Abstract
PyMOL is often used to generate images of biomolecular structures. Hundreds of parameters in PyMOL provide precise control over the appearance of structures. We developed 241 Python functions—called “shortcuts”—that extend and ease the use of PyMOL. A user runs a shortcut by entering its name at the PyMOL prompt. We clustered the shortcuts by functionality into 25 groups for faster look‐up. One set of shortcuts generates new styles of molecular representation. Another group saves files with time stamps in the file names; the unique filenames avoid overwriting files that have already been developed. A third group submits search terms in the user's web browser. The help function prints the function's documentation to the command history window. This documentation includes the PyMOL commands that the user can reuse by copying and pasting onto the command line or into a script file. The shortcuts should save the average PyMOL user many hours per year searching for code fragments in their computer or on‐line.
Statement for Lay Public
Computer‐generated images of protein structures are vital to the interpretation of and communication about the molecular structure of proteins. PyMOL is a popular computer program for generating such images. We made a large collection of macros or shortcuts that save time by executing complex operations with a few keystrokes.
Keywords: macros, molecular artwork, molecular graphics, molecular illustration, molecular visualization, PyMOL scripting language, Python scripting, scientific illustration, shortcuts, visual narratives
Abbreviations
- GUI
graphical user interface
- PDB
Protein Data Bank
1. INTRODUCTION
Static, high‐quality images of molecular models are valuable for illustrating important structural insights into biological molecules and dominate the visual representation of molecular structures in scientific publications.1 Many of the most effective images require a large amount of time and technical knowledge to produce.2 There is interest in techniques that save time, enhance the quality of the image, or do both. One obvious approach to saving time is to make macros or functions to carry out recurring tasks.3 These functions can have short names that are fast to enter. Functions can also take arguments on the command line. We call these functions “shortcuts”. Shortcuts spare the user of the need to enter multiple lines of code. Shortcuts also keep the user's fingers on the keyboard. Shortcuts are several times faster to enter than navigating pull‐down menus.4 Shortcuts also extend the lengths of time between interruptions to use the mouse. Removal of the dominant hand from the keyboard to use the mouse interrupts typing and often the flow of thoughts. The above advantages of shortcuts can improve productivity when using molecular graphics programs.
We developed an extensive collection of shortcuts to bring their benefits to the users of the molecular graphics program PyMOL, which has more than 100,000 users.5 PyMOL is popular for preparing static images for publication and for making molecular movies because it has over 600 parameters that provide exquisite control over the appearance of the image.2, 6 The shortcuts are stored in one file, pymolshortcuts.py. This file is read during the start‐up of PyMOL after adding one line to the pymolrc file, the start‐up file for PyMOL. This simple edit makes the shortcuts available all of the time. The script file has 12,814 lines of code and comments, but its reading during PyMOL's startup causes no noticeable delay. Each shortcut is a Python function with a document string that explains how to use the shortcut. The user enters help (shortcut name) to display the documentation in the command history window. The documentation includes the corresponding PyMOL Macro Language code (pml) that the users can copy into a script file or paste onto the command line at the PyMOL prompt. Earlier, we introduce eighteen shortcuts in two sets8: one set displayed molecules in standard orientations and the other set made styles of molecular representation that are not available through a single command in the pull‐down menus or on the command line in PyMOL. Here, we introduce 23 new categories of shortcuts. Selected groups are discussed below. These shortcuts can help both novice and expert users work more efficiently with PyMOL.
2. RESULTS
We developed a set of shortcuts for more efficient use of PyMOL by both novice and expert users. The shortcuts were organized into 25 sets. Some groups of shortcuts address many common stumbling blocks in the use of PyMOL that can consume a significant amount of time. Other groups of shortcuts launch web searches and external programs from within PyMOL. The library's construction is described followed by explanations of several select categories of shortcuts that should be of wide interest. The current list of the shortcuts by category is supplied in the README.md file of the project's GitHub site http://github.com/MooersLab/pymolshortcuts and can displayed in PyMOL's command history window by entering the shortcut SC.
