Abstract
Multidrug resistance in pathogenic bacteria is an increasing problem in patient care and public health. Molecular nanomachines (MNMs) have the ability to open cell membranes using nanomechanical action. We hypothesized that MNMs could be used as antibacterial agents by drilling into bacterial cell walls and increasing susceptibility of drug-resistant bacteria to recently ineffective antibiotics. We exposed extensively drug-resistant Klebsiella pneumoniae to light-activated MNMs and found that MNMs increase the susceptibility to Meropenem. MNMs with Meropenem can effectively kill K. pneumoniae that are considered Meropenem-resistant. We examined the mechanisms of MNM action using permeability assays and transmission electron microscopy, finding that MNMs disrupt the cell wall of extensively drug-resistant K. pneumoniae, exposing the bacteria to Meropenem. These observations suggest that MNMs could be used to make conventional antibiotics more efficacious against multi-drug-resistant pathogens.
Keywords: molecular nanomachines, nanomechanical action, light activation, antimicrobial, antimicrobial resistance, multidrug resistance, extensive drug resistance
Multi-drug-resistant (MDR) pathogens are an increasing problem worldwide. Annually, 700 000 deaths are attributed to MDR and antimicrobial-resistant (AMR) strains of common bacterial infections. This number, if current trends in the use of antibiotics continue, is projected to increase beyond 10 million annual deaths by 2050.1 MDR infections create an increasingly large burden in healthcare and preventative practices.2 In their 2013 antibiotic-resistant threat report, the Centers for Disease Control and Prevention (CDC) listed 18 MDR and AMR pathogens that require immediate attention. Carbapenem-resistant Enterobacteriaceae (CRE) were identified as one of three pathogens at the highest threat level, demanding urgent action.3 Recognizing the global impact of MDR and AMR pathogens on patient care, the World Health Organization (WHO) put forth a Global Action Plan (GAP) in 2015 to ensure continued success in effective treatment and prevention of these infectious diseases.4 In 2017, the WHO also identified CRE as one of three carbapenem-resistant pathogens in their highest priority category (Priority 1: Critical) for research and development of new antibiotics, again highlighting the urgent need for solutions to counter pathogens resistant to last resort antibiotics.5
Klebsiella pneumoniae belongs to the family of Enterobacteriaceae and is one of the most important causes of nosocomial infections worldwide.6 This Gram-negative opportunistic pathogen colonizes the human intestine and is of great clinical importance, especially among very sick patients.7K. pneumoniae causes various healthcare-associated infections, including pneumonia, bloodstream infections, urinary tract infections, wound or surgical site infections, and meningitis.8−10 Over the last few decades, MDR K. pneumoniae infections have rapidly increased in hospital settings, making first-line antibiotics vastly ineffective. The emergence of carbapenem-resistant strains of K. pneumoniae as a major nosocomial infection has raised many concerns as antibiotic treatment options available against this pathogen are very limited.11−13 With the rapid emergence of resistance to conventional antibiotics that were once considered wonder drugs, there is an emergent need for the development of new unconventional antibiotic agents that can effectively counter MDR pathogens.
Molecular nanomachines (MNMs) are synthetic organic nanomolecules that have a rotor component with light-induced actuation (motorization) that rotates unidirectionally relative to a stator (Figure 1a).14−16 These MNMs can disrupt synthetic lipid bilayers and cell membranes with their rapid rotational movement. In the absence of light, MNMs diffuse into bilayers and display little or no toxicity.16−18 The mechanism for light-induced unidirectional rotation of MNMs can be explained as follows: (1) upon photoexcitation of the central twisted double bond, it will move into an excited state, wherein the rotor and stator are orthogonal to each other; (2) normally relaxation could occur in either direction, but since there is a neighboring stereogenic center at the methyl group, the two ways that it can rotate are diastereomeric and therefore different in energy, so relaxation occurs through the preferred lower energy twist direction that completes a half turn; (3) the molecule finds itself in a sterically encumbered twist state, and the rotor and stator will thermally slide past each other to a lower energy state; (4) then a second photoexcitation followed by (5) a thermally induced twist puts the molecule back in the original state.16−18 Recently, ultraviolet-light-activated MNMs were shown to use nanomechanical action to drill into cell membranes, creating pores in targeted cancer cells and causing cell death.17 Light-activated fast motor, MNM 1 (Figure 1b), was shown to cause cell necrosis in human prostate adenocarcinoma cells (PC-3) and mouse embryonic fibroblast cells (NIH 3T3). MNMs have various properties dependent on their steric structure and attached functional groups. They can be modified to give them specific properties and functions. Light-activated MNM 1 rotates ∼2–3 million revolutions per second and is considered a fast motor. Light-activated MNM 2 is a slow motor, rotating only ∼1.8 revolutions per hour and is a nanomechanical control for MNM 1. MNM 3 is similar to MNM 1 but with a triphenylphosphonium (TPP) cation attached to its stator portion. TPP targets eukaryotic mitochondria, causing MNM 3 to accumulate within the mitochondria.19 MNMs can also have peptide appendages for specific cell adhesion. Nanomechanical action of fast motor MNMs makes them potential broad-spectrum antibacterials. We hypothesized that MNM 1 can disrupt bacterial cell walls and act as a potent nanomechanical antibacterial agent either alone or facilitating the action of conventional antimicrobials.
Figure 1.
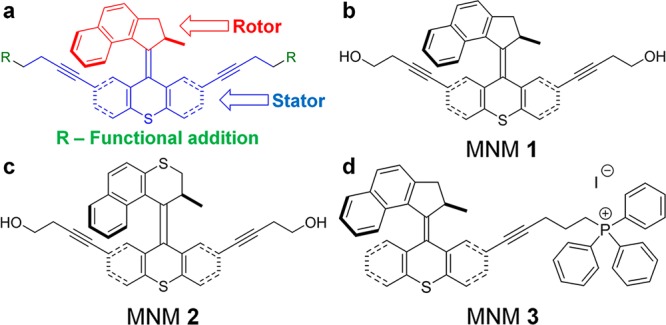
Molecular nanomachine structures. (a) Representative MNMs, illustrating the rotor portions (red), which rotate upon light activation relative to the stator portion (blue). R groups (green) are functional molecules that can be added to provide increased solubility, fluorophores for tracking, or serve as recognition sites for cellular targeting. (b) MNM 1 is a fast motor with a unidirectional rotor activated by 365 nm light. (c) MNM 2 is the corresponding slow motor that serves as a control. (d) MNM 3 is a fast motor similar to MNM 1 but with a triphenylphosphonium (TPP) cation attached to the stator portion. TPP targets eukaryotic mitochondria, causing MNM 3 to accumulate within mitochondria. This served as a control to demonstrate eukaryotic cell targeting of MNMs.
Among various AMR mechanisms used by MDR K. pneumoniae to resist carbapenems, the loss of cell wall outer membrane porins and production of K. pneumoniae carbapenemase (KPC) confer the highest levels of carbapenem resistance.20−23 The cell wall outer membrane (OM) lacking porins acts as a mechanical barrier that prevents carbapenem to permeate the OM and reach its target site, penicillin-binding proteins (PBP) in the periplasmic space.24 We explore the use of light-activated MNM 1 nanomechanical properties to drill pores and disrupt the cell wall in MDR K. pneumoniae to allow carbapenem to traverse the cell wall OM and cause bacterial cell death. One of the effective ways to kill AMR and MDR bacteria is to increase the intracellular concentration of antibiotics. Biological and chemical approaches have been shown to modulate metabolic pathways to help increase the effective concentrations of antibiotics within bacteria.25,26 In this study, we assay the use of a nanomechnical approach to disrupt the K. pneumoniae cell wall and increase the effective concentrations of antibiotics.
Here, we use an extensively drug-resistant (ψkp6) and an antibiotic-sensitive (ψkp7) strain of K. pneumoniae to first show that light-activated MNM 1 using its nanomechanical action can display antibacterial properties irrespective of pathogen antibiotic susceptibility profiles. Then we show that light-activated MNM 1 in combination with Meropenem has the ability to make an extensively drug-resistant K. pneumoniae susceptible to Meropenem at subtherapeutic concentrations. Our results indicate that light-activated MNM 1 uses its nanomechanical action to assist in bypassing the cell wall OM-induced antibacterial resistance posed by K. pneumoniae. Thus, MNM 1, together with antibiotics like Meropenem, is shown as a potent antibacterial agent with the potential to effectively counter the increasing problem of multidrug resistance not only in K. pneumoniae but also in many other MDR pathogens.
