Abstract
This article is written for the 2020 tool issue of Protein Science. It briefly introduces the widely used antimicrobial peptide database, initially online in 2003. After a description of the main features of each database version and some recent additions, the focus is on the peptide design parameters for each of the four unified classes of natural antimicrobial peptides (AMPs). The amino acid signature in AMPs varies substantially, leading to a variety of structures for functional and mechanistic diversity. Also, Nature is a master of combinatorial chemistry by deploying different amino acids onto the same structural scaffold to tune peptide functions. In addition, the single‐domain AMPs may be posttranslationally modified, self‐assembled, or combined with other AMPs for function. Elucidation of the design principles of natural AMPs will facilitate future development of novel molecules for various applications.
Keywords: antimicrobial peptides, classification, database tool, peptide design, peptide signature
1. INTRODUCTION
Peptides are small proteins (usually <100 amino acids) that play a variety of functions in biological systems. Antimicrobial peptides (AMPs) are universal players in innate immunity to protect the host from infection.1, 2, 3 The common denominator for AMPs is their antimicrobial activity against various pathogens, including bacteria, fungi, viruses, and parasites. This property forms the basis for developing AMPs into new antibiotics to meet the challenge of drug resistance.4, 5 These molecules are implicated in male fertility, Alzheimer's disease, and cancer metastasis, to list just a few.6, 7, 8, 9 AMPs may also determine dog hair color, regulate insect sleep, and shape host microbiota.10, 11, 12
Although lysozyme was discovered in 1922, the interest in AMPs was not reignited until the 1980s.13, 14, 15 Since then, the number of AMPs discovered per year increased from ~50 in the 1990s to ~100 in the 2000s.16 With more and more such peptides discovered, we constructed the antimicrobial peptide database (APD) to help manage such information.17 In this article, I will first introduce the different versions of the APD. Then, I will describe the design parameters for the major classes of AMPs based on a unified peptide classification (UC) system.18 Since the article is prepared for Protein Science, it is proper to ask the following questions. What amino acids (aa) are frequently utilized (or abundant) in natural AMPs? What is the parameter space for the major kinds of structural scaffolds in nature? What are the major activities of these peptides? Is there any characteristic amino acid signature for AMPs with a known mechanism of action? Understanding Nature's design principles of AMPs can inspire the construction of new molecules to benefit our society.
2. THE DATABASE TOOL
2.1. The first version of the APD
The first version of our database, with 525 peptide entries, was online in August 2003, and a paper was published in Nucleic Acid Research.17 This APD programmed a powerful search engine for antibacterial, antifungal, antiviral, and anticancer activities of AMPs as well as toxic effects on mammalian cells (mainly hemolysis). While antimicrobial activity is usually represented by minimal inhibitory concentrations (MIC), it is also estimated by the diffusion method in the literature. This makes it difficult to combine the activity into a data set to quantitatively predict whether a peptide is an AMP. Hemolysis is often evaluated by incubating the peptide with red blood cells for 2 hr. This may underestimate hemolysis. It is recommended to incubate peptides up to 1,000 μg/mL with blood cells for 18 hr at 37°C. To further complicate the picture, other blood cells (e.g., chicken, cattle, and sheep) are also used. The combination of antimicrobial activity and toxicity information gives users a clue whether a particular peptide preferentially kills pathogens. Additional clue could be obtained by conducting statistical analysis of peptides. The APD reveals a higher hydrophobic content (Pho) for toxic AMPs than nontoxic peptides.17 The hydrophobic amino acids defined in the APD include Leu, Ile, Val, Phe, Met, Ala, Cys, and Trp. This database observation is in line with the results from human cathelicidin studies, laying the basis for improving peptide cell selectivity.19 The physical basis for peptide selective killing is usually attributed to the membrane differences between bacteria (rich in phosphatidylglycerols) and mammalian cells (rich in phosphocholines). The first version of the APD also programmed a rule‐based prediction interface. Importantly, the database is able to provide users with five most similar sequences in the APD. Such a similarity can inspire functional characterization of new peptides.
2.2. The second‐version APD2
The APD was updated and expanded to the APD2 during 2007–2008.20 We tried to collect every peptide described as “antimicrobial peptides” in the literature even though there were no activity data in some cases. The peptide entries increased to 1,228 in the APD2. Also, the peptide search functions increased from 5 to 9. Users can search peptides with anti‐HIV, anti‐Gram‐positive (G+) only, and anti‐Gram‐negative (G−) bacteria only. Importantly, we also demonstrated the use of the database for peptide design. This was initiated by identifying the frequently occurring amino acids (~10%). We found it feasible to use Gly, Leu, and Lys to design AMPs with antimicrobial activity against bacteria.20 This led to our subsequent development of the database filtering technology for designing DFTamP1 against methicillin‐resistant Staphylococcus aureus (MRSA) USA300.21 Recently, we have extended this technology to in vitro and in vivo filtering, generating new ideas for peptide design.22
2.3. The third‐version APD3
The database was further expanded into APD3 during 2010–2015. The APD3 classifies the peptide sources into six life kingdoms: bacteria, archaea, protists, fungi, plants, and animals. Based on the existence of α‐helix and β‐sheet, the 3D structures of AMPs are classified into α, β, αβ, and non‐αβ families.23 Figure 1 gives examples for each structural family. We also proposed a UC method.18 With the use of genomic and proteomic approaches for peptide discovery, it became necessary to define a set of criteria for registration of peptides into the database. Thus, since 2011, the APD3 has mainly registered natural peptides with demonstrated antimicrobial activity.24 Our practice led to a clean data set (2,619 entries reported in the APD3) for classification, prediction, and design of AMPs.
Figure 1.
