Abstract
In the Special Issue on Tools for Protein Science in 2018, we presented Molstack: a concept of a cloud‐based platform for sharing electron density maps and their interpretations. Molstack is a web platform that allows the interactive visualization of density maps through the simultaneous presentation of multiple datasets and models in a way that allows for easy pairwise comparison. We anticipated that the users of this conceptually simple platform would find many different uses for their projects, and we were not mistaken. We have observed researchers use Molstack to present experimental evidence for their models in the form of electron density maps, omit maps, and anomalous difference density maps. Users also employed Molstack to present alternative interpretations of densities, including rerefinements and speculative interpretations. While we anticipated these types of projects to be the main use cases, we were pleased to see Molstack used to display superpositions of different models, as a tool for story‐driven presentations, and for collaboration as well. Here, we present developments in the platform that were driven by user feedback, highlight several cases that used Molstack to enhance the publication, and discuss possible directions for the platform.
Keywords: data interpretation, validation, visualization, cryo‐EM, crystallography
1. INTRODUCTION
The accurate presentation and dissemination of experimental data is a cornerstone of scientific research and facilitates the validation of interpretation. Issues related to reproducibility are not restricted to pitfalls in the experiment alone but can quite often arise from an ambiguous interpretation of the results. Therefore, it is crucial that the authors of a study can posit a uniform explanation and presentation of their findings that will hold up under scrutiny from others and current knowledge in the concerned field.
In macromolecular crystallography, the result of a diffraction experiment is an electron density map. The structural model is an interpretation of that map. The process of map interpretation and modeling amino acid residues, water molecules, crystallization agents, and ligands has been considerably streamlined through automation, but structural errors continue to persist. The macromolecular model from the Protein Data Bank (PDB) is often treated as a ground truth but should be treated as an interpretation of the underlying experimental data. The interpretation can be complicated not only by the intrinsic nature of densities that result from the averaging of multiple states but also by human error and cognitive biases. Reviewers may sometimes have access only to validation reports and not models themselves;1 therefore, presenting an electron density map in a clear and unambiguous fashion is paramount for the review process. The release of a deposited structure and the publication of a paper constitute the points at which the coordinates and structure factors for a given PDB deposit become available for the public and are available for scrutiny. The detection of errors in a structure can lead to the redeposition of the structure and sometimes even retraction of the paper.2 The ripple effect of continued reliance on an erroneous finding can have tremendous ramifications, especially in translational research,2 as the impact of structural biology research on biomedical research disciplines such as drug discovery3, 4, 5 cannot be underestimated. The consistent interpretations of experimental data, which should conform to chemical knowledge, depend on the quality of tools available to inspect, validate, and correct structural models.
The vast increase in the number of computer programs to aid in each step of the structure determination process has helped to reduce the opportunities for human error and improve reproducibility.6, 7, 8, 9, 10, 11, 12, 13, 14
In the Special Issue on Tools for Protein Science in 2018, we presented Molstack: a concept of a cloud‐based platform for sharing electron density maps and their interpretations.15 Two years after the initial release of the Molstack server, we present new developments to Molstack that were driven by user feedback, highlight several cases that used Molstack to enhance the publication, and discuss possible future directions for the platform.
2. RESULTS
For a detailed description of the Molstack platform, we refer the readers to the previous publication.15 Shortly, Molstack is a rich internet application to disseminate structural data. It is a hosted platform that provides authoring tools and a web viewer to present and verify the models and maps. For convenience, we would like to provide a brief description of major concepts and terminology used here such as projects, stacks, and views. First, the Molstack project is a set of maps and models organized into groups that are displayed individually in two separate but synchronized viewports (Figure 1). We refer to these groups as stacks. In each viewport, the user viewing the project can select one stack to be displayed and compare two stacks side‐by‐side. The projects can have defined views. Views allow the project creator to define coordinates and scenes at which the user viewing the project should focus his/her attention. The views can be accompanied by a description, and together, they form a narrative for the project.
