Abstract
We present a GPU implementation of the continuous constant pH molecular dynamics (CpHMD) based on the most recent generalized Born implicit-solvent model in the pmemd engine of the Amber molecular dynamics package. To test the accuracy of the tool for rapid pKa predictions, a series of 2-ns single-pH simulations were performed for over 120 titratable residues in 10 benchmark proteins that were previously used to test the various continuous CpHMD methods. The calculated pKa’s showed a root-mean-square deviation of 0.80 and correlation coefficient of 0.83 with respect to experiment. 90% of the pKa’s were converged with estimated errors below 0.1 pH units. Surprisingly, this level of accuracy is similar to our previous replica-exchange simulations with 2 ns per replica and an exchange attempt frequency of 2 ps−1 (Huang, Harris and Shen, J Chem Info Model, 2018). Interestingly, for the linked titration sites in two enzymes, although residue-specific protonation state sampling in the single-pH simulations was not converged within 2 ns, the protonation fraction of the linked residues appeared to be largely converged, and the experimental macroscopic pKa values were reproduced to within 1 pH unit. Comparison with replica-exchange simulations with different exchange attempt frequencies showed that the splitting between the two macroscopic pKa’s is underestimated with frequent exchange attempts such as 2 ps−1, while single-pH simulations overestimate the splitting. The same trend is seen for the single-pH vs. replica-exchange simulations of a hydrogen-bonded aspartyl dyad in a much larger protein. A 2-ns single-pH simulation of a 400-residue protein takes about one hour on a single NVIDIA GeForce RTX 2080 graphics card, which is over 1000 times faster than a CpHMD run on a single CPU core of a high-performance computing cluster node. Thus, we envision that GPU-accelerated continuous CpHMD may be used in routine pKa predictions for a variety of applications, from assisting MD simulations with protonation state assignment to offering pH-dependent corrections of binding free energies and identifying reactive hot spots for covalent drug design.
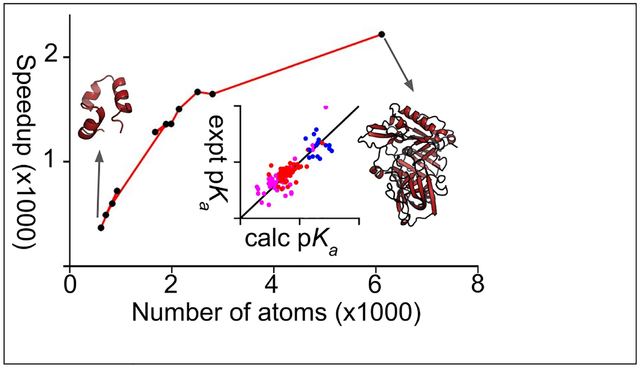
Graphical Abstract
INTRODUCTION
Molecular dynamics (MD) simulations are widely used in structure-based drug design and to understand biophysical processes on a molecular level,1,2 but one weakness of the most common simulations is that they require the protonation states of the titratable sites to be fixed. Typically, the protonation states of these residues are set to match those of model compounds representing single amino acids in water at pH 7, but these states may not be correct, as the pKa of a titratable site in the protein environment may differ significantly from the model pKa in solution. Sometimes, structure-based pKa prediction tools, such as the empirical method Propka3 and the Poisson-Boltzmann solver based methods APBS-PDB2PQR,4 H++,5 and DelPhiPKa,6 are used to obtain estimates of the protonation states prior to running simulations; however, these tools do not directly account for the dynamic flexibility of the protein, which may lead to incorrect assignment of protonation states. Most importantly, even if the initial assignment is correct, protonation states may change in the course of conformational dynamics, as demonstrated in pH-dependent protein folding,7,8 protein-ligand binding,9–11 enzyme catalysis,12 and ion/substrate transport across the membrane.13,14
One solution to the above problems is to use constant pH molecular dynamics (CpHMD) methods to determine protonation states on the fly during the MD simulation.15–20 CpHMD methods treat the pH as a macroscopic thermodynamic variable of the simulation, similar to how temperature and pressure are commonly handled in MD simulations. By linking the protonation states of the titratable residues to the solute conformation, the effects of conformational flexibility on the pKa estimates are explicitly taken into account. Several CpHMD methods have been developed in the past, and they generally fall into two categories: The Monte-Carlo based methods,15,16,18,20 which maintain a constant pH by periodically attempting Metropolis Monte Carlo moves to protonate or deprotonate titratable sites and continuous CpHMD,17,21–26 which is rooted in the λ-dynamics method for free energy calculations27 and treats the protonation states of the titratable sites as continuous dynamic variables of the system. Continuous CpHMD lets systems escape local energy minima by allowing transient access to partially protonated states which are unphysical and must be discarded from the final pKa calculations. In contrast, Monte-Carlo based CpHMD allows only fully protonated or deprotonated states, but as a result they tend to converge more slowly. Additionally, the proper way to extend these methods to fully explicit solvent is unclear.19,20 A more extensive discussion of the various CpHMD methods can be found in the reviews.28,29
Both Monte-Carlo and continuous CpHMD simulations have historically required some enhanced sampling technique, such as temperature-30,31 or pH-based replica exchange,22,32 to obtain converged pKa estimates. However, at the present time, replica exchange is not necessarily attractive for computation on Graphics Processing Units (GPUs), as in some molecular dynamics packages, including Amber, each replica requires a separate graphics card, and to ensure frequent exchange between neighboring replicas, a large number of parallel replicas are sometimes needed. Such simulations would require a relatively expensive GPU cluster and could prevent the method from being widely used.
Here we present the first implementation of the generalized Born (GB) based continuous CpHMD method in the GPU-accelerated pmemd program33 of the Amber molecular dynamics (MD) package.34 The performance of the code was benchmarked in pKa predictions for Asp, Glu, and His sidechains of 10 small and medium sized proteins. Considering that common users may not have access to a GPU node equipped with multiple GPU cards, single pH titration simulations were used. We restricted the simulation length to 2 ns to facilitate comparison with our previous work, where the pKa’s were calculated using pH replica-exchange (pH-REX) titration simulations of 2 ns per replica on the CPUs.35 We note that even though the time scale for conformational rearrangements in implicit solvent is much shorter than in explicit solvent, 2 ns may be insufficient for conformational relaxation to specific pH conditions, particularly for buried residues and those undergoing strong electrostatic or hydrogen-bond interactions. However, the objective of this work is to demonstrate the tool not for constant pH simulations but for pKa predictions given a user-specified simulation time, e.g, 2 ns. Encouragingly, despite the short simulation length, the calculated pKa values of ten benchmark proteins agree well with our previous pH-REX results and experimental data. Interestingly, although the microscopic pKa’s for the linked residues in two enzymes were not converged, the single-pH simulations were able to reproduce the experimentally observed macroscopic pKa’s. On a single NVIDIA 2080 graphics card, a 2-ns single-pH simulation of a 400-residue protein takes about one hour. Thus, we envision the GPU-accelerated continuous CpHMD tool will be routinely applied to predict pKa’s on commodity hardware.
METHODS AND PROTOCOLS
The Compute Unified Device Architecture (CUDA) code of the GB molecular dynamics method in the pmemd.cuda program33 of Amber (version 2018)34 was modified. The energies, forces, and other numbers computed with the code matched those given by our CPU implementation, subject to the precision differences between the GPU and CPU code. As in the CPU implementation, the code is currently limited to using the GB-Neck2 model36 for both conformational and titration dynamics. Although the method does not impose a limit on the total number of titration sites, the maximal number of titratable sites is currently set to an arbitrary number 1000 (Asp/Glu has 2 titratable groups due to double-site titration), which however can be changed in the future.
We performed single-pH simulations on 11 proteins: the 36-residue villin headpiece subdomain (HP36, pdbid 1VII), 45-residue binding domain of 2-oxoglutarate dehydrogenase multi-enzyme complex (BBL, pdbid 1W4H), 56-residue N-terminal domain of ribosomal protein L9 (NTL9, pdbid 1CQU), 56-residue turkey ovomucoid third domain (OMTKY, pdbid 1OMU), 105-residue reduced form of human thioredoxin (pdbid 1ERT), 129-residue hen egg-white lysozyme (HEWL, pdbid 2LZT), 143-residue hyperstable Δ+PHS variant of staphylococcal nuclease (SNase, pdbid 3BDC), 124-residue ribonuclease A (RNase A, pdbid 7RSA), 155-residue E. coli ribonuclease H (RNase HI, pdbid 2RN2), 185-residue oxidized form of Bacillus circulans xylanase (xylanase, pdbid 1BCX), and 389-residue unbound β-secretase 1 catalytic domain (BACE1, pdbid 1SGZ). These proteins have been previously used to validate our CPU implementation of the same continuous CpHMD method.35
The initial structures for the simulations were taken from our previous paper.35 We started from the PDB coordinates by adding acetylated N terminus and amidated C terminus caps, building any disulfide bonds, and adding hydrogens with the CHARMM program (version c42a1).37 The protonation states of the titratable residues were set so that Asp/Glu were deprotonated and His/Lys/Arg/Cys/Tyr were protonated. The structures then underwent 50 steps of steepest descent minimization in GBSW implicit solvent38 with a harmonic force constant of 50 kcal/mol/Å2 applied to each heavy atom. Next, dummy atoms were added to the Asp/Glu residues, and the structure was minimized for 10 steps of steepest descent and 10 steps of Newton-Raphson minimization. These final structures were then converted to the structure files with the Leap utility in Amber.34
All simulations used the Amber ff14SB force field39 and the GB-Neck2 implicit-solvent model.36 All bonds containing hydrogens were constrained with the SHAKE algorithm,40 the salt concentration was set to 0.15 M, and a 2 fs timestep was used. For the 2 proteins without His residues (HP36 and NTL9), 14 single-pH simulations were run with pH 1–7.5 in 0.5 unit increments, and for the other proteins 18 simulations were run with pH 1–9.5 in 0.5 unit increments. Each simulation lasted 2 ns except for BACE1 simulations which were extended to 10 ns each. The CpHMD settings and options were the same as in our previous replica-exchange simulations,35 except that the latter also employed a pH replica-exchange protocol, in which exchanges between adjacent pH replicas were attempted every 250 MD steps (exchange attempt frequency of 2 ps−1). In the current work, we performed additional replica-exchange simulations for SNase and RNase H with exchanges attempted every 500 and 1000 MD steps which correspond to the exchange attempt frequencies of 1 ps−1 and 0.5 ps−1, respectively. Larger pH ranges were used in the replica-exchange simulations. For the 2 proteins without His, 16 replicas were used with pH 0–7.5 in increments of 0.5 units, and for the other proteins 20 replicas with pH ranging from 0–9.5 in increments of 0.5 units were used.