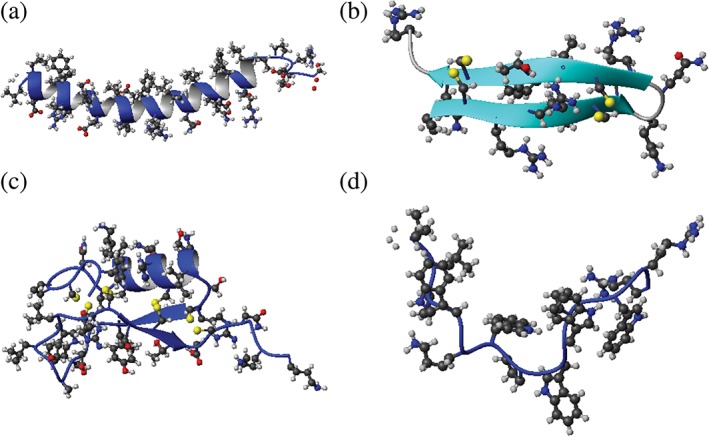
Representative structures of antimicrobial peptides. Shown are the ribbon diagrams of (a) α‐helical LL‐37, (b) β‐sheet gomesin, (c) the oyster defensin with a mixed α and β structure, and (d) indolicidin with a non‐αβ structure. The PDB IDs are http://firstglance.jmol.org/fg.htm?mol=2K6O,60 http://firstglance.jmol.org/fg.htm?mol=1KFP,61 http://firstglance.jmol.org/fg.htm?mol=2B68,62 and http://firstglance.jmol.org/fg.htm?mol=1G89,63 respectively. Structures are determined by solution NMR, and the first structure in the ensemble is used here. Side chains are represented in ball and stick (sulfur, yellow; oxygen, red; carbon, black; and nitrogen, blue). Disulfide bonds are represented by pairs of yellow balls
2.4. The current status
While we aim to develop this database into a new version, here we highlight some recent additions to the APD. We have annotated additional search functions, including antitoxin, anti‐MRSA, anti‐inflammatory, and ion channel inhibition. An increase in peptide search function in the APD is depicted in Figure 2. At present, there are 24 searchable peptide functions/activities, such as antibacterial, antiviral, anticancer, and wound healing. The APD also annotated the peptide information based on the mechanism of action of peptides. It is evident that AMPs use a variety of mechanisms to attack pathogens beyond membranes (Figure 3). Because frog peptides are over 1,000 in the APD, we further annotated these peptides based on their geographic origins. It is interesting to note that AMPs from different continents vary in amino acid composition profile (i.e., AMP signature) (Figure 4). For instance, AMPs from South America possess the highest alanine, while AMPs from Europe are highest in leucine. The biological significance of such differences requires further studies. Will different pathogens in a defined ecological system be the major player in shaping the AMP signature in frogs?
Figure 2.
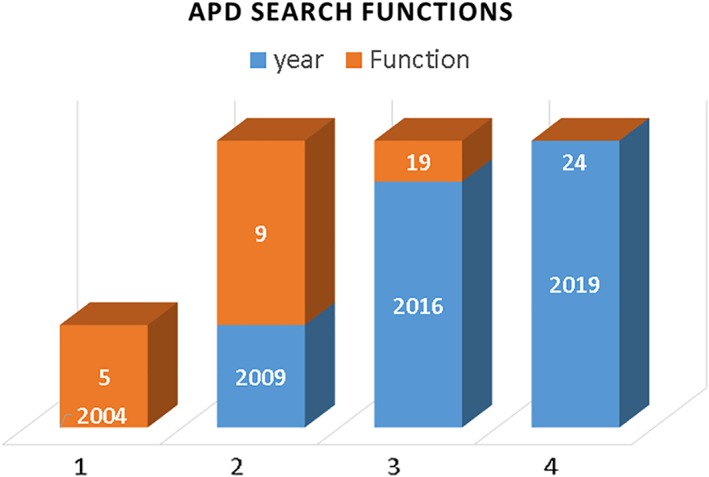
Expansion of search functions for peptide activity such as antibacterial, antiviral, anticancer, antitoxin, and anti‐inflammatory: 5 in the antimicrobial peptide database (APD), 9 in the APD2, 19 in the APD3, and 24 in the current APD (http://aps.unmc.edu/AP)
Figure 3.
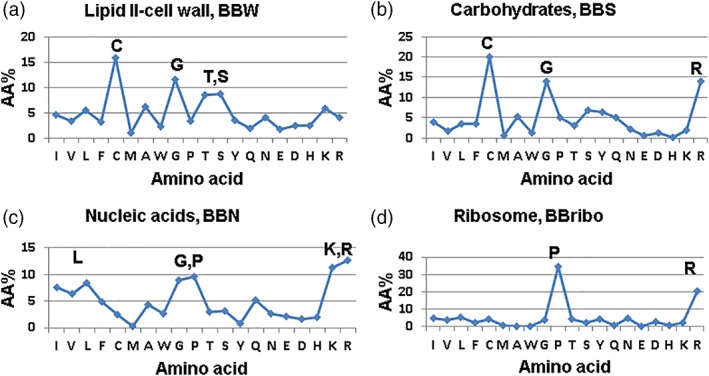
Amino acid signatures of antimicrobial peptides that inhibit different molecular targets: (a) cell wall, (b) carbohydrate, (c) nucleic acid, and (d) ribosome
Figure 4.
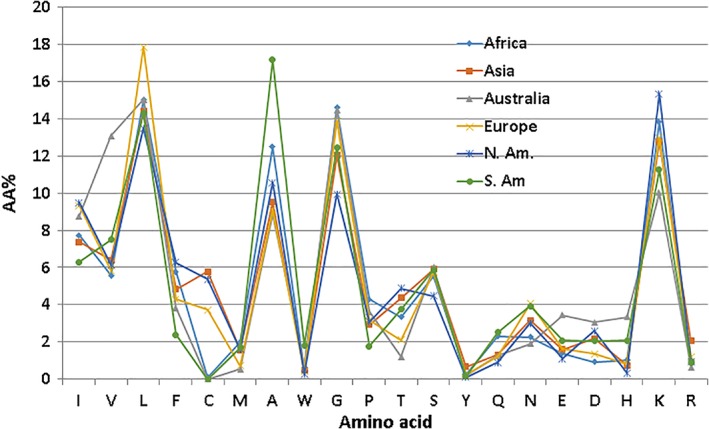
Amino acid signatures of frog antimicrobial peptides from different continents, including Africa, Asia, Australia, Europe, and North and South America
2.5. Why use the APD rather than other databases?
The interest in AMPs stimulated the construction of other databases. Table 1 documents the timeline of web‐accessible databases for AMPs. Since these databases have been discussed elsewhere,25 here I only mention the data in select databases. While CAMP contains 5,398 predicted sequences,26 DBAASP hosts over 10,000 synthetic peptides, including some inactive ones.27 DRAMP has 14,739 peptides from patents and also includes some natural peptides without any activity data.28 While such data can be useful for different purposes, these databases are not used here for the following reasons: first, without activity data, predicted peptides may not be true AMPs. Second, the inclusion of thousands of synthetic/predicted peptides can mask or distort the picture for natural AMPs. Third, databases for a single kingdom or special class of peptides do not have a complete data set for natural AMPs. The natural AMPs in the APD were accumulated and manually curated in the past 16 years. A new peptide must meet a set of criteria (known activity, known sequence, less than 200 amino acids, and from natural sources) in order to be accepted into the APD.24 As of June 2019, the APD hosted 3,081 peptides mainly from natural sources. On average, these peptides have a peptide length of 32.9 and a net charge of +3.3. This set of peptide data is most suitable for this study.
Table 1.
