Abstract
A highly reducing polyketide synthase gene cluster from the Magnaporthe oryzae genome was previously identified to produce phytotoxic compounds including pyriculol. A homologous gene cluster was found in Neurospora crassa through bioinformatics analysis. Heterologous expression of this cluster led to the production of salicylic aldehyde sordarial and related intermediates. A series of combinatorial gene expression experiments established the set of genes required to produce sordarial, and the likely biosynthetic pathway.
Graphical Abstract
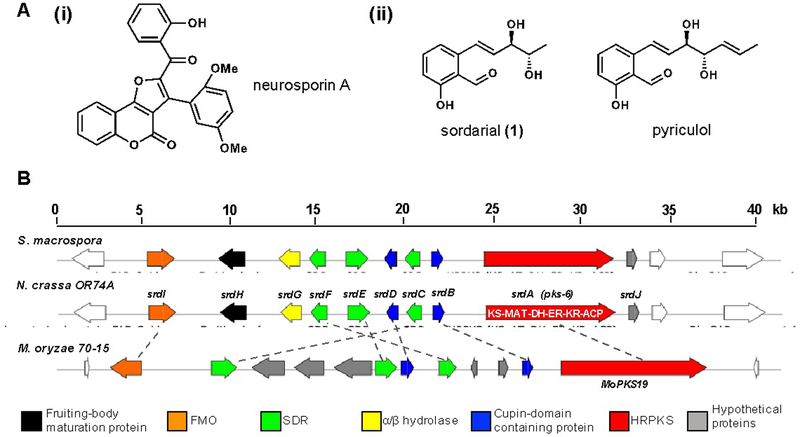
Natural products possess a wide range of bioactivities: some are medicinally valuable for pharmaceutical purposes,1 while others produced by plant pathogens can have a detrimental impact on the ecological system.2–3 Among the large number of fungal phytotoxins, one of the most well-known and economically damaging is pyriculol produced by Magnaporthe oryzae (Figure 1A), responsible in causing the rice-blast disease.4–5 Pyriculol is a salicylic aldehyde proposed to derive from a polyketide origin. Recently, the gene encoding a polyketide synthase, MoPKS19, in M. oryzae was identified to be responsible in the biosynthesis of pyriculol and related compounds.6 The ΔMoPKS19 knockout strain was reported to be absent of phytotoxicity.6 MoPKS19 resides in a gene cluster that contains additional biosynthetic enzymes found in fungal PKS pathways, including oxidoreductases, cupin-domain containing proteins, etc. (Figure 1B). The roles of most of the other enzymes in this cluster are unknown.
Figure 1.
Biosynthetic gene cluster of interest and related compounds. (A) Structure of (i) neurosporin A, (ii) salicylaldehyde polyketide compounds sordarial and pyriculol. (B) Comparison between N. crassa OR74A pks-6 gene cluster (renamed srd cluster) and its homologous clusters in S. macrospora and M. oryzae 70–15. The core pks genes in three clusters are >50% identical with the same domain organization KS-MAT-DH-ER-KR-ACP. Domain abbreviations: KS: ketosynthase; MAT: malonyl-CoA ACP acyltransferase; DH: dehydratase; ER: enoylreductase; KR: ketoreductase; and ACP: acyl-carrier protein.
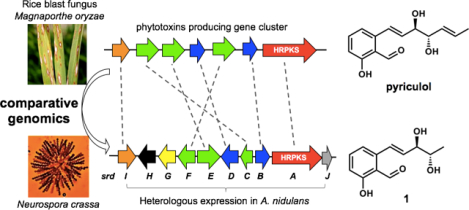
Identification of the pyriculol biosynthetic pathway, in particular MoPKS19, allows systematic scanning of sequenced fungi to locate similar gene clusters. This can enable the prediction of whether a fungus can produce compounds structurally related to the phytotoxin. Using this comparative genomic approach, homologs to MoPKS19 can be found in many fungi. Perhaps the most surprising fungus to contain a similar gene cluster is Neurospora crassa, the first fungi to be genome sequenced7 and a model research organism for fungal biology.8 The cluster (named srd in this work) is anchored by pks-6 (Figure 1B) together with a similar set of post-PKS tailoring enzymes. This N. crassa cluster is well-conserved in several fungi, including a cluster of unknown function in Sordaria macrospora (Figure 1B, Table S1). The pks genes in these clusters are highly homologous to each other, with pks-16 having 95% and 54% identity to S. macrospora pks (XM_003351980.1) and MoPKS19 in M. oryzae 70–15, respectively. The highly reducing polyketide synthase (HRPKS) encoded by pks-6 contains the typical domains KS, MAT, DH, ER, KR and ACP arranged in a linear fashion. Genes flanked by pks-6 were also found to have high identities with those in the S. macrospora cluster. Together two clusters of 10 genes have identical organizations (Figure 1B). Bioinformatics analysis revealed that predicted genes outside of boundaries have low or no identity, thereby establishing putative boundaries of the 10-gene cluster.