2.1. Library composition and assembly
The shortcuts were derived from the following sources: (a) past needs of lab staff and students, (b) posts in the PyMOL mailing list, and (c) questions about PyMOL in Stack Overflow (https://stackoverflow.com). The shortcuts were assembled in a single file, pymolshortcuts.py. Each shortcut has a document string that provides a brief description, usage, examples, more details, the corresponding pml code with one command per line, the pml code with all commands on one line for easy reuse on the command line, and the corresponding Python code (Figure 1). The compound command with all code on one line is useful for recycling the code in horizontal scripting and partly compensates for the absence of support in PyMOL for recycling useful code fragments.8
Figure 1.

The document string for the CBSS shortcut. The documentation includes the corresponding PyMOL scripting language code with one command per line or with all commands on a single line. The latter format enables horizontal scripting where all of the commands are on one line to ease iterative refinement of the parameter settings without leaving the PyMOL GUI and with minimal use of the mouse.8 The documentation also includes the Python version of the code for reuse in Python scripts. This function depends on Jared Sampson's 2014 colorbindfriendly.py script, which is copyrighted. The documentation in the script includes copyright and permission statements; these are not shown here for brevity
2.2. New styles of molecular representation
One set of shortcuts apply styles of molecular representation that are not available through existing commands in the pull‐down menus or on the command line. These shortcuts can be applied to the structure of any biological macromolecule.
A popular representation style is the black and white cartoon (e.g., shortcut: BW) (Figure 2a). These cartoons can give simple images for inclusion in documents where the cost of printing a large number of color figures is prohibitive or where color may be distracting or otherwise inappropriate or less effective. Examples of such documents include course syllabi, workshop handouts, organizational logos, review articles, book chapters, fliers, and exams.
Figure 2.
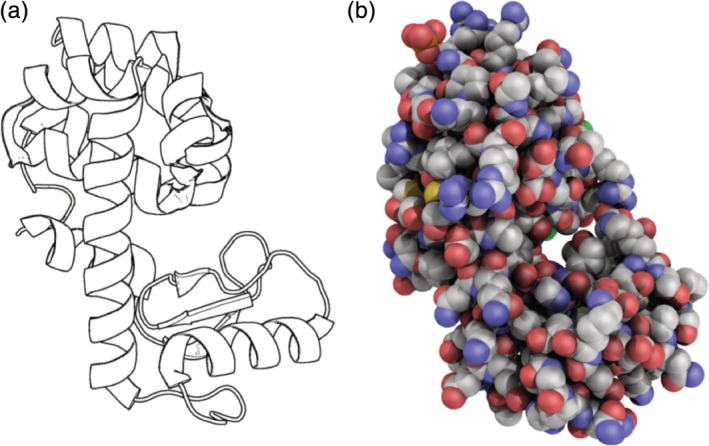
Examples of supplemental molecular representations that are generated from shortcuts. (a) Application of the black‐and‐white cartoon shortcut BW applied to wildtype T4 lysozyme (PDB‐ID 3fa0). (b) Application of the ambient occlusion shortcut (AO) to wildtype T4 lysozyme (PDB‐ID 3fa0)
Another popular representation style is the photorealistic efffect via ambient occlusion applied to van der Waals sphere representations (Figure 2b).9 This shortcut uses ray tracing to generate the shadows that give the photorealistic effect. Movement of the molecular object with the mouse destroys the ray tracing; the effect is restored by rerunning the shortcut AO. The shortcut AOBW gives a grayscale version of the photorealistic effect that is suitable for book chapters.
The shortcut gscale will convert any images into the proper grayscale values by element type. The RGB values for each element on a 0 to 1 scale were converted to grayscale with the formula Y = 0.2126 * R + 0.7152 * G + 0.0722 * B.