Timeline of web accessible databases dedicated to AMPs (accessed June 2019)a
| Database | Peptides | Web site (http://) | Yearb |
|---|---|---|---|
| APD | AMPs from bacteria, archaea, protists, fungi, plants, and animals | http://aps.unmc.edu/AP/ | 2004; 2009; 2016 |
| Peptaibol | Fungal peptides | http://peptaibol.cryst.bbk.ac.uk/home.shtml | 2004 |
| Cybase | Cyclic peptides | http://www.cybase.org.au/ | 2006; 2008 |
| Defensins | Defensins | http://defensins.bii.a-star.edu.sg/ | 2007 |
| BACTIBASE | Bacteriocins | http://bactibase.hammamilab.org/main.php | 2007; 2010 |
| PhytAMP | Plant AMPs | http://phytamp.pfba-lab-tun.org/main.php | 2009 |
| CAMP | Predicted and patented AMPs | http://www.camp.bicnirrh.res.in/ | 2010; 2014; 2016 |
| YADAMP | AMPs | http://yadamp.unisa.it/ | 2012 |
| DADP | Frog peptides | http://split4.pmfst.hr/dadp/ | 2012 |
| THIOBASE | Bacterial thiopeptides | http://db-mml.sjtu.edu.cn/THIOBASE/ | 2012 |
| AVPpred | Antiviral peptides | http://crdd.osdd.net/servers/avppred/ | 2012 |
| MilkAMP | Milk peptides | http://milkampdb.org/home.php | 2014 |
| DBAASP | AMPs | http://dbaasp.org/home.xhtml | 2014; 2016 |
| BaAMPs | Anti‐biofilm AMPs | http://www.baamps.it/ | 2015 |
| DRAMP | AMPs | http://dramp.cpu-bioinfor.org/ | 2016 |
| AntiTbPdb | Anti‐TB peptides | https://webs.iiitd.edu.in/raghava/antitbpdb/ | 2018 |
| dbAMP | AMPs | http://140.138.77.240/~dbamp/index.php | 2019 |
Adapted and updated based on the original list of the APD “links.”
Years for the publication of different versions of each database.
3. NATURE'S DESIGN PRINCIPLES OF ANTIMICROBIAL PEPTIDES
3.1. Structural diversity of single‐domain AMPs
Nature uses two types of machines to make peptides: ribosome biosynthesis and multiple enzyme systems. To be more complete, the APD includes both types of peptides. Most of the AMPs are gene encoded (97%) in the APD and are the focus of this article. There are only 23 enzyme‐made peptide antibiotics (database search using “nonribosomally synthesized” in the name field), such as gramicidin and daptomycin. Because there are different classification systems for AMPs from different kingdoms, we proposed a UC method to facilitate data organization in the database.18 This method does not depend on molecular source, activity, or 3D structures. In this approach, peptides are separated into four classes based on chain connection patterns. Class I (UCLL) consists of linear peptides (Figure 5a). Some AMPs do not work alone and require the combination of two independent chains (UCLL2, where 2 indicates two chains). AMPs in the second class (UCSS) are required to form at least one bond between different amino acid side chains (Figure 5b–g). While the third peptide class (UCSB) contains a covalent bond between side chain and backbone Figure 5h,i), the fourth class (UCBB) forms a peptide bond between the N‐ and C‐termini of peptides (Figure 5j,k). The APD has peptide examples for all these classes. Each class can be further divided based on bond type and peptide chain number. In the following, I describe the parameter space defined by each major class of peptides that differ in chain‐connection pattern. The peptide parameters include length, net charge, hydrophobic content, and amino acid composition, which constitute a useful starting point for designing peptides with a defined structure and activity.
Figure 5.
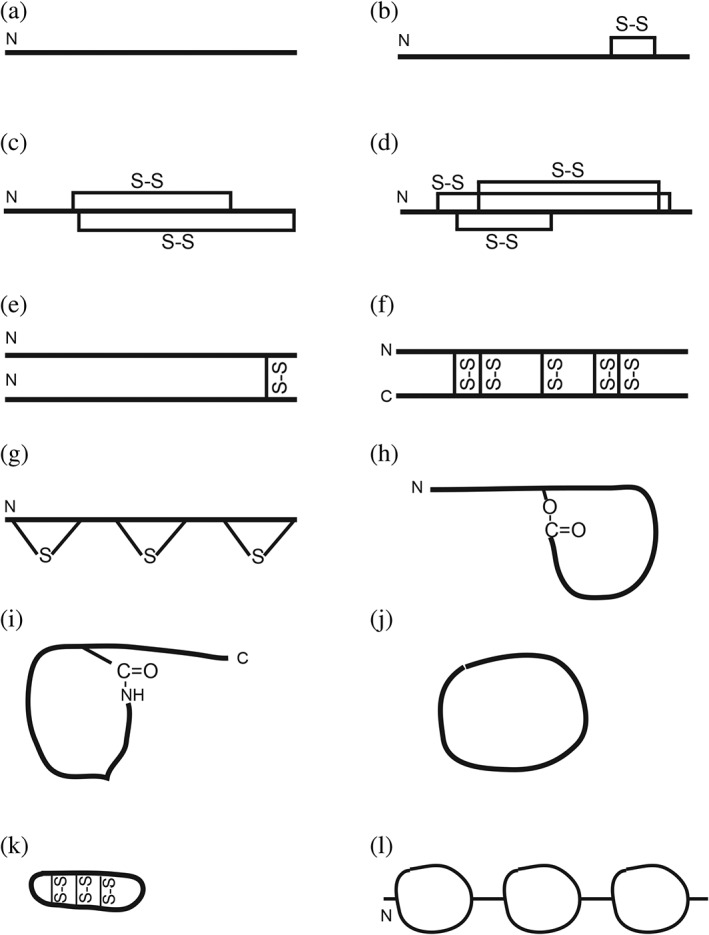
Scaffolds of antimicrobial peptides based on the unified peptide classification (UC) scheme18
3.1.1. UCLL
Linear peptides (Figure 5a) frequently become amphipathic (the segregation of hydrophobic and hydrophilic side chains) after binding to bacterial membranes. Typical examples are magainins, cecropins, and human cathelicidin LL‐37. The amphipathic helix model (Figure 1a) is so popular that it becomes a token for cationic AMPs.5, 29 However, such peptides can use different amino acids depending on biological sources. This can be seen from Table 2 that summarizes the parameters for known helical peptides from different sources. The abundant amino acids (~8% or above) are Ile, Leu, Ala, and Lys in insect AMPs, but Leu, Ala, Gly, and Lys in amphibian AMPs. In fish, phenylalanines are more abundant, while arginines are preferred over lysines. Histidines are also abundant and can play unique biological functions such as pH‐dependent membrane permeation.30 Note that there is an increase in cationic amino acids in helical peptides from reptiles and mammals since both lysine and arginine are abundant (~30% combined). One possibility is to increase the neutralizing capability of these peptides to tame inflammation. In the major antibacterial peptide of human cathelicidin LL‐37, Arg23 plays an important role in combating pathogenic bacteria and Zika viruses.31, 32 These examples illustrate that Nature is able to achieve functional diversity by deploying different amino acids on the same helix model (Table 2). Peptide length is another important parameter for peptide design. With a decrease in peptide length, there is a clear increase in Phe and Leu in α‐helical peptides (Figure 6a,b). Such an increase may be necessary to retain membrane‐binding ability of short helical peptides. For comparison, we only found an increase in the Cys content for β‐sheet‐containing AMPs (Figure 6c).