Results and Discussion
Characterization of Optimum Conditions for MNM Light Activation against K. pneumoniae
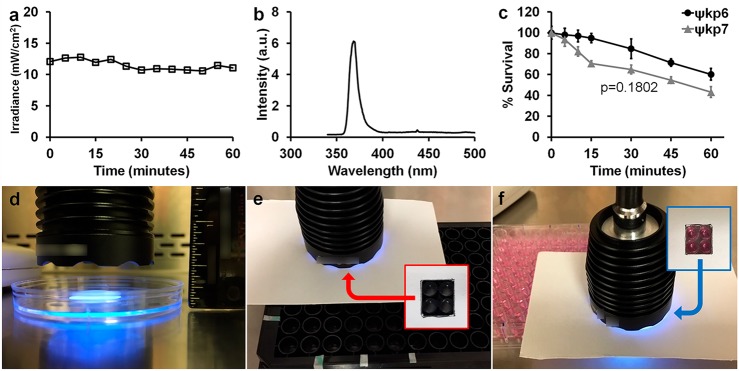
The irradiance of the 365 nm light-emitting diode (LED) light source (Sunlite Eagle 8WFP UV365 LED) used to activate the MNM was within a constant energy output range of 10.5 to 12 mW/cm2 over 60 min of exposure, measured at a constant distance (Figure 2a). It had a narrow wavelength spectrum of 360–376 nm, with a peak intensity at 368 nm (Figure 2b). Any bactericidal effects caused by the heat generated from the light source was controlled by the used of no MNM and slow MNM controls. Under these conditions, we assayed the bactericidal effects of the light source on an extensively drug-resistant K. pneumoniae (ψkp6) and an antibiotic-sensitive K. pneumoniae (ψkp7). The ψkp6 and ψkp7 antibiotic susceptibilities were characterized against several antibiotics using microdilution assays (Table 1). With 5 min of light exposure, we observed a viability reduction of 3% in ψkp6 and 6.5% in ψkp7. With 10 min of light exposure, it was 4% in ψkp6 and 18% in ψkp7, and at 60 min, 40% in ψkp6 and 55% in ψkp7 (Figure 2c). The overall bactericidal effects of 356 nm light on ψkp6 and ψkp7 were not significantly different (p = 0.1802). Therefore, a 5 min light activation time was chosen to minimize the effects of 365 nm light on K. pneumoniae. For viability assays, 120–240 μL volumes of bacterial cultures were exposed to light directly placed above it at a distance of 1.3 cm (Figure 2d). For permeability and toxicity assays, the light source was directly placed above the 96-well plate at a distance of 0.65 cm from the culture or media (Figure 2e,f).
Figure 2.
Characterization of 365 nm light source used to activate molecular nanomachines. (a) Irradiance of the 365 nm light source remained within a constant range of 10.5 to 12 mW/cm2 measured over 1 h. (b) Range of wavelengths emitted by the 365 nm light source and their relative intensities with peak light intensity at a 368 nm wavelength. (c) Bactericidal effect of the 365 nm light source on K. pneumoniae over 60 min of light exposure. ψkp6 (AR-0666), an extensively drug-resistant strain of K. pneumoniae; ψkp7 (NIH-1), an antibiotic-sensitive strain of K. pneumoniae. Percent survival was calculated by dividing the CFU/mL at each time point by the starting CFU/mL. A Mann–Whitney test was used to compare the survival of ψkp6 (AR-0666) and ψkp7 (NIH-1) strains (p = 0.1802). (d) Light source placed directly above bacterial cultures at a constant distance of 1.3 cm for the duration of light exposure. (e,f) Light source placed directly above the 96-well plate to only expose four wells, as shown by the insets.
Table 1. Antibiotic Susceptibilities of K. pneumoniae Strains ψkp7 (NIH-1) and ψkp6 (AR-0666).
| |
ψkp7a |
ψkp6b |
|||||
|---|---|---|---|---|---|---|---|
| antibiotic | class | MICc (μg/mL) | MBC99d (μg/mL) | ASTe | MIC (μg/mL) | MBC99 (μg/mL) | AST |
| Meropenem | carbapenem | 0.0625 | 0.0625 | S(−) | 16 | 16 | R(+) |
| tetracycline | tetracycline | 4 | 8 | S(−) | 256 | 256 | R(+) |
| gentamicin | aminoglycoside | 1 | 1 | S(−) | >512 | >512 | R(+) |
| amikacin | aminoglycoside | 1 | 1 | S(−) | >512 | >512 | R(+) |
| streptomycin | aminoglycoside | 4 | 4 | S(−) | 4 | 4 | S(−) |
| hygromycin | aminoglycoside | 64 | 64 | S(−) | 32 | 32 | S(−) |
| kanamycin | aminoglycoside | 2 | 2 | S(−) | >512 | >512 | R(+) |
| spectinomycin | aminoglycoside | 32 | 64 | S(−) | 64 | >512 | R(+) |
| rifampin | rifamycins | 32 | 32 | R(+) | 16 | 16 | R(+) |
| isoniazid | isonicotinate | >128 | >128 | R(+) | >128 | >128 | R(+) |
| ampicillin | penicillin | >512 | >512 | R(+) | >512 | >512 | R(+) |
| vancomycin | glycopeptide | >128 | >128 | R(+) | >128 | >128 | R(+) |
ψkp7 strain (NIH-1), carbapenemase nonproducing (KPC negative), antibiotic-sensitive strain obtained from NIH.
ψkp6 strain (AR-0666), carbapenemase producing (KPC positive), extensively drug-resistant strain obtained from the CDC.
Minimal inhibitory concentration (MIC), the lowest concentration needed to inhibit bacterial growth determined by colony-forming units (CFU).
Minimal bactericidal concentration (MBC99), the lowest concentration needed to kill 99% of the bacteria determined by CFU.
Antibiotic susceptibility testing (AST): S(−), sensitive; R(+), resistant.
Light-Activated MNM 1 Causes Reduced Bacterial Viability through Its Fast Rotational Movement in K. pneumoniae
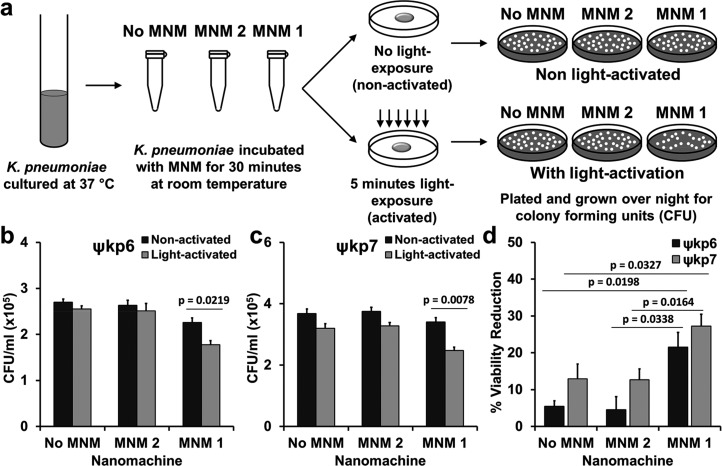
To characterize the antibacterial properties of MNM 1, we exposed the extensively drug-resistant (ψkp6) and the antibiotic-sensitive (ψkp7) K. pneumoniae strains to 10 μM of MNM 1 (fast motor), MNM 2 (slow motor) control, and to the MNM solvent of 0.1% dimethyl sulfoxide (DMSO) control (no MNM), with 5 min of 365 nm light activation (Figure 3a). DMSO solvent was used so that the MNMs remain soluble in media, and DMSO at a concentrations of 0.1% has no effects on cell viability.17 The only significant reduction in colony-forming unit (CFU) counts was observed in light-activated MNM 1 for both ψkp6 and ψkp7 (p = 0.0219 and 0.0078, respectively) (Figure 3b,c). The percent viability reduction of ψkp6 exposed to light-activated MNM 1 was 21.3%, significantly higher than that of the no MNM (DMSO) control (5.4%) and MNM 2 control (4.6%) (Figure 3b). Similarly, the percent viability reduction of ψkp7 exposed to light-activated MNM 1 was 27.2%, significantly higher than that of the no MNM control (12.9%) and MNM 2 control (12.7%) (Figure 3c). No toxicity or bactericidal effects were observed when 10 μM of non-light-activated MNM 1 was exposed to either ψkp6 or ψkp7. These results show that high-speed rotation of light-activated MNM 1 caused nanomechanical damage to K. pneumoniae irrespective of their antibiotic susceptibility, causing a significant relative reduction in viability (14–17%). In contrast, neither the light-activated MNM 2 nor the nonactivated MNM 1 caused a significant reduction in viability. Our results showed no significant difference in the viability reduction observed in K. pneumoniae irrespective of their antibiotic sensitivity profiles. This suggests that antimicrobial resistance (AMR) mechanisms of this extensively drug-resistant strain have little or no effect on the nanomechanical action of light-activated MNM 1.
Figure 3.