To calculate pKa’s, the probability of deprotonation (unprotonated fraction) was fit to the generalized Henderson-Hasselbalch (HH) equation, as in our previous work.35 For simplicity, the word generalized will be omitted in later discussions. The statistical errors in the pKa’s were estimated from the covariances of the fit parameters. For the macroscopic pKa of histidine, the total unprotonated fraction was used in the fitting. For the pKa’s of HID and HIE, the fractions of the respective tautomers were used (see more explanation in the footnote of Table 1).
Table 1:
Calculated pKa’s of the model alanine pentapeptides from the GPU single-pH simulations compared to the CPU replica-exchange results and experiment
| GPU | CPUc | Exptd | ||
|---|---|---|---|---|
| Fita | Bootstrapb | |||
| Asp | 3.7±0.03 | 3.7±0.10 | 3.8±0.05 | 3.67±0.04 |
| Glu | 4.2±0.02 | 4.2±0.06 | 4.1±0.05 | 4.25±0.05 |
| His | 6.3±0.03 | 6.3±0.10 | 6.4±0.03 | 6.54±0.04 |
| HIDe | 7.0±0.03 | 7.0±0.08 | 7.0±0.05 | n/d |
| HIEf | 6.4±0.03 | 6.4±0.11 | 6.5±0.02 | n/d |
pKa’s and errors from the fitting of all data points (see main text).
pKa’s and errors from a bootstrap procedure (see main text).
pKa’s from our previous replica-exchange simulations with errors calculated as the standard deviations of the pKa’s from five sets of replica-exchange simulations.35
NMR-determined pKa’s by Thurlkill et al.41 These pKa’s were used as the reference (model) values in our simulations.
The pKa of HID is associated with HIP ⇋ HID.
The pKa of HIE is associated with HIP ⇋ HIE.
RESULTS AND DISCUSSION
Titration simulations of model peptides.
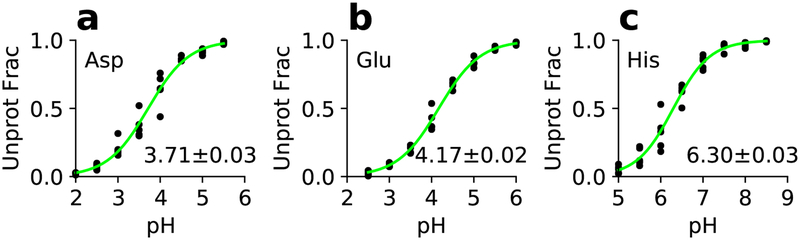
First, we verified that the GPU-based single-pH titration simulations with the model parameters from our previous work35 can reproduce the reference experimental pKa’s of model alanine pentapeptides, CH3CO-Ala-Ala-X-Ala-Ala-NH2, where X represents Asp, Glu, or His. These reference pKa’s are also referred to elsewhere as model or solution pKa’s. For each pentapeptide, 5 independent 2-ns titration simulations were performed at each of the 8 evenly spaced pH conditions with an interval of 0.5 units, in the pH ranges 2–5.5 for Asp, 2.5–6 for Glu, and 5–8.5 for His. Fig. 1 displays the unprotonated fraction versus pH (titration data) for each residue and the least-square fit to the HH equation. Fitting of all 40 data points for each peptide returned a pKa of 3.71 for Asp, 4.17 for Glu, and 6.33 for His. These pKa’s are within 0.1 units of the previous CPU-based replica-exchange pKa’s and within 0.2 units of the experimental values (Table 1). This level of agreement is acceptable and therefore we decided that we could use the previously derived model parameters for the titration simulations of proteins.
Figure 1: Simulated titration data for the model alanine pentapeptides in solution.
Unprotonated fraction as a function of pH for Asp (a), Glu (b), and His (c). Green curves are the best fits to the HH equation. The calculated pKa’s based on all five sets of titration data and the fitting errors are shown.
Error estimates for the model pKa’s.
From the least-square fit of five titration datasets (40 data points) to a single HH equation, an error for the pKa, i.e., fitting parameter, can be obtained. This fitting error is below 0.03 for model Asp, Glu, and His, demonstrating a high precision of the pKa (Fig. 1). We were also interested in the error (reproducibility) of the calculated pKa given only one titration dataset (one data point for each pH). To address this question, a bootstrap method was used, where the unprotonated fractions at different pH were combined to generate every possible combination out of 58 virtual sets of titration data, where 5 is the number of simulations per pH and 8 is the number of pH conditions. Thus, 58 pKa’s were obtained, from which an average and standard deviation were calculated (Figure S3). As expected, the bootstrap procedure gave identical pKa’s but larger errors compared to those obtained from the best fits of all five datasets (Table 1). The bootstrap errors are only somewhat larger than the standard deviations from the CPU replica-exchanges simulations,35 which is surprising, as previous work based on the continuous CpHMD in CHARMM,21,30 which uses the GBSW model38 and CHARMM22 protein force field42 as well as the discrete CpHMD in Amber, which employs the GB-OBC model43 (igb=2)31,32 and Amber99SB force field44 indicated that the single-pH model titration data are very noisy, with errors much larger than those from the replica-exchange simulations. We will continue the discussion of pKa convergence and error in the context of protein titration simulations.
Timing of the GPU-accelerated simulations.
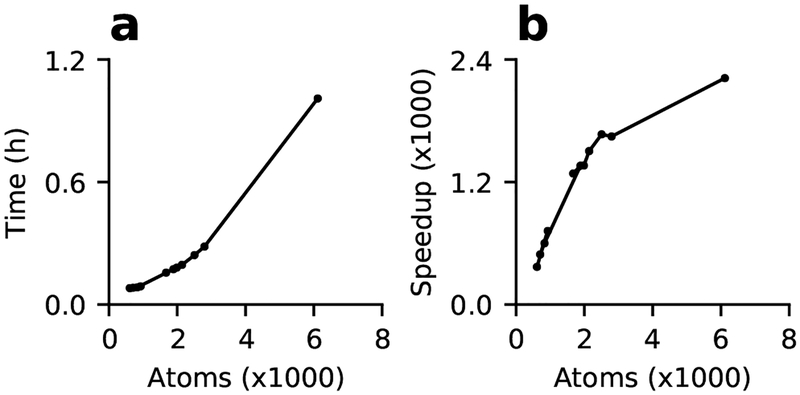
To demonstrate the feasibility of GPU CpHMD titration for routine use on a single desktop equipped with a graphics card, we examined the wall clock time of 2-ns simulations of the benchmark proteins on a Nvidia GeForce RTX 2080 graphics card. The GPU wall clock time increases with the system size, from 5 minutes for the 36-residue HP39 to 17 minutes for the 185-residue xylanase, and about one hour for the 389-residue BACE1 (Fig. 2a). Compared to a single core on the AMD Opteron 6276 processor cluster node, the wall-clock speedup on the GPU increases with system size and reaches about 2400 times. Thus, CpHMD simulations no longer require a high-performance computing cluster and are significantly more cost effective on commodity GPUs.
Figure 2: Timing of the GPU-based titration simulations.
(a) The time (in hours) required to run a 2-ns single-pH simulation on a single NVIDIA Geforce 2080 graphics card for the 11 proteins discussed in the main text. (b) GPU speedup relative to one core on the AMD Opteron 6276 processor node with 64 cores. The simulations ran on 2–8 cores and the time was extrapolated to one core.
We compared the speed of the GPU CpHMD titration with the GPU-accelerated GBSW-based continuous CpHMD21,30 in the CHARMM-OpenMM interface.45 The latter implementation was reported to output a trajectory of about 81 ns/day for SNase on a Nvidia GeForce GTX 780 Ti graphics card.45 Using our implementation, SNase ran at 250 ns/day on an RTX 2080 card. Considering the difference in single-precision floating point operations per second (FLOPS) between 780 Ti and 2080 cards (5.3 vs. 10.1) and the Amber benchmark data for GB simulations on various graphics cards (http://ambermd.org/GPUPerformance.php), we estimated the speed of our GPU implementation to be slightly faster than the implementation in the CHARMM-OpenMM interface.
Protein titration simulations: overall comparison to the replica-exchange simulations and experiment.