Table 2.
Helical peptides (UCLL) from different sourcesa
| Source | Count | Length | Net charge | Pho | Abundant AA (>8%) |
|---|---|---|---|---|---|
| Insects | 9 | 11–34 | +1−+9 | 47–71 | I 10.5%, L 18%, A 14%, K 17.4% |
| Amphibians | 116 | 8–34 | 0−+7 | 31–75 | L 15.4%, A 14.1%, G 12.6%, K 11.9% |
| Fish | 21 | 12–44 | 0−+10 | 15–50 | I 10.1%, F 8.6%, G 12.8%, H 9.5%, R 9.3% |
| Reptiles | 9 | 22–38 | +3−+15 | 32–50% |
L 9.1%, F 9.4%, K 17%, R 12.3% |
| Mammals | 14 | 17–49 | +3−+11 | 3–52% | I 8.2%, L 8,6%, K 14.1%, R 15.1% |
Note that, although we only provided abundant amino acids here for AMPs (<50 aa), readers may refer to the APD to view the whole amino acid signatures or for updated ones. This applies to all the tables in this study.
Figure 6.
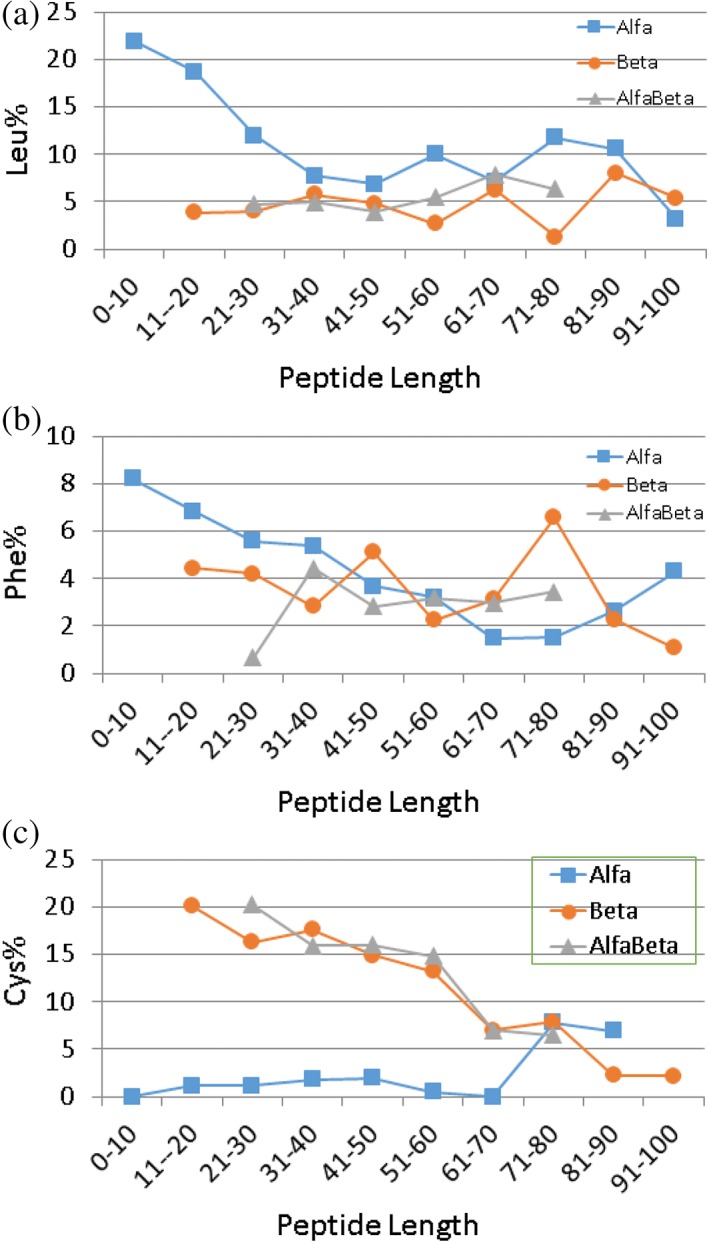
Length‐dependent amino acid use in (a) α‐helical, (b) β‐sheet, and (c) αβ‐mixed AMPs. Other amino acids do not show any clear trend and are therefore not presented here
Scientists working on insects first discovered proline‐rich AMPs.33 Other well‐known examples are Trp‐rich, Arg‐rich, and His‐rich AMPs.34, 35 The word “rich” has different meanings in different articles. If we define 30% as rich in the APD, there are also other less popular members, including Asp‐rich, Ser‐rich, Lys‐rich, Ala‐rich, Phe‐rich, and Leu‐rich. Such peptides can have unique functions and properties. For example, metals modulate the activity of His‐rich peptides.34, 36
3.1.2. UCSS
Two types of sidechain bonding are common in AMPs. The first subclass one‐chain peptides (UCSS1a) contain at least one disulfide bond (Figure 5b–f), while the second subclass (UCSS1b) shares the thioether bond (Figure 5g). The cysteine‐containing peptides are very diverse in source and structure. Peptides in this subclass can be further splitted into families based on the number of S—S bonds. The classic examples are linear AMPs that form a two‐stranded β‐sheet stabilized by 1–2 disulfide (S—S) bonds (Figure 5c). The two disulfide bonds (yellow) in spider gomesin can be seen in Figure 1b. Both human α‐ and β‐defensins consist of three S—S bonds that stabilize the β‐sheet (Figure 5d). Defensins are reported to contain a γ‐core motif that can instruct the search of potentially new AMPs.37 The oyster defensin contains four pairs of S—S bonds (Figure 1c). In plants, up to six S—S bonds have been found in snakins.38 Table 3 summarizes the APD‐derived parameter space for these peptides. In terms of 3D structure, the majority of S—S linked peptides form a β‐sheet (Figure 1bc). Both Cys and Gly are abundant amino acids. There is a shift in the use of basic amino acids from mainly arginine for AMPs with two S—S bonds to similar contents of Lys and Arg in plant AMPs with 4–6 S—S bonds (Table 3).
Table 3.