Viability reduction of K. pneumoniae with light-activated molecular nanomachines. (a) Experimental setup for bacterial viability reduction assays. A log growth phase culture of K. pneumoniae incubated with no MNM (dimethyl sulfoxide (DMSO)), MNM 2, or MNM 1 for 30 min, activated with 365 nm light for 5 min and plated for CFU/mL counts. (b) Extensively drug-resistant strain of K. pneumoniae (ψkp6, AR-0666) exposed to no MNM (DMSO), 10 μM of MNM 2, or 10 μM of MNM 1. Comparison of CFU/mL of K. pneumoniae after MNM exposure, without and with light activation. (c) Antibiotic-sensitive strain of K. pneumoniae (ψkp7, NIH-1) exposed to no MNM (DMSO), 10 μM of MNM 2, or 10 μM of MNM 1. Comparison of CFU/mL of K. pneumoniae after MNM exposure, without and with light activation. Results presented are means and standard error from four replicates for each group. The p values are from an unpaired two-tailed Student’s t test.
Light-Activated MNM 1 Causes Cell Wall Inner and Outer Membrane Disruptions in K. pneumoniae
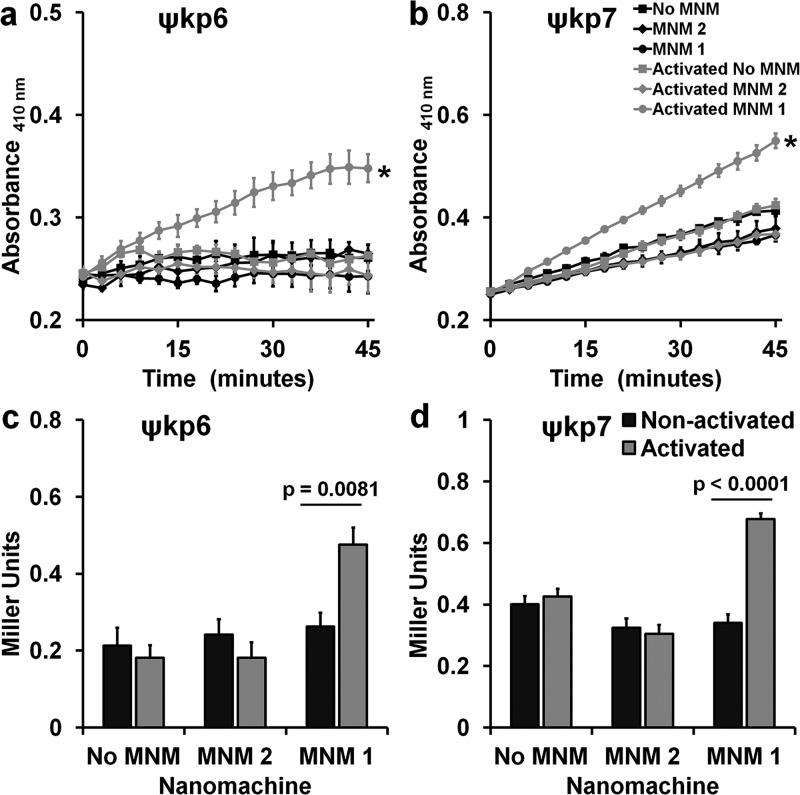
To confirm that the viability reduction observed in K. pneumoniae is a result of cell wall disruptions caused by the fast drilling action of light-activated MNM 1, we carried out three assays to characterize the cell wall inner membrane permeability, outer membrane permeability, and cell membrane integrity. Cell wall inner membrane permeability of K. pneumoniae exposed to no MNM (DMSO control), 10 μM of MNM 2, or 10 μM of MNM 1 was determined using o-nitrophenyl-β-d-galactoside, which is a substrate to cytoplasmic β-galactosidase that would leak through the cell wall inner membrane when disrupted. In both the extensively drug-resistant (ψkp6) and the antibiotic-sensitive (ψkp7) K. pneumoniae, light-activated MNM 1 showed a significant increase in the β-galactosidase activity, which was represented by an increase in absorbance at a 410 nm wavelength (Figure 4a,b). This was in contrast to both the light-activated MNM 2 and the nonactivated MNM 1 that did not cause a significant increase in absorbance at 410 nm. To further characterize these differences, we calculated the differences in β-galactosidase activity at 30 min postexposure in Miller units.27 In both ψkp6 and ψkp7, light-activated MNM 1 showed a significant increase in inner membrane permeability compared to that in nonactivated MNM 1, MNM 2, and no MNM (DMSO) control (Figure 4c,d). These results indicate that, upon light activation, MNM 1 causes nanomechanical damage to the K. pneumoniae cell wall inner membrane, allowing the leakage of cytoplasmic β-galactosidase enzyme.
Figure 4.
Cell wall inner membrane permeability with and without light activation of molecular nanomachines. Cell wall inner membrane permeability of K. pneumoniae exposed to no MNM (DMSO), 10 μM of MNM 2, or 10 μM of MNM 1 determined by cytoplasmic β-galactosidase activity using o-nitrophenyl-β-d-galactoside (ONPG) as the substrate, measured with an increase in absorbance at 410 nm. (a,c) Extensively drug-resistant strain of K. pneumoniae (ψkp6, AR-0666) exposed to MNM. (a) Comparison in absorbance at 410 nm of K. pneumoniae with ONPG after MNM exposure, without (nonactivated) and with light activation (activated). (b,d) Antibiotic-sensitive strain of K. pneumoniae (ψkp7, NIH-1) exposed to MNMs. (b) Comparison in absorbance at 410 nm of K. pneumoniae with ONPG after MNM exposure, without (nonactivated) and with light activation (activated). (c,d) ONPG assay at 30 min with Miller calculation for inner membrane permeability of K. pneumoniae exposed to no MNM (DMSO control), 10 μM of MNM 2, or 10 μM of MNM 1. Comparison of inner membrane permeability of K. pneumoniae after MNM exposure, without and with light activation. Results presented are means and standard error from four replicates for each group. (a–c) Values of *p < 0.05 are from a one-way ANOVA. (c,d) Values of p are from an unpaired two-tailed Student’s t test.
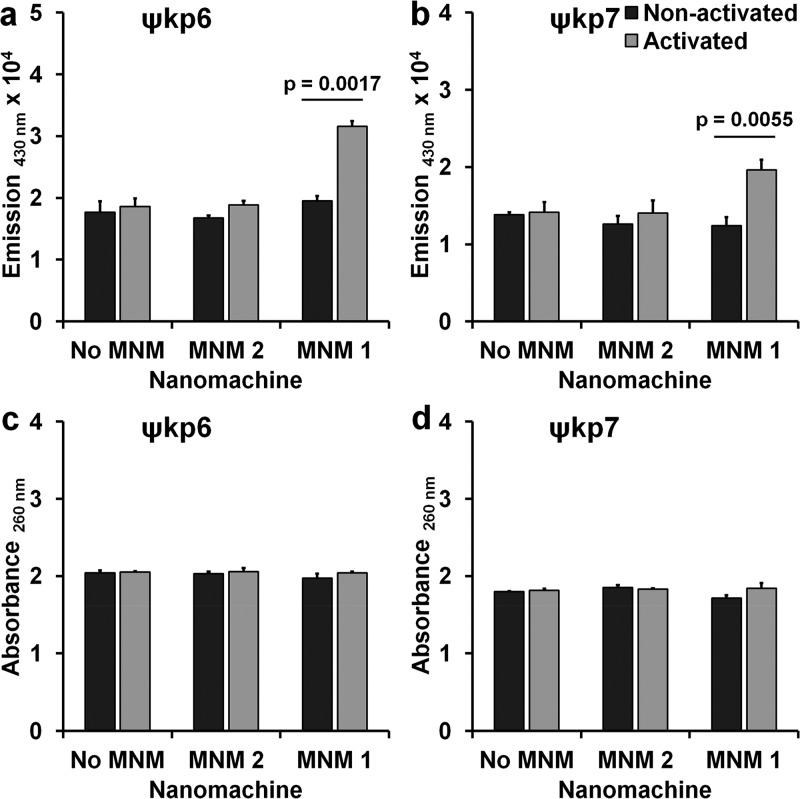
We then studied the ability of light-activated MNM 1 to permeabilize the cell wall outer membrane using an N-phenyl-1-naphthylamine (NPN) uptake assay.28 In both ψkp6 and ψkp7, light-activated MNM 1 showed a significant increase in NPN partitioning to the cell wall outer membrane, which was represented by an increase in emission at a 430 nm wavelength (Figure 5a,b). This was in contrast to both the light-activated MNM 2 and the nonactivated MNM 1 that did not cause a significant increase in emission at 430 nm. These results indicate that light-activated MNM 1 causes disruptions in the cell wall outer membrane, allowing the uptake of NPN.
Figure 5.
