scholar]
- Rodrigo A. C.; Barnard A.; Cooper J.; Smith D. K. Self-Assembling Ligands for Multivalent Nanoscale Heparin Binding. Angew. Chem., Int. Ed. 2011, 50, 4675–4679. 10.1002/anie.201100019. [DOI] [PubMed] [Google Scholar]
- Montalvo G. L.; Zhang Y.; Young T. M.; Costanzo M. J.; Freeman K. B.; Wang J.; Clements D. J.; Magavern E.; Kavash R. W.; Scott R. W.; Liu D.; Degrado W. F. De novo Design of Self-Assembling Foldamers that Inhibit Heparin-Protein Interactions. ACS Chem. Biol. 2014, 9, 967–975. 10.1021/cb500026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fechner L. E.; Albanyan B.; Vieira V. M. P.; Laurini E.; Posocco P.; Pricl S.; Smith D. K. Electrostatic Binding of Polyanions Using Self-Assembled Multivalent (SAMul) Ligand Displays-Structure-Activity Effects on DNA/Heparin Binding. Chem. Sci. 2016, 7, 4653–4659. 10.1039/C5SC04801J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.; Guo W.; Ding Y.; Cheng H.; Wei H. Modulating Luminescence of Tb3+ with Biomolecules for Sensing Heparin and Its Contaminant OSCS. Biosens. Bioelectron. 2016, 86, 858–863. 10.1016/j.bios.2016.07.085. [DOI] [PubMed] [Google Scholar]
- Zhong Z.; Anslyn E. V. A Colorimetric Sensing Ensemble for Heparin. J. Am. Chem. Soc. 2002, 124, 9014–9015. 10.1021/ja020505k. [DOI] [PubMed] [Google Scholar]
- Välimäki S.; Beyeh N. K.; Linko V.; Ras R. H. A.; Kostiainen M. A. A Supramolecular Host–Guest Complex for Heparin Binding and Sensing. Nanoscale 2018, 10, 14022–14030. 10.1039/C8NR03132K. [DOI] [PubMed] [Google Scholar]
- Ding Y.; Shi L.; Wei H. A “turn on” fluorescent probe for heparin and its oversulfated chondroitin sulfate contaminant. Chem. Sci. 2015, 6, 6361–6366. 10.1039/C5SC01675D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa Y.; Hayashida R.; Seki T.; Anzai J. Fluorometric Determination of Heparin Based on Self-Quenching of Fluorescein-Labeled Protamine. Talanta 2008, 76, 736–741. 10.1016/j.talanta.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Chen L. J.; Ren Y. Y.; Wu N. W.; Sun B.; Ma J. Q.; Zhang L.; Tan H.; Liu M.; Li X.; Yang H. B. Hierarchical Self-Assembly of Discrete Organoplatinum(II) Metallacycles with Polysaccharide via Electrostatic Interactions and Their Application for Heparin Detection. J. Am. Chem. Soc. 2015, 137, 11725–11735. 10.1021/jacs.5b06565. [DOI] [PubMed] [Google Scholar]
- Chan C. W.; Smith D. K. Pyrene-Based Heparin Sensors in Competitive Aqueous Media-the Role of Self-Assembled Multivalency (SAMul). Chem. Commun. 2016, 52, 3785–3788. 10.1039/C6CC00163G. [DOI] [PubMed] [Google Scholar]
- Hawkins M. J.; Soon-Shiong P.; Desai N. Protein Nanoparticles as Drug Carriers in Clinical Medicine. Adv. Drug Delivery Rev. 2008, 60, 876–885. 10.1016/j.addr.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Kratz F. Albumin as a Drug Carrier: Design of Prodrugs, Drug Conjugates and Nanoparticles. J. Controlled. Release 2008, 132, 171–183. 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Taguchi K.; Chuang V. T. G.; Maruyama T.; Otagiri M. Pharmaceutical Aspects of the Recombinant Human Serum Albumin Dimer: Structural Characteristics, Biological Properties, and Medical Applications. J. Pharm. Sci. 2012, 101, 3033–3046. 10.1002/jps.23181. [DOI] [PubMed] [Google Scholar]
- Nosrati H.; Sefidi N.; Sharafi A.; Danafar H.; Manjili H. K. Bovine Serum Albumin (BSA) Coated Iron Oxide Magnetic Nanoparticles as Biocompatible Carriers for Curcumin-Anticancer Drug. Bioorg. Chem. 2018, 76, 501–509. 10.1016/j.bioorg.2017.12.033. [DOI] [PubMed] [Google Scholar]