A recent genetics report associated pks-6 to the biosynthesis of furanocoumarin neurosporin A (Figure 1A), a metabolite produced by N. crassa for chemo-resistance against predation by arthropod fungivores.9 However, based on the gene cluster organization and predicted gene functions, we reason this cluster is unlikely to be involved in neurosporin A biosynthesis, but instead should produce compounds similar to pyriculol. N. crassa has not been classified as a plant pathogen, with the exception of a recent report showing host-pathogen interaction with Scot pine (Pinus sylvestris) cultured in laboratory conditions.10 Therefore, we set out to identify the metabolite encoded in this cluster in N. crassa and determine if it is structurally related to the phytotoxin pyriculol.
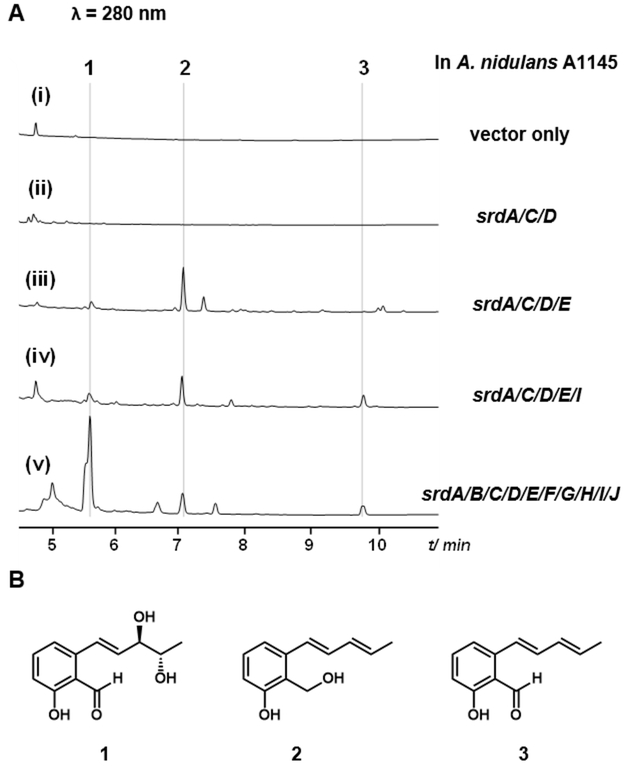
We used a fungal heterologous overexpression system to examine the natural product encoded in the srd gene cluster. The host is an Aspergillus nidulans A1145 strain that can be transformed and maintain three expression vectors (pYTU, pYTP and pYTR).11 A combination of four A. nidulans-adapted promoters (glaA, amyB, gpdA and trpC) with three fungal selection markers (pyrG, pyroA and riboB) allows starch-inducible or constitutive expression of up to twelve genes (Table S2). We first introduced all ten of the genes into A. nidulans. Analysis of the organic extract following three days of culturing revealed the emergence of three metabolites 1–3 (Figure 2A, v) compared to the vector only control (Figure 2A, i). UV-absorption analysis showed that all three compounds have extensive π-electron system, with 3 being the most conjugated (λmax = 371 nm) (Figure S2). Large scale fermentation was carried out in starch-supplemented glucose minimum media, and the three compounds were purified by chromatography methods.
Figure 2.