Cell wall outer membrane permeability and cell membrane integrity with and without light activation of molecular nanomachines. (a,b) Cell wall outer membrane permeability assay of K. pneumoniae exposed to no MNM DMSO, 10 μM of MNM 2, or 10 μM of MNM 1. Outer membrane permeability determined by the increase in fluorescence due to the partitioning of phenylnaphthylamine (NPN) into the cell wall outer membrane, measured by the increase in emission at 430 nm. Comparison of emission at 430 nm of K. pneumoniae with NPN after MNM exposure, without (nonactivated) and with light activation (activated). (a) Extensively drug-resistant strain of K. pneumoniae (ψkp6, AR-0666). (b) Antibiotic-sensitive strain of K. pneumoniae (ψkp7, NIH-1). (c,d) Cell membrane integrity assay of K. pneumoniae exposed to no MNM (DMSO), 10 μM of MNM 2, or 10 μM of MNM 1. Disruptions in cell membrane integrity determined by cytoplasmic release of DNA and RNA, measured with an increase in absorbance at 260 nm. Comparison of absorbance at 260 nm of K. pneumoniae after MNM exposure, without (nonactivated) and with light activation (activated). (c) Extensively drug-resistant strain of K. pneumoniae (ψkp6). (d) Antibiotic-sensitive strain of K. pneumoniae (ψkp7). Results presented are means and standard error from four replicates for each group. The p values are from an unpaired two-tailed Student’s t test.
To further characterize the extent of the cell wall damage caused by light-activated MNM 1, we assayed the leakage of cytoplasmic constituents using absorbance at 260 nm that detects DNA and RNA in the supernatant.29 Our studies showed that there was no significant difference in absorbance at 260 nm or relative changes with light-activated MNM 1, MNM 2, or with no MNM control in both the K. pneumoniae strains ψkp6 and ψkp7 (Figure 5c,d). These results indicate that cell membrane damage caused by light-activated MNM 1 was not large enough to allow the leakage of cytoplasmic DNA or RNA.
Our K. pneumoniae permeability assays indicate that light-activated MNM 1 is able to damage both the inner and outer membrane of the cell wall. This allowed smaller molecules such as enzymes and fluorescent dyes to cross the cell wall but not larger molecules such as DNA or RNA. The nanomechanical action of MNM 1 was not affected by antibiotic-resistant mechanisms because it caused cell wall damage to both K. pneumoniae strains alike.
Light-Activated MNM 1 Combined with Meropenem To Make an Extensively Drug-Resistant K. pneumoniae More Sensitive to Meropenem
Carbapenems are last resort antibiotics used in clinical settings against Gram-negative pathogens. Carbapenem antibiotics cause bactericidal effects through PBPs with the inhibition of cell wall synthesis.30 Loss of cell wall outer membrane porins is known to contribute to carbapenem resistance in K. pneumoniae by acting as a physical barrier preventing carbapenem antibiotics from reaching their target sites in the periplasmic space.31 As light-activated MNM 1 caused cell wall damage to K. pneumoniae irrespective of its antibiotic-resistant profile, we hypothesized that MNM 1 will synergize with currently ineffective carbapenem antibiotics to make them more effective.
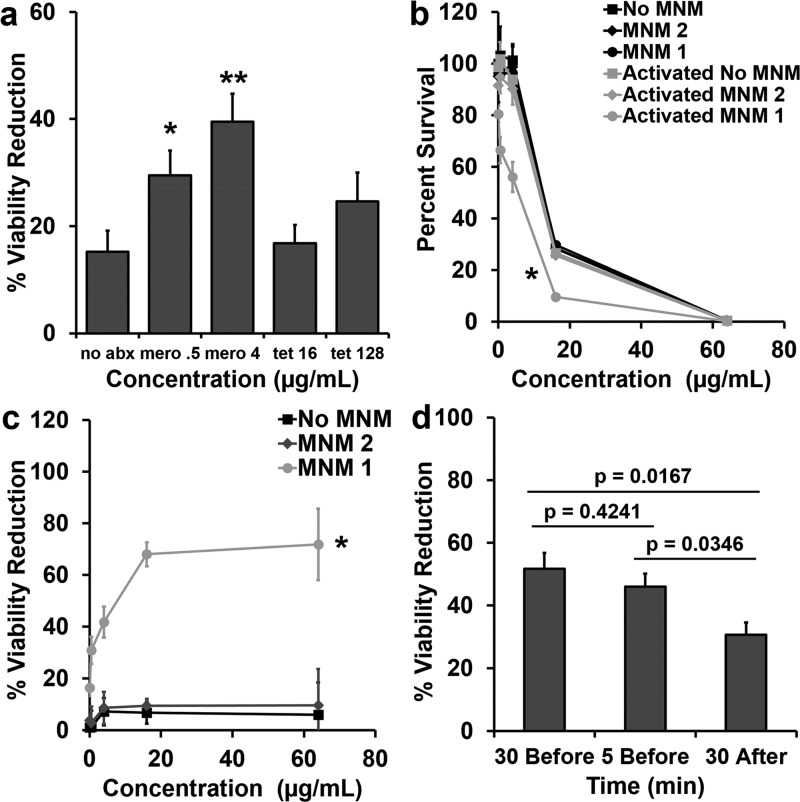
To test this hypothesis, we used light-activated MNM 1 with Meropenem and tetracycline (control) at subtherapeutic concentrations against the extensively drug-resistant K. pneumoniae strain (ψkp6). In contrast to carbapenems, tetracycline antibiotics are protein synthesis inhibitors that prevent the initiation of translation by binding to the 30S ribosomal subunit.32 We assayed Meropenem at concentrations of 0.5 and 4 μg/mL and tetracycline at 16 and 128 μg/mL. These concentrations were lower than the MIC and MBC99 against ψkp6 (Table 1). Light-activated MNM 1 and Meropenem at concentrations of 0.5 and 4 μg/mL showed significant reduction in ψkp6 viability compared to that with nonactivated MNM 1 with the same concentrations of Meropenem (p = 0.0455 and 0.0095, respectively) (Figure 6a). Light-activated MNM 1 and tetracycline at concentrations of 16 and 128 μg/mL did not show a significant reduction in ψkp6 viability.
Figure 6.
Viability reduction of K. pneumoniae with Meropenem and light-activated molecular nanomachines. Viability reduction of extensively drug-resistant K. pneumoniae (ψkp6) exposed to antibiotics (Meropenem or tetracycline), and no MNM (DMSO), 10 μM of MNM 2, or 10 μM of MNM 1. (a) Percent viability reduction of K. pneumoniae exposed to light-activated MNMs with no antibiotics (no abx), 0.5 μg/mL Meropenem (mero 0.5), 4 μg/mL Meropenem (mero 4), 16 μg/mL tetracycline (tet 16), or 128 μg/mL tetracycline (tet 128). (b) Percent survival of K. pneumoniae exposure to different concentrations of Meropenem and MNM without (nonactivated) and with light activation (activated). (c) Percent viability reduction of light-activated with no MNM, MNM 2, and MNM 1 compared to nonactivated controls with different concentrations of Meropenem (0.5–64 μg/mL). (d) Percent viability reduction of K. pneumoniae with light-activated MNM 1 with 4 μg/mL Meropenem added 30 min before light activation, 5 min before light activation, or 30 min after light activation. Percent viability reduction was calculated by comparing light-activated groups with nonactivated groups. Results presented are means and standard error from three replicates for each group. The p values are from an unpaired two-tailed Student’s t test, compared to no abx group; *p < 0.05, **p < 0.01.
When we used various doses of Meropenem (0.5–64 μg/mL) in combination with 10 μM of light-activated MNM 1 to study the dose-dependent combined effects of the combined therapy in reducing bacterial viability; as was expected, higher concentrations of Meropenem alone showed increased reductions in viability, with 16 and 64 μg/mL of Meropenem showing 70 and 98%, respectively (Figure 6b). Without light activation, MNM 1 or 2 did not have any additional viability reduction in K. pneumoniae (Figure 6b). However, when MNM 1 was light-activated for 5 min in combination with Meropenem, it caused a significant reduction in bacterial viability (p < 0.05), shifting the survival curve to the left (Figure 6b). At subtherapeutic concentrations of Meropenem (4 μg/mL), light-activated MNM 1 caused a 41.7% relative reduction in viability, and at 64 μg/mL of Meropenem, the relative reduction in viability was 72% (Figure 6c). These results indicate that Meropenem when combined with light-activated MNM 1 acts to reduce bacterial viability in an extensively drug-resistant K. pneumoniae strain that is otherwise resistant to Meropenem.
To further characterize the mechanism of interactions between Meropenem and light-activated MNM 1, we added Meropenem 30 min before, 5 min before, and 30 min after light activation. Our results show that the presence of Meropenem during light activation of MNM 1 showed higher viability reduction in ψkp6 (30 min before, 51.7% and 5 min before, 46.0%), compared to that when added after light activation (30 min after, 30.7%) (Figure 6d). This suggests that there is a temporal relationship between Meropenem and light-activated MNM 1, and perhaps the cell wall damage or perturbation caused by MNM 1 is a transient effect. Although we characterized the temporal aspect of the mechanistic relationship between Meropenem and MNM 1, it still needs more careful characterization. However, the MNMs alone, disrupting cell walls, do result in bacterial death, albeit slower than in the presence of antibiotics.