- Elsadek B.; Kratz F. Impact of Albumin on Drug Delivery — New Applications on the Horizon. J. Controlled Release 2012, 157, 4–28. 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- Auvinen H.; Zhang H.; Nonappa; Kopilow A.; Niemelä E. H.; Nummelin S.; Correia A.; Santos H. A.; Linko V.; Kostiainen M. A. Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv. Healthcare Mater. 2017, 6, 1700692. 10.1002/adhm.201700692. [DOI] [PubMed] [Google Scholar]
- Pitek A. S.; Jameson S. A.; Veliz F. A.; Shukla S.; Steinmetz N. F. Serum Albumin ‘Camouflage’ of Plant Virus Based Nanoparticles Prevents Their Antibody Recognition and Enhances Pharmacokinetics. Biomaterials 2016, 89, 89–97. 10.1016/j.biomaterials.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele E.; Spinelli G. P.; Miele E.; Tomao F.; Tomao S. Albumin-Bound Formulation of Paclitaxel (Abraxane® ABI-007) in the Treatment of Breast Cancer. Int. J. Nanomed. 2009, 4, 99–105. 10.2147/ijn.s3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socinski M. A.; Bondarenko I.; Karaseva N. A.; Makhson A. M.; Vynnychenko I.; Okamoto I.; Hon J. K.; Hirsh V.; Bhar P.; Zhang H.; Iglesias J. L.; Renschler M. F. Weekly Nab-Paclitaxel in Combination with Carboplatin Versus Solvent-Based Paclitaxel Plus Carboplatin as First-Line Therapy in Patients with Advanced Non–Small-Cell Lung Cancer: Final Results of a Phase III Trial. J. Clin. Oncol. 2012, 30, 2055–2062. 10.1200/JCO.2011.39.5848. [DOI] [PubMed] [Google Scholar]
- Kan M.; Wang F.; Xu J.; Crabb J.; Hou J.; McKeehan W. An Essential Heparin-Binding Domain in the Fibroblast Growth Factor Receptor Kinase. Science 1993, 259, 1918–1921. 10.1126/science.8456318. [DOI] [PubMed] [Google Scholar]
- Kostiainen M. A.; Hardy J. G.; Smith D. K. High-Affinity Multivalent DNA Binding by Using Low-Molecular-Weight Dendrons. Angew. Chem., Int. Ed. 2005, 44, 2556–2559. 10.1002/anie.200500066. [DOI] [PubMed] [Google Scholar]
- Kostiainen M. A.; Szilvay G. R.; Smith D. K.; Linder M. B.; Ikkala O. Multivalent Dendrons for High-Affinity Adhesion of Proteins to DNA. Angew. Chem., Int. Ed. 2006, 45, 3538–3542. 10.1002/anie.200504540. [DOI] [PubMed] [Google Scholar]
- Kostiainen M. A.; Szilvay G. R.; Lehtinen J.; Smith D. K.; Linder M. B.; Urtti A.; Ikkala O. Precisely Defined Protein-Polymer Conjugates: Construction of Synthetic DNA Binding Domains on Proteins by Using Multivalent Dendrons. ACS Nano 2007, 1, 103–113. 10.1021/nn700053y. [DOI] [PubMed] [Google Scholar]
- Miyadera T.; Kosower E. M. Receptor Site Labeling Through Functional Groups. 2. Reactivity of Maleimide Groups. J. Med. Chem. 1972, 15, 534–537. 10.1021/jm00275a024. [DOI] [PubMed] [Google Scholar]
- Ghosh S. S.; Kao P. M.; McCue A. W.; Chappelle H. L. Use of Maleimide-Thiol Coupling Chemistry for Efficient Syntheses of Oligonucleotide-Enzyme Conjugate Hybridization Probes. Bioconjugate Chem. 1990, 1, 71–76. 10.1021/bc00001a009. [DOI] [PubMed] [Google Scholar]
- Gindy M. E.; Ji S.; Hoye T. R.; Panagiotopoulos A. Z.; Prud’homme R. K. Preparation of Poly(ethylene glycol) Protected Nanoparticles with Variable Bioconjugate Ligand Density. Biomacromolecules 2008, 9, 2705–2711. 10.1021/bm8002013. [DOI] [PubMed] [Google Scholar]