Heterologous reconstitution of the srd biosynthetic gene cluster produced sordarial 1. (A) HPLC analysis (λ = 280 nm) of A. nidulans expressing different srd genes. (i) negative control with expression vector only; (ii) srdA/C/D; (iii) srdA/C/D/E; (iv) (iii) srdA/C/D/E/I; (v) the entire srd cluster. (B) Structures of 1, 2 and 3 as confirmed through purification and NMR analysis.
As we predicted from bioinformatic comparison to the pyriculol cluster, NMR structural characterization revealed 1–3 to be a series of compounds structurally related to the salicylic aldehyde pyriculol (Figure 2). The 13C-NMR data of 1 agrees with sordarial data reported in literature, and the optical rotation data [α] +10.10 (c 2.0, CH3OH) matches that of the reported (3’R, 4’S) enantiomer (Table S3).12 Hence, 1 is identified to be sordarial previously isolated from S. macrospora,13 Gelasinospora heterospora,12 and Gelasinospora longispora.12 The production of 1 from the 10-gene cluster is consistent with S. macrospora being a known producer of 1. 2 is identified as 1-hydroxy-2-hydroxymethyl-3-pent-1,3-dienylbenzene, a known pentadienyl-substituted salicylic alcohol previously isolated from Deuteromycete strain HA-45–93,14 whereas 3 is elucidated to be a new compound. The large scale fermentation reached titers of 41 mg, 10.7 mg and 8.6 mg in 8L culture for 1–3 respectively. The NMR spectra of 3 are almost superimpossible with those of 2 (Figures S4–S8), showing resonances of a 1,2,3-trisunstituted phenyl and a conjugated diene moiety. A key difference is the replacement of the hydroxymethyl group in 2 (δH 4.76, s; δC 56.4) by an aldehyde in 3 (δH 10.38, s; δC 196.9), This observation postulates a pentadienyl-substituted salicylic aldehyde structure for 3 (Figure 2B), which was confirmed by the HRSIMS (Figure S1), IR (Figure S3) and 2D-NMR spectra (Figures S4−S8). Assignment of the NMR data for 3 is listed in Table S3.
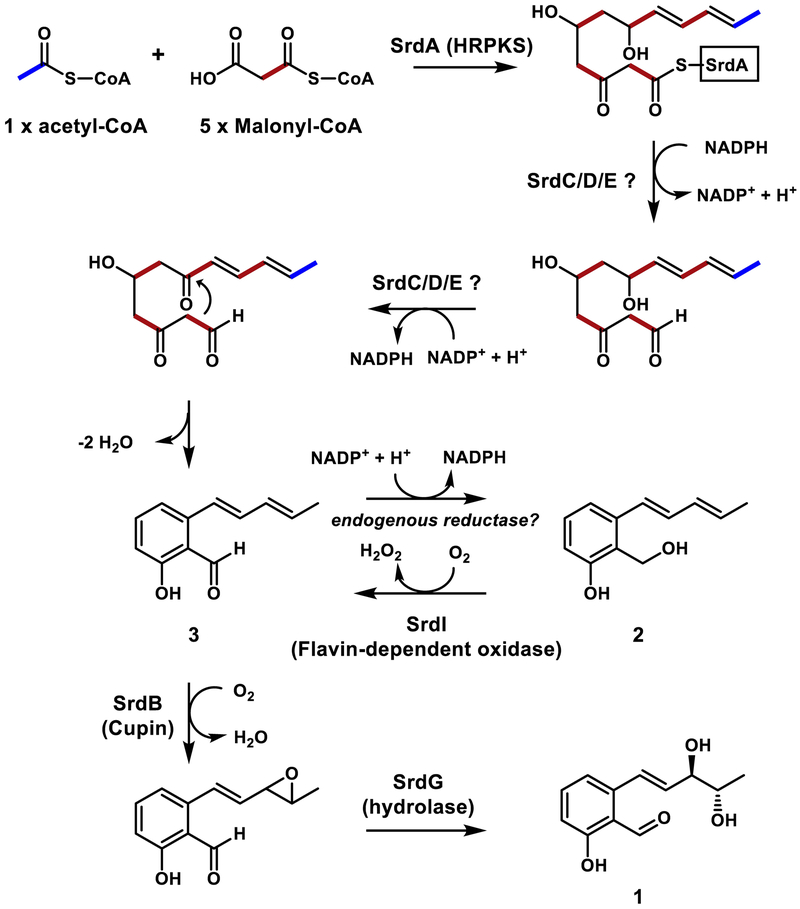
Based on the structures of 1–3, we reason sordarial (1) is the final product of the gene cluster and 2–3 are intermediates that accumulated in the heterologous host. The anti-diol in 1 could derive from the corresponding epoxide which can be oxidized from 3 (Figure 3). To examine the minimum number of genes required to produce the salicylic structure, we expressed the PKS srdA with different combinations of pathway genes, except the srdH, which does not encode a biosynthetic enzymes. Most binary and tertiary combinations with srdA failed to yield any products, including srdA with srdC and srdD (Figure 2A, ii). The minimal construct that afforded a detectable metabolite is the combination of PKS (srdA), short-chain dehydrogenases (SDR, srdC and srdE) and a cupin-domain containing oxidoreductase (srdD), which produced 2. Upon introduction of a flavin-binding oxidoreductase srdI into the above strain, we observed the production of compound 3 together with 2. This suggests the SrdI is involved in oxidizing the alcohol group in 2 to the corresponding aldehyde. This result is consistent with a similar study in the pyriculol pathway: the SrdI homolog encoded by MoC190XR1 was shown through knockout studies to oxidize dihydroxypyriculol to pyriculol in M. oryzae.6 The ΔMoC19OXR1 mutant strain only produced dihydroxypyriculol.6 This oxidation to afford salicylic aldehyde is critical for the activity of pyriculol.