Figure 1.
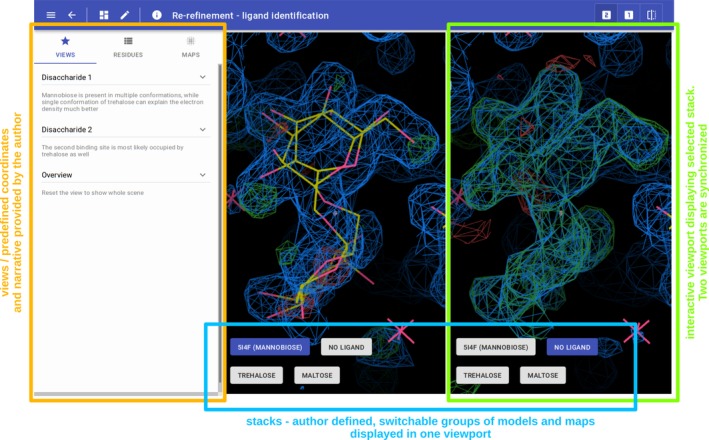
A Molstack project annotated with the description of individual parts
The Molstack platform allows users to create and share Molstack projects. The platform offers a “hands‐off” approach, and users are allowed to combine any type of models and maps into individual stacks and define views as they see fit. This flexibility allows the Molstack platform to host projects that can be used for distinct purposes.
2.1. New features
Since the initial release, we have removed reported bugs and improved the interface and responsiveness/performance of the platform. Here, we would like to present several user‐facing improvements and new features that we introduced to enhance the usability of the platform.
2.1.1. Collections
One of the early requested features was the ability to group several related Molstack projects together into a single collection. Therefore, we have implemented interfaces for the creation of a collection and selection of the projects that should be grouped, the presentation of a collection, and searching/browsing of public collections (Figure 2). The implementation of the collections offers different opportunities to organize projects into logical groups. We allow users to create collections that comprise their own and any public projects, and the collections honor the public visibility settings for individual projects (i.e., projects that are private or hidden will not be displayed as part of the collection even when the collection is publicly visible). Similar to an individual project, a collection can have three visibility settings: private, hidden, or public. The collections are shareable using long and short identifiers as described later and discoverable using the search feature. Some collections are highlighted in the published use cases, such as grouping projects related to a single publication, project, or presentation.
Figure 2.
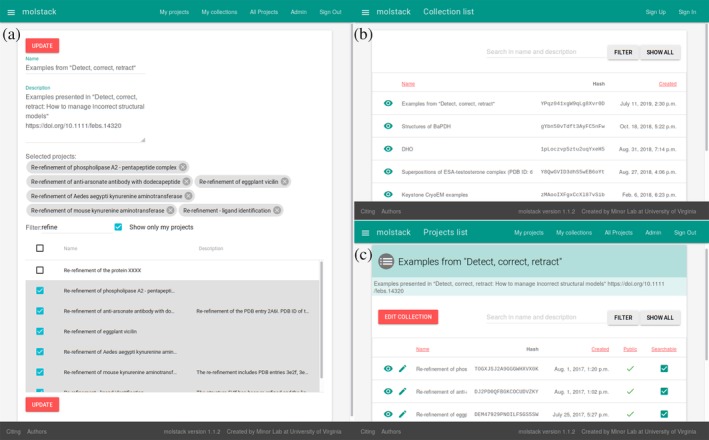
The interfaces related to managing collections of the projects. (a) Collection description and selection of the projects. (b) List and search of publicly visible collections. (c) View of the collection displaying constituent projects
2.1.2. Search
The increasing number of projects has made it more difficult to browse and manage projects. We have introduced a free text search of projects' names and descriptions in all lists, including both lists of public projects and private ones. The same search mechanism is used for the collections.
