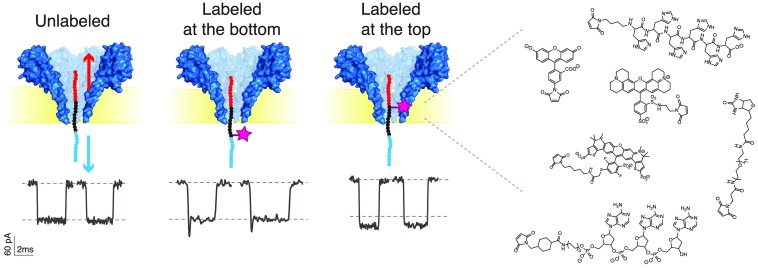
Abstract
While DNA sequencing is now amply available, fast, and inexpensive, protein sequencing remains a tremendous challenge. Nanopores may allow for developing a protein sequencer with single-molecule capabilities. As identification of 20 different amino acids currently presents an unsurmountable challenge, fingerprinting schemes are pursued, in which only a subset of amino acids is labeled and detected. This requires modification of amino acids with chemical structures that generate a distinct nanopore ionic current signal. Here, we use a model peptide and the fragaceatoxin C nanopore to characterize six potential tags for a fingerprinting approach using nanopores. We find that labeled and unlabeled proteins can be clearly distinguished and that sensitive detection is obtained for labels with a spectrum of different physicochemical properties such as mass (427–1275 Da), geometry, charge, and hydrophobicity. Additionally, information about the position of the label along the peptide chain can be obtained from individual current-blockade event features. The results represent an important advance toward the development of a single-molecule protein-fingerprinting device with nanopores.
Keywords: nanopore, protein fingerprinting, amino acid labeling, protein analysis, biological nanopores, single-molecule protein sequencing
DNA, RNA, proteins, and metabolites form a complex network of interactions that determines the phenotype of cells.1,2 To obtain a broad understanding of a biological system, we need a comprehensive approach that integrates genomics, transcriptomics, proteomics, and metabolomics.3−7 Recent technological developments have mostly focused on the study of genomes, making DNA sequencing fast, cheap, and ubiquitous.8,9 The study of other -omics, especially proteomics, however, still remains costly and time-consuming.10−12
One of the main challenges in proteomics is the lack of sensitive techniques that allow for detection of proteins present in low abundance, because, unlike for DNA, there is no biochemical method to amplify proteins present in a sample.13,14 Several antibody-based methods are commonly used for protein analysis. These methods are cost-effective; however, they have several well-known limitations. For example, there is a limited availability of antibodies, and they often lack the high specificity needed for complex samples. Single-molecule techniques offer ultimate sensitivity and hold great promise for single-cell protein analysis.15 Nanopores, in particular, have demonstrated to be ultrasensitive biosensors,16 capable of successfully sequencing biopolymers such as DNA.17,18 In a nanopore sensor, an insulating membrane made of a lipid bilayer or a solid-state membrane separates two compartments filled with an electrolyte. A nanometer-sized pore is made within the membrane, by inserting a single protein pore into a lipid bilayer or drilling a pore in a solid-state membrane using an electron beam. When a voltage is applied across the membrane, an ionic current flows through the nanometer-sized aperture. Molecules passing or translocating through the pore modulate the ionic current, which provides the basic sensor signal. For DNA translocation, fine changes within the current blockade signal were found to correlate to the sequence of the DNA passing through the pore,19,20 paving the way to DNA sequencing at the single-molecule level.
At first glance one might think that nanopore-based DNA sequencing can be straightforwardly modified to also allow for protein sequencing, but there are multiple challenges.15 While DNA is composed of only four different bases, proteins contain over 20 different amino acids, presenting a very significant technical hurdle for protein sequencing. In recent years, the groups of Joo and Marcotte proposed an alternative simpler idea, namely, protein fingerprinting, in which proteins can be identified if a subset of amino acids is labeled and read.21,22 Joo and colleagues proposed protein identification through detection of the sequence of cysteines and lysines along the peptide chain, a sequence that then is compared to a protein database for protein identification.21 Nanopores may offer an attractive technique for the implementation of such protein fingerprinting, thanks to their high sensitivity and time resolution. For such a nanopore protein fingerprinting method, cysteines, lysines, or other amino acids should be modified with labels that can produce a distinct modulation in the current while the linearized protein traverses the nanopore. The identification of such chemical modifications has so far not been realized using nanopores.
Here, we study the detection of six potential sequencing tags on a model peptide, as measured with the fragaceatoxin C (FraC) nanopore.23,24 We attach different chemical groups to a single cysteine in the central part of the peptide and measure their effect on the nanopore signals. We show that labels in the range between 427 and 1275 Da can be clearly and reproducibly detected. Information about the position of the label can be extracted from the individual event characteristics. The relative current blockade correlates with label properties such as geometry and molecular weight, and the translocation time is found to be proportional to the calculated tag charge and size. The results represent an important advance toward the development of a single-molecule protein-fingerprinting device with nanopores. Beyond protein fingerprinting, the identification of single amino acid modifications is also of great important for other biotechnology applications such as the detection of post-translational modifications (PTMs).