The ability to color molecular surfaces of proteins by biophysical properties with shortcuts is added with the timcolor and yrb shortcuts. The latter colors that surface to emphasize likely sites of protein–protein intermolecular interaction.10
2.3. Searches of web sites launched from inside PyMOL
This set of shortcuts was stimulated by the need to search the PyMOL Wiki for examples of using of a PyMOL function when that function's in‐line documentation lacks an example of use. For example, the documentation for the command isomesh (used to display electron density maps) has the syntax of the command defined but lacks examples of usage that are easier for beginners to understand. The user enters PW isomesh on the command line, and then the user's default web browser opens a new tab and retrieves a PyMOL Wiki webpage that describes the command isomesh. The user does not have to open their web browser, find the PyMOL wiki main page, and navigate to the webpage about the isomesh page. The entry of the function and its arguments on the command line is faster than using the link to the PyMOL Wiki in PyMOL's help pull‐down menu in the GUI. These time savings are compounded when multiple searches of the PyMOL Wiki need to be made during a work session in PyMOL.
The search of several select sites can be launched in parallel after entering the shortcuts and their search terms on the command line with each command separated by a semicolon after the search term. A tab for each website will open in the browser, and the searches will be run in parallel. The running of searches in parallel compounds the time saved. The shortcut multiple search of core sites (MC) will send the search term to arXiv, bioRxiv, chemRxiv, GitHub, PubMed, and Research Gate. The user can edit the code in the pymolshortcuts.py file to exclude a website, or they can use one of these functions as a template to write a new function that omits one or more of these websites. Likewise, the shortcut multisearch of all sites (MA) will send the search phrase to all of the searchable websites in the pymolshortcuts.py script.
2.4. Saving files with time stamps in the filename
There is no automated version control for files saved by PyMOL. Old versions of a file are overwritten by default. A common solution to this problem is to save the new file with a different filename; however, this solution becomes annoying to implement when repeated many times during the creation of an image. We address this issue with shortcuts that save the file with the date and time to the nearest second appended to the filename stem. The time‐stamp makes the filename unique and avoids the problem of overwriting of earlier versions of the file. The filename extensions is used by PyMOL to determine the file type to be saved (Table 1). These shortcuts start with the letter “s” and are followed by the corresponding file extension. For example, the command spse scene will save the current scene to disk as a session file with the filename sceneY19m11d04h11m50s10.pse.
Table 1.
PyMOL file type, extensions, and the corresponding shortcut to save a file with date and time
| Category | Description | Extension | Shortcut |
|---|---|---|---|
| Molecular structures | Protein Data Bank | .pdb | spdb |
| Protein Data Bank xml | .pdb | spdb | |
| PDB, Q‐charge, and Radius | .pqr | spqr | |
| Sybyl file format | .mol | smol | |
| Sybyl file format | .mol2 | smol2 | |
| SD File | .sdf | ssdf | |
| Python Pickle file | .pkl | spkl | |
| Python Pickle file | .pkla | spkla | |
| Intermediate Data Text Format | .idtf | sidtf | |
| Wavefront mesh file | .obj | sobj | |
| Molecular Operating Environment | .moe | smoe | |
| Maestro file | .mae smae | ||
| Crystallographic Information File | .cif | scif | |
| Macromodel | .mmd | smmd | |
| Macromodel | .smmod | smmod | |
| XYZ, binary format | .pmo | spmo | |
| Alignments | aln | .aln | saln |
| fasta | .fasta | sfasta | |
| Output | Output data file | .out | sout |
| Output data file | .dat | sdat | |
| Volume Data | CCP4 | .ccp4 | sccp4 |
| Geometry | Virtual Reality Modeling Language 2 | .vrml2 | svrml2 |
| POV‐ray tracing | .pov | spov | |
| VRML 2 | .wrl | swrl | |
| Wavefront Material | .mtl | smtl | |
| Collada | .dae | sdae | |
| Sessions | PyMOL Session | .pse | spse |
| PyMOL Session | .psw | spsw | |
| Image | Portable Network Graphic | .png | spng |
These shortcuts require the datetime module that does not come with PyMOL. Versions of incentive PyMOL less than 2.0 are difficult to augment with external modules. Users of these earlier versions of PyMOL can use the script pymolshortcutNoDateTime.py that lacks the functions that call datetime. This second script is available at the same GitHUb site.