In the future, we plan to expand the search to view and stack descriptions and to index all chemicals present in the uploaded files. The chemical search has been suggested by several users as a crucial feature for the more widespread use of alternative and speculative interpretations. However, using chemical identities poses a significant technical challenge. Molstack accepts all types of models without ligand curation at the expense of model consistency. Searches using simple methods relying on the ligand identifiers are not robust enough. For example, when depositing a speculative interpretation that is not deposited elsewhere, the user may use any identifier for the ligand that is not related to the ligands already present in the PDB. When implemented, the search using chemicals will most probably use a substructure search such as the one used by PubChem.16
2.1.3. Short URLs/public sharing/privacy
The main aspect of Molstack is the ability to share the density maps and their interpretations. Currently, Molstack's projects and collections have three types of user‐configurable visibility: private, hidden, and public. Private projects are accessible only to the person who created the project. Access to the project requires proper login credentials. The private setting is intended for work‐in‐progress projects or to limited presentations by the project owner.
Hidden projects are viewable by anyone who has access to the link. These projects are not displayed in publicly accessible searches or indexed by search engines. This mechanism is similar to “link sharing” options available through cloud storage such as Google Drive or Microsoft OneDrive. The links used by Molstack use long, case‐sensitive, 20‐character, unique randomized IDs, and it is virtually impossible at this time to guess or scan more than 7 x 1035 identifiers to access project without explicit knowledge of the identifier. The hidden setting is intended for limited sharing collaborators to share progress, receive feedback, and so on. Once the link is shared, it can be accessed without the need for a Molstack account. The links to the hidden projects have the form of https://molstack.bioreproducibility.org/project/view/JJISBFEC2K6OU51GOETK/ and https://molstack.bioreproducibility.org/collection/view/YPqz041xgW9qLg8Xvr0D/ for hidden collections.
The public setting will make projects visible on the list of public projects, accessible from a search, and indexable by search engines. For more convenient sharing the public projects can be accessed using short ID and shortened link. The short IDs are four‐character long unique strings. Because short IDs are easier to guess (~14 x 106 identifiers to check), they are only used for projects that are fully public. The shortened links have a form of https://molstack.bioreproducibility.org/p/kdZe/ and https://molstack.bioreproducibility.org/c/AnAP/ for projects and collections, respectively.
On top of that, to ease the use of the identifiers, we have added a search box in the landing page that accepts short project and collection identifiers that can be used to directly jump to the requested project/collection.
Currently, we recommend using the shortened links when referring to Molstack's projects or collections in the publications, as they are directly clickable or easy to copy to manually navigate to a specific webpage. However, in places where the links will be too long to provide (as in tables with multiple projects) we recommend using four‐letter short IDs.
2.1.4. Map contouring and navigation
One of the commonly requested features by users was the ability to change the contouring level of maps. This was also requested by the reviewers of several of our papers where Molstack was used to present data. For the initial Molstack release, we had adopted a policy that the map contouring levels are fixed and defined by the project creator. This was to ensure that the project is viewed as intended by the creator. However, as Molstack is currently used mostly for presenting the evidence for the model, that is, being evaluated during the review process, the adjustable map contouring levels are beneficial for thorough verification. We have decided to revert from the previous policy and implement the interface for dynamically changing the contour levels. Due to the potential complexity of Molstack's projects and the number of maps, this was no trivial task. Single‐view software tools such as Coot7 or NGL17 on the RCSB PDB18 website (https://rcsb.org) allow for the selection of a map that is “scrolled” and has its contouring level adjusted. This works well with single views and a limited (two or three) number of maps, but with Molstack, we expect projects to have multiple maps that need to be synchronized not only across the two views but also across multiple stacks. Therefore, we have opted for a solution that all maps across all stacks have their contouring level adjusted simultaneously when “scrolled,” with an optional locking mechanism and the ability to reset to original values intended by the project creator (Figure 3).
Figure 3.