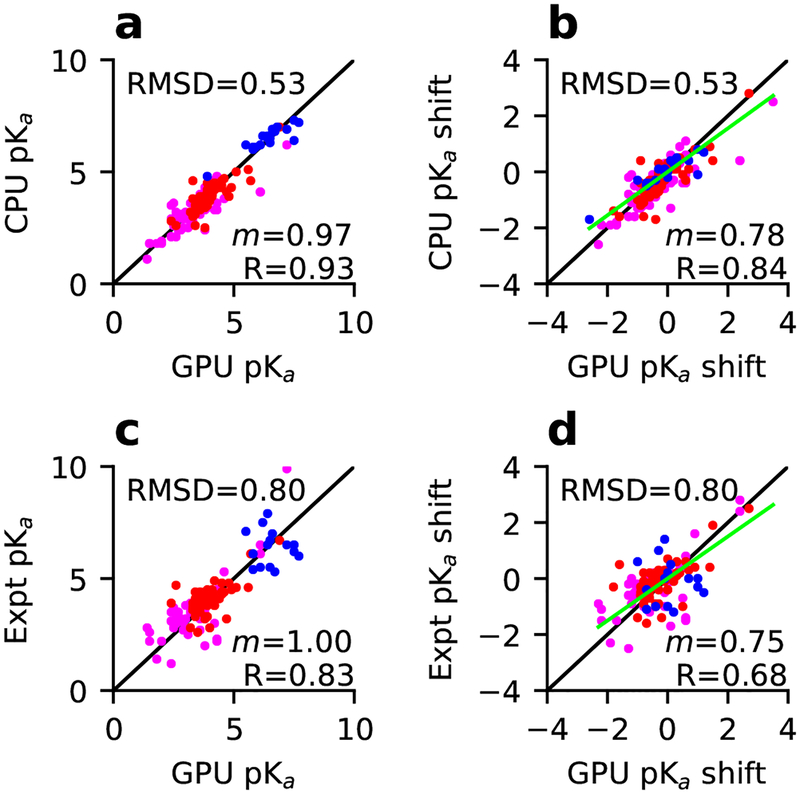
We performed single-pH titration simulations of 10 benchmark proteins, comprising 4 small proteins and 6 enzymes, on a single GPU card. Each protein was run for 2 ns at single pH values between 1 and 7.5/9.5, with a pH spacing of 0.5 units. We compared the calculated pKa’s to our previous replica-exchange simulations on CPUs,35 which used the same amount of sampling (2 ns per replica, with replicas separately by 0.5 pH units) (Table 2). Assuringly, the pKa’s are very similar, with a root-mean-square deviation (RMSD) of 0.54, linear regression slope (m) of 0.97 and Pearson’s correlation coefficient (R) of 0.93 (Fig. 3a). The comparisons for Asp, Glu, and His pKa’s are given in Table 3. In terms of the pKa shifts relative to the model values, a comparison between the GPU single-pH simulations and CPU replica-exchange simulations gives m of 0.78 and R of 0.84.
Table 2:
Protein pKa’s calculated from the GPU single-pH simulations compared to the CPU replica-exchange simulations and experimenta
| Residue | Expt | REX | Single | Residue | Expt | REX | Single | Residue | Expt | REX | Single | Residue | Expt | REX | Single |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBL | HP36 | OMTKY | HEWL | ||||||||||||
| Asp129 | 3.9 | 2.8 | 3.3(0.09) | Asp44 | 3.1 | 2.1 | 2.4(0.06) | Asp8 | 2.7 | 2.5 | 2.9(0.08) | Glu7 | 2.6 | 3.5 | 3.5(0.04) |
| Glu141 | 4.5 | 4.1 | 3.9(0.04) | Glu45 | 4.0 | 3.7 | 3.8(0.02) | Glu11 | 4.1 | 3.9 | 4.1(0.02) | His15 | 5.5 | 6.5 | 6.5(0.03) |
| His142 | 6.5 | 6.9 | 7.2(0.01) | Asp46 | 3.5 | 3.6 | 3.2(0.03) | Glu20 | 3.2 | 3.7 | 3.8(0.05) | Asp18 | 2.8 | 1.1 | 1.4(0.02) |
| Asp145 | 3.7 | 2.6 | 2.5(0.06) | Glu72 | 4.4 | 4.1 | 4.1(0.05) | Asp28 | 2.3 | 3.6 | 4.3(0.05) | Glu35 | 6.1 | 4.6 | 5.7(0.06) |
| Glu161 | 3.7 | 3.3 | 3.6(0.07) | Glu44 | 4.8 | 4.6 | 4.5(0.02) | Asp48 | 1.4 | 1.8 | 1.8(0.02) | ||||
| Asp162 | 3.2 | 3.2 | 2.6(0.06) | RNase A | His53 | 7.5 | 6.6 | 6.2(0.07) | Asp52 | 3.6 | 3.3 | 3.8(0.03) | |||
| Glu164 | 4.5 | 4.0 | 4.1(0.10) | Glu2 | 2.8 | 3.2 | 3.2(0.09) | Asp66 | 1.2 | 2.1 | 2.4(0.15) | ||||
| His166 | 5.4 | 6.0 | 5.8(0.04) | Glu9 | 4.0 | 3.4 | 3.8(0.03) | RNase H | Asp87 | 2.2 | 1.8 | 2.0(0.17) | |||
| His12 | 6.2 | 6.4 | 7.5(0.04) | Glu6 | 4.5 | 4.3 | 4.9(0.29) | Asp101 | 4.5 | 4.8 | 4.3(0.06) | ||||
| NTL9 | Asp14 | 2.0 | 2.4 | 3.8(0.08) | Asp10* | 6.1 | 4.8 | 7.1(0.18) | Asp119 | 3.5 | 2.4 | 2.7(0.10) | |||
| Asp8 | 3.0 | 2.1 | 2.6(0.04) | Asp38 | 3.5 | 2.8 | 2.6(0.08) | Glu32 | 3.6 | 3.2 | 3.3(0.09) | ||||
| Glu17 | 3.6 | 3.4 | 3.5(0.04) | His48 | 6.0 | 7.2 | 7.7(0.06) | Glu48 | 4.4 | 2.5 | 3.8(0.12) | SNase | |||
| Asp23 | 3.1 | 2.9 | 2.9(0.08) | Glu49 | 4.7 | 2.6 | 2.6(0.05) | Glu57 | 3.2 | 4.1 | 4.7(0.06) | His8 | 6.5 | 6.5 | 6.4(0.06) |
| Glu38 | 4.0 | 3.6 | 3.6(0.09) | Asp53 | 3.9 | 4.3 | 3.8(0.02) | Glu61 | 3.9 | 2.8 | 2.4(0.05) | Glu10 | 2.8 | 3.7 | 4.0(0.02) |
| Glu48 | 4.2 | 3.8 | 4.0(0.06) | Asp83 | 3.5 | 2.9 | 2.4(0.07) | His62 | 7.0 | 6.9 | 6.6(0.07) | Asp19* | 2.2 | 1.8 | 1.5(0.17) |
| Glu54 | 4.2 | 3.8 | 3.9(0.02) | Glu86 | 4.1 | 3.5 | 3.4(0.03) | Glu64 | 4.4 | 3.1 | 3.4(0.04) | Asp21* | 6.5 | 4.1 | 6.1(0.17) |
| His105 | 6.7 | 6.3 | 6.5(0.07) | Asp70* | 2.6 | 2.6 | 1.7(0.17) | Asp40 | 3.9 | 2.8 | 3.2(0.09) | ||||
| Thioredoxin | Glu111 | 3.5 | 3.5 | 3.6(0.03) | His83 | 5.5 | 6.2 | 6.1(0.05) | Glu43 | 4.3 | 3.7 | 3.7(0.10) | |||
| Glu6 | 4.8 | 3.9 | 4.8(0.11) | His119 | 6.1 | 6.1 | 5.8(0.03) | Asp94 | 3.2 | 3.2 | 3.0(0.07) | Glu52 | 3.9 | 3.9 | 3.8(0.04) |
| Glu13 | 4.4 | 4.4 | 4.5(0.03) | Asp121 | 3.1 | 3.5 | 2.4(0.02) | Asp102 | <2.0 | 3.4 | 3.5(0.15) | Glu57 | 3.5 | 3.4 | 3.5(0.02) |
| Asp16 | 4.0 | 4.0 | 4.1(0.06) | Asp108 | 3.2 | 3.1 | 3.0(0.04) | Glu67 | 3.8 | 4.5 | 4.2(0.01) | ||||
| Asp20 | 3.8 | 2.9 | 3.0(0.01) | Xylanase | His114 | <5.0 | 7.0 | 6.8(0.05) | Glu73 | 3.3 | 3.9 | 3.8(0.04) | |||
| Asp26 | 9.9 | 6.2 | 7.2(0.27) | Asp5 | 3.0 | 3.3 | 4.2(0.06) | Glu119 | 4.1 | 3.9 | 3.8(0.04) | Glu75 | 3.3 | 2.6 | 3.4(0.22) |
| His43 | n/d | 6.1 | 5.9(0.03) | Asp12 | 2.5 | 2.5 | 3.1(0.06) | His124 | 7.1 | 6.2 | 5.5(0.06) | Asp77 | <2.2 | 1.9 | 2.0(0.07) |
| Glu47 | 4.1 | 4.3 | 4.4(0.04) | Glu79 | 4.6 | 5.1 | 5.6(0.10) | His127 | 7.9 | 6.6 | 6.4(0.02) | Asp83 | <2.2 | 2.1 | 2.6(0.31) |
| Glu56 | 3.3 | 4.5 | 3.9(0.21) | Asp84 | <2.0 | 3.4 | 3.9(0.08) | Glu129 | 3.6 | 4.6 | 3.3(0.05) | Asp95 | 2.2 | 4.3 | 4.3(0.11) |
| Asp58 | 5.3 | 3.8 | 4.6(0.25) | Asp102 | <2.0 | 3.3 | 4.3(0.08) | Glu131 | 4.3 | 4.3 | 4.5(0.05) | Glu101 | 3.8 | 3.5 | 3.9(0.08) |
| Asp60 | 2.8 | 3.6 | 2.5(0.11) | Asp107 | 2.7 | 3.1 | 3.6(0.14) | Asp134 | 4.1 | 4.3 | 4.3(0.12) | His121 | 5.2 | 6.8 | 6.7(0.05) |
| Asp61 | 4.2 | 4.6 | 4.1(0.05) | Asp120 | 3.2 | 4.0 | 3.6(0.06) | Glu135 | 4.3 | 4.2 | 4.2(0.04) | Glu122 | 3.9 | 3.0 | 3.3(0.11) |
| Asp64 | 3.2 | 3.1 | 2.8(0.03) | Asp122 | 3.6 | 3.4 | 3.4(0.07) | Glu147 | 4.2 | 3.9 | 3.7(0.09) | Glu129 | 3.8 | 4.5 | 4.2(0.05) |
| Glu68 | 4.9 | 4.3 | 4.2(0.05) | His150 | <2.3 | 4.8 | 3.9(0.09) | Asp148 | <2.0 | 2.4 | 2.8(0.05) | Glu135 | 3.8 | 4.2 | 3.9(0.05) |