Peptide parameters for constructing AMPs in the UCSS class
| Peptide | Count | Length (aa) | Net charge | Pho (%) | Abundant amino acids |
|---|---|---|---|---|---|
| Rana peptides | 445 | 10–49 | 0−+10 | 25–75 | L 12.7%, A 9.9%, G 11.0%, K 14.8% |
| Plant 2S‐S α | 3 | 33–37 | +2−+7 | 21–22 | C 11.4%, G 14.3%, Q 8.6%, E 10%, R 28.6% |
| Saposin‐like 3S‐S α | 12 | 73–88 | −11−+11 | 33–52 | I 8.1%, L 10.2%, K 9.9% |
| Plant 2S‐S β | 10 | 20–44 | +1−+9 | 16–38 | C 14.9%, G 10.6%, R 17.7% |
| Plant 3S‐S β | 11 | 27–76 | −1−+5 | 33–48 | C 17%, G 14.7%, T 8.2% |
| Plant 4S‐S β | 78 | 40–92 | 0−+12 | 26–45 | C 16.2%, G 9.5%, S 7.5%, K 7.9%, R 7.4% |
| Plant 5S‐S β | 8 | 41–66 | −2−+6 | 28–46 | C 20.9%, G 11.4%, (K = R 6.9%) |
| Plant 6S‐S β | 5 | 63–79 | +6− +10 | 31–35 | C 17.6%, G 9.4%, S 8.5%, K 9.4%, R 8.5% |
| α‐Defensin β | 44 | 29–47 | −2−+9 | 28–56 | C 18.3%, G 10%, R 18.5% |
| β‐Defensin αβ | 87 | 25–102 | −1−+12 | 39–53 | C 13.9%, G 10.7%, R 10.4% |
| Lantibiotics | 69 | 18–44 | −2−+8 | 31–57 | C 13.9%, G 9.2%, T 12.4%, S 11.7% |
Of note, a disulfide bond can also stabilize a helical peptide. A total of 445 frog AMPs (10–45 aa) contain a Rana box39 usually at the C‐termini (Figure 5b). However, they share the identical four abundant amino acids (Leu, Ala, Gly, and Lys) with those linear amphibian peptides without a disulfide bond (UCLL1, 558 entries), indicative of a similar potential in forming helical conformations. Although there are only a few examples, two disulfide bonds can also stabilize a helical hairpin‐like structure in plant AMPs.40 Such plant peptides appear to have a rather unique amino acid composition since glutamins and glutamates are both abundant (Table 3). There are also saposin‐like peptides (~80 AA residues), which consist of multiple helices stabilized by three pairs of S—S bonds. In these peptides, amino acids Ile, Leu, and Lys are abundant. Also, residues Val, Cys, Glu, and Asp are above 7%.
A special construct was also found in a frog, where a single cysteine in distinctin forms a disulfide bond with another copy of the same chain (Figure 5e). This homodimer then dimerizes into a helix bundle structure to increase peptide stability to proteases.41 Other AMPs with an odd number of cysteines may form intermolecular disulfide bonds as well. Five intermolecular S—S bonds have been found in rattusin (Figure 5f), a β‐sheet peptide.42 Over 10 disulfide‐bridged homodimers exist in the APD. These examples substantially enriched the disulfide‐bonded scaffolds for AMPs (Figure 5b–f).
Nature has further expanded its defense arsenal by exerting environment‐dependent host defense. A typical example is that the reduced human β‐defensin 143 is more active under reduced conditions. However, one should not generalize this observation as reduction of the S—S bonds can reduce the activity of other AMPs such as mollusca myticin C.44
Lantibiotics, made by Gram‐positive bacteria, constitute another family of the UCSS peptides. Nisin, a typical example, has been utilized by many countries as a food preservative. This success stimulated the isolation and characterization of many more lantibiotics. These heavily posttranslationally modified peptides share the common thioether bonds between cysteine and serine/threonine.45 The abundance of Cys, Thr, and Ser in these peptide sequences (Table 3) is necessary for the formation of up to seven such ring structures.46 These rings (Figure 5g) are essential for structural rigidity, peptide stability, and antibacterial activity.45 A series of posttranslational enzymes/neutralizing proteins is involved in the processing of a translated peptide, covering dehydration, ring formation, transport, and immunity protein to avoid self‐killing. In this sense, the “innate immune” concept extends to bacteria, which produce all kinds of bacteriocins with structures far more complex than those found in eukaryotes.
3.1.3. UCSB
The UCSB class is of particular interest because several peptide antibiotics in this class (e.g., daptomycin and colistin) are already in clinical use. In the current APD, the UCSB class has 31 members. Nearly all of these peptides originate from bacteria. Based on the bond type from sidechain to backbone, these peptides can be further classified into two families: lactone and lactam. While lactone contains an ester bond, lactam forms an amide bond. Table 4 provides the parameter space for both lactone and lactam peptides. It is evident that these two types of AMPs use different abundant amino acids. A detailed analysis of lactones revealed a dominance of the 4, 6, and 9 aa rings formed between a OH sidechain of Thr and the carboxylic group of the C‐terminus (Figure 5h). D‐amino acids are frequently incorporated to facilitate the ring formation. Also, an acyl chain may be attached to enhance the activity (e.g., fusaricidin and daptomycin). In two cases, an eight aa ring structure is formed between a Tyr OH and the peptide C‐terminus.
Table 4.
Classification of the UCSB peptides and parameter space of lactone and lactam
| Bond type | Peptide count | Length | Net charge | Pho | Abundant AA |
|---|---|---|---|---|---|
| Lactone | 13 | 6–13 | −3−+4 | 15–71% | V 12.5%; T 14.16%, K 9.16% |
| Lactam | 16 | 10–21 | −2−+6 | 20–66% | V 8.4%, F 8.1%, G 17.9%, |
In contrast, lactam peptides usually form a bond between the N‐terminus NH2 and the sidechain of Glu7/8 or Asp8/9 (Figure 5i). In the APD, the observed bonding pairs are C1‐D9, G1‐D8, G1‐E8, and G1‐E7.23 When it is a C1‐D9 ring, additional disulfide bonds can occur. These peptides may fold into a lasso structure, where the tail can enter the macrolactam ring and is locked there via steric hindrance.47 However, there is an exception. In polymyxins, the ring occurs between a sidechain NH2 of 2,4‐diaminobutanoic acid at position 4 and the carboxylic C‐terminus.
3.1.4. UCBB
Circular AMPs have been discovered in bacteria, plants, and animals. These peptides have a circular structure due to the formation of a peptide bond between the amino‐ and carboxylic termini (Figure 5j). The smallest circular AMP from bacteria consists of only six amino acids, while the largest one, also from bacteria, has 82 aa. The formation of the small ring requires the incorporation of D‐amino acids.