Results and Discussion
Distinguishing Labeled and Unlabeled Peptides Using the FraC Nanopore
Nanopore protein fingerprinting requires the electrical detection of chemical modifications on specific amino acids. We sought to characterize the blockade levels produced by different chemical labels in a well-defined manner. In order to avoid bandwidth-related distortions of the signal, we designed a method to obtain long translocation times. We used a 30 amino acid long peptide, containing 10 glutamates at the N-terminus, and 10 arginines at the C-terminus, which at neutral pH features a strongly negatively charged N-terminus and a strongly positively charged C-terminus. Upon applying a negative bias to the trans side of the nanopore setup (Figure 1), the peptide is dragged into the nanopore with its positive end entering first. When the negative region subsequently enters the pore, the electrophoretic force pulls the negatively charged end of the peptide in the opposite direction, thus stretching the peptide and stalling the molecule at the point where the forces in both directions equilibrate. The “tug-of-war” created thus allows for long observation times where the center part of the peptide is probed in the pore constriction.25 A similar peptide was previously used by Asandei etal. to study the effect of pH in peptide–nanopore interactions.26
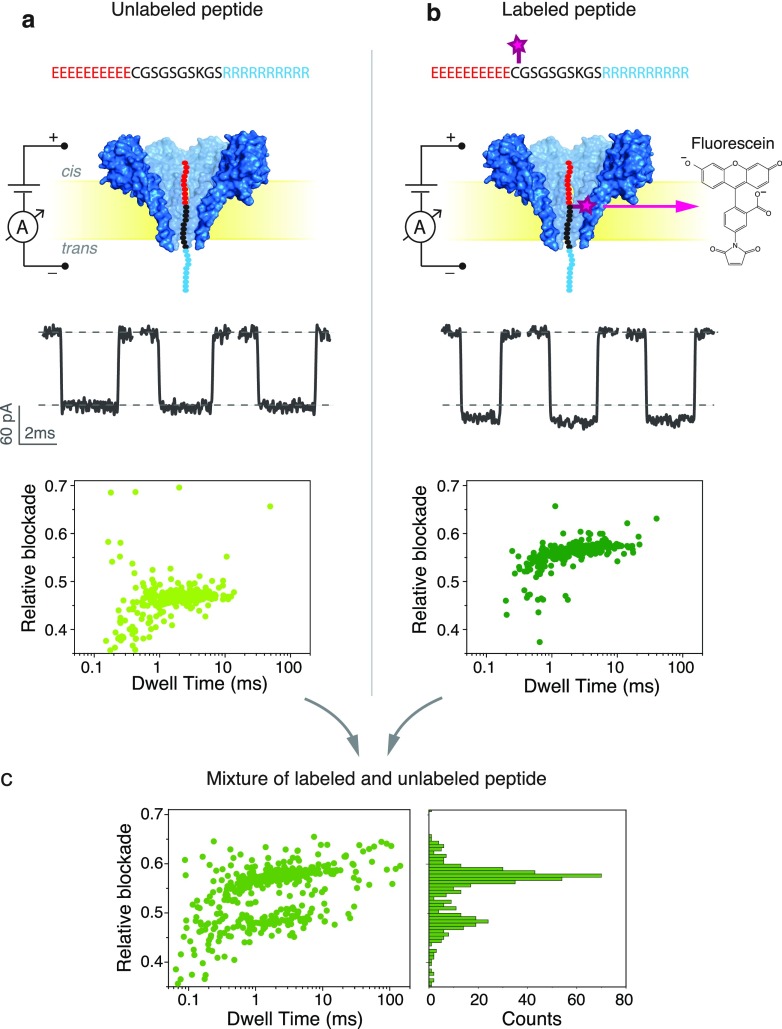
Figure 1.
Analysis of labeled and unlabeled peptides with FraC. From top to bottom: Schematic representation, typical events, and scatter plot of relative blockade vs dwell time of the unlabeled model peptide (a) and for the fluorescein-labeled model peptide (b) through the FraC nanopore. (c) Scatter plot of relative blockade vs dwell time (left) and relative blockade histogram (right) of a mixture containing labeled and unlabeled peptide. All measurements were done using a buffer containing 1 M NaCl, 10 mM Tris, and 1 mM EDTA at pH 7.5. The peptides were added to the cis compartment, and recordings were performed at a bias of −90 mV. Data were recorded at a sampling frequency of 500 kHz and further low-pass filtered at 10 kHz. FraC is shown as a surface representation from the PDB structure 4TSY.
Figure 1 and Figure S1 illustrate the typical nanopore experiments. All our experiments were performed using buffer containing 1 M NaCl, 10 mM Tris, and 1 mM EDTA at pH 7.5. We use the wild-type type I FraC nanopore, which most likely describes octameric nanopores,27 and the peptide described above as our substrate. Our model peptide is added to the cis compartment at concentrations between 0.1 and 0.5 μM. Negative voltages are applied to the trans compartment for all of our measurements to avoid gating that is observed in FraC under positive bias. The measurements presented here are performed under a voltage bias of −90 mV, unless stated otherwise.
Well-defined events are consistently observed when the peptide is present in the cis compartment (Figure 1a and Figure S1). Notably, the relative current blockade is well reproducible from pore to pore with an average relative blockade of 0.47 ± 0.03 (error is standard deviation from N = 3 experiments) at −90 mV, as measured in three independent experiments. The relative blockade is defined as the ratio between the current blockade (ΔI = IOpenPore – IBlockade) and the open pore current (IOpenPore). Long translocation times of 4.2 ± 0.6 ms are observed. The translocation time is observed to decrease with increasing bias (Figure S1), indicating that the peptide exits the pore to the trans side of the chamber.28,29 A detailed characterization of the translocation behavior of this model peptide through the FraC nanopore can be found elsewhere.25
To probe the effect of an added chemical label to the peptide, we first used maleimide chemistry to label the cysteine in position C11, i.e., near the N-terminus, with a fluorescein dye (Figure 1b). Fluorescein maleimide is a small molecule with a molecular weight of only 427 Da. Our labeled peptides were HPLC-purified, and the labeling was verified using MALDI-TOF mass spectrometry (Figure S2). We find that our samples are nearly 100% labeled. We performed measurements of the unlabeled and the fluorescein-labeled peptide, as shown in Figure 1.