Figure 3.
Proposed biosynthesis pathway of sordarial by the srd pathway from Neurospora crassa.
Figure 3 shows a proposed biosynthetic pathway for sordarial (1). The most interesting aspect of this pathway is formation of an aromatic product from an HRPKS. Typically in fungi, aromatic polyketides are synthesized by nonreducing polyketide synthases (NRPKSs) that do not have reductive domains such as KR, DH and ER.15 In order to form the aromatic phenol from a β-reduced polyketide chain, re-oxidation of the chain with redox enzymes may be required. This is consistent with the requirement of a number of redox enzymes (SrdC, D and E) to form the first isolable intermediate 2. SrdA can synthesize a reduced polyketide chain from one molecule of acetyl-CoA and five molecules of malonyl-CoA. The polyketide chain is then reductively released as an aldehyde. The remaining enzymes then oxidize one of the hydroxy groups to facilitate the intramolecular aldol condensation, followed by dehydration to yield the salicylic aldehyde 3. This aldehyde can undergo facile reduction by endogenous reductases to yield the alcohol 2.16 The flavin-dependent SrdI is responsible to re-oxidized the alcohol 2 back to 3. SrdI and related enzymes in other salicylaldehyde pathways therefore counteract against the propensity of the aldehydes to be reduced under physiological conditions. 3 is then selectively epoxidized by SrdB to yield the epoxide, which can be hydrolyzed stereoselectively by the hydrolase SrdG to give the final product 1.
Initial interest of sordarial bioactivity stemmed from its immunosuppressant activity.12,17 Subsequently, other salicylaldehyde polyketides differing in substituted chain lengths have been reported with phytotoxic bioactivity. Sordarial and pyriculol share strong structural similarity, with pyriculol containing an additional olefin presumably arise from a longer polyketide product. Such structural similarity suggests potential phytotoxic activities of sordarial as well. The salicylic aldehyde moiety is common to a panel of phytotoxic fungal secondary metabolite from Ascochyta sp.18 and Magnaporthe sp.,6 and was reported to be essential for plant pathogenesis. From various studies including chemical modification of agropyrenol produced by Ascochyta agropyrina and genetic manipulation efforts in M. oryzae,6 it is known that any modification of the diol of the 3′, 4′-dihydroxypentenyl side chain at C-2 as well as the aldehyde group at C-1 led to strong reduction or total loss of phytotoxicity. N. crassa is widely used as a model host in various fungal biology research and is generally accepted as a non-pathogenic fungus.10 Our results here suggest that sordarial could be produced from the srd cluster under certain environmental triggers. Future in planta studies with N. crassa can be crucial in evaluating the conditions upon which this cluster is expressed and the phytotoxin produced.