Most of the plant circular peptides are cyclotides (~30 aa). It appears that different activities of these peptides result from their capability of recognizing phosphatidylserine.48 In addition, cyclotides contain three S—S bonds to further stabilize the structure, making them very stable. The high stability of the cyclotide has been exploited as a template for developing medicine. Peptide sequence motifs have been grafted to the loop region of cyclotides to inhibit protein–protein association involved in cancer.48, 49
In animals, small circular proteins, called θ‐defensins (Figure 5k), have been found in monkeys.50 The parameter ranges for these circular AMPs are summarized in Table 5. They possess a constant length of 18. However, the amino acid use is rather biased. While isoleucine and valine are major hydrophobic amino acids, arginine is the dominant basic one. Up to date, the following 12 amino acids are absent in these minidefensins, including Met, Ala, Trp, Pro, Ser, Tyr, Gln, Asn, Glu, Asp, His, and Lys. The circular structure is remarkably stable with three S—S bonds across the two β‐strands (Figure 5k).
Table 5.
Peptide parameter space for circular peptides (UCBB)
| Source | Count | Size | Net charge | Pho | Abundant AA (>8%) |
|---|---|---|---|---|---|
| Bacteria | 30 | 6–82 | −2−+7 | 36–100% | I, 10.4 L 10.3, A 20.0; G 12.1 |
| Plants | 155 | 8–37 | −2−+4 | 32–57% | C 20.14%, G 11.76%, T 8.14%, and S 8.29% |
| Animals | 15 | 18 | +3−+6 | 55–66% | I 8.14%, V 8.51%, C 33.33%; G 12.22%; R 25.55% |
3.2. Multiple‐domain AMPs
As discussed above, most of the AMPs consist of a single sequence domain (Model 1: A). Some can also be composed of multiple domains. Such peptides can be searched in the APD name field by entering “modular design”. A total of 14 such AMPs have been annotated in the APD, mostly crustacean peptides. The structures of shrimp penaeidins comprise an N‐terminal Pro‐rich domain and a C‐terminal Cys‐rich domain (Model 2: A + B).
With the increase in polypeptide length, AMPs fall into the category of antimicrobial proteins (>100 aa). A longer sequence enables functional programming of additional peptide domains. The recent discovery of MjPen‐II with 117 aa51 has expanded the construction of shrimp penaeidins to three different sequence domains: Ser‐rich + Pro‐rich + Cys‐rich (Model 3: A + B + C, Figure 5l). Some antimicrobial chemokines are known to have a folded domain and a disordered region, conferring both antimicrobial and immune modulation capabilities.52
3.3. Activity signature of antimicrobial peptides
AMPs are moonlighting molecules with multiple functions. Table 6 lists the abundant amino acids in peptides with antibacterial, antifungal, antiviral, antiparasitic, and anticancer activities. Interestingly, these peptides tend to have similar abundant amino acids, implying a common molecular design. It is interesting to note that there is a different requirement for AntiG+ and AntiG− AMPs (G+ = Gram‐positive and G− = Gram‐negative). For 21 AntiG+ helical AMPs (UCLL1), the average pho and net charge are 53.3% and + 1.1, respectively. These are 47.8% and + 2.7 for 15 AntiG− helical AMPs (<50 aa). Thus, AntiG+ helical AMPs tend to have a higher hydrophobic content and lower cationicity, whereas AntiG− helical AMPs appear to have a lower hydrophobic content and higher cationicity. The hydrophobic content has a shortcoming because equal weights for small and large hydrophobic amino acids mask their difference in hydrophobicity. To gain additional insight, Figure 7 plotted the amino acid signatures for AntiG+ and AntiG− helical AMPs. The hydrophobic profiles are drastically different. In particular, there are 18.29% Leu and 9.27% Ala for AntiG+ AMPs, but 10.72% Leu and 16.95% Ala for AntiG− peptides, indicative of the requirement of high hydrophobicity to kill G+ pathogens. This finding is in line with our peptide engineering results based on the only human cathelicidin.53 This plot also contains detailed information on basic amino acids. While the levels of arginines in these two groups of AMPs are more or less similar, the AntiG− group possesses a much higher content of lysines than the AntiG+ group. However, arginine is preferred in certain cases. In 2003, we found antiviral peptides have a higher content of arginine.17 Years later, we had a chance to apply this finding to peptide design by converting lysines to arginines and obtained peptides with improved antiviral activity against human immune deficient virus type 1.54 A detailed analysis of AMPs from different structural classes revealed that, except for antibacterial peptides, arginine is preferred in β‐sheet peptides, whereas lysine is preferably associated with helical AMPs (Table 6).
Table 6.
Abundant amino acids in AMPs with different activitiesa
| Target pathogen (peptide count) | α | β | αβ | Non‐αβ |
|---|---|---|---|---|
| G+ bacteria (520) | L, A, G, K (52) | C, G, S, K (9) | C, G (15) | C, G, T, S, K (6) |
| G− bacteria (297) | L, A, G, K (30) | C, G, K (9) | I, S, T, K (4) | G, S, K (4) |
| Fungi (1106) | L, A, G, K (191) | C, G, R (43) | C, G, K, R (51) | W, P, R (4) |
| Viruses (188) | V, L, G, K (45) | C, G, R (29) | C, G (15) | W, P, R (1) |
| Parasites (113) | L, A, G, K (34) | C, G, R (5) | C, K (12) | P, S, R (2) |
| Cancer (227) | L, A, G, K (80) | C, G, R (7) | C, G, S, K (12) | I, G, S, T, K (1) |
Abundant ~8%.
Figure 7.