We find that the current levels from the unlabeled and labeled peptides are clearly separated (Figure 1a,b). The unlabeled peptide produced a relative blockade of 0.47 ± 0.01, while the labeled peptide produced a relative blockade of 0.57 ± 0.01. These values correspond to the mean and standard deviation derived from a Gaussian fit of the relative blockade histograms. The clear increase in the relative blockade upon labeling is consistent with an additional blockade by the added volume due to the presence of the fluorescein tag. Control experiments with a mixture of labeled and unlabeled peptides confirmed the presence of two populations with blockade levels of 0.46 ± 0.01 and 0.56 ± 0.01, very close to the values obtained from the independent measurements (Figure 1c). We conclude that the fluorescein-labeled peptides can be readily distinguished from their unlabeled counterpart. The difference in the blockade levels is so clear and reproducible that, while nanopore data typically rely on data comparison in stochastic scatter plots of hundreds of events, we can even distinguish labeled from unlabeled molecules one by one from their current levels in individual events (Figure 1a,b).
Identifying the Most Sensitive Region of the Model Peptide in the FraC Pore
The fact that we observed events with a sizable (>1 ms) translocation time indicates that the peptide is, as designed, stalled in the pore at the point where the forces pulling in both directions are equal. Our previous experiments and simulations confirmed the mechanism of such peptide stalling in the FraC nanopore.25 During this stalling, a particular region of the peptide will stay closest to the pore constriction at the applied voltage of −90 mV. This portion of the peptide represents the most sensitive sensing region of our peptide–nanopore system, and its identification is therefore important for testing the tags for fingerprinting. Hence, we tested three different variants of our model peptide in which the cysteine is placed at different positions along the central part of the peptide, namely, at position 11, 15, or 20 as counted from the N-terminus (Figure 2). Notably, a similar approach was used in the early days of DNA sequencing, where a DNA homopolymer was trapped in the alpha-hemolysin nanopore, and single bases were changed sequentially at different positions to find the region closest to the pore constriction.19
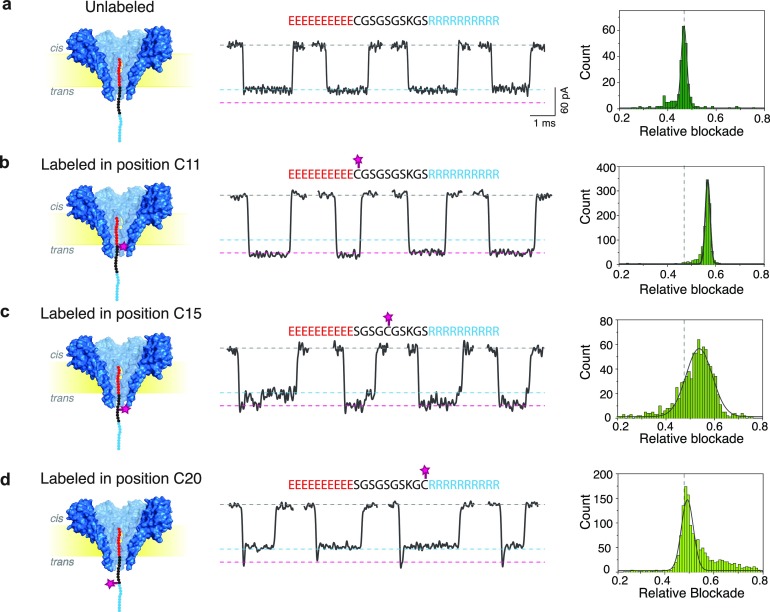
Figure 2.
Peptide labeled at different positions through FraC. From left to right: Schematic, current traces, and relative blockade histograms for unlabeled peptide (a) and for fluorescein-labeled peptide at positions C11 (b), C15 (c), and C20 (d). All labeled peptide samples were HPLC purified and verified using mass spectrometry. Measurements were done in a buffer containing 1 M NaCl, 10 mM Tris, and 1 mM EDTA at pH 7.5. Peptides were added to the cis compartment and measured at −90 mV.
The three different peptide variants were labeled with fluorescein maleimide, HPLC-purified, and mass spectrometry-verified as shown in Figure S2. The three samples were measured with the FraC nanopores, as shown in Figure 2. For reference, the relative blockade observed for an unlabeled peptide is displayed as well in Figure 2a (derived from the scatter plots presented in Figure 1a). The unlabeled peptide produced relative current blockades of 0.47 ± 0.01, while we consistently observed a larger relative blockade of 0.57 ± 0.01 for the peptide labeled with fluorescein in position C11, as discussed in the previous section. With the peptide labeled with fluorescein in position C15, however, pronounced current fluctuations were observed and events often contained a lower current level in the last fraction of the event (Figure 2c). As a consequence, a broad population is observed in the relative blockade histogram with a mean of 0.54 and a larger standard deviation of 0.05. Finally, when the label was placed in position C20, a blockade level of 0.48 ± 0.02 was observed, i.e., virtually no increase in the relative blockade compared to the unlabeled peptide but merely leading to a weak tail on the right-hand side of the histogram. Most interestingly, while the average current level of the events corresponded to that of the unlabeled peptide for most of the event duration, a clear spike in the current was observed at the start of most of these events (81%) (Figure 2d).
These results suggest that when the label is placed at position C11, it remains in the proximity of the pore constriction during the entire duration of the event. As a consequence, a well-defined increase in the blockade is observed. On the other hand, when the label is placed in position C15, fluctuations in the current level are observed, indicating that the label occupies the pore constriction only for a fraction of the time of the event duration. Finally when the label is placed in position C20, it appears to be too far from the nanopore constriction for most of the event duration, while it is only temporarily observed at the beginning of the event as the label moves fast through the nanopore. From examining these three positions that were tested, we thus conclude that the label remains the closest to the nanopore constriction when it is placed in position C11, and hence we identify this as the most sensitive region in our model peptide in the FraC system. The results indicate that both the relative blockade and individual event characteristics can be used to extract information on the position of the label in the peptide system.