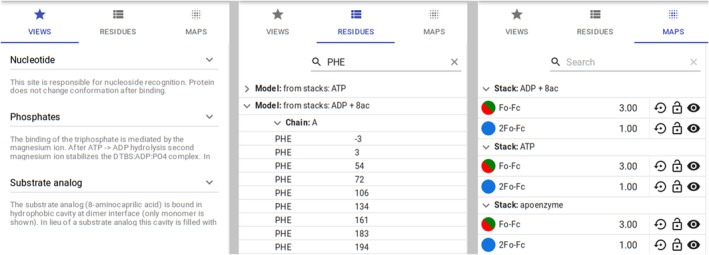
The tabs of the viewer interface for defined views (left), model navigation (middle), and managing maps contouring levels (right). The views panel allows fast navigation between the points of interest defined by the project creator. The residues panel allows independent navigation using the list of residues and other fragments from the uploaded models as points of reference. The interface allows users to filter the residues by name and residue number (shown here for Phe). The maps panel allows management of contouring levels. Components in each individual row are (from left to right): map color, map name, current RMSD level used for contouring, reset to original value, lock map RMSD level, hide map
To improve usability, we have implemented an interface to navigate the molecule. The users may click on individual residues to center the view on a specific part of the molecule. Although this feature was implemented mostly to make it easier for the project creators to define views, it is also useful for people viewing the project to select different parts of the molecule to inspect. The presence of multiple stacks, multiple models per stack, and the possibility that the same model is present in multiple stacks complicates the organization of the models. We display unique models and label them according to stacks in which they are present. To ease the navigation of larger macromolecules, we allow the residue list to be narrowed to specific residue IDs (three‐letter codes) and residue numbers.
2.2. Published use cases
During the last 2 years, Molstack users created various projects and utilized the platform to highlight different aspects of their data and models. We have reviewed the published cases and categorized them according to common usage patterns.
As expected, most of the users use Molstack projects to show the evidence for their interpretations: original, rerefinements, and potential alternative interpretations that are not deposited elsewhere. In X‐ray crystallography, one usually provides different types of omit maps (for a review of different types of omit maps, see work by Shabalin et al.19) to demonstrate the significance of an electron density map and to prove that the region of interest is not biased by the model. If some heavier atoms (usually Z >= 15) are present and the diffraction experiment is well‐designed and executed,20 the experimental evidence for the presence of these atoms may include anomalous difference density maps. Less frequently, Molstack is used for the comparison of different structures either by showing structures individually in different stacks or by showing a superposition of multiple models. Several projects provide the narrative and/or a rationale of the interpretation in the form of view descriptions. Finally, some of the authors used collections to group projects together, especially when the number of related projects was high.
PyrC 21—Molstack was used to present 2mF o ‐DF C and omit maps validating the modeling of ligands in the active sites of dihydroorotase (DHO) from Yersinia pestis. Moreover, anomalous electron density maps calculated from data collected at different wavelengths are used as evidence for the presence of zinc in the active site. The authors also used a Molstack collection to gather all previously determined structures of DHOs from Homo sapiens, Porphyromonas gingivalis, Salmonella enterica, Escherichia coli, Bacillus anthracis, and Aquifex aeolicus and coherently present and analyze them. Molstack was also used to present a superposition of the active sites of these DHOs to highlight biologically relevant molecules bound in the active site.
GHPR 22—Molstack was used to visualize the active sites for glyoxylate/hydroxypyruvate reductase (GHPR) structures that had cofactors and/or ligands bound in those regions. The authors used Molstack to present structures that they have determined, structures determined by others, and structures that they reinterpreted. Descriptions of the views were used to provide the narrative and discussion about the observed map features and the rationale for the reinterpretations. Collection grouped related projects.
Testosterone 23—Molstack was used in three different ways: (a) to present omit maps for the modeling of testosterone in its two binding sites (interestingly, multiple stacks were used to present maps at different contouring levels); (b) to visualize superpositions of the equine serum albumin (ESA)–testosterone complex and selected complexes of serum albumin (SA) with compounds known to bind in the testosterone binding sites; and (c) to visualize conformational changes to SA through superpositions of the ESA–testosterone complex with ligand‐free structures of human serum albumin and ESA.