2.5. Shortcuts to text editors
PyMOL provides a simple text editor that can be found under the File: Edit pymolrc pull‐down menu. This editor has syntax highlighting for the PyMOL Macro Language but does not support snippet libraries. In contrast, most text editors support snippet libraries. A set of shortcuts opens externally installed text editors from within PyMOL by using the subprocess module that is part of the Python interpreter that is inside of PyMOL. The user edits the file paths to the text editors at line 500 in the pymolshortcuts.py script if the editors are not installed in the Applications folder on a Mac (template file‐paths for other systems are provided). The supported full‐featured text editors for programmers include emacs, vim, Sublime Text, Visual Studio Code, TextMate (Mac OS only), and Atom. The supported simpler text editors that are more intuitive for nonprogrammers include gedit, jedit, Notepad++, and BBedit. These editors provide intelligent autocompletion, libraries of code snippets, and other advanced editing features that are not available in PyMOL's built‐in text editor. In addition, a shortcut opens the PDBeditor.jar program for efficient editing of PDB files.7 The editor shortcuts eliminate searches for the start‐up icon of the editing program. These small time savings add up.
2.6. Multi‐model display and removal
The display of symmetry mates and conformers in multi‐conformer models can be laborious because the default commands are hard to use to give the desired outcome or the command to display all of the conformers is hard to remember. For example, the default display of symmetry mates available under the “A” pull‐down in the internal GUI is often not very useful because the symmetry operations are applied without regard to the boundaries of the unit cell (Figure 3a). Instead, the user usually wants the symmetry mates inside one unit cell displayed (Figure 3b). If the user is interested in the shape the solvent channels, these can be visualized with a 2 × 2 × 1 array (Figure 3c) or a 3 × 3 × 1 array of unit cells (Figure 3d). The supercell.py script by Thomas Holder https://pymolwiki.org/index.php/Supercell will do this, but the user must enter multiple parameters on the command line. To simplify the use of this script, we wrote shortcuts for all combinations involving two or three unit cells in each direction.
Figure 3.
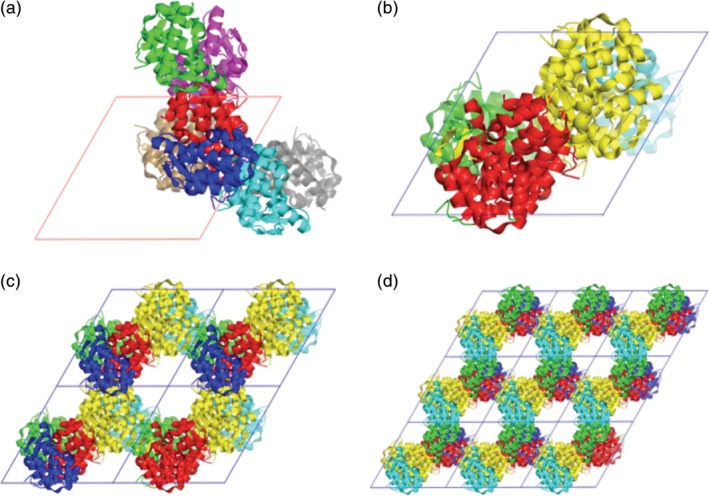
Displays of symmetry mates of bacteriophage T4 lysozyme (PDB‐ID 3Fa0) in space group P3221 without (a) and with shortcuts (b–d). The views are down the c‐axis. Each symmetry mate has a different color. (a) Symmetry mates generated inPyMOL with the Action:generate:symmetry mates cascade of commands by using the internal GUI. (B) The shortcut sc111 generated one unit cell filled with symmetry mates. (c) The shortcut sc221 shortcut generated a 2 × 2 × 1 array of unit cells. (d) The shortcut sc331 shortcut generated a 3 × 3 × 1 array of unit cells
The supercell.py script generates molecular objects for each symmetry mate. Each object has a menu bar in the internal GUI. When finished with the symmetry mates, the user is left with dozens of molecular objects to be removed manually one‐by‐one. The user often restarts PyMOL out of frustration by the tedium of this task. The shortcut rmsc removes the supercell and all of the symmetry mates.