Ultrastructural Observations Show Light-Activated MNM 1 and Meropenem Destroy Extensively Drug-Resistant K. pneumoniae
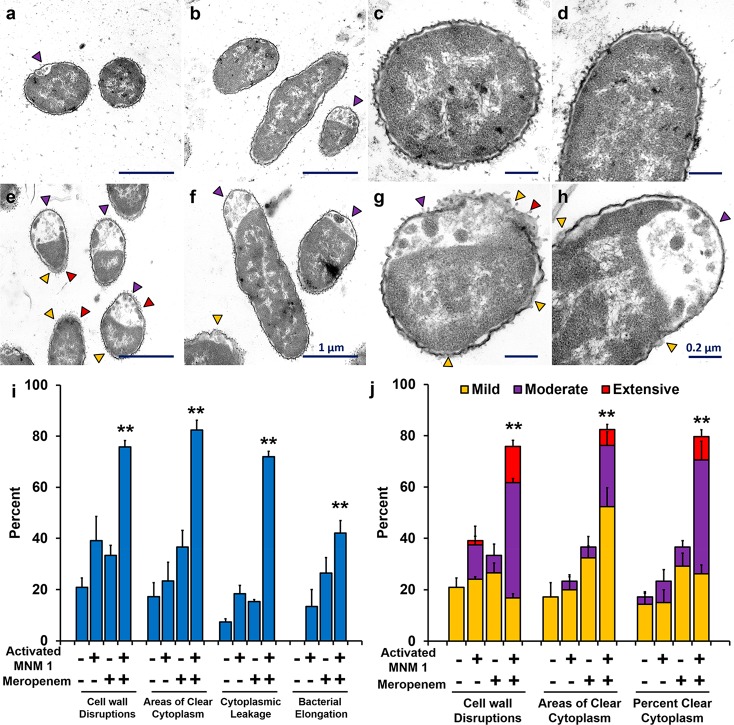
To further confirm the combined action between light-activated MNM 1 and 4 μg/mL of Meropenem, we exposed ψkp6 to MNM 1 with and without Meropenem and light activation and observed under transmission electron microscopy (TEM) (Figure 7). The ψkp6 exposed to Meropenem and nonactivated MNM 1 showed minimal ultrastructural and morphological changes (Figure 7a–d). In contrast, ψkp6 exposed to Meropenem with light-activated MNM 1 showed distinct ultrastructural and morphological changes, many of which can possibly be attributed to changes with Meropenem alone (Figure 7e–h).33,34 These observations included cell wall disruptions (yellow arrowhead), areas of clear cytoplasm (purple arrowhead), areas of cytoplasmic leakage (red arrowhead), and bacterial elongation. These observations were quantified in 60–80 ψkp6 per group (Figure 7i,j). Compared to the control groups, ψkp6 exposed to light-activated MNM 1 and Meropenem showed the presence of significantly higher cell wall disruptions, cytoplasmic clearance, and cytoplasmic leakage (p > 0.005) (Figure 7i). The extent of the ultrastructural damage caused was further quantified as mild, moderate, and extensive. The light-activated MNM 1 and Meropenem showed a significantly higher, moderate, and extensive ultrastructural damage in ψkp6 compared to that of the control groups (Figure 7j). These TEM observations confirm our viability reduction results, in which light-activated MNM 1 made subtherapeutic concentrations of Meropenem effective against the extensively drug-resistant K. pneumoniae strain (ψkp6).
Figure 7.
Cell wall disruptions and changes in K. pneumoniae exposed to Meropenem and light-activated molecular nanomachines observed through transmission electron microscopy. (a–d) Representative TEM images of K. pneumoniae incubated with 4 μg/mL Meropenem and 10 μM of nonactivated MNM 1 for 2.5 h. (e–h) Representative TEM images of K. pneumoniae incubated with 4 μg/mL Meropenem and 10 μM of light-activated MNM 1 for 2.5 h (30 min prior to 5 min of 395 nm light activation and 2 h postlight activation). (a,e) Cross section of bacilli at 20 000× magnification. (b,f) Longitudinal section of bacilli at 20 000× magnification. (c,g) Cross section of bacilli at 60 000× magnification. (d,h) Longitudinal section of bacilli at 60 000× magnification. (a–h) Purple arrowheads show areas of cytoplasmic clearance. Yellow arrowheads show areas of cell wall disruptions. Red arrowheads show areas of cytoplasmic leakage. Scale bar for a,b and e,f is 1 μm, as shown in panel f. Scale bar for c,d and g,h is 0.2 μm, as shown in panel h. (i,j) Selected ultrastructural changes observed in 60–80 K. pneumoniae in groups with exposure to Meropenem and MNM 1 were documented and quantified. These ultrastructural changes included (i) presence of cell wall disruptions (p = 0.0007), areas of cytoplasmic clearance (p = 0.0002), cytoplasmic leakage (p < 0.0001), and bacterial elongation (p = 0.0022). (j) Extent of damage observed in K. pneumoniae categorized as mild, moderate, or extensive for ultrastructural change: cell wall disruptions (p = 0.0004), number of areas of clear cytoplasm (p = 0.0002), and percent clear cytoplasm (p = 0.0003). Results presented are the percentage of bacilli number in each exposure group. One-way ANOVA was used to compare the differences in means of each group; **p < 0.01.
We used TEM to confirm the combined action of light-activated MNM 1 with Meropenem and showed that, upon light activation, the extensively drug-resistant K. pneumoniae undergoes pathological and morphological changes such as cytoplasmic clearance and bacterial elongation that are associated with the action of Meropenem. The significance of this finding is that light-activated MNM 1 was able to make a subtherapeutic concentration effective again. To study the mechanism of action between Meropenem and MNM 1, we examined the temporal effects of Meropenem addition (Figure 7i). We show that it is important that Meropenem be present during light activation of MNM 1, as the cell wall disruptions could be transient.
Light-Activated MNM 1 Does Not Cause Cytotoxicity in J774A.1 Macrophage Cells
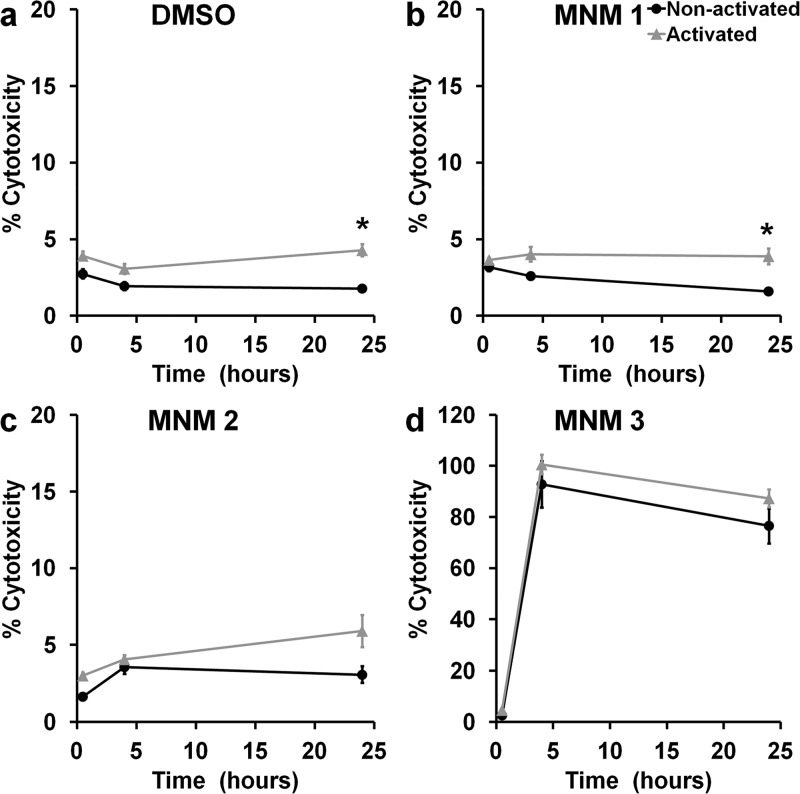
The use of a broad-spectrum nanomechanical antibiotic carries with it the concern of nonspecific damage or associated cytotoxicity to adjacent host cells. To characterize the cytotoxic effects of light-activated MNM 1 (fast motor) on mammalian cells, we used J774A.1 macrophages and exposed them to various concentrations of MNM (0.5–100 μM) and observed them for up to 24 h postexposure (Figure 8). We assayed 0.1% DMSO (solvent) in media and MNM 2 (slow motor) as negative controls and MNM 3 (fast motor with TPP, targeting mitochondria) as a positive control for mammalian cell toxicity to perform lactate dehydrogenase (LDH) cytotoxicity assays (Figure 8). At 24 h postexposure, percent cytotoxicity observed was as follows: 1% DMSO without light exposure = 1.8% and with light exposure = 4.3% (Figure 8a); 100 μM of MNM 1 without light activation = 1.6% and with light activation = 3.9% (Figure 8b); 100 μM of MNM 2 without light activation = 3.1% and with light activation = 5.9% (Figure 8c); and 100 μM of MNM 3 without light activation = 76.5% and with light activation = 87.2% (Figure 8d). There was no statistical significance between nonactivated MNM 1 and MNM 2 or no MNM (DMSO) control. This shows that even at a 10× concentration (100 μM) used against K. pneumoniae, MNM 1 does not display any cytotoxicity in macrophages. However, when exposed to light, both the no MNM (DMSO) control and MNM 1 showed an increase in cytotoxicity (p < 0.005), showing the cytotoxic effects of 365 nm light on mammalian cells.