Exploring Diverse Labels Using the Peptide-FraC System
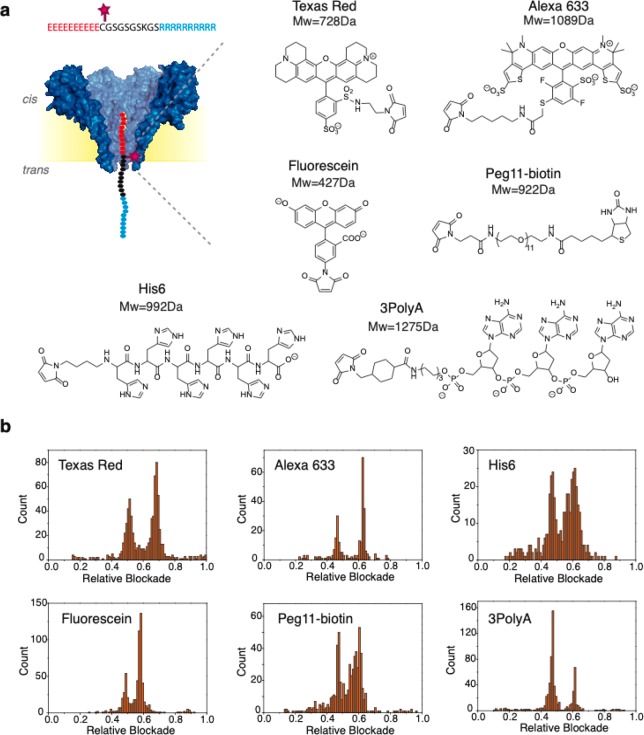
Protein fingerprinting requires the use of multiple different chemical tags attached to different amino acids. The development of efficient chemical procedures to label different amino acids is still a subject of active research in chemistry.30 Here, we focus on maleimide chemistry due to specificity, availability of labels, and simplicity of the reaction conditions. We proceeded to label the peptide in position C11 with a variety of tags as shown in Figure 3a. In ascending order of their molecular weight, the six labels studied were fluorescein (427 Da), Texas Red (728 Da), PEG11-biotin (922 Da), His6 (992 Da), Alexa633 (1089 Da), and 3polyA (1275 Da). We note that these are relatively large tags, 2–7 times larger than the largest amino acid (tryptophan, 204 Da), with different physicochemical properties. Fluorescein, Texas Red, and Alexa633 are fluorescent dyes. Their hydrophobic nature potentially allows specific binding modes to the inner lumen of the pore, but might also promote protein aggregation and reduce protein solubility in a fingerprinting approach, in which a protein should be labeled at every 5–10 amino acids. The hydrophilic tags tested were 3polyA, His6, and PEG11-biotin. 3polyA is a small oligonucleotide containing three adenine bases and a maleimide coupling group at the 5′-end. Due to the three phosphate groups, this tag has a net negative charge. His6 is a stretch of six histidines with a maleimide at the end. At the pH of our measurements (pH 7.5), this tag can carry a neutral or single negative charge, given by its carboxyl group. By lowering the pH to values closer to the pKa of histidine the number of charges of this tag can be modified. Lastly, the PEG11-biotin structure is composed of 11 PEG units. PEG is known to bind positive ions such as Na+, which is present in our buffer. According to previous studies, a PEG molecule of 11 units can bind one Na+ ion at most, and therefore this tag is likely to have a neutral or single positive charge.31,32
Figure 3.
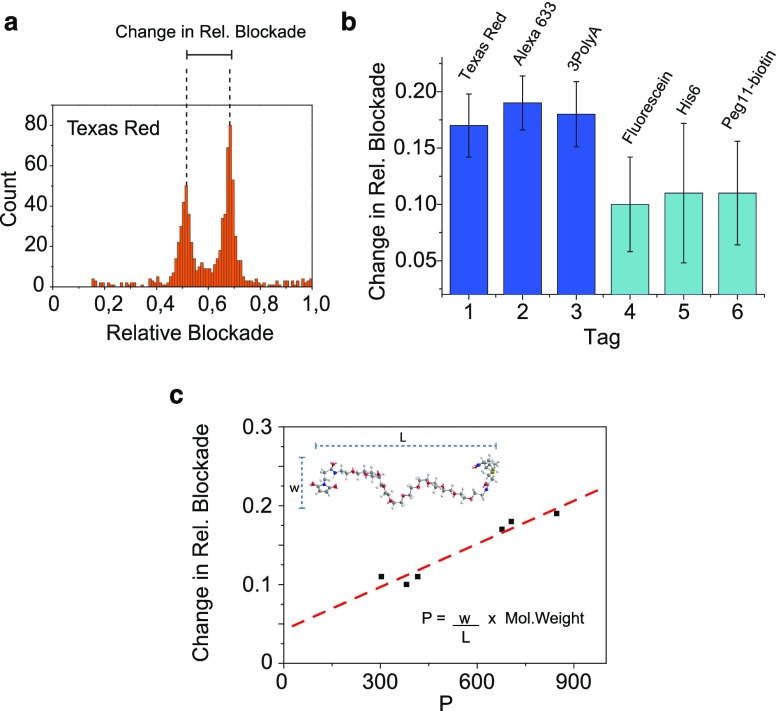
(a) Different chemical structures used to label the peptide at position C11. The six structures correspond to Texas Red (728 Da), Alexa633 (1089 Da), 3polyA (1275 Da), fluorescein (427 Da), PEG11-biotin (922 Da), and His6 (992 Da). (b) Histograms of the relative blockade of unlabeled and labeled peptide mixtures. Two peaks can be observed in each of the histograms. The first peak corresponds to the unlabeled peptide, and the second peak corresponds to the labeled peptide with each of the tags. These measurements were performed in buffer containing 1 M NaCl, 10 mM Tris, and 1 mM EDTA at pH 7.5. Peptide mixtures were added in the cis chamber and measured at −90 mV. Data were recorded at 500 kHz and low-pass filtered with a Gaussian filter at 10 kHz.