Several structural biology methods rely on ensembles of structures due to insufficient data. These methods include NMR, SAXS, and EM. The alternative conformers are displayed with the show all_states command, which is hard to remember when used infrequently. We wrote shortcuts to display (e.g., nmr) and later remove the ensemble of models (nmroff).
2.7. Examples of molecules in standard orientation
One of the most critical decisions in the analysis of a new protein structure is selecting the orientation with which to display the model of the protein molecule. This orientation is often used to compare all future structures of the protein. This orientation minimizes the overlap of secondary structure elements. Of course, this orientation can be rotated by x degrees about a vector normal to the page and remain equivalent. One may decide that a particular rotation is the one of the greatest biological relevance with regards to the protein's function. The settings for this standard orientation can be saved and reused with isomorphous structures. The user can store these settings mapped to a function name to always have these standard orientations available. PyMOL's get_view function returns 7 lines of code for one orientation, which takes up too much space when used on the command line. We developed the roundview.py script to return the settings in a compact format on one line.8 We have mapped the roundview function to the shortcut rv. We provide shortcuts to several protein molecules in standard orientations. These shortcuts can be used as templates for making views of new molecules in a standard orientation.
2.8. Examples of complex displays
Molecular images of protein–protein interfaces, protein‐nucleic acid complexes, ligand‐binding sites, metal binding sites, and base‐stacking in nucleic acids can involve the creation of dozens of hydrogen bonds and coordinate covalent bonds to metals (Figure 4). Each of these bonds is a separate molecular object that can be specified with the PyMOL scripting language called pml code. The pml code for making these complex images can be displayed in the command history window by entering the help command and the name of the shortcut. This code can be recycled to make similar figures with new molecules.
Figure 4.
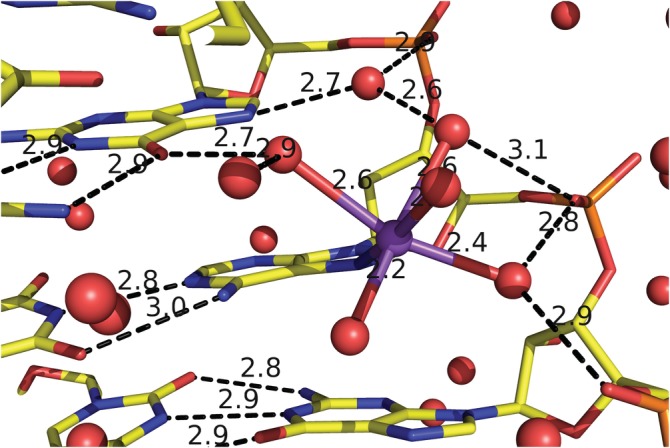
Complex figure with manually identified hydrogen bonds and the coordinate covalent bonds between a sodium, waters and adenine in the major groove of a RNA double helix (PDB‐ID 3ND4)
3. DISCUSSION
We present a collection of 241 shortcuts that help the user be more productive while working in PyMOL by eliminating many navigation steps that require the use of the mouse and by helping the user avoid tempting distractions outside of PyMOL. Illusions of progress from multi‐tasking and frequent external distractions hinder the productivity of many workers. Many of the shortcuts call external programs or websites and some send commands or even search terms. The user does not have to spend time outside of PyMOL finding an external program to open nor waiting for it to launch. The shortcuts enable the user's fingers to remain on the keys longer because the need to use the mouse has been reduced. The use of the mouse to manipulate the camera in the viewport of PyMOL is inevitable, but any tool that reduces the use of the mouse will accelerate the work. The opening of the external programs and websites from within PyMOL saves the user time and allows the user's attention to switch quickly back to the task at hand in PyMOL until a natural break in the work occurs. Without the shortcuts, the user has to make excursions outside of PyMOL that can take a long time if the user becomes distracted by other programs or searches on the internet. Keeping the user's work context anchored in PyMOL's GUI helps the user complete the task that is at hand.