Figure 8.
Cytotoxicity of molecular nanomachine treatment for J774A.1 macrophages. Percent cytotoxicity of macrophage cells measured with an LDH assay at 0.5, 4, and 24 h exposed to 100 μM MNM without or with light activation. Percent cytotoxicity was calculated using a low control (natural cell death) (0%) and a high control (Triton-x induced cell death) (100%) (a) without MNM (1% DMSO) (p = 0.0282), (b) with 100 μM of MNM 1 (fast motor) in 1% DMSO (p = 0.0428), (c) with 100 μM of MNM 2 (slow motor) in 1% DMSO (p = 0.1971), (d) with 100 μM of MNM 3 (fast motor with TPP, targeting mitochondria) in 1% DMSO (p = 0.8748). The DMSO concentration was 1% because 100 μM MNM was assayed. Results presented are mean and standard error from four replicates; *p < 0.05.
Light-Activated MNM 1 Assists Meropenem in Killing an Extensively Drug-Resistant K. pneumoniae
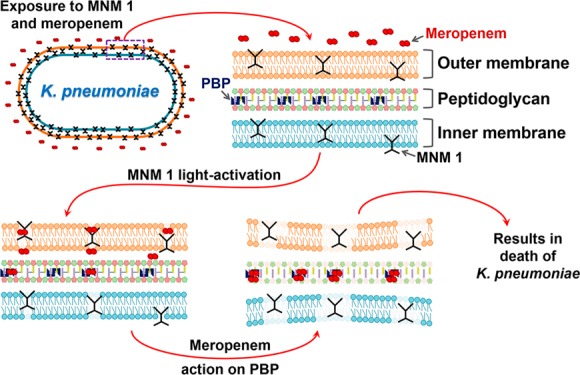
Meropenem-resistant K. pneumoniae uses different mechanisms to prevent Meropenem from reaching PBP within peptidoglycan in the periplasmic space. One such resistant mechanism is a cell wall outer membrane lacking porins that keep Meropenem out of the bacteria.24 When MNM 1 is activated with 365 nm light, it rotates unidirectionally at 3 MHz to drill pores into the cell wall of K. pneumoniae through its nanomechanical action. These pores allow Meropenem to travel across the cell wall outer membrane and reach PBP. This causes the destruction of the peptidoglycan layer, destabilizing the bacterial cell wall and leading to the death of K. pneumoniae. This synergistic mechanism between light-activated MNM 1 and Meropenem allows subtherapeutic concentrations of Meropenem to kill Meropenem-resistant extensively drug-resistant K. pneumoniae. In the current study, we observed an approximately 10-fold increase in efficacy using MNM, possibly due to the extensive resistance mechanisms present in the strain. Efficacy might be improved by further optimizing concentrations of MNM and Meropenem used as well as combining this approach with strategies to block resistance mechanisms, but current data show promise for use of MNM to improve the utility of conventional antimicrobials.
There are a few limitations in this study. MNM 1 has a nonspecific action, without any specific binding affinity to K. pneumoniae, and toxicity was examined solely in a cell line rather than in a whole animal. When MNM 1 was previously used to target and permeabilize cancer cells, they had short sequence peptides that allowed selective binding and high cell specificity. Targeting specific bacterial receptors using ligands can increase the specificity to the pathogen.35 Several ligands including asialo-GM1, asialo-GM2, and GM2 have been shown to have specificity to K. pneumoniae and can be attached to the MNM stator to increase their specificity and efficacy.36−38
Another concern of the nonspecific nature of MNM 1 is the toxicity and possible damage it can cause to surrounding host cells during light activation. In order to address this issue, we looked at the cytotoxicity of MNM 1 in macrophages (Figure 8). We only observed a 1.6% cytotoxicity with 100 μM (10× more) MNM 1. However, with 365 nm light exposure, the macrophage cytotoxicity increased to 3.9–5.9% (p < 0.05), highlighting the concerns with this use of 365 nm light. Similarly, longer exposures display more bacterial toxicity due to light alone, reducing the observation of the effects of the MNM. This issue caused us to limit our MNM 1 activation time to 5 min because 365 nm light displayed higher bactericidal effects over longer exposure times (Figure 2c). We are in the process of developing 405 nm light-activated MNMs that will be safer and should allow longer activation times without as much killing due to the light itself.
A wavelength of 365 nm has a relatively low penetration in host organs and tissue. This currently limits the use of MNMs for the potential treatment of deep tissue infections, as MNMs will not be activated effectively. We are also exploring the synthesis of next generation MNMs that are activated with longer wavelengths (>700 nm) in the near-infrared (NIR) region. This will greatly increase the ability of MNM activation in much deeper host targets and also allow the activation of MNM for longer times to achieve a far superior antimicrobial efficacy, without any associated harmful effects on the host. However, the energies at these wavelengths are much lower. We have been exploring two-photon NIR, although the potential depth may be somewhat limited, this approach would allow very precise targeting within tissues.18
Our current study characterizes the use of light-activated MNM 1 as an effective nanomechanical antibacterial agent against extensively drug-resistant K. pneumoniae. In addition to its ability to counter antibacterial resistance, light-activated MNM 1 has several potential therapeutic applications. It can be used to treat skin infections and wound infections by direct light activation at the site of infection, and its broad-spectrum activities make it possible to treat urinary tract infections through a catheter device for light delivery. Light-activated MNM 1 also has the potential to disrupt biofilms on indwelling prosthetic devices by direct application of MNM 1 and light activation at the site of infection. This strategy could allow antibiotic treatment to be more efficacious against biofilm-forming pathogens.
Conclusions
In this study, we show that light-activated MNM 1 displays antibacterial properties against K. pneumoniae that is not diminished even in an extensively drug-resistant strain (Figure 3). This is because bacterial antimicrobial resistance mechanisms are not developed against a nanomechanical agent that disrupts bacterial cell walls. We have shown the ability of light-activated MNM 1 to disrupt cell walls by its nanomechanical action, using K. pneumoniae cell wall IM and OM permeability assays (Figures 4 and 5). The ability to use nanomechanical force to disrupt bacterial cell walls is a characteristic feature of MNMs with the potential of many therapeutic applications and has not been characterized before. With only 5 min of MNM 1 light activation, we observed 14–17% in viability reduction of K. pneumoniae. Next, we show that light-activated MNM 1 can combine with Meropenem at subtherapeutic concentrations to be effective against an extensively drug-resistant K. pneumoniae strain (Figure 6). The ability to help otherwise ineffective antibiotics to be efficacious is another distinctive aspect of light-activated MNM 1. The use of MNM 1 in combination with other conventional antibiotics allows the potential to recycle many currently available antibiotics against MDR pathogens. We plan to evaluate MNM 1 in a range of Gram-positive and Gram-negative MDR pathogens as well as other antimicrobials to better understand how broad their utility will be.
Methods
Bacterial Strains
Two clinical strains of K. pneumoniae were used: an extensively drug-resistant K. pneumoniae, AR-0666 (ψkp6) obtained from the CDC, and a KPC-negative antibiotic-sensitive strain, NIH-1 (ψkp7) obtained from the National Institutes of Health (NIH).
Synthesis of Molecular Machines
The molecular motors 1 and 2 were freshly prepared according to our previous protocols.17,18 The molecular motor 3 is newly designed and synthesized as described in the Supporting Information.
Molecular Nanomachines
MNM 1 is a fast motor with a rotor that rotates at (2–3) × 106 revolutions per second relative to its stator (Figure 1b). MNM 2 is a slow motor that rotates about 1.8 revolutions per hour (Figure 1c). MNM 2 served as a negative control. MNM 3 is MNM 1 attached to the TPP cation at the stator (Figure 1d). TPP targets eukaryotic mitochondria and was used to demonstrate eukaryotic cell targeting of MNMs.