In summary, using genome mining and heterologous expression we identified and isolated three compounds 1-3 that are structurally related to the phytotoxin pyriculol. Compound 1 is the known sordarial, while 2 and 3 are likely biosynthetic intermediates. The assignment of 1 to the pks-6 containing gene cluster in N. crassa suggests that this well-characterized model fungus has the biosynthetic potential to yield phytotoxin-like compounds.
EXPERIMENTAL SECTION
General Experimental Procedures:
Optical rotations were analyzed using a Rudolph Research Autopol III automatic polarimeter. UV and mass spectra were obtained using a Shimadzu 2020 EV LC–MS (Kinetex 1.7 μm C18 100 Å, LC Column 100 × 2.1 mm) using positive-and negative-mode electrospray ionization with a linear gradient of 5–95% acetonitrile MeCN–H2O with 0.5% formic acid in 15 min followed by 95% MeCN for 3 min with a flow rate of 0.3 mL/min. Column chromatography was performed using a CombiFlash system using a HP CL18 reverse phase column. HPLC fractionation used a semi-preparative C18 column of Kinetics New column. IR spectra were obtained using the PerkinElmer® Frontier FIR spectrometer, annotated by the PerkinElmer® Spectrum software suite. 1D and 2D NMR spectra were obtained on Bruker AV500 spectrometers at the UCLA Molecular Instrumentation Center. High resolution mass spectra were obtained from Agilent 6530 Q-TOF ESI with a 1260 Infinity LC with Autosampler at the UCLA Molecular Instrumentation Center.
Fungal Strain and Culture Media:
Neurospora crassa OR74A was purchased from the ARS Culture Collection (NRRL A-2983). Neurospora crassa was grown on Difco PDA (potato dextrose agar) Plates from BD biosciences and cultured for extraction of gDNA using the Zymo ZR Fungal /Bacterial DNA Microprep™ kit. Genomic sequence of N. crassa was obtained from online sources.7 Aspergillus nidulans A1145 ΔSTΔEM was used for heterologous expression of genes from N. crassa.19
Plasmids Construction for Heterologous Expression:
Escherichia coli strain XL1 Blue was used for plasmid propagation. Saccharomyces cerevisiae strain BJ5464 was used for homologous recombination of DNA fragments to assemble the vectors used in heterologous expression (list of plasmids see Table S2). Plasmids pYTU, pYTP, pYTR were used as fungal expression vectors to insert genes which contain auxotrophic fungal markers for uracil (pyrG), pyridoxine (pyroA) and riboflavin (riboB), respectively. For detailed plasmid cloning procedures, please see Supplementary Information.
Extraction and Isolation:
To purify 1−3 for structure characterization, the A. nidulans transformants were cultured in 8L liquid CD-ST medium and shaken at 28 °C for 4 days. Culture was filtered to separate the supernatant and the mycelium. Ethyl acetate was used to extract the secondary metabolites from the supernatant and acetone was used for the mycelium extraction. The organic extracts were combined and concentrated in vacuo. The concentrated organic extract was subjected to column chromatography using a CombiFlash® system eluted with a gradient wash of hexane/acetone (9/1–1:1, v/v) to yield three fractions Frs. 1–3 as guided by UV characteristics. Frs. 1–3 were further purified by HPLC with a semipreparative C18 column (Kinetics New column, 5μm, 10 × 250 mm) eluting with MeCN/H2O (45:55, v/v, flow rate, 3.0 mL/min) to furnish 3 (8.6 mg, tR = 35 min), 2 (10.7 mg, tR = 24 min), and 1 (41 mg, tR = 18 min). Compound 3, pale yellow powder; IR νmax 3022, 1737, 1640, 1600, 1569, 1451, 1327, 1310, 1276, 1236, 1194, 1168, 987, 805, 750, 720, 534, 469 cm−1; 1H and 13C NMR data, see Table S3; HRESIMS [M + H]+ m/z 189.0942 (calcd for C12H13O2, 189.0916).