Figure 3b shows the six relative blockade histograms observed for peptides labeled with each of the six tags mentioned above. Example event traces for each of the labels can be found in Figure S3. In each of these measurements, labeled and unlabeled peptides were measured as a mixture, where the unlabeled peptide was acting as a reference. Hence, two peaks can be observed in each of the histograms. The first peak near 0.45–0.50 corresponds to the unlabeled peptide, while the second peak near 0.56–0.66 corresponds to the labeled peptide for each of the tags. The change in relative blockade caused by each label, calculated by the difference in relative blockade between the labeled and unlabeled peptide, is shown in Figure 4a. Only based on the change in relative blockade, the tags can be categorized into two groups: Texas Red, Alexa633, and 3polyA cause a large change in relative blockade of 0.17–0.19, while fluorescein, His6, and Peg11-biotin cause a smaller change of 0.10–0.11 (Figure 4b). As a control, we also attempted to measure the free tags in solution, but translocation of these small groups occurred too fast for reliable detection, verifying the virtue of our tug-of-war approach.
Figure 4.
(a) Relative blockade histogram for the mixture of unlabeled peptide and peptide labeled with Texas Red showing the change in relative blockade. (b) Shift in relative blockade measured for each label. Texas Red, Alexa633, and 3polyA cause a larger change in relative blockade. (c) Correlation between the change in relative blockade and parameter P = M × w/L, which characterizes the label’s geometry (see inset) and molecular weight (R2 = 0.92).
Interestingly, the increase in relative blockade produced by a particular tag (Figure 4) does not correlate well with its molecular weight (R2 = 0.16), as shown in Figure S4. For example, while Texas Red has a lower molecular weight than PEG11-biotin (728 Da vs 922 Da, respectively), it generates a larger blockade. The poor correlation is potentially related to their different geometry. While PEG has a linear and very flexible structure, Texas Red is a more rigid extended structure comprising multiple tightly packed aromatic rings. Indeed, one can imagine that a larger number of ions is blocked by Texas Red when this label resides in the constriction as compared to the linear structure of PEG that most probably extends partially out of the constriction area. To find an alternative figure of merit that takes into consideration both the molecular weight and shape, we define a phenomenological parameter P = S x M, where M is its molecular weight of the label and S is a shape factor calculated as the ratio between the width (w) of the molecule and its length (L). To calculate the length and width of the labels, an energy minimization of each structure is done using molecular mechanics at the MM2 level in ChemDraw 3D. This process returns the energy-minimized conformation of the molecule. The parameter L is then calculated as the length of the molecule across the longest axis, and the width is measured perpendicular to the length (Figure 4c inset, Figure S5). Figure 4c shows the increase in relative blockade measured for each label vs the calculated P value. A strong correlation (R2 = 0.92) is observed between the increase in relative blockade and parameter P, indicating that not only the molecular weight of the label but also its geometry affect the amount of current blocked by a label.
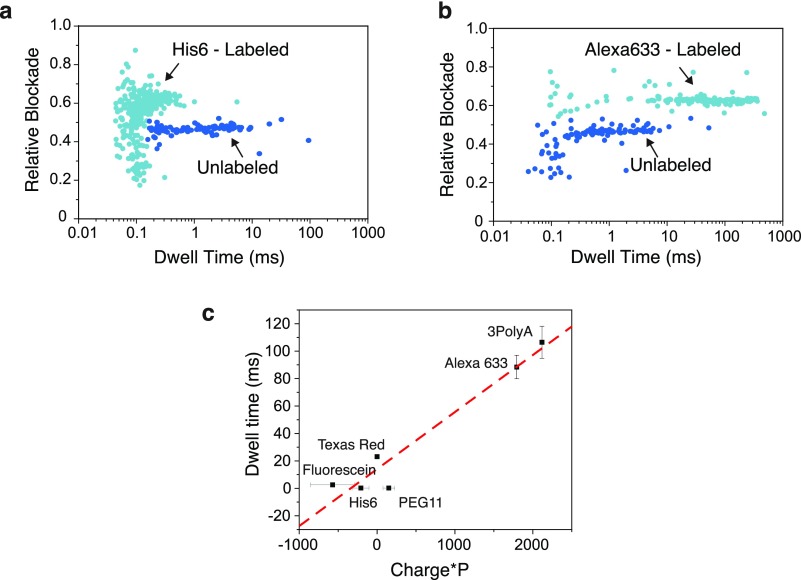
Different chemical labels not only cause a measurably different increase in the relative blockade but also influence the translocation time of the peptide; see Figure 5, where panels a and b compare the scatter plots of relative blockade vs dwell time for the peptide labeled with His6 and with Alexa633, respectively. The dwell time is observed to be significantly different, with a mean translocation time of 29 ms for Alexa633 vs 0.22 ms for His6. We find that two main properties of the tags have an effect on the translocation time, namely, charge and size. Charges present in the labels can act to increase the electrophoretic force pulling toward the cis or the trans opening of the pore, thus increasing or decreasing the translocation time, respectively. Control experiments with peptides containing a constant number of 10 positive charges in the C-terminus but varying number of negative charges in the N-terminus (10, 12, or 14) showed that the translocation dwell times increased with the increasing number of negative charges, as expected (Figure S6). To account for size, we again take into consideration the size (MW) and the geometry of the label through the parameter P. Figure 5c shows the scatter diagrams of the dwell time vs the net charge multiplied by P for each of the labels. A strong correlation between these two characteristics is observed with R2 = 0.93.