Minimal Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC99) of Antibiotics in K. pneumoniae
Log-phase K. pneumoniae cultures [(4–5) × 105 CFU/mL] grown in Mueller–Hinton broth were exposed to antibiotics for 16 h in 96-well plates in triplicate. Serial dilutions (1:2) of each antibiotic were assayed in a microdilution assay against K. pneumoniae. A PerkinElmer EnVision microplate reader was used to measure culture optical density at 600 nm. After antibiotic exposure, bacterial cultures were plated for CFL/mL counts. The MIC and MBC99 values were calculated relative to the starting CFU/mL. MIC was defined as the minimal concentration of antibiotic needed to inhibit the growth of the starting culture of bacteria (≤100%). MBC99 was defined as the minimal concentration of the antibiotic needed to kill 99% of the starting culture of bacteria (≤1%).
K. pneumoniae Viability Reduction Assay
Log-phase K. pneumoniae cultures [(2–4) × 105 CFU/mL] grown in Lysogeny broth were exposed to MNM in triplicate. The concentration of MNMs used was 10 μM in 0.1% DMSO. K. pneumoniae cultures were incubated with MNMs for 30 min prior to 5 min of 365 nm light activation. The 365 nm light source was placed directly above the cultures at a constant distance of 1.3 cm (Figure 1d). After light exposure, bacterial cultures were plated for CFL/mL counts.
Inner Membrane Permeability Assay
K. pneumoniae [(2–4) × 105 CFU/mL] was washed once with 10 mM sodium phosphate (pH 7.4) and resuspended in the same buffer containing 1.5 mM o-nitrophenyl-β-d-galactoside.27 Cultures were incubated with 10 μM MNMs in a black 96-well plate with clear bottoms with 100 μL of K. pneumoniae in four replicates. MNMs were light-activated for 5 min with the light source placed directly above the 96-well plate (Figure 2e). The production of o-nitrophenol was monitored at an absorbance of 410 nm every 3 min for 45 min postlight exposure. Miller calculation was used to determine the inner membrane permeability.
Outer Membrane Permeability Assay
K. pneumoniae [(2–4) × 105 CFU/mL, 100 μL] was incubated with 10 μM MNM in a black 96-well plate with clear bottoms for 30 min and then light-activated for 5 min in four replicates (Figure 2e). After light activation, 10 mM 1-N-phenylnaphthylamine was mixed and incubated for 30 min. The fluorescence intensity due to the partitioning of NPN into the OM was measured with a microplate reader fluorescence spectrophotometer with an excitation wavelength of 350 nm and an emission wavelength of 430 nm.
Cell Membrane Integrity Assay
Similar to the OM permeability assay, K. pneumoniae was exposed to MNMs and light-activated for 5 min in four replicates. These cultures were spun down at 10 000 rpm, and the supernatant was placed in a 96-well plate. The release of cytoplasmic constituents of the cell was monitored using the absorbance at 260 nm.29
MNM and Meropenem Combined Assay
Similar to viability reduction assays, ψkp6 cultures [(2–4) × 105 CFU/mL] were incubated with 10 μM of MNM and Meropenem for 30 min and activated with 365 nm light for 5 min in triplicate. Different concentrations of Meropenem (0,5, 4, 16, and 64 μg/mL) were used with 10 μM of MNM. Tetracycline (16 and 128 μg/mL) was used as an antibiotic control with MNM. These cultures were then plated for CFU/mL counts.
Transmission Electron Microscopy
Log-phase ψkp6 (5 × 106 CFU/mL) was exposed to 10 μM of MNM 1 and 4 μg/mL of Meropenem with and without light activation for TEM. The four exposure groups were (a) MNM 1 only, without light activation; (b) MNM 1 only, with light activation; (c) MNM 1 with Meropenem, without light activation; and (d) MNM 1 with Meropenem, with light activation. Postexposure, K. pneumoniae was incubated with Meropenem for an additional 2 h. Then the mixture was fixed with 4% formaldehyde, 2.5% glutaraldehyde, and 1% acrolein. After three washes, the mixture was embedded in Epon 812 resin and stained with 5% uranyl acetate. The embedded samples were sectioned into grids and imaged with a JEOL 1200 TEM. Cell wall disruptions, cytoplasmic clearance, cytoplasmic leakage, and bacterial elongation were quantified using 60–80 ψkp6 for each group.
Macrophage Cytotoxic Assay
A LDH cytotoxicity colorimetric assay kit (Biovision, #K311) was used to measure the cytotoxicity of MNMs at different concentrations (0.5, 1, 10, 50, and 100 μM) in a J774A.1 macrophage cell line. J774A.1 cells (5 × 105 cells/mL) grown in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum were incubated with MNMs in a 96-well plate (100 μL in each well) and exposed to 5 min of 365 nm light (Figures 2f and 8d). Cytotoxicity of MNMs with and without light activation was measured at 0.5, 4, and 24 h postexposure. DMSO and MNM 2 were used as negative controls. MNM 3 was used as a cell-targeted positive control.
Statistical Analyses
All experiments were done with at least three replicates (n ≥ 3). The number of replicates used in each experiment is stated in the figure legend of each experiment. Prism GraphPad was used to perform the two-tailed unpaired Student’s t test statistical analyses to compare the means of two exposure groups. For comparison among three or more groups, analysis of variance (ANOVA) was used. A Mann–Whitney U test was used to compare different survival plots. Means and standard errors are presented in each of the graphs plotted in Microsoft Excel. A value of p < 0.05 was defined as statistically significant.
Acknowledgments
This work was supported in part by grants from the Discovery Institute, the Welch Foundation, NIH R01 Grant AI104960, BBSRC, and the Royal Society University Research Fellowship. We thank Drs. Dustin K. James and Preeti Sule for help coordinating this work, Dr. Riti Sharan for her assistance in antibiotic MIC assays in K. pneumoniae strains, Dr. Kristen Maitland for help with characterizing the light source, and Dr. Stanislav Vitha and Richard Littleton at the Texas A&M University Microscopy Imaging Center (MIC) for assistance with transmission electron microscopy.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.9b07836.
Detailed synthesis information and validation of the structure for the newly designed molecular motor 3; 1H, 13C, DEPT-135 13C, 1H–13C HSQC, and 31P NMR spectra are provided, validating that the correct structure was obtained after synthesis (PDF)
Author Contributions
& T.G. and R.S.G. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance 2016, 1–84. [Google Scholar]
- Brown E. D.; Wright G. D. Antibacterial Drug Discovery in the Resistance Era. Nature 2016, 529, 336–343. 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- CDC . Antibiotic Resistant Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, 2013; pp 1–112.
- WHO . Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; pp 1–19.
- WHO . Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017; pp 1–87.
- Han J. H.; Goldstein E. J.; Wise J.; Bilker W. B.; Tolomeo P.; Lautenbach E. Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae in a Network of Long-Term Acute Care Hospitals. Clin. Infect. Dis. 2016, 64, ciw856. 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrie C. L.; Mirceta M.; Wick R. R.; Edwards D. J.; Thomson N. R.; Strugnell R. A.; Pratt N. F.; Garlick J. S.; Watson K. M.; Pilcher D. V.; McGloughlin S. A.; Spelman D. W.; Jenney A. W. J.; Holt K. E. Gastrointestinal Carriage Is a Major Reservoir of Klebsiella pneumoniae Infection in Intensive Care Patients. Clin. Infect. Dis. 2017, 65, 208–215. 10.1093/cid/cix270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Facility Guidance for Control of Carbapenem-Resistant Enterobacteriaceae; Centers for Disease Control and Prevention: Atlanta, GA, 2015; pp 1–20.