| Glu70 | 4.6 | 5.0 | 5.1(0.02) | His157 | 6.5 | 7.3 | 7.5(0.03) | Glu154 | 4.4 | 3.8 | 3.6(0.05) | ||||
| Glu88 | 3.7 | 3.8 | 3.3(0.04) | Glu173 | 6.7 | 7.0 | 6.9(0.13) | all | mud | 0.64 | 0.60 | ||||
| Glu95 | 4.1 | 3.5 | 3.9(0.02) | BACE1 | rmsd | 0.87 | 0.80 | ||||||||
| Glu98 | 3.9 | 3.9 | 3.6(0.07) | Asp32* | 5.2 | 4.2 | 6.6(0.25) | ||||||||
| Glu103 | 4.4 | 4.7 | 4.6(0.01) | Asp228* | 3.5 | 2.4 | <1 | ||||||||
Experimental pKa’s were taken from our previous work.35 The REX data were taken from our previous replica-exchange simulations with an exchange attempt frequency (EAF) of 2 ps−1.35 The errors in the pKa’s estimated from the least-square fits are given in parentheses. The statistics, mean unsigned deviation (mud) and root-mean-square deviation (rmsd) from the experimental values are calculated for residues with NMR determined pKa’s. As such the two BACE1 residues are excluded. The linked residues are indicated by * and their pKa’s were obtained from the best fits to the coupled titration model (Equation 1).
Figure 3: Comparison of the pKa’s and pKa shifts from the GPU single-pH simulations to the CPU replica-exchange simulations or experiment.
The GPU pKa estimates compared to (a) CPU and (b) experiment and the corresponding plots ((c) and (d)) for the pKa shifts. The model pKa’s from Thurlkill et al.41 were used for calculating the shifts. Data for Asp, Glu, and His resides are colored magenta, dark red, and blue, respectively. The diagonal (y = x) and linear regression lines are colored black and green, respectively. RMSD, regression slope (m) and Pearson’s correlation coefficient (R) are shown. The regression was performed with a zero intercept (R only changed in the second decimal point). In c and d, only pKa’s with NMR determined pKa’s are included. In d, the data for Asp26 in thioredoxin are hidden for clarity (the experimental pKa shift > 4).
Table 3:
Comparisons of Asp, Glu, and His pKa’s from the GPU single-pH simulations and CPU replica-exchange simulations and experimenta
| Res | N | maxd | mud | rmsd | R |
|---|---|---|---|---|---|
| Single vs. REX | |||||
| Asp | 48 | 2.0 | 0.49 | 0.66 | 0.84 |
| Glu | 54 | 1.3 | 0.28 | 0.42 | 0.84 |
| His | 18 | 1.1 | 0.35 | 0.44 | 0.90 |
| Single (REX) vs. experiment | |||||
| Asp | 42 | 2.7 (2.1) | 0.69 (0.75) | 0.92 (1.06) | 0.78 (0.71) |
| Glu | 54 | 2.1 (1.2) | 0.45 (0.54) | 0.61 (0.70) | 0.64 (0.53) |
| His | 18 | 1.6 (1.5) | 0.90 (0.67) | 1.04 (0.81) | −0.08 (0.16)b |
The maximum unsigned deviation (maxd), mean unsigned deviation (mud), root-mean-square deviation (rmsd), and Pearson’s correlation coefficient (R) are listed. The replica-exchange (REX) statistics for His is taken from our previous work,35 and those for Asp and Glu are recalculated. Only residues with NMR determined pKa’s are included.
See main text for discussion.
A comparison between the pKa’s calculated form the single-pH simulations and experiment yields a RMSD of 0.80, m of 1.0 and R of 0.83 (Fig. 3c). The comparisons for Asp, Glu, and His pKa’s are given in Table 3. A comparison of the corresponding pKa shifts to experiment yields m of 0.75 and R of 0.68 (Fig. 3d). These statistics are comparable to those for our previous replica-exchange simulations, which had a RMSD of 0.87, R of 0.81 for pKa’s, and R of 0.61 for pKa shifts.35 This finding is surprising, as previous work based on both continuous30 and discrete31,32 constant pH MD showed that the pKa accuracies from single-pH simulations are much lower than those from temperature or pH replica-exchange simulations. We will continue the discussion later.
Analysis of the pKa errors by residue types.
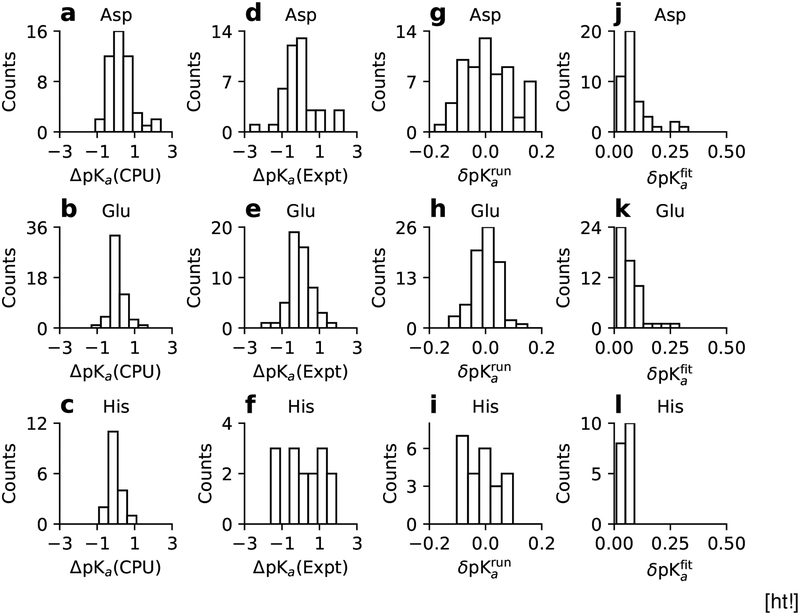
Now we examine the calculated pKa’s of Asp, Glu, and His individually by calculating the comparison statistics (Table 3) and by plotting the histograms of the pKa differences between the GPU single-pH and CPU replica-exchange simulations or experiment (Fig. 4). Assuringly, the histograms of the differences between the single-pH and replica-exchange pKa’s for Asp, Glu, and His are peaked around 0 and span a range of −1 to 1 unit, with Asp showing a few data counts between 1–2. Both the comparison statistics and histograms of the differences between the single-pH and experimental pKa’s (or pKa errors) indicate differences among the three types of residues.
Figure 4:
(a-c) Comparisons of the Asp, Glu, and His pKa’s from the GPU single-pH and CPU replica-exchange simulations(ΔpKa(CPU) = GPU pKa−CPU pKa). (d-f) Comparisons of the single-pH results and experiment (ΔpKa(Expt) = GPU pKa−Expt pKa). (g-i) Histograms of the convergence error estimates (δpKarun) obtained from the difference between the 2-ns and 1.5-ns pKa’s. Asp26 of Thioredoxin (δpKarun =−0.34) falls off the plot. (j-l) Histograms of the statistical error estimates from the pKa fitting. The linked residues, Asp19/Asp21 in SNase and Asp10/Asp79 in RNase H, are excluded in the error estimates.