PyMOL plugins often provide a GUI to external programs. The GUI interface is usually accessed via a cascading pull‐down menu. Many cursor actions with the mouse are required to gain access to the functions in the pull‐down menu. While pull‐down menus are more intuitive than the command line and are very helpful for demonstrating the features of a plugin, shortcuts are faster because a single action is required to execute the function of interest. In addition, the shortcut can be iterated in a simple loop entered interactively on the command line or called from a pml script file. In contrast, executing the action in a GUI with a mouse cursor becomes laborious, error‐prone, and tedious after the third iteration. The ability to call shortcuts from script files means that they can support reproducible research practices as long as the pymolshortcuts.py file is archived with the script file. Speed of execution, ease of iteration, and support for reproducible research are three advantages of shortcuts.
The use of shortcuts benefits new structural biologists by habituating them to the use of the command line. This benefit can have a spin‐off effect because comfort with the command line is good preparation for doing other scientific computing tasks. There has been rapid growth in the number of computer programs available for structural biology and structural bioinformatics. Most of these programs lack GUIs during the developmental phase because GUIs are time‐consuming to assemble. The new software may only be accessible via the command line for several years while the core functions are being optimized. To be competitive, future structural biologists need to be comfortable working with the command line to take advantage of software that currently lack GUIs. The script PyMOL helps build comfort with the command line because it provides motivational examples of how a lot can be accomplished by a few keystrokes. These shortcuts also reinforce the efforts of Software Carpentry to help biologists and other scientists become competent with using scientific software from the command line.11
The shortcuts presented here have several new capabilities compared to the aliases presented earlier.8 First, the shortcuts can take arguments on the command line. Second, each shortcut, as a Python function, has documentation (a so‐called docstring) that can be printed to the command history window in PyMOL by using the help function. Third, the shortcuts can open external applications. Fourth, multiple searches of the internet can be launched from the PyMOL command line. Fifth, the command to open a script file with a full‐featured text editor can now be launched from within PyMOL. These editors allow access to libraries of code fragments or snippets in the PyMOL Macro language. These snippets enable faster assembly of scripts. Sixth, image processing software can now open images saved from PyMOL to enable faster and more interactive editing of figures. Seventh, the number of shortcuts has been extended from 18 to 241 and from two categories to 25 categories. The shortcuts presented here represent a large advance over the aliases that were described in an earlier publication.8
4. MATERIALS AND METHODS
4.1. Script creation
The script startupAliases.py 8 was used as a template to make the pymolshortcuts.py script. The new script has with 25 new categories of shortcuts. Each shortcut name has several alphanumeric characters. No new special characters are used in the name of the shortcut to avoid long reaches from the home keys on the keyboard and to avoid conflicts with existing key bindings to keyboard shortcuts.
Each shortcut is a Python function that has an associated pymol.cmd method that maps the function name to a new command that can be invoked in PyMOL by entering the name of the function at the prompt on one of the two command lines inside PyMOL. Each function has a document string that is accessible within PyMOL with the help command. The document string contains a description of the shortcut, one or more examples of its usage, the corresponding PyMOL Macro Language code as a vertical script, the same as a horizontal script to ease copying and pasting the code on the command line, and as the corresponding Python code. The shortcuts depends only on modules that are shipped with the Python interpreter embedded in PyMOL with the exception of the timestamp related shortcuts, which depend on the module datetime. This module is installed inside PyMOL versions >2.0 by entering the following command at the PyMOL prompt conda install datetime. The the pip module can be used to install the datetime module in the Python interpreter used to build the open‐source versions of PyMOL. The module is not easy to add to the incentive versions of PyMOL that are earlier than version 2.0. The PyMOL Wiki has three webpages that describe how to install the Open Source PyMOL on Windows, Mac, and Linux. The alternate version of the script pymolshortcutsNoDateTime.py does not call the datetime module.