Figure 5.
Scatter plots of the peptide labeled with (a) His6 and (b) Alexa633. Faster translocation times are observed in peptides labeled with His6 compared to Alexa633. (c) Plot of dwell time vs net charge × P, where P = M × w/L (see Figure 4). A correlation (R2 = 0.93) is observed between these parameters. The errors in the x-axis for fluorescein, His6, and PEG11 represent the range of values that the parameter can take due to the possible charged states of the molecules.
Toward Fingerprinting: Resolving Labels during Peptide Translocation
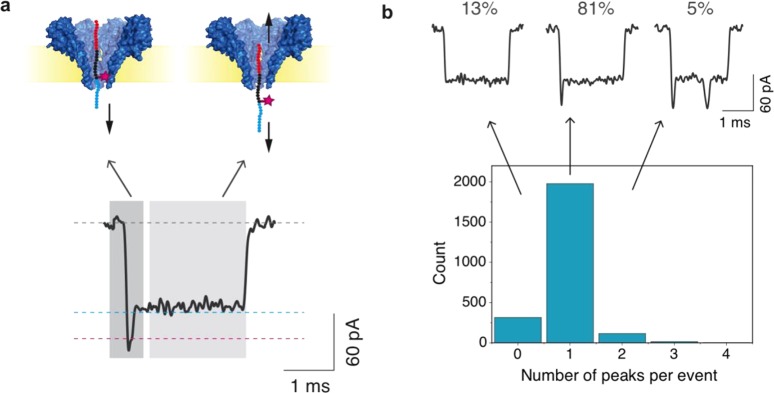
Above, we demonstrated the successful detection of chemical labels in a model peptide, where a tug-of-war mechanism stalled the central part of the peptide close to the pore constriction. While this represents an important step forward to fingerprint proteins, peptides will be translocating at a high speed in a more generalized nanopore fingerprinting scheme, and label may escape detection due to an overly short detection time. However, our data show that we can detect single labels “on the fly” as a short dip in the current. As shown in Figures 6a and 2d, we observed a clear spike in the first fraction of the event when measuring a peptide with a label in position C20. We quantified the number of peaks in each of the events using a MATLAB script. As shown in Figure 6b, 1978 out of a total of 2427 events, i.e., 81%, contained a clear single dip in the current. Only 315 events (13%) contained no distinguishable peak, and 134 events showed more than one peak (6%). The high percentage of events containing a single peak (81%), even without any further optimization, is encouraging for the realization of a generalized fingerprinting scheme. The small percentage of events in which no peak is visible is likely due to fast translocations of the label through the pore, which therefore occasionally escapes detection due to the finite time resolution. This percentage is expected to be reduced significantly if an enzyme, such as ClpX, is used to enable controlled protein translocation through the nanopore as previously shown by Nivala etal.33 ClpX is a robust enzyme that can process modified substrates and can significantly reduce the translocation time of polypeptides.34 Finally, event traces with more than one peak may be attributed to multiple readings of the label due to thermal motion of the peptide. Again, using a processing enzyme to control the translocation speed of the peptide through the pore would reduce the occurrence of these events.
Figure 6.
(a) Typical event observed for fluorescein-labeled peptide in position 20C. A decrease in current is observed in the first fraction of the event as the labeled portion of the peptide moves through the FraC pore. (b) Histogram of the number of peaks observed per event. Data were recorded at −90 mV using a sampling frequency of 500 kHz. Event traces were low-pass filtered at 5 kHz.
Conclusion
Using a bipolar peptide and the FraC nanopore as a model system, we studied the characteristics of different chemical labels and explored their potential for a nanopore fingerprinting approach. Six different labels were characterized in terms of their current blockade and translocation time. We observed a correlation between the translocation time of the peptide-tag system and the charge and size of the tags. Furthermore, we could successfully interpret the current blockade generated by a particular label if information about the geometry and molecular weight of the label is available. Our study indicates that it is possible to label amino acids with multiple distinguishable labels. We explored different positions along the peptide sequence to find the most sensitive region and showed that information about the position of the label can be derived from their relative blockade and event characteristics. Moreover, spikes in the current traces could be reproducibly observed, consistent with a fingerprinting scheme where a polypeptide sequentially translocates through the pore. Altogether, our results are very promising for a protein fingerprinting approach in which different chemical tags are used to label and recognize different amino acids.
Methods/Experimental
Peptide Design and Synthesis
The peptides used in this work were the model peptide C11 with sequence EEEEEEEEEECGSGSGSKGSRRRRRRRRRR (HPLC purity = 95.8%, MW = 3678.9 Da), model peptide with C15 with sequence EEEEEEEEEESGSGCGSKGSRRRRRRRRRR (HPLC purity = 95.1%, MW = 3678.9 Da), and model peptide with C20 with sequence EEEEEEEEEESGSGSGSKGCRRRRRRRRRR (HPLC purity = 95.7%, MW = 3678.9 Da). Peptides were synthesized by Biomatik Corporation (Cambridge, CA, USA). The synthesis was performed using standard solid-phase methods, and the peptides were further purified using reverse-phase HPLC and analyzed by mass spectrometry (Biomatik). Peptides were kept lyophilized or, when necessary, aliquoted in LC-MS grade water to a final concentration of 10 mg/mL at −20 °C.
Chemical Tags Containing a Maleimide Group
The maleimide-containing molecules used as tags were fluorescein-5-maleimide (Thermo), Texas Red C2 maleimide (Thermo), Alexa Fluor 633 C5 maleimide (Thermo), EZ-link maleimide-PEG11-biotin (Thermo), Histidine6 maleimide (Biomatik), 5′-maleimide 3PolyA (Biosynthesis).