The distribution of the pKa errors for His is broad and essentially flat, in contrast with those for Asp and Glu, which are narrower and peaked around 0. The rmsd of the His pKa’s with respect to experiment is 1.04, larger than the rmsd of 0.92/0.61 for Asp/Glu pKa’s. Significantly, there is essentially no correlation (correlation coefficient of −0.08 in Table 3) between the calculated and experimental pKa’s for His, while the R value for Asp/Glu is 0.78/0.64. The performance of the single-pH simulations for His pKa’s is slightly worse than the previous replica-exchange simulations (R of 0.16 and RMS error of 0.81).35 Close examination revealed that salt bridges involving His are more persistent in single-pH as compared to replica-exchange simulations. In our previous work,35 we found that the original GB-Neck2 parameters gave significantly higher pKa’s for His involved in electrostatic interactions with acidic groups. Close examination revealed overstabilization of salt bridges involving His. Therefore, we decreased the imidazole hydrogen Born radius from 1.3 to 1.17 Å, following a similar modification of the Born radii of the guanidinium hydrogens of Arg which weakened the salt-bridge strength to a similar value as in explicit solvent.36 Our modification of the His Born radius brought nearly all calculated His pKa’s down and closer to experiment; however, for partially buried His involved in strong attractive electrostatic interactions, the pKa’s remained somewhat overestimated. We suggested that the major source of error is the underestimation of desolvation energy, which results in an underestimated pKa downshift (see a detailed analysis and discussion in our previous work35). This is clearly an area that requires further testing and improvement.
Although Asp and Glu differ in only one methylene group, the histograms of the pKa errors show noticeable differences. Most of the pKa errors for Glu are within ±0.5 units and do not extend beyond ±1; however, there are several larger pKa errors (above 2 units in magnitude) for Asp and the histogram appears to be somewhat skewed to the left (more negative errors). The sidechain of Asp is shorter than that of Glu by one methylene group and as a result, the carboxylate oxygen can form a hydrogen bond with its own or a neighbor backbone amide group. These sidechain-to-backbone hydrogen bonds were observed in our simulations with both Amber and CHARMM force fields. The hydrogen bond formation or breakage perturbs the protonation state of Asp and contributes to the larger fluctuation in the model peptide titration (Table 1) and perhaps also the protein titration simulations (Fig. 4a). In the parameterization of the GB-Neck2 model,36 the strengths of the Glu· · · Lys and Glu· · · Arg salt bridges were examined; however, the strengths of the Asp· · · Lys and Asp· · · Arg salt bridges were not. As the structural differences in Asp and Glu lead to changes in the interactions formed by the sidechains, perhaps a slight adjustment is needed for the Asp parameters, which are currently the same as for Glu.
Most pKa’s are converged within 2 ns but some require longer sampling time.
To assess the convergence of protonation-state sampling and pKa’s, we examined the running estimates of the unprotonated fractions at different pH and the running estimates of the pKa values (Fig. S6). The unprotonated fractions of a majority of the 120 residues with experimental values and the corresponding pKa’s appear to stabilize within 2 ns. The convergence behavior seems comparable to the previous replica-exchange simulations,35 consistent with the model peptide titration data discussed earlier. We note that pKa, which is calculated by fitting unprotonated fractions from all pH conditions, converges more quickly than unprotonated fractions. For example, the pKa of Asp162 in BBL is converged, although the unprotonated fraction at one of the pH conditions appears to slightly increase between 1 ns and 2 ns (Fig. S6). To quantify the convergence of the calculated pKa’s, we plotted the histograms of the convergence error estimates, obtained as the pKa differences between the 2-ns and 1.5-ns pKa simulations for Asp, Glu, and His (Fig. 4g–i). The errors for Glu and His pKa’s fall in the range of ±0.1. Except Asp19/Asp21 in SNase, Asp10/Asp70 in RNase H, and Asp26 in thioredoxin, the errors for Asp pKa’s fall in the range of ±0.2. The slower convergence of the Asp pKa’s compared to Glu pKa’s is consistent with the larger deviations between the calculated and experimental pKa’s of Asp. Curiously, the unprotonated fractions of Asp19/Asp21 in SNase and Asp10/Asp70 in RNase H are not converged and their titration data do not follow a single sigmoidal trend. We will come back to the discussion of these residues.
Higher mixed fractions are correlated with insufficient sampling.
One potential drawback of continuous CpHMD is the presence of mixed states, defined here as λ values between 0.2 and 0.8. Although mixed states are necessary and their presence can accelerate sampling, they are unphysical and a high occupancy of them may lower the accuracy of a CpHMD simulation. We were puzzled that the previous replica-exchanges simulations35 showed somewhat higher mixed fractions than the continuous CpHMD simulations in CHARMM.30 In contrast, the mixed fractions in the current single-pH simulations are very low, below 10% at all pH for a majority of residues (Fig. S6), consistent with the CpHMD simulations in CHARMM with a lower replica exchange frequency30 (see later discussion). A few residues which showed a mixed fraction above 30% are: Asp26 of thioredoxin, Glu79/Asp84/His150 of xylanase, His114 of RNase H, and His12/His48 of RNaseA, and none of them displayed converged protonation-state sampling. The worst case is His150 of xylanase, which had a mixed fraction of 35 to 55% at pH below 6. His150 is deeply buried, with an experimental pKa below 2.3, compared to the estimate of 3.9 from our 2-ns simulations. 2 ns is likely too short to complete the conformational rearrangement accompanying the protonation of His150. While the present data show that the mixed fractions are correlated with insufficient sampling, it remains to be seen if they decrease in much longer trajectories.
Statistical error estimates for the calculated pKa’s.
Unlike replica-exchange titration, single-pH simulations are independent of each other and therefore the pKa error obtained in the least-square fit of one set of titration data may be a good representation of the statistical error. To test this hypothesis, we performed an additional three sets of single-pH simulations at 9 evenly spaced pH conditions between 5 and 9 for RNase A to explore the statistical errors of the His pKa’s. Table 4 lists the calculated pKa’s and least-square fitting errors based on the original set of titration data and the pKa’s and errors from the bootstrap procedure based on a total of 39 virtual sets of titration data. The calculated pKa’s deviate by 0.1 units at most and the fitting errors using one dataset are similar and highly correlated with the bootstrap errors. Although this test is not a rigorous proof, it suggests that the fitting error derived from a single set of titration simulations can be an approximate representation of the statistical error. The ease of obtaining error estimates is an advantage for single-pH over replica-exchange simulations, for which deriving error estimates is less straightforward. We list the pKa fitting errors for all titratable residues in Table 2. The corresponding histograms are given in Figure 4j–l. Except for a handful of residues with fitting errors around 0.2–0.3, the errors are smaller than 0.1, demonstrating that the statistical noise in the calculated pKa’s from the single-pH simulations is generally low.
Table 4:
Error analysis for the pKa’s of histidines in RNase A.
| Residue | Fita | Bootstrapb |
|---|---|---|
| H12 | 7.5±0.11 | 7.5±0.08 |
| H48 | 7.6±0.14 | 7.7±0.13 |
| H105 | 6.7±0.04 | 6.8±0.06 |
| H119 | 5.6±0.07 | 5.7±0.06 |
pKa’s and fitting errors based on the first set of simulations.
pKa’s and errors estimated from the bootstrap method using three sets of simulations.
Single-pH simulations could not converge microscopic pKa’s but captured macroscopic pKa’s of linked residues in SNase and RNase H.
A close examination of the residues that do not display a single sigmoidal trend, Asp19/Asp21 of SNase and Asp10/Asp70 of RNase H, revealed linked (coupled) titration, i.e., protonation of one residue is linked to the protonation of the other. As a result, the titration plot of one residue shows evidence of pH-dependent titration of the other residue and the transition from the fully protonated to the fully deprotonated persists over a very wide pH range (>4 pH units for Asp19/Asp21 in RNase and Asp10/Asp70 in RNase H, see Fig. 5 and Fig. 6). Interestingly, although the residue-specific unprotonated state fractions of of these linked residues are not converged (Fig. S6), the convergence of the total unprotonated fractions of the two linked residues is much better (Fig. S4 and S5).
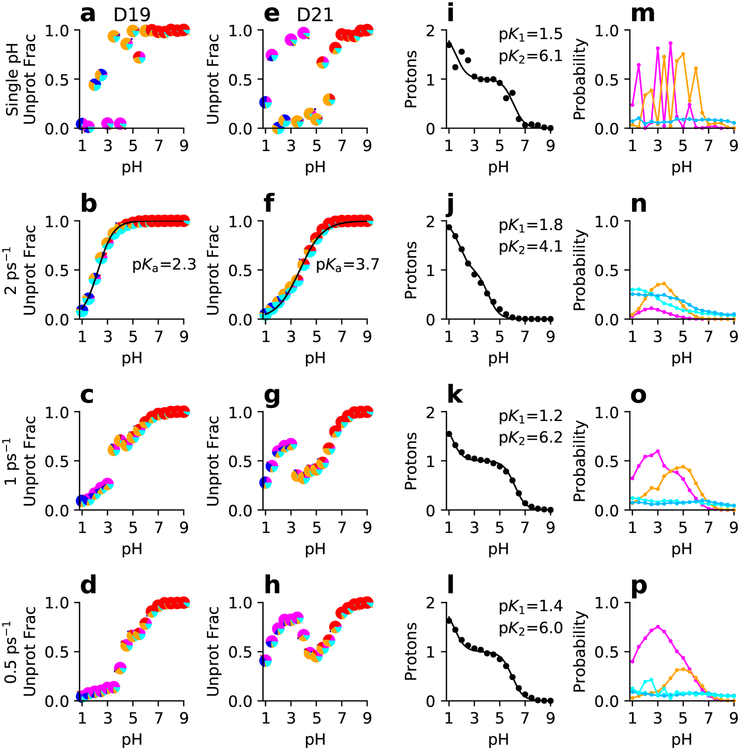
Figure 5: Titration data of the linked residues D19 and D21 in SNase from single-pH and replica-exchange simulations.
From top to bottom rows, single-pH and replica-exchange simulations with different exchange attempt frequencies are shown. (a-h) Unprotonated fraction as a function of pH and best fit to the residue-specific HH equation for D19 (a-d) and D21 (e-h). Each data point is a pie chart showing the probability of the four protonation states: D19H/D21H (blue), D19−/D21− (red), D19H/D21− (magenta), and D19(−)/D21H (orange). Cyan indicates that one of the residues is in a mixed state. (i-l) Total number of protons as a function of pH and best fit to the linked-titration model. (m-p) Probabilities of the singly-protonated (magenta for D19H/D21− and orange for D19−/D21H) and mixed states (light cyan for D19 titration and dark cyan for D21 titration) as functions of pH. The macroscopic experimental pKa’s are 2.2 and 6.5 for D19 and D21, respectively.
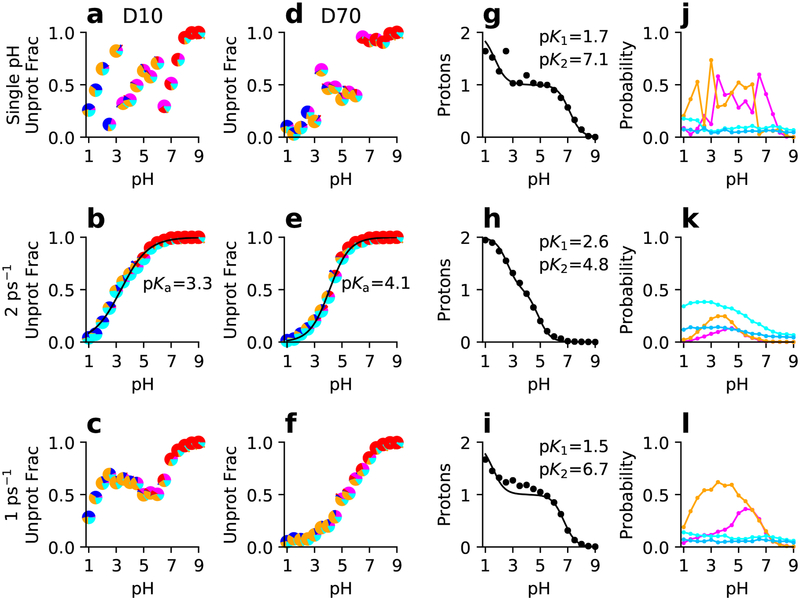
Figure 6: Simulated titration of the linked residues D10 and D70 in RNase H from single-pH and replica-exchange simulations.
From top to bottom rows, single-pH and replica-exchange simulations with different exchange attempt frequencies are shown. (a-f) Unprotonated fraction as a function of pH and best fit to the residue-specific HH equation for D10 (a-c) and D70 (d-f). Each data point is a pie chart showing the probability of the four protonation states: D10H/D70H (blue), D10−/D70− (red), D10H/D70− (magenta), and D10(−)/D70H (orange). Cyan indicates that one of the residues is in a mixed state. (g-i) Total number of protons as a function of pH and best fit to the linked-titration model. (j-l) Probabilities of the singly-protonated (magenta for D10H/D70− and orange for D10−/D70H) and mixed states (light cyan for D10 titration and dark cyan for D70 titration) as functions of pH. The experimental macroscopic pKa’s are 6.1 and 2.6 for D10 and D70, respectively.
For linked titration, the residue-specific microscopic pKa’s cannot be experimentally determined. Rather, the signals, e.g., NMR chemical shifts, are fit to a coupled two-proton model, which results in two macroscopic pKa’s representing the stepwise titration events.46 To obtain macroscopic pKa’s from simulations, the average number of protons bound to the two residues (Nprot) at different pH values can be fit to an analogous statistical mechanics model,24,47,48
| (1) |
where the denominator is the partition function, and the fitting parameters, pK1 and pK2, are the macroscopic pKa’s. The resulting macroscopic pKa’s for the linked residues in SNase are 1.5 and 6.1 (Fig. 5i) and for those in RNase H are 1.7 and 7.1 (Fig. 6i). Note, linked titration events can also be studied by a decoupled-site model representing the sum of two HH equations.49,50
To identify which residue should be assigned the higher pKa, we can observe which residue becomes protonated first as pH decreases. For SNase, since the occupancy of Asp19−/Asp21H is higher than Asp19H/Asp21− at pH above 4.5, i.e., Asp21 protonates first (Fig. 5m), Asp21 has the higher pKa. Thus, we assigned Asp19 and Asp21 the pKa’s of 1.5 and 6.1, in excellent agreement with the respective experimental assignment of 2.2 and 6.5. Similarly for RNase H, since the occupancy of Asp10H/Asp70− is higher than Asp10−/Asp70H at pH above 6 (Fig. 5j), we assigned Asp10 and Asp70 the pKa’s of 7.1 and 1.7, in good agreement with the respective experimental assignment of 6.1 and 2.6.
Comparison of single-pH and replica-exchange simulations with an exchange attempt frequency of 2 ps−1.
Interestingly, the residue-specific titration data of Asp19/Asp21 in SNase (Fig. 5b and f) and Asp10/Asp70 in RNase H (Fig. 5b and e) from our previous replica-exchange simulations did not offer noticeable evidence of coupling. A comparison to the single-pH titration data reveals that the occupancies of the singly protonated states are much lower while those of the mixed states are much higher, up to 30% for SNase and up to 45% for RNase H in the pH range where two singly protonated states seem to compete in the single-pH simulations. The decreased amount of coupling seen in the replica-exchange simulations is consistent with the underestimation of the higher pKa and the splitting between the low and high pKa observed in experiment. Since the fraction of mixed states was much higher in the replica-exchange compared to single-pH simulations, we hypothesized that the underestimation of pKa splitting could be due to insufficient sampling time for the conformation to relax to the specific pH conditions in the replica-exchange simulations. In these simulations,35 the exchange attempt frequency (EAF) was every 250 MD steps (or 2 ps−1), rather than every 500 (1 ps−1) or 1000 MD steps (0.5 ps−1), as used in our previous CHARMM-based CpHMD.30 Our decision to increase EAF was inspired by the reported observation that high EAF improved pKa estimates in the discrete-CpHMD simulations in Amber.32 We will come back to this discussion in Concluding Remarks.
A high frequency of exchange attempts may not allow sufficient conformational relaxation.
To test if the lack of conformational relaxation is responsible for the decreased amount of coupling in our previous replica-exchange simulations, we performed additional replica-exchange simulations with EAFs of 1 ps−1 and 0.5 ps−1 for SNase and EAF of1 ps−1 for RNase H. As seen from Fig. 5 and 6, at the lower EAFs, the occupancies of the singly protonated states increase to similar levels as in the single-pH simulations, while the mixed fractions decrease to below 10% as in the single-pH simulations. Importantly, the higher macroscopic pKa’s increase by about 2 units, to similar values as from the single-pH simulations of SNase and RNase H. The increased amount of coupling is also reflected in the noisier residue-specific titration data, which no longer conform to the one-proton HH equation. Additionally, the solvent-accessible surface areas (SASA) and number of hydrogen bonds of the two residues predicted by simulations with the lower EAFs of 1 and 0.5 ps−1 agree more closely with the single-pH results than the simulations with the high EAF of 2 ps−1 (Fig. S1 and Fig. S2), which offers a direct piece of evidence for the insufficient conformational relaxation for the coupled residue pairs with the EAF of 2 ps−1. Interestingly, for both proteins, the nucleophile (residue with the lower pKa) has a larger SASA and number of hydrogen bonds compared to the proton donor (higher pKa), in agreement with the findings from CpHMD simulations using the CHARMM force field and the GBSW model.51
Having demonstrated that a high EAF may lead to insufficient conformational relaxation for linked residues, we wondered if the pKa’s of other independent residues are affected as well. A comparison of the pKa’s from the SNase simulations with different EAFs and single pH values (no exchange) showed that, although the differences are much smaller than for the couple residues, the pKa’s from the two slower exchange (EAF of 1 and 0.5 ps−1) simulations agree with each other and with the single-pH simulations much better as compared to the fast exchange (EAF of 2 ps−1) simulations (Table S1). A similar trend is observed for the RNase H simulations (Table S1). These data are consistent with the findings in regards to the linked titration and support the notion that an EAF of 2 ps−1 may not allow sufficient conformational relaxation. In other words, the relaxation time for adapting to changes of protonation states is likely above 0.5 ps for the tested proteins.
Hydrogen-bonded dyad in BACE1 requires enhanced sampling or significantly longer simulations.
To further test the ability of single-pH simulations to quantitatively reproduce the experimental apparent pKa’s of coupled sites in enzymes, we performed titration simulations of BACE1, which is more than three times as large as HEWL, a frequently used benchmark protein for pKa calculations,16,32 and more than twice as large as xylanase, the largest protein among the 10 benchmark proteins studied here. Most importantly, the aspartyl dyad of BACE1 (Asp32 and Asp228) is subject to much stronger hydrogen bonding than the linked residues in SNase and RNase H.51 Our previous replica-exchange simulations with an EAF of 2 ps−1 35 and the hybrid-solvent continuous CpHMD in CHARMM12 correctly predicted the pKa order of the dyad but underestimated the two macroscopic pKa’s and magnitude of their splitting. The current single-pH simulations also reproduced the experimental pKa order; however, compared to the experimental data, the macroscopic pKa assigned to Asp32 is too high by 1.4 units, and Asp228, which is the dyad residue with the lower pKa did not protonate even at pH 1 (the lowest simulation pH). The latter may be attributed to a persistent hydrogen bond between the carboxylate oxygens of the dyad, whereby Asp228 was exclusively acting as a hydrogen bond acceptor (data not shown). The overestimation of the higher pKa and underestimation of the lower pKa, i.e, overestimation of the pKa splitting, is consistent with the single pH simulations of the linked titration in SNase and RNase H. To attempt to break the hydrogen bond, we prolonged the single-pH simulations to 10 ns; however, while the pKa of Asp32 remained largely unchanged, Asp228 remained deprotonated in the entire pH range (Figure S6). This data suggests that for buried, linked residues with strong hydrogen bonding, pH replica-exchange or significantly longer simulation length is required. Detailed exploration of linked titration involving strong hydrogen bonds is a topic of our future work.
Comparison of the 2-ns and 10-ns single-pH simulations of BACE1.
Although no experimental titration data are available for BACE1 except for the dyad pKa’s, comparison of the 2-ns and 10-ns single-pH simulations may allow us to further assess the convergence behavior of pKa calculations for larger proteins. Out of a total of 50 titratable residues, the majority showed negligible pKa changes when extending the simulations from 2 to 10 ns; however, 10 residues showed pKa changes of 0.5–1.5 units (Table S1). The second largest change is for Asp83; its pKa increased from 2.2 based on the 2-ns simulations to 3.4 based on the 10-ns simulations. The latter is closer to the pKa of 5.3 from the replica-exchange simulations (Table S1). Trajectory analysis revealed that in the first 2 ns, Asp83 sometimes formed a salt bridge with Arg96 at pH above 3, and as the simulation continued, the salt bridge became less frequently sampled. This trend continued throughout the 10-ns simulations, which explains why the calculated pKa shows an increasing trend and it is not converged at 10 ns (Figure S6). This data suggests that for Asp83, a single-pH simulation requires longer than 10 ns to converge the pKa, and replica-exchange simulations are effective in breaking the salt bridge and accelerating convergence.
The largest change is for His360, which has a pKa of 4.7 based on the 2-ns simulations and the pKa increased by 1.5 units to 6.2 based on the 10-ns simulations. Trajectory analysis showed that although starting out as largely buried, His360 became exposed to solvent in the prolonged simulations, which resulted in the pKa increase. Curiously, the replica-exchange simulations gave a pKa of 4.8, similar to the 2-ns single pH simulations. We hypothesize that His360 did not have sufficient time to become solvent exposed at low pH conditions due to the frequent exchange attempts, and that is why the calculated pKa is similar to that from the 2-ns simulations. Another question is whether His360 becoming exposed to solvent is realistic. Our extensive replica-exchange simulations based on a hybrid-solvent CpHMD method in CHARMM indicated that this is not the case.10,12 Although the GB-Neck2 model has been demonstrated to enable accurate folding of a set of small proteins,36 explicit solvent representation remains more accurate and yields pKa’s in better agreement with experiment.18,22 Thus, prolonged CpHMD simulations may worsen the pKa calculations as the conformational states deviate further from the crystal structure, as demonstrated by a previous discrete CpHMD simulation.18 Future work may shed more light on these issues.
CONCLUDING REMARKS
We presented and validated a GPU implementation of the generalized Born continuous CpHMD in the Amber pmemd.cuda program. A 2-ns single-pH simulation of a 400-residue protein runs in about 1 hour on a single NVIDIA Geforce 2080 graphics card, which represents a three-orders-of-magnitude speedup compared to the wall-clock time on a single CPU core of a high-performance computing cluster node (AMD Opteron 6276). Most calculated pKa’s of Asp, Glu, and His sidechains of the 10 proteins from 2-ns single-pH simulations were converged and in close agreement (RMSD of 0.54) with our previous replica-exchange simulations on CPUs.35 Compared to the experimental pKa’s, the single-pH simulations gave a RMSD of 0.80 units, which is slightly lower than the RMSD of 0.87 from our previous replica-exchange simulations. Surprisingly, the errors in the protein pKa’s from the single-pH simulations were generally small (below 0.2 units). This finding is in stark contrast to the reported data from the continuous CpHMD in CHARMM21,30,52 and the discrete CpHMD in Amber,31,32 which found that errors from single-pH simulations were much larger than those from replica-exchange simulations. Perhaps the most unexpected finding is that the single-pH simulations were able to correctly predict the pKa order of the linked residues in SNase and RNase H and reproduce the experimental macroscopic pKa’s to about 1 pH units, although the pKa splitting was overestimated. By contrast, our previous replica-exchange simulations, which used an exchange attempt frequency (EAF) of 2 ps−1 35 underestimated the pKa splitting and gave a significant amount of mixed states which were largely absent in the single-pH simulations. Additional replica-exchange simulations with less frequent exchange attempts gave similar pKa’s and conformational behavior as the single-pH simulations, suggesting that an EAF of 2 ps−1 may not allow sufficient conformational relaxation at specific pH conditions. A more extensive study will be carried out in the future to further investigate the topic.
We suggest that the above finding does not contradict a previous study32 using the GB-OBC43 based discrete CpHMD, which found that an EAF of 50 ps−1 significantly improved the pKa of a salt-bridged residue (Asp66 in HEWL), which did not titrate with an EAF of 0.5 ps−1. While our single-pH simulations did not encounter this issue (Table 2), likely because salt bridges are significantly weakened in the newer GB-Neck2 model,36 we suggest that conformational relaxation time for buried residues, such as the aforementioned linked pairs, is much longer than solvent-exposed ones. As a result, too frequent exchanges may not allow internal residues to adjust themselves and the local environment to a change in pH. This may explain why the pKa’s of buried residues are more sensitive to the EAF, while pKa’s of solvent-exposed residues are not. For example, a deeply buried Asp26 in thioredoxin has a calculated pKa of 6.2 from replica-exchange and 7.2 from single-pH simulations. The latter is in better agreement with the experimental pKa of 9.9.
It is indisputable that enhanced sampling in either temperature30,31 or pH space22,32 accelerates protonation-state sampling and thereby the convergence of calculated pKa’s. The analysis of the pKa results from the 2-ns, 10-ns single-pH and replica-exchange simulations of BACE1 confirmed this point and further explored the topic of replica-exchange frequency. Although the pKa order of the catalytic dyad, Asp32/Asp228, was correctly predicted, Asp228 did not protonate at pH 1 (the lowest simulation pH), due to a persistent hydrogen bond, which requires enhanced sampling or significantly longer sampling time to break. As a result, the extent of coupling and pKa splitting were overestimated. By contrast, the extent of coupling and pKa splitting were underestimated in the replica-exchange simulations with an EAF of 2 ps−1. Thus, our future work will explore the usage of either much longer single-pH simulations or an optimum EAF to make the most accurate and efficient predictions of pKa’s for tough cases, i.e., buried, salt-bridged, and linked titratable residues.
A most recent study53 demonstrated the capability of replica-exchange GBNeck2-CpHMD for accurate prediction of downshifted Cys and Lys pKa’s. Work is underway to benchmark the performance using single-pH simulations. Another area of interest is the improvement of His pKa’s, which have the largest errors in the current dataset. Although there remains much room for improvement, our present data are encouraging and demonstrate that single-pH simulations may be routinely performed on a desktop computer equipped with a single GPU card for a variety of applications, from assisting MD stimulations with protonation state assignment to offering pH-dependent corrections of binding free energies10 and nucleophilic hot spots for covalent drug design.53
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge National Institutes of Health (R01GM098818 and R01GM118772) and National Science Foundation (CBET143595) for funding.
Footnotes
SOFTWARE AVAILABILITY
The software will be distributed as a part of the Amber molecular dynamics package (Amber20).
Supporting Information Available
A supplemental table, additional analysis for linked titration, and residue-specific titration plots as well as convergence analyses for all residues of the 11 proteins are included.
References
- (1).Frenkel D; Smit B Understanding Molecular Simulation, 2nd ed.; Academic Press: London, 2002. [Google Scholar]
- (2).Durrant JD; McCammon JA Molecular Dynamics Simulations and Drug Discovery. BMC Biol. 2011, 9, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Søndergaard CR; Mats HM Olsson MR; Jensen JH Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput 2011, 7, 2284–2295. [DOI] [PubMed] [Google Scholar]
- (4).Baker NA; Sept D; Joseph S; Holst MJ; McCammon JA Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Anandakrishnan R; Aguilar B; Onufriev AV H++ 3.0: Automating pK Prediction and the Preparation of Biomolecular Structures for Atomistic Molecular Modeling and Simulations. Nucleic Acids Res 2012, 40, W537–W541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang L; Zhang M; Alexov E DelPhiPKa Web Server: Predicting pKa of Proteins, RNAs and DNAs. Bioinformatics 2016, 32, 614–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Khandogin J; Chen J; Brooks III CL Exploring Atomistic Details of pH-Dependent Peptide Folding. Proc. Natl. Acad. Sci. USA 2006, 103, 18546–18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yue Z; Shen J pH-Dependent Cooperativity and Existence of a Dry Molten Globule in the Folding of a Miniprotein BBL. Phys. Chem. Chem. Phys 2018, 20, 3523–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ellis CR; Tsai C-C; Hou X; Shen J Constant pH Molecular Dynamics Reveals pH-Modulated Binding of Two Small-Molecule BACE1 Inhibitors. J. Phys. Chem. Lett 2016, 7, 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Harris RC; Tsai C-C; Ellis CR; Shen J Proton-Coupled Conformational Allostery Modulates the Inhibitor Selectivity for β-Secretase. J. Phys. Chem. Lett 2017, 8, 4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Henderson JA; Harris RC; Tsai C-C; Shen J How Ligand Protonation State Controls Water in Protein-Ligand Binding. J. Phys. Chem. Lett 2018, 9, 5440–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ellis CR; Shen J pH-Dependent Population Shift Regulates BACE1 Activity and Inhibition. J. Am. Chem. Soc 2015, 137, 9543–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Huang Y; Chen W; Dotson DL; Beckstein O; Shen J Mechanism of pH-Dependent Activation of the Sodium-Proton Antiporter NhaA. Nat. Commun 2016, 7, 12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yue Z; Chen W; Zgurskaya HI; Shen J Constant pH Molecular Dynamics Reveals How Proton Release Drives the Conformational Transition of a Transmembrane Efflux Pump. J. Chem. Theory Comput 2017, 13, 6405–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Baptista AM; Teixeira VH; Soares CM Constant-pH Molecular Dynamics using Stochastic Titration. J. Chem. Phys 2002, 117, 4184–4200. [Google Scholar]
- (16).Mongan J; Case DA; McCammon JA Constant pH Molecular Dynamics in Generalized Born Implicit Solvent. J. Comput. Chem 2004, 25, 2038–2048. [DOI] [PubMed] [Google Scholar]
- (17).Lee MS; Salsbury FR Jr.; Brooks CL III Constant-pH Molecular Dynamics using Continuous Titration Coordinates. Proteins 2004, 56, 738–752. [DOI] [PubMed] [Google Scholar]
- (18).Swails JM; York DM; Roitberg AE Constant pH Replica Exchange Molecular Dynamics in Explicit Solvent Using Discrete Protonation States: Implementation, Testing, and Validation. J. Chem. Theory Comput 2014, 10, 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lee J; Miller BT; Damjanović A; Brooks BR Constant pH Molecular Dynamics in Explicit Solvent with Enveloping Distribution Sampling and Hamiltonian Exchange. J. Chem. Theory Comput 2014, 10, 2738–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Radak BK; Chipot C; Suh D; Jo S; Jiang W; Phillips JC; Schulten K; Roux B Constant-pH Molecular Dynamics Simulations for Large Biomolecular Systems. J. Chem. Theory Comput 2017, 13, 5933–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Khandogin J; Brooks CL III Constant pH Molecular Dynamics with Proton Tautomerism. Biophys.J 2005, 89, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wallace JA; Shen JK Continuous constant pH Molecular Dynamics in Explicit Solvent with pH-Based Replica Exchange. J. Chem. Theory Comput 2011, 7, 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Donnini S; Tegeler F; Groenhof G; Grubmüller H Constant pH Molecular Dynamics in Explicit Solvent with λ-Dynamics. J. Chem. Theory Comput 2011, 7, 1962–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wallace JA; Shen JK Charge-Leveling and Proper Treatment of Long-Range Electrostatics in All-Atom Molecular Dynamics at Constant pH. J. Chem. Phys 2012, 137, 184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Goh GB; Knight JL; Brooks III CL Constant pH Molecular Dynamics Simulations of Nucleic Acids in Explicit Solvent. J. Chem. Theory Comput 2012, 8, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Huang Y; Chen W; Wallace JA; Shen J All-Atom Continuous Constant pH Molecular Dynamics With Particle Mesh Ewald and Titratable Water. J. Chem. Theory Comput 2016, 12, 5411–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kong X; Brooks CL III λ-dynamics: A New Approach to Free Energy Calculations. J. Chem. Phys 1996, 105, 2414–2423. [Google Scholar]
- (28).Wallace JA; Shen JK Predicting pKa Values with Continuous Constant pH Molecular Dynamics. Methods Enzymol 2009, 466, 455–475. [DOI] [PubMed] [Google Scholar]
- (29).Chen W; Morrow BH; Shi C; Shen JK Recent Development and Application of Constant pH Molecular Dynamics. Mol. Simul 2014, 40, 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Khandogin J; Brooks CL III Toward the Accurate First-principles Prediction of Ionization Equilibria in Proteins. Biochemistry 2006, 45, 9363–9373. [DOI] [PubMed] [Google Scholar]
- (31).Meng Y; Roitberg AE Constant pH Replica Exchange Molecular Dynamics in Biomolecules Using a Discrete Protonation Model. J. Chem. Theory Comput 2010, 6, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Swails JM; Roitberg AE Enhancing Conformation and Protonation State Sampling of Hen Egg White Lysozyme Using pH Replica Exchange Molecular Dynamics. J. Chem. Theory Comput 2012, 8, 4393–4404. [DOI] [PubMed] [Google Scholar]
- (33).Götz AW; Williamson MJ; Xu D; Poole D; Grand SL; Walker RC Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput 2012, 8, 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Case DA; Ben-Shalom IY; Brozell SR; Cerutti DS; Cheatham T III; Cruzeiro VWD; Darden TA; Duke RE; Ghoreishi D; Gilson MK; Gohlke H; Goetz AW; Greene D; Harris R; Homeyer N; Izadi S; Kovalenko A; Kurtzman T; Lee TS; LeGrand S; Li P; Lin C; Liu J; Luchko T; Luo R; Mermelstein DJ; Merz KM; Miao Y; Monard G; Nguyen C; Nguyen H; Omelyan I; Onufriev A; Pan F; Qi R; Roe DR; Roitberg A; Sagui C; Schott-Verdugo S; Shen J; Simmerling CL; Smith J; Salomon-Ferrer R; Swails J; Walker RC; Wang J; Wei H; Wolf RM; Wu X; Xiao L; York DM; Kollman PA AMBER 2018 2018. [Google Scholar]
- (35).Huang Y; Harris RC; Shen J Generalized Born Based Continuous Constant pH Molecular Dynamics in Amber: Implementation, Benchmarking and Analysis. J. Chem. Inf. Model 2018, 58, 1372–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Nguyen H; Roe DR; Simmerling C Improved Generalized Born Solvent Model Parameters for Protein Simulations. J. Chem. Theory Comput 2013, 9, 2020–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Brooks BR; Brooks CL III; Mackerell AD Jr.; Nilsson L; Petrella RJ; Roux B; Won Y; Archontis G; Bartels C; Boresch S; Caflisch A; Caves L; Cui Q; Dinner AR; Feig M; Fischer S; Gao J; Hodoscek M; Im W; Kuczera K; Lazaridis T; Ma J; Ovchinnikov V; Paci E; Pastor RW; Post CB; Pu JZ; Schaefer M; Tidor B; Venable RM; Woodcock HL; Wu X; Yang W; York DM; Karplus M CHARMM: the biomolecular simulation program. J. Comput. Chem 2009, 30, 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Im W; Lee MS; Brooks CL III Generalized Born Model with a Simple Smoothing function. J. Comput. Chem 2003, 24, 1691–1702. [DOI] [PubMed] [Google Scholar]
- (39).Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ryckaert JP; Ciccotti G; Berendsen HJC Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- (41).Thurlkill RL; Grimsley GR; Scholtz JM; Pace CN pK Values of the Ionizable Groups of Proteins. Protein Sci 2006, 15, 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).MacKerell AD Jr.; Bashford D.; Bellott M; Dunbrack RL Jr.; Evanseck JD; Field MJ; Fischer S; Gao J; Guo H; Ha S; Joseph-McCarthy D; Kuchnir L; Kuczera K; Lau FTK; Mattos C; Michnick S; Ngo T; Nguyen DT; Prodhom B; Reiher WE III; Roux B; Schlenkrich M; Smith JC; Stote R; Straub J; Watanabe M; Wiórkiewicz-Kuczera J; Yin D; Karplus M All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- (43).Onufriev A; Bashford D; Case A, Exploring D Protein Native States and Large-Scale Conformational Changes with a Modified Generalized Born model. Proteins 2004, 55, 383–394. [DOI] [PubMed] [Google Scholar]
- (44).Hornak V; Okur A; Rizzo RC; Simmerling C HIV-1 protease flaps Spontaneously Open and Reclose in Molecular Dynamics Simulations. Proc. Natl. Acad. Sci. USA 2006, 103, 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Arthur EJ; B. CL III Efficient Implementation of Constant pH Molecular Dynamics on Modern Graphics Processors. J. Comput. Chem 2016, 37, 2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Castañeda CA; Fitch CA; Majumdar A; Khangulov V; Schlessman JL; García-Moreno E, Molecular B Determinants of the pKa Values of Asp and Glu residues in Staphylococcal Nuclease. Proteins 2009, 77, 570–588. [DOI] [PubMed] [Google Scholar]
- (47).Ullmann GM Relations Between Protonation Constants and Titration Curves in Polyprotic Acids: a Critical View. J. Phys. Chem. B 2003, 107, 1263–1271. [Google Scholar]
- (48).Ellis CR; Tsai C-C; Lin F-Y; Shen J Conformational Dynamics of Cathepsin D and Binding to a Small-molecule BACE1 Inhibitor. J. Comput. Chem 2017, 38, 1260–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Onufriev A; Case DA; Ullmann GM A Novel View of pH Titration in Biomolecules. Biochemistry 2001, 40, 3413–3419. [DOI] [PubMed] [Google Scholar]
- (50).Webb H; Tynan-Connolly BM; Lee GM; Farrell D; O’Meara F; Søndergaard CR; Teilum K; Hewage C; McIntosh LP; Nielsen JE Remeasuring HEWL pKa Values by NMR Spectroscopy: Methods, Analysis, Accuracy, and Implications for Theoretical pKa Calculations. Proteins 2011