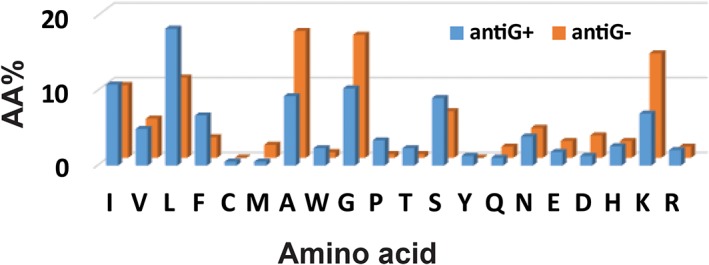
Amino acid (aa) signatures for helical AMPs (<50 aa) against Gram‐positive (G+) or Gram‐negative (G−) bacteria. It is remarkable that leucine (blue column) is higher in antimicrobial peptides against G+ bacteria, but alanine, glycine, and lysine are higher in antimicrobial peptides against G− bacteria (gold columns)
3.4. Mechanistic signatures of antimicrobial peptides
The AMPs are often generalized as amphipathic, cationic, and membrane targeting. A characteristic feature of these peptides is their rapid killing by both the peptides synthesized in L‐ or D‐amino acids.1 Recent studies have expanded this view because AMPs can also inhibit protein synthesis. A typical feature of these peptides is slow killing and different MIC values of the L‐ and D‐forms of the peptides.33 Interestingly, our database analysis of AMPs with a defined mechanism reveals distinct amino acid signatures (Figure 3). While peptides inhibiting cell wall synthesis are abundant in Cys, Gly, Thr, and Ser, ribosome inhibitory peptides are rich in proline and arginine. Such amino acids are required for a defined structure that recognizes its molecular target. For example, a specific sequence of Pro‐rich peptides can fit into the exit tunnel of ribosomes to block the release of the synthesized peptides.55
4. CONCLUDING REMARKS
This article introduces the APD tool to the Protein Science community. It also summarizes peptide design parameters for each class. A careful annotation of the AMP sequences in the APD is a prerequisite for us to classify natural AMPs. Typically, AMPs comprise a single domain. They can adopt a variety of structures for different functions/activities (Figures 1 and 5). Alternatively, the same helix or cyclotide fold can be utilized widely in Nature. The peptide activity may also be modulated by amino acid changes, posttranslational modifications,56 and environmental conditions.43 Some AMPs do not work alone, and two or more peptide chains need be combined for activity. In addition, different AMPs can work together to show a synergistic effect, which may be important for persistent efficacy in innate immune systems. Our understanding of the Nature's construction principles of AMPs herein can guide the design of a new generation of peptides for a variety of potential applications.57 One can also mimic multiple‐domain AMPs. A smart design is to combine AMPs with a pathogen‐targeting motif to increase peptide specific binding to pathogens and to minimize potential off‐target binding.58 Attempts are also made to induce AMPs to combat pathogen infection.59 The success of this method depends on a thorough understanding of the biological system (e.g., patients) and elegant control of AMPs at a defined site and time. Use of probiotic bacteria to express needed bacteriocins may offer an alternative approach.12
In summary, Nature has designed a variety of structural scaffolds for host defense. They can have different activities and be induced via different pathways to curb pathogen infection. In addition, the same scaffold can be tuned at the amino acid level by species from different kingdoms to fulfill the defense requirement in a defined ecological niche. The APD provides a useful platform for classifying, decoding, and designing AMPs.
ACKNOWLEDGMENTS
This study was supported by the NIH/NIAID grant R01AI105147 and the Nebraska POC award to GW. The author acknowledges the IT services of the University of Nebraska Medical Center (UNMC) for keeping the database in the running state for 16 years.
Wang G. The antimicrobial peptide database provides a platform for decoding the design principles of naturally occurring antimicrobial peptides. Protein Science. 2020;29:8–18. 10.1002/pro.3702
Correction added on August 16, 2019 after first online publication: minor typographical errors have been corrected throughout the article.
Funding information University of Nebraska Medical Center; NIH, Grant/Award Number: R01AI105147
REFERENCES
- 1. Boman HG. Antibacterial peptides: Basic facts and emerging concepts. J Intern Med. 2003;254:197–215. [DOI] [PubMed] [Google Scholar]
- 2. Hoffmann JA, Hetru C. Insect defensins: Inducible antibacterial peptides. Immunol Today. 1992;13:411–415. [DOI] [PubMed] [Google Scholar]
- 3. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. Pillars article: The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996. 86: 973‐983. J Immunol. 2012;188:5210–5220. [PubMed] [Google Scholar]
- 4. Haney EF, Hancock RE. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers. 2013;100:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. [DOI] [PubMed] [Google Scholar]
- 6. Soscia SJ, Kirby JE, Washicosky KJ, et al. The Alzheimer's disease‐associated amyloid beta‐protein is an antimicrobial peptide. PLoS One. 2010;5:e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang J, Cheng M, Law IKM, Ortiz C, Sun M, Koon HW. Cathelicidin suppresses colon cancer metastasis via a P2RX7‐dependent mechanism. Mol Ther Oncolytics. 2019;12:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weber G, Chamorro CI, Granath F, et al. Human antimicrobial protein hCAP18/LL‐37 promotes a metastatic phenotype in breast cancer. Breast Cancer Res. 2009;1:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu H, Dong J, Gu Y, et al. The novel human beta‐defensin 114 regulates lipopolysaccharide(LPS)‐mediated inflammation and protects sperm from motility loss. J Biol Chem. 2013;288:12270–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Candille SI, Kaelin CB, Cattanach BM, et al. A β‐defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Toda H, Williams JA, Gulledge M, Sehgal A. A sleep‐inducing gene, nemuri, links sleep and immune function in Drosophila. Science. 2019;363:509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hols P, Ledesma‐García L, Gabant P, Mignolet J. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 2019;27:690–702. [DOI] [PubMed] [Google Scholar]
- 13. Steiner H, Hultmark D, Engström A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. [DOI] [PubMed] [Google Scholar]
- 14. Ganz T, Selsted ME, Szklarek D, et al. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: Isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang G. Database‐guided discovery of potent peptides to combat HIV‐1 or superbugs. Pharmaceuticals. 2013;6:728–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z, Wang G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004;32:D590–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang G. Improved methods for classification, prediction, and design of antimicrobial peptides. Methods Mol Biol. 2015;1268:43–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X, Li Y, Han H, Miller DW, Wang G. Solution structures of human LL‐37 fragments and NMR‐based identification of a minimal membrane‐targeting antimicrobial and anticancer region. J Am Chem Soc. 2006;128:5776–5785. [DOI] [PubMed] [Google Scholar]
- 20. Wang G, Li X, Wang Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mishra B, Wang G. Ab initio design of potent anti‐MRSA peptides based on database filtering technology. J Am Chem Soc. 2012;134:12426–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mishra B, Narayana JL, Lushinikova T, Wang XQ, Wang G. Low cationicity is important for systemic in vivo efficacy of database‐derived peptides against drug‐resistant Gram‐positive pathogens. Proc Natl Acad Sci U S A. 2019;116:13517–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang G. Antimicrobial peptides: Discovery, design and novel therapeutic strategies. 2nd ed. Wallingford, England: CABI, 2017. [Google Scholar]