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the NIH 1R35GM118056 to Y.T. Chemical characterization studies were supported by shared instrumentation grants from the NSF (CHE-1048804) and the NIH NCRR (S10RR025631). Z. Z. is supported by UCLA Undergraduate Research Fellowship Program (URFP). Y.-S.H. is supported by NIH Biotechnology Training in Biomedical Sciences and Engineering (T32GM067555). We thank Dr. Man-Cheng Tang for help in gene cluster annotation. We thank Leibniz Hang for assistance with HRMS analysis, and Rachel Knapp and Prof. Neil Garg for assistance with IR analysis of compounds.
Footnotes
Supporting Information
Experimental details, spectroscopic and computational data. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Newman DJ; Cragg GM J. Nat. Prod 2016, 79, 629–661. [DOI] [PubMed] [Google Scholar]
- (2).Möbius N; Hertweck C Curr. Opin. Plant. Biol 2009, 4, 390–8. [DOI] [PubMed] [Google Scholar]
- (3).Dayan FE; Owens DK; Duke SO Pest. Manag. Sci 2012, 68, 519–528. [DOI] [PubMed] [Google Scholar]
- (4).Ebbole DJ Ann. Rev. Phytopathol 2007, 45, 437–456. [DOI] [PubMed] [Google Scholar]
- (5).Kim JC; Min JY; Kim HT; Cho KY; Yu SH Biosci. Biotechnol. Biochem 1998, 62, 173–174. [DOI] [PubMed] [Google Scholar]
- (6).Jacob S; Grötsch T; Foster AJ; Schüffler A; Rieger PH; Sandjo LP; Liermann JC; Opatz T; Thines E Microbiology, 2017, 163, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Galagan JE et al. , Nature, 2003, 422, 859–868. [DOI] [PubMed] [Google Scholar]
- (8).Davis RH; Perkins DD Nat. Rev. Genet 2002, 3, 397–403. [DOI] [PubMed] [Google Scholar]
- (9).Zhao Y; Ding J; Yuan W; Huang J; Huang W; Wang Y; Zheng W Environ. Microbiol 2017, 19, 3920–3929. [DOI] [PubMed] [Google Scholar]
- (10).Kuo HC; Hui S; Choi J; Aseigbu FO; Valkonen JP; Lee YH Sci. Rep 2014, 4, 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sato M; Yagishita F; Mino T; Uchiyama N; Patel A; Chooi YH; Goda Y; Xu W; Noguchi H; Yamamoto T; Hotta K; Houk KN; Tang Y; Watanabe K Chembiochem 2015, 16, 2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fujimoto H, Sumino M, Nagano J, Natori H, Okuyama E, Yamazaki M Chem. Pharm. Bull 1999, 47, 71–6. [DOI] [PubMed] [Google Scholar]
- (13).Bouillant ML; Favre-Bonvin J; Salin N; Bernillon J Phytochemistry 1988, 27, 1517–1519. [Google Scholar]
- (14).Gehrt A, Erkel G, Anke H, Anke T, Sterner O Nat. Prod. Lett 1997, 9, 259–264. [Google Scholar]
- (15).Chooi Y-H; Tang Y J. Org. Chem 2012, 77, 9933–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).(a) Guillén P; Guis M; Martínez-Reina G; Colrat S; Dalmayrac S; Deswarte C; Bouzayen M; Roustan JP; Fallot J; Pech JC; Latché A Plant. J 1998, 16, 335–343. [DOI] [PubMed] [Google Scholar]; (b) Yamauchi Y; Hasegawa a.; Taninaka A; Mizutani M; Sugimoto J. Biol. Chem 2011, 286, 6999–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Fujimoto H, Asai T, Kim Y-P, Ishibashi M Chem. Pharm. Bull 2006, 54, 550–553. [DOI] [PubMed] [Google Scholar]
- (18).Cimmino A; Zonno MC; Andolfi A; Troise C; Motta A; Vurro M, Evidente AJ Agric. Food. Chem 2013, 61, 1779–1783. [DOI] [PubMed] [Google Scholar]
- (19).Liu N, Hung Y, Gao S-S, Hang L, Zou Y, Chooi Y-H *, Tang Y Org. Lett 2017, 19, 3560–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.