- Simons S. S. Jr. Selective Covalent Labeling of Cysteines in Bovine Serum Albumin and in Hepatoma Tissue Culture Cell Glucocorticoid Receptors by Dexamethasone 21-Mesylate. J. Biol. Chem. 1987, 262, 9669–9675. [PubMed] [Google Scholar]
- Janatova J.; Fuller J. K.; Hunter M. J. Heterogeneity of Bovine Albumin with Respect to Sulfhydryl and Dimer Content. J. Biol. Chem. 1968, 243, 3612–3622. [PubMed] [Google Scholar]
- Hirayama K.; Akashi S.; Furuya M.; Fukuhara K. Rapid Confirmation and Revision of the Primary Structure of Bovine Serum Albumin by ESIMS and FRIT-FAB LC MS. Biochem. Biophys. Res. Commun. 1990, 173, 639–646. 10.1016/S0006-291X(05)80083-X. [DOI] [PubMed] [Google Scholar]
- Majorek K. A.; Porebski P. J.; Dayal A.; Zimmerman M. D.; Jablonska K.; Stewart A. J.; Chruszcz M.; Minor W. Structural and Immunologic Characterization of Bovine, Horse, and Rabbit Serum Albumins. Mol. Immunol. 2012, 52, 174–182. 10.1016/j.molimm.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.; Chen J.; Wu L.; Li W.; Chen J.; Cheng H.; Pan J.; Cai B. Hyaluronic Acid-Coated Bovine Serum Albumin Nanoparticles Loaded with Brucine as Selective Nanovectors for Intra-Articular Injection. International Journal of Nanomedicine 2013, 8, 3843–3853. 10.2147/IJN.S50721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almajano M. P.; Gordon M. H. Synergistic Effect of BSA on Antioxidant Activities in Model Food Emulsions. J. Am. Oil Chem. Soc. 2004, 81, 275–280. 10.1007/s11746-004-0895-6. [DOI] [Google Scholar]
- van der Gucht J.; Spruijt E.; Lemmers M.; Cohen Stuart M. A. Polyelectrolyte Complexes: Bulk Phases and Colloidal Systems. J. Colloid Interface Sci. 2011, 361, 407–422. 10.1016/j.jcis.2011.05.080. [DOI] [PubMed] [Google Scholar]
- Voets I. K.; de Keizer A.; Cohen Stuart M. A. Complex Coacervate Core Micelles. Adv. Colloid Interface Sci. 2009, 147–148, 300–318. 10.1016/j.cis.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Etrych T.; Leclercq L.; Boustta M.; Vert M. Polyelectrolyte Complex Formation and Stability When Mixing Polyanions and Polycations in Salted Media: A Model Study Related to the Case of Body Fluids. Eur. J. Pharm. Sci. 2005, 25, 281–288. 10.1016/j.ejps.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Sun W.; Mao S.; Mei D.; Kissel T. Self-Assembled Polyelectrolyte Nanocomplexes between Chitosan Derivatives and Enoxaparin. Eur. J. Pharm. Biopharm. 2008, 69, 417–425. 10.1016/j.ejpb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Makhro A.; Huisjes R.; Verhagen L. P.; del Mar Mañú-Pereira M.; Llaudet-Planas E.; Petkova-Kirova P.; Wang J.; Eichler H.; Bogdanova A.; van Wijk R.; et al. Red Cell Properties after Different Modes of Blood Transportation. Front Physiol. 2016, 7, 288–308. 10.3389/fphys.2016.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges J.; Trinh Q. D.; Jonas L.; Budäus L.; Larbig R.; Schlomm T.; Karakiewicz P. I.; Heinzer H.; Huland H.; Graefen M.; Steuber T. Influence of Low-Molecular-Weight Heparin Dosage on Red Blood Cell Transfusion, Lymphocele Rate and Drainage Duration After Open Radical Prostatectomy. Eur. J. Surg. Oncol. 2012, 38, 1082–1088. 10.1016/j.ejso.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Evans B. C.; Nelson C. E.; Yu S. S.; Beavers K. R.; Kim A. J.; Li H.; Nelson H. M.; Giorgio T. D.; Duvall C. L. Ex Vivo Red Blood Cell Hemolysis Assay for the Evaluation of pH-responsive Endosomolytic Agents for Cytosolic Delivery of Biomacromolecular Drugs. J. Vis. Exp. 2013, e50166 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.