MBL 24—Molstack was used to present omit maps for ligands in nine rerefined and redeposited metallo‐β‐lactamase (MBL) structures and to compare structures and maps before and after rerefinement. The authors used a collection to group all rerefined projects together.
T6ODM 25—Molstack was used to present omit maps for the active site of thebaine 6‐O‐demethylase (T6ODM) and additional densities that were assigned as unknown atoms (UNX). Molstack was also used to present an interpretation of the additional density as a putative modification of a lysine residue that may have reacted with 2‐oxoglutarate, a crystallization agent, to produce a saccharopine. This is an example of a more “speculative” interpretation, which, while probable, was not checked by other means such as mass spectrometry. As the modification was an artifact of crystallization and was not affecting the structure of the protein, the authors decided to deposit the model with UNX at this site and provided their additional interpretation using Molstack. View descriptions provided a narrative and explanation of the visualized features.
DJ‐1 26—Molstack was used to present omit maps for the covalent modification of a catalytically active cysteine residue by iodoacetic acid and present the unmodified protein.
hARD 27—Moltstack was used to present an omit map for the substrate analog (selenomethionine) in the structure of human acireductone dioxygenase (hARD).
Detect, correct, retract 28—Molstack was used to visualize the original and rerefined models and maps for seven structures that contained errors ranging from significant steric clashes to incorrect ligand assignment.
CheckMyBlob 29—Cases that were identified by CheckMyBlob, a machine learning algorithm for recognizing density blobs, as incorrectly modeled ligands are visualized with Molstack. Four different structures were rerefined and redeposited, and Molstack was used to present the evidence for the corrected assignment in the form of omit maps. View descriptions provide a narrative for the projects. All projects related to this work are grouped using a collection.
2HADH 30—Molstack visualizations of active site ligands accompany a broad array of phylogenetic, kinetic, and structural data for 2‐hydroxyacid dehydrogenases (2HADHs) in the 2HADH knowledgebase.
2.3. Other use cases
2.3.1. Sharing/discussion between collaborators
Through personal communication with our colleagues, we have learned that they find Molstack to be quite useful for sharing data with collaborators. They noted that Molstack has an advantage over other solutions such as sharing files or static pictures in that it offers an immediate interactive experience in the browser without the need to set up specialized software. While this is not an issue when sharing files with crystallographers, using Molstack simplifies sharing with nonstructural biologists, who may not always have specialized software or training to use it. In our own experience, Molstack substantially simplifies local and remote collaborations.
2.3.2. Cryo‐EM maps
While initially developed for X‐ray crystallography, Molstack has a hands‐off approach to handling files. One can upload any type of map in CCP4/MRC format and any type of model in PDB/mmCIF format. This allows Molstack to be a versatile tool for the presentation of maps from cryo‐EM experiments. While we have not yet seen Molstack to be used in such a way, we have demonstrated Molstack's practicality for cryo‐EM by creating several demonstration projects that can be accessed here: https://molstack.bioreproducibility.org/c/n7cu.
2.3.3. External component for presentation
We are actively exploring the applicability of a Molstack viewer to other projects as an embeddable component. For example, we have developed a server for real‐space analysis of independent components in a collection of maps (https://mapica.minorlab.org) that uses the Molstack viewer for comparison of different map components extracted from analyzed files. Using the Molstack viewer in different projects allows us to better define the functionality and crystallize an API of the component.
3. DISCUSSION
The last 2 years of continuous development and multiple submitted projects gave us ample opportunity to evaluate, discuss, and refine the idea of Molstack as a sharing platform. We were able to identify four major areas on which we would like to focus: evidence presentation and validation, alternative interpretations, storytelling, and collaboration. As we have demonstrated above, Molstack can already be effectively used in all these areas, but it can be substantially strengthened by the development of area‐specific features. For example, we would like to implement the calculation and presentation of validation metrics, such as clash‐scores or density correlation. This will allow for an independent comparison of different models when presenting alternative interpretations or provide additional, numerical evidence for the modeled features. Storytelling will greatly benefit from the ability to modify the representation of the molecules and provide additional visualization of different elements such as atom interactions or rudimentary functions for aligning macromolecules. To do this, we are evaluating the use of alternative viewers such as NGL Viewer31 that have better visualization capabilities. Finally, collaboration can be enhanced by implementing group projects where multiple users can edit projects, discuss the changes, and reference specific parts of the macromolecule.