Peptide Labeling and Purification
Polypeptides containing a cysteine residue were labeled using maleimide chemistry. For labeling, the final peptide concentration was 1 mg/mL, and an excess of tag from 10:1 was used. An exception was 3PolyA, where a ratio of 2:1 was used due to the low amounts of tag available. Labeling proceeded at 4 °C overnight in 1× phosphate-buffered saline at pH 7. Labeling was done with degassed buffers and under nitrogen to prevent cysteine oxidation. For synthetic peptides, no significant difference in labeling was observed if cysteines were reduced previous to labeling, and therefore tris(2-carboxyethyl)phosphine (Sigma-Aldrich) was not necessary. Labeled peptides were purified using reverse-phase chromatography. For that an Agilent 1260 Infinity HPLC system was used with a Waters CSH C18 column as the stationary phase and a mobile phase consisting of a gradient of acetonitrile and water with 0.1% trifluoroacetic acid (TFA). HPLC fractions were collected and analyzed using MALDI-TOF (Autoflex Speed). The matrix consisted of 10 mg/mL α-cyano-4-hydroxycinnamic acid and 0.2% TFA.
Electrical Recording in Planar Lipid Membranes
Electrical recording were performed using planar lipid membranes as described before.35,36 Briefly, a 25-μm-thick Teflon film (Goodfellow Corporation, PA, USA) containing an orifice of approximately 70 μm separates the cis and trans compartments. To form the membranes, 10 μL of 5% hexadecane in pentane is added to the Teflon film and the pentane is allowed to evaporate. The reservoirs are filled with buffer and 10 μL of 10 mg/mL 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids) in pentane. Membranes were spontaneously formed using the Montal–Mueller method.37 Ag/AgCl electrodes are placed in each compartment, with the ground electrode in the cis side. WT FraC oligomers are added to the cis side of the chamber.23,24 Upon pore insertion, the pore is characterized by measuring traces at different voltages and taking an IV curve. The substrate was added to the cis side of the chamber and measured at −90 mV.
Data Acquisition and Analysis
Nanopore recordings were collected using a patch-clamp amplifier (Axopatch 200B, Molecular Devices, USA) at a filtering frequency of 100 kHz. The data were digitized using an Axon Digidata 1550B digitizer at a sampling frequency of 500 kHz. The signal was low-pass filtered at 10 kHz and processed using a Matlab script (Transalyzer).38 Event traces were filtered at 5 kHz for display.
Acknowledgments
We would like to thank Niels van den Broek, Robert Cordfunke, Mike Filius, Marek Noga, Sonja Schmid, and Chun Heung Wong for fruitful discussions. This work was supported by the European Research Council Advanced Grant SynDiv (No. 669598) to C.D., by The Netherlands Organisation for Scientific Research (NWO/OCW) through the NanoFront and BaSyC grants, and by the Foundation for Fundamental Research on Matter (Vrije Programma SMPS) to C.J., C.D., R.E., P.R.B., and G.M.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.9b05156.
Example traces of the translocation of unlabeled peptides, MS spectra of the purified peptides, example event traces of peptides with the six different labels, the P parameter calculated for each label, and control experiments with peptides containing different numbers of negative amino acids (PDF)
The authors declare the following competing financial interest(s): L.R.P., C.D., and C.J. are co-founders and shareholders of Bluemics B.V., a company engaged in the development of nanopore sensors for protein analysis.
Supplementary Material
References
- Kim M.; Rai N.; Zorraquino V.; Tagkopoulos I. Multi-Omics Integration Accurately Predicts Cellular State in Unexplored Conditions for Escherichia Coli. Nat. Commun. 2016, 7, 13090. 10.1038/ncomms13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin S.; Vondriska T. M. Genomes, Proteomes, and the Central Dogma. Circ.: Cardiovasc. Genet. 2011, 4, 576. 10.1161/CIRCGENETICS.110.957795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K. J.; Snyder M. P. Integrative Omics for Health and Disease. Nat. Rev. Genet. 2018, 19, 299–310. 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M.; Mann M. From Genomics to Proteomics. Nature 2003, 422, 193–197. 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- Karahalil B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. 10.2174/0929867323666160926150617. [DOI] [PubMed] [Google Scholar]
- Haider S.; Pal R. Integrated Analysis of Transcriptomic and Proteomic Data. Curr. Genomics 2013, 14, 91–110. 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Beyer A.; Aebersold R. On the Dependency of Cellular Protein Levels on MRNA Abundance. Cell 2016, 165, 535–550. 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Wetterstrand K. A.DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) www.genome.gov/sequencingcostsdata (accessed Jul 2, 2018).