- 24. Wang G, Li X, Wang Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang G. Database resources dedicated to antimicrobial peptides In: Chen C‐Y, Yan X, Jackson CR, editors. Antimicrobial resistance and food safety: Methods and techniques. Boston: Academic Press, 2015; p. 365–384. [Google Scholar]
- 26. Waghu FH, Barai RS, Gurung P, Idicula‐Thomas S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016;44:D1094–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pirtskhalava M, Gabrielian A, Cruz P, et al. DBAASP v.2: An enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016;44:D1104–D1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fan L, Sun J, Zhou M, et al. DRAMP: A comprehensive data repository of antimicrobial peptides. Sci Rep. 2016;6:24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yount NY, Weaver DC, Lee EY, et al. Unifying structural signature of eukaryotic α‐helical host defense peptides. Proc Natl Acad Sci U S A. 2019;116:6944–6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mihailescu M, Sorci M, Seckute J, et al. Structure and function in antimicrobial piscidins: Histidine position, directionality of membrane insertion, and pH‐dependent permeabilization. J Am Chem Soc. 2019;141:9837–9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He M, Zhang H, Li Y, et al. Cathelicidin‐derived antimicrobial peptides inhibit Zika virus through direct inactivation and interferon pathway. Front Immunol. 2018;9:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang G, Narayana JL, Mishra B, et al. Design of antimicrobial peptides: Progress made with human cathelicidin LL‐37. Adv Exp Med Biol. 2019;1117:215–240. [DOI] [PubMed] [Google Scholar]
- 33. Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects: Structure and function. Dev Comp Immunol. 1999;23:329–344. [DOI] [PubMed] [Google Scholar]
- 34. Gusman H, Lendenmann U, Grogan J, Troxler RF, Oppenheim FG. Is salivary histatin 5 a metallopeptide? Biochim Biophys Acta. 2001;1545:86–95. [DOI] [PubMed] [Google Scholar]
- 35. Zanetti M, Gennaro R, Romeo D. Cathelicidins: A novel protein family with a common proregion and a variable C‐terminal antimicrobial domain. FEBS Lett. 1995;37:1–5. [DOI] [PubMed] [Google Scholar]
- 36. Kim SY, Zhang F, Gong W, et al. Copper regulates the interactions of antimicrobial piscidin peptides from fish mast cells with formyl peptide receptors and heparin. J Biol Chem. 2018;293:15381–15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yeaman MR, Yount NY. Unifying themes in host defence effector polypeptides. Nat Rev Microbiol. 2007;5:727–740. [DOI] [PubMed] [Google Scholar]
- 38. García‐Olmedo F, Molina A, Alamillo JM, Rodríguez‐Palenzuéla P. Plant defense peptides. Biopolymers. 1998;47:479–491. [DOI] [PubMed] [Google Scholar]
- 39. Conlon JM, Kolodziejek J, Nowotny N. Antimicrobial peptides from Ranid frogs: Taxonomic and phylogenetic markers and a potential source of new therapeutic agents. Biochim Biophys Acta. 2004;1696:1–14. [DOI] [PubMed] [Google Scholar]
- 40. Ryazantsev DY, Rogozhin EA, Dimitrieva TV, et al. A novel hairpin‐like antimicrobial peptide from barnyard grass (Echinochloa crusgalli L.) seeds: Structure‐functional and molecular‐genetics characterization. Biochimie. 2014;99:63–70. [DOI] [PubMed] [Google Scholar]
- 41. Batista CV, Scaloni A, Rigden DJ, et al. A novel heterodimeric antimicrobial peptide from the tree‐frog Phyllomedusa distincta . FEBS Lett. 2001;494:85–89. [DOI] [PubMed] [Google Scholar]
- 42. Min HJ, Yun H, Ji S, et al. Rattusin structure reveals a novel defensin scaffold formed by intermolecular disulfide exchanges. Sci Rep. 2017;7:45282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schroeder BO, Wu Z, Nuding S, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β‐defensin 1. Nature. 2011;469:419–423. [DOI] [PubMed] [Google Scholar]
- 44. Pallavicini A, Costa Mdel M, Gestal C, et al. High sequence variability of myticin transcripts in hemocytes of immune‐stimulated mussels suggests ancient host‐pathogen interactions. Dev Comp Immunol. 2008;32:213–226. [DOI] [PubMed] [Google Scholar]
- 45. Jack RW, Sahl HG. Unique peptide modifications involved in the biosynthesis of lantibiotics. Trends Biotechnol. 1995;13:269–278. [DOI] [PubMed] [Google Scholar]
- 46. Garg N, Tang W, Goto Y, Nair SK, van der Donk WA. Lantibiotics from Geobacillus thermodenitrificans . Proc Natl Acad Sci U S A. 2012;109:5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hegemann JD, Zimmermann M, Xie X, Marahiel MA. Lasso peptides: An intriguing class of bacterial natural products. Acc Chem Res. 2015;48:1909–1919. [DOI] [PubMed] [Google Scholar]
- 48. Troeira Henriques S, Craik DJ. Cyclotide structure and function: The role of membrane binding and permeation. Biochemistry. 2019;56:669–682. [DOI] [PubMed] [Google Scholar]
- 49. Camarero JA, Campbell MJ. The potential of the cyclotide scaffold for drug development. Biomedicine. 2019;7:E31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tang YQ, Yuan J, Osapay G, et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha‐defensins. Science. 1999;286:498–502. [DOI] [PubMed] [Google Scholar]
- 51. An MY, Gao J, Zhao XF, Wang JX. A new subfamily of penaeidin with an additional serine‐rich region from kuruma shrimp (Marsupenaeus japonicus) contributes to antimicrobial and phagocytic activities. Dev Comp Immunol. 2016;59:186–198. [DOI] [PubMed] [Google Scholar]
- 52. Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. 2014;7:545–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang X, Mishra B, Lushnikova T, Narayana JL, Wang G. Amino acid composition determines peptide activity spectrum and hot‐spot‐based design of merecidin. Adv Biosyst. 2018;2:1700259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang G, Waston K, Peterkofsky A, Buckheit R Jr. Identification of novel human immunodeficiency virus type 1 inhibitory peptides based on the antimicrobial peptide database. Antimicrob Agents Chemother. 2010;54:1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Graf M, Mardirossian M, Nguyen F, et al. Proline‐rich antimicrobial peptides targeting protein synthesis. Nat Prod Rep. 2017;34:702–711. [DOI] [PubMed] [Google Scholar]
- 56. Wang G. Post‐translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr Biotechnol. 2012;1:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptide as antibiotics: Design and application strategies. Curr Opin Chem Biol. 2017;38:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone‐guided "smart" antimicrobial peptide. Antimicrob Agents Chemother. 2006;50:3651–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nylén F, Bergman P, Gudmundsson GH, Agerberth B. Assays for identifying inducers of the antimicrobial peptide LL‐37. Methods Mol Biol. 2017;1548:271–281. [DOI] [PubMed] [Google Scholar]
- 60. Wang G. Structures of human host defense cathelicidin LL‐37 and its smallest antimicrobial peptide KR−12 in lipid micelles. J Biol Chem. 2008;283:32637–32643. [DOI] [PubMed] [Google Scholar]
- 61. Mandard N, Bulet P, Caille A, Daffre S, Vovelle F. The solution structure of gomesin, an antimicrobial cysteine‐rich peptide from the spider. Eur J Biochem. 2002;269:1190–1198. [DOI] [PubMed] [Google Scholar]
- 62. Gueguen Y, Herpin A, Aumelas A, et al. Characterization of a defensin from the oyster Crassostrea gigas . J Biol Chem. 2006;281:313–323. [DOI] [PubMed] [Google Scholar]
- 63. Rozek A, Friedrich CL, Hancock RE. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry. 2000;39:15765–15774. [PubMed] [Google Scholar]