We look forward to new projects being submitted to Molstack and new, innovative approaches to present one's data. We are excited that Molstack is gaining user attraction, growing, and maturing from a side project used to augment other research into an independent project. We hope that we will be able to continue Molstack's development in the outlined directions and serve the biomedical community with innovative tools for data dissemination.
4. MATERIALS AND METHODS
The Molstack platform has been implemented using the Django framework with a PostgreSQL database backend. User authentication with ORCID is based on the OAuth2 protocol. Electron density maps are generated from structure factors using the FFT program from CCP4 and a grid sampled at 1/2.5 of the maximum resolution. Uploaded models are processed using mmLib.32
The interactive parts of the website (project editing and viewing) are built using the React JavaScript framework. The current version uses the Uglymol33 viewer (https://uglymol.github.io/).
Molstack is currently optimized for desktop presentation and has been tested on current versions of Google Chrome (versions 75, 74, 73 on Linux, Windows, macOS and version 75 on Android), Mozilla Firefox (66, 65 on Linux, Windows, MacOS), Microsoft Edge 18 (on Windows) and Apple Safari 12.1 (on MacOS).
ACKNOWLEDGMENTS
The authors thank Ivan Shabalin, Mateusz Czub, David Cooper, Ewa Niedzialkowska, and Marcin Cymborowski for valuable discussions and testing the software. Funding for this research was provided by NIH BD2K grant HG008424, GM117325, GM117080, and GM132595. Funding was also provided by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201200026C.
Porebski PJ, Bokota G, Venkataramany BS, Minor W. Molstack: A platform for interactive presentations of electron density and cryo‐EM maps and their interpretations. Protein Science. 2020;29:120–127. 10.1002/pro.3747
Funding information National Human Genome Research Institute, Grant/Award Number: HG008424; National Institute of Allergy and Infectious Diseases, Grant/Award Number: HHSN272201200026C; National Institute of General Medical Sciences, Grant/Award Numbers: GM117080, GM117325, GM132595
Contributor Information
Przemyslaw J. Porebski, Email: pjp2b@virginia.edu.
Wladek Minor, Email: wladek@iwonka.med.virginia.edu.
REFERENCES
- 1. Baker EN, Dauter Z, Einspahr H, Weiss MS. In defence of our science—Validation now! Acta Cryst. 2010;D66:115. [DOI] [PubMed] [Google Scholar]
- 2. Minor W, Dauter Z, Helliwell JR, Jaskolski M, Wlodawer A. Safeguarding structural data repositories against bad apples. Structure. 2016;24:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng H, Handing KB, Zimmerman MD, Shabalin IG, Almo SC, Minor W. X‐ray crystallography over the past decade for novel drug discovery—Where are we heading next? Expert Opin Drug Discov. 2015;10:975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grabowski M, Chruszcz M, Zimmerman MD, Kirillova O, Minor W. Benefits of structural genomics for drug discovery research. Infect Disord Drug Targets. 2009;9:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Westbrook JD, Burley SK. How structural biologists and the Protein Data Bank contributed to recent FDA new drug approvals. Structure. 2019;27:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheldrick GM. A short history of SHELX. Acta Crystallogr. 2008;A64:112–122. [DOI] [PubMed] [Google Scholar]
- 7. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams CJ, Headd JJ, Moriarty NW, et al. MolProbity: More and better reference data for improved all‐atom structure validation. Protein Sci. 2018;27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. Auto‐rickshaw: An automated crystal structure determination platform as an efficient tool for the validation of an X‐ray diffraction experiment. Acta Crystallogr D Biol Crystallogr. 2005;61:449–457. [DOI] [PubMed] [Google Scholar]
- 10. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Otwinowski Z, Minor W. Processing of X‐ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. [DOI] [PubMed] [Google Scholar]
- 12. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL‐3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Cryst. 2006;D62:859–866. [DOI] [PubMed] [Google Scholar]
- 13. Adams PD, Baker D, Brunger AT, et al. Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems. Annu Rev Biophys. 2013;42:265–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kabsch W. XDS. Acta Crystallogr. 2010;D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porebski PJ, Sroka P, Zheng H, Cooper DR, Minor W. Molstack‐interactive visualization tool for presentation, interpretation, and validation of macromolecules and electron density maps. Protein Sci. 2018;27:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S, Thiessen PA, Bolton EE, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlić A, Rose PW. NGL viewer: Web‐based molecular graphics for large complexes. Bioinformatics. 2018;34:3755–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shabalin IG, Porebski PJ, Minor W. Refining the macromolecular model ‐ achieving the best agreement with the data from X‐ray diffraction experiment. Cryst Rev. 2018;24:236–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Handing KB, Niedzialkowska E, Shabalin IG, Kuhn ML, Zheng H, Minor W. Characterizing metal‐binding sites in proteins with X‐ray crystallography. Nat Protoc. 2018;13:1062–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipowska J, Miks CD, Kwon K, et al. Pyrimidine biosynthesis in pathogens—Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae. Int J Biol Macromol. 2019;136:1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kutner J, Shabalin IG, Matelska D, et al. Structural, biochemical, and evolutionary characterizations of glyoxylate/hydroxypyruvate reductases show their division into two distinct subfamilies. Biochemistry. 2018;57:963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Czub MP, Venkataramany BS, Majorek KA, et al. Testosterone meets albumin—The molecular mechanism of sex hormone transport by serum albumins. Chem Sci. 2019;10:1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raczynska JE, Shabalin IG, Minor W, Wlodawer A, Jaskolski M. A close look onto structural models and primary ligands of metallo‐β‐lactamases. Drug Resist Updat. 2018;40:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kluza A, Niedzialkowska E, Kurpiewska K, et al. Crystal structure of thebaine 6‐O‐demethylase from the morphine biosynthesis pathway. J Struct Biol. 2018;202:229–235. [DOI] [PubMed] [Google Scholar]
- 26. Mussakhmetov A, Shumilin IA, Nugmanova R, et al. A transient post‐translational modification of active site cysteine alters binding properties of the parkinsonism protein DJ‐1. Biochem Biophys Res Commun. 2018;504:328–333. [DOI] [PubMed] [Google Scholar]
- 27. Miłaczewska A, Kot E, Amaya JA, et al. On the structure and reaction mechanism of human acireductone dioxygenase. Chemistry. 2018;24:5225–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wlodawer A, Dauter Z, Porebski PJ, et al. Detect, correct, retract: How to manage incorrect structural models. FEBS J. 2018;285:444–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kowiel M, Brzezinski D, Porebski PJ, Shabalin IG, Jaskolski M, Minor W. Automatic recognition of ligands in electron density by machine learning. Bioinformatics. 2019;35:452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matelska D, Shabalin IG, Jabłońska J, et al. Classification, substrate specificity and structural features of D‐2‐hydroxyacid dehydrogenases: 2HADH knowledgebase. BMC Evol Biol. 2018;18:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rose AS, Hildebrand PW. NGL viewer: A web application for molecular visualization. Nucleic Acids Res. 2015;43:W576–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Painter J, Merritt EA. mmLib Python toolkit for manipulating annotated structural models of biological macromolecules. J Appl Cryst. 2004;37:174–178. [Google Scholar]
- 33. Wojdyr M. UglyMol: A WebGL macromolecular viewer focused on the electron density. J Open Source Softw. 2017;2:350. [Google Scholar]