4.2. Accessibility
The files pymolshortcuts.py and pymolshortcutsNoDateTime.py are available for download from a dedicated webpage on github: https://github.com/MooersLab/PyMOLshortcuts. The README.md file of the github site contains instructions for installing the script and lists of the shortcuts by topic and their descriptions.
CONFLICT OF INTEREST
The author declares no conflict of interest.
ACKNOWLEDGMENTS
I thank the 275 graduate students in the OUHSC Graduate Program in Biomedical Sciences who provided invaluable feedback during nine or more hours of training per year in the use of PyMOL over the past eleven years. I thank Ann West for the opportunity to present an early version of this work at the 2018 Oklahoma Structural Biology Symposium.
Funding was provided by grants from the Presbyterian Health Foundation (PIs: Mooers; Wu and Mooers; Jankencht and Mooers), NSF (NSF MCB‐1616845), and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103640 (PI: Ann West).
Research reported in this publication was also supported in part by a 2018‐2019 Warren Delano Memorial Open‐Source PyMOL Fellowship, which is administered by Schrodinger Inc., the maintainers of PyMOL.
The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Mooers BHM. Shortcuts for faster image creation in PyMOL. Protein Science. 2020;29:268–276. 10.1002/pro.3781
Funding information Directorate for Biological Sciences, Grant/Award Number: MCB‐1616845; National Institute of General Medical Sciences, Grant/Award Number: 20 GM103640
REFERENCES
- 1. Goodsell D, Jenkinson J. Molecular illustration in research and education: past, present, and future. J Mol Biol. 2018;430:3969–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mura C, McCrimmon C, Vertrees J, Sawaya M. An Introduction to Biomolecular Graphics. PLoS Comput Biol. 2010;6:e1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sweigart A. Automate the boring stuff with Python: practical programming for total beginners. San Francisco, CA, USA: No Starch Press, 2015. [Google Scholar]
- 4. Omanson R, Miller C, Young E, Schwantes D. Comparison of mouse and keyboard efficiency. Proc Human Factors Ergon Soc Ann Meet. 2010;54:600–604. [Google Scholar]
- 5. Delano W. Use of PyMOL as a communications tool for molecular science. Abstr Paper Am Chem Soc. 2004;228:U228–U230. [Google Scholar]
- 6. Hodis E, Schreiber G, Rother K, Sussman J. eMovie: a storyboard‐based tool for making molecular movies. Trends Biochem Sci. 2007;32:199–204. [DOI] [PubMed] [Google Scholar]
- 7. Lee J, Kim S‐H. PDB Editor: a user‐friendly Java‐based Protein Data Bank file editor with a GUI. Acta Crystallogr D Biol Crystallogr. 2009;65(4), 399–402. 10.1107/s090744490900451x [DOI] [PubMed] [Google Scholar]
- 8. Mooers B. Simplifying and enhancing the use of PyMOL with horizontal scripts. Protein Sci. 2016;25:1873–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tarini M, Cignoni P, Montani C. Ambient occlusion and edge cueing for enhancing real time molecular visualization. IEEE Trans Vis Comput Graph. 2006;12:1237–1244. [DOI] [PubMed] [Google Scholar]
- 10. Hagemans D, Vanbelzen I, Moránluengo T, Rüdiger S. A script to highlight hydrophobicity and charge on protein surfaces. Front Mol Biosci. 2015;2:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson G. Software carpentry: getting scientists to write better code by making them more productive. Comput Sci Eng. 2006;8:66–69. [Google Scholar]