- Gomez-Simmonds A.; Uhlemann A. C. Clinical Implications of Genomic Adaptation and Evolution of Carbapenem-Resistant Klebsiella pneumoniae. J. Infect. Dis. 2017, 215, S18–S27. 10.1093/infdis/jiw378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung P. Y. The Emerging Problems of Klebsiella pneumoniae Infections: Carbapenem Resistance and Biofilm Formation. FEMS Microbiol Lett. 2016, 363, fnw219. 10.1093/femsle/fnw219. [DOI] [PubMed] [Google Scholar]
- Hauck C.; Cober E.; Richter S. S.; Perez F.; Salata R. A.; Kalayjian R. C.; Watkins R. R.; Scalera N. M.; Doi Y.; Kaye K. S.; Evans S.; Fowler V. G. Jr.; Bonomo R. A.; van Duin D. Spectrum of Excess Mortality Due to Carbapenem-Resistant Klebsiella pneumoniae Infections. Clin. Microbiol. Infect. 2016, 22, 513–519. 10.1016/j.cmi.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price L. S.; Poirel L.; Bonomo R. A.; Schwaber M. J.; Daikos G. L.; Cormican M.; Cornaglia G.; Garau J.; Gniadkowski M.; Hayden M. K.; Kumarasamy K.; Livermore D. M.; Maya J. J.; Nordmann P.; Patel J. B.; Paterson D. L.; Pitout J.; Villegas M. V.; Wang H.; Woodford N.; Quinn J. P. Clinical Epidemiology of the Global Expansion of Klebsiella pneumoniae Carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone A.; Giannella M.; Fortini D.; Giordano A.; Meledandri M.; Ballardini M.; Venditti M.; Bordi E.; Capozzi D.; Balice M. P.; Tarasi A.; Parisi G.; Lappa A.; Carattoli A.; Petrosillo N. High Rate of Colistin Resistance Among Patients With Carbapenem-Resistant Klebsiella pneumoniae Infection Accounts for an Excess of Mortality. Clin. Microbiol. Infect. 2013, 19, E23–E30. 10.1111/1469-0691.12070. [DOI] [PubMed] [Google Scholar]
- Watson M. A.; Cockroft S. L. Man-Made Molecular Machines: Membrane Bound. Chem. Soc. Rev. 2016, 45, 6118–6129. 10.1039/C5CS00874C. [DOI] [PubMed] [Google Scholar]
- Xu T.; Gao W.; Xu L. P.; Zhang X.; Wang S. Fuel-Free Synthetic Micro-/Nanomachines. Adv. Mater. 2017, 29, 1603250. 10.1002/adma.201603250. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez V.; Chiang P. T.; Chen F.; Ruan G.; Marti A. A.; Kolomeisky A. B.; Wang G.; Tour J. M. Unimolecular Submersible Nanomachines. Synthesis, Actuation, and Monitoring. Nano Lett. 2015, 15, 8229–8239. 10.1021/acs.nanolett.5b03764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez V.; Chen F.; Nilewski L. G.; Duret G.; Aliyan A.; Kolomeisky A. B.; Robinson J. T.; Wang G.; Pal R.; Tour J. M. Molecular Machines Open Cell Membranes. Nature 2017, 548, 567–572. 10.1038/nature23657. [DOI] [PubMed] [Google Scholar]
- Liu D.; Garcia-Lopez V.; Gunasekera R. S.; Greer Nilewski L.; Alemany L. B.; Aliyan A.; Jin T.; Wang G.; Tour J. M.; Pal R. Near-Infrared Light Activates Molecular Nanomachines to Drill into and Kill Cells. ACS Nano 2019, 13, 6813–6823. 10.1021/acsnano.9b01556. [DOI] [PubMed] [Google Scholar]
- Zielonka J.; Joseph J.; Sikora A.; Hardy M.; Ouari O.; Vasquez-Vivar J.; Cheng G.; Lopez M.; Kalyanaraman B. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem. Rev. 2017, 117, 10043–10120. 10.1021/acs.chemrev.7b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit H.; Queenan A. M.; Anderson G. J.; Domenech-Sanchez A.; Biddle J. W.; Steward C. D.; Alberti S.; Bush K.; Tenover F. C. Novel Carbapenem-Hydrolyzing Beta-Lactamase, KPC-1, From a Carbapenem-Resistant Strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Moland E.; Hanson N. D.; Herrera V. L.; Black J. A.; Lockhart T. J.; Hossain A.; Johnson J. A.; Goering R. V.; Thomson K. S. Plasmid-Mediated, Carbapenem-Hydrolysing Beta-Lactamase, KPC-2, in Klebsiella pneumoniae Isolates. J. Antimicrob. Chemother. 2003, 51, 711–714. 10.1093/jac/dkg124. [DOI] [PubMed] [Google Scholar]
- Woodford N.; Tierno P. M. Jr.; Young K.; Tysall L.; Palepou M. F.; Ward E.; Painter R. E.; Suber D. F.; Shungu D.; Silver L. L.; Inglima K.; Kornblum J.; Livermore D. M. Outbreak of Klebsiella pneumoniae Producing a New Carbapenem-Hydrolyzing Class A Beta-Lactamase, KPC-3, in a New York Medical Center. Antimicrob. Agents Chemother. 2004, 48, 4793–4799. 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M. Has the Era of Untreatable Infections Arrived?. J. Antimicrob. Chemother. 2009, 64, i29–i36. 10.1093/jac/dkp255. [DOI] [PubMed] [Google Scholar]
- Doumith M.; Ellington M. J.; Livermore D. M.; Woodford N. Molecular Mechanisms Disrupting Porin Expression in Ertapenem-Resistant Klebsiella and Enterobacter spp. Clinical Isolates From the UK. J. Antimicrob. Chemother. 2009, 63, 659–667. 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- Peng B.; Su Y. B.; Li H.; Han Y.; Guo C.; Tian Y. M.; Peng X. X. Exogenous Alanine and/or Glucose Plus Kanamycin Kills Antibiotic-Resistant Bacteria. Cell Metab. 2015, 21, 249–262. 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Allison K. R.; Brynildsen M. P.; Collins J. J. Metabolite-Enabled Eradication of Bacterial Persisters by Aminoglycosides. Nature 2011, 473, 216–220. 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith K. L.; Wolf R. E. Jr. Measuring Beta-Galactosidase Activity in Bacteria: Cell Growth, Permeabilization, and Enzyme Assays in 96-Well Arrays. Biochem. Biophys. Res. Commun. 2002, 290, 397–402. 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- Helander I. M.; Mattila-Sandholm T. Fluorometric Assessment of Gram-Negative Bacterial Permeabilization. J. Appl. Microbiol. 2000, 88, 213–219. 10.1046/j.1365-2672.2000.00971.x. [DOI] [PubMed] [Google Scholar]
- Graca da Silveira M.; Vitoria San Romao M.; Loureiro-Dias M. C.; Rombouts F. M.; Abee T. Flow Cytometric Assessment of Membrane Integrity of Ethanol-Stressed Oenococcus oeni Cells. Appl. Environ. Microbiol. 2002, 68, 6087–6093. 10.1128/AEM.68.12.6087-6093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering R. C. Jr.; Eliopoulos G. M.; Sentochnik D. E. The Carbapenems: New Broad Spectrum Beta-Lactam Antibiotics. J. Antimicrob. Chemother. 1989, 24, 1–7. 10.1093/jac/24.suppl_A.1. [DOI] [PubMed] [Google Scholar]
- Domenech-Sanchez A.; Martinez-Martinez L.; Hernandez-Alles S.; del Carmen Conejo M.; Pascual A.; Tomas J. M.; Alberti S.; Benedi V. J. Role of Klebsiella pneumoniae OmpK35 Porin in Antimicrobial Resistance. Antimicrob. Agents Chemother. 2003, 47, 3332–3335. 10.1128/AAC.47.10.3332-3335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I.; Roberts M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol Mol. Biol. Rev. 2001, 65, 232–260. 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veras D. L.; de Souza Lopes A. C.; Vaz da Silva G.; Araujo Goncalves G. G.; de Freitas C. F.; de Lima F. C. G.; Vieira Maciel M. A.; Feitosa A. P. S.; Alves L. C.; Brayner F. A. Ultrastructural Changes in Clinical and Microbiota Isolates of Klebsiella pneumoniae Carriers of Genes Bla SHV, Bla TEM, Bla CTX-M, or Bla KPC When Subject to Beta-Lactam Antibiotics. Sci. World J. 2015, 2015, 1–13. 10.1155/2015/572128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavuzzi A. M. L.; Alves L. C.; Veras D. L.; Brayner F. A.; Lopes A. C. S. Ultrastructural Changes Caused by Polymyxin B and Meropenem in Multiresistant Klebsiella pneumoniae Carrying blaKPC-2 Gene. J. Med. Microbiol. 2016, 65, 1370–1377. 10.1099/jmm.0.000367. [DOI] [PubMed] [Google Scholar]
- Brown L.; Wolf J. M.; Prados-Rosales R.; Casadevall A. Through the Wall: Extracellular Vesicles in Gram-Positive Bacteria, Mycobacteria and Fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly H.; Keisari Y.; Crouch E.; Sharon N.; Ofek I. Recognition of Bacterial Surface Polysaccharides by Lectins of the Innate Immune System and Its Contribution to Defense Against Infection: The Case of Pulmonary Pathogens. Infect. Immun. 2008, 76, 1322–1332. 10.1128/IAI.00910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.; Brooks T. Common Oligosaccharide Moieties Inhibit the Adherence of Typical and Atypical Respiratory Pathogens. J. Med. Microbiol. 2004, 53, 833–840. 10.1099/jmm.0.45643-0. [DOI] [PubMed] [Google Scholar]
- Krivan H. C.; Roberts D. D.; Ginsburg V. Many Pulmonary Pathogenic Bacteria Bind Specifically to the Carbohydrate Sequence GalNAc Beta 1–4Gal Found in Some Glycolipids. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 6157–6161. 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.