- Goodwin S.; McPherson J. D.; McCombie W. R. Coming of Age: Ten Years of Next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S.; Kulej K.; Garcia B. A. Why Proteomics Is Not the New Genomics and the Future of Mass Spectrometry in Cell Biology. J. Cell Biol. 2017, 216, 21–24. 10.1083/jcb.201612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A. A Cast of Thousands. Nat. Biotechnol. 2003, 21, 213. 10.1038/nbt0303-213. [DOI] [PubMed] [Google Scholar]
- Mitchell P. In the Pursuit of Industrial Proteomics. Nat. Biotechnol. 2003, 21, 233–237. 10.1038/nbt0303-233. [DOI] [PubMed] [Google Scholar]
- Millioni R.; Tolin S.; Puricelli L.; Sbrignadello S.; Fadini G. P.; Tessari P.; Arrigoni G. High Abundance Proteins Depletion vs Low Abundance Proteins Enrichment: Comparison of Methods to Reduce the Plasma Proteome Complexity. PLoS One 2011, 6, e19603 10.1371/journal.pone.0019603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Fonslow B. R.; Shan B.; Baek M.-C.; Yates J. R. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem. Rev. 2013, 113, 2343–2394. 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Pérez L.; Joo C.; Dekker C. Paving the Way to Single-Molecule Protein Sequencing. Nat. Nanotechnol. 2018, 13, 786–796. 10.1038/s41565-018-0236-6. [DOI] [PubMed] [Google Scholar]
- Dekker C. Solid-State Nanopores. Nat. Nanotechnol. 2007, 2, 209–215. 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- Jain M.; Olsen H. E.; Paten B.; Akeson M. The Oxford Nanopore MinION: Delivery of Nanopore Sequencing to the Genomics Community. Genome Biol. 2016, 17, 239. 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D.; Akeson M.; Branton D. Three Decades of Nanopore Sequencing. Nat. Biotechnol. 2016, 34, 518–524. 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart D.; Heron A. J.; Mikhailova E.; Maglia G.; Bayley H. Single-Nucleotide Discrimination in Immobilized DNA Oligonucleotides with a Biological Nanopore. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 7702–7707. 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E. A.; Derrington I. M.; Pavlenok M.; Niederweis M.; Gundlach J. H. Nucleotide Discrimination with DNA Immobilized in the MSPA Nanopore. PLoS One 2011, 6, e25723 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Docter M.; van Ginkel J.; de Ridder D.; Joo C. Single-Molecule Protein Sequencing through Fingerprinting: Computational Assessment. Phys. Biol. 2015, 12, 055003 10.1088/1478-3975/12/5/055003. [DOI] [PubMed] [Google Scholar]
- Swaminathan J.; Boulgakov A. A.; Marcotte E. M. A Theoretical Justification for Single Molecule Peptide Sequencing. PLoS Comput. Biol. 2015, 11, e1004080 10.1371/journal.pcbi.1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Willems K.; Soskine M.; Wloka C.; Maglia G. Electro-Osmotic Capture and Ionic Discrimination of Peptide and Protein Biomarkers with FraC Nanopores. Nat. Commun. 2017, 8, 935. 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C.; Mutter N. L.; Soskine M.; Maglia G. Alpha-Helical Fragaceatoxin C Nanopore Engineered for Double-Stranded and Single-Stranded Nucleic Acid Analysis. Angew. Chem., Int. Ed. 2016, 55, 12494–12498. 10.1002/anie.201606742. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Restrepo-Pérez L.; Soskine M.; Maglia G.; Joo C.; Dekker C.; Aksimentiev A. Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate. ACS Nano 2019, 13, 2398–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asandei A.; Chinappi M.; Kang H. K.; Seo C. H.; Mereuta L.; Park Y.; Luchian T. Acidity-Mediated, Electrostatic Tuning of Asymmetrically Charged Peptides Interactions with Protein Nanopores. ACS Appl. Mater. Interfaces 2015, 7, 16706–16714. 10.1021/acsami.5b04406. [DOI] [PubMed] [Google Scholar]
- Huang G.; Voet A.; Maglia G. FraC Nanopores with Adjustable Diameter Identify the Mass of Opposite-Charge Peptides with 44 Da Resolution. Nat. Commun. 2019, 10, 835. 10.1038/s41467-019-08761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movileanu L.; Schmittschmitt J. P.; Martin Scholtz J.; Bayley H. Interactions of Peptides with a Protein Pore. Biophys. J. 2005, 89, 1030–1045. 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesemans A.; Soskine M.; Maglia G. A Protein Rotaxane Controls the Translocation of Proteins Across a ClyA Nanopore. Nano Lett. 2015, 15, 6076–6081. 10.1021/acs.nanolett.5b02309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez E. T.; Swaminathan J.; Marcotte E. M.; Anslyn E. V.. Solution-Phase and Solid-Phase Sequential, Selective Modification of Side Chains in KDYWEC and KDYWE as Models for Usage in Single-Molecule Protein Sequencing. New J. Chem. 2017, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan M. J.; Agnes G. R. Poly(Ethylene Glycol) Doubly and Singly Cationized by Different Alkali Metal Ions: Relative Cation Affinities and Cation-Dependent Resolution in a Quadrupole Ion Trap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2002, 13, 177–186. 10.1016/S1044-0305(01)00350-6. [DOI] [PubMed] [Google Scholar]
- Wong S. F.; Meng C. K.; Fenn J. B. Multiple Charging in Electrospray Ionization of Poly(Ethylene Glycols). J. Phys. Chem. 1988, 92, 546–550. 10.1021/j100313a058. [DOI] [Google Scholar]
- Nivala J.; Marks D. B.; Akeson M. Unfoldase-Mediated Protein Translocation through an α-Hemolysin Nanopore. Nat. Biotechnol. 2013, 31, 247–250. 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ginkel J.; Filius M.; Szczepaniak M.; Tulinski P.; Meyer A. S.; Joo C. Single-Molecule Peptide Fingerprinting. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 201707207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglia G.; Heron A. J.; Stoddart D.; Japrung D.; Bayley H. Analysis of Single Nucleic Acid Molecules with Protein Nanopores. Methods Enzymol. 2010, 475, 591–623. 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsmann T.; Heimburg T.; Keyser U.; Mahendran K. R.; Winterhalter M. Protein Reconstitution into Freestanding Planar Lipid Membranes for Electrophysiological Characterization. Nat. Protoc. 2015, 10, 188–198. 10.1038/nprot.2015.003. [DOI] [PubMed] [Google Scholar]
- Montal M.; Mueller P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. U. S. A. 1972, 69, 3561–3566. 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesa C.; Dekker C. Data Analysis Methods for Solid-State Nanopores. Nanotechnology 2015, 26, 1–7. 10.1088/0957-4484/26/8/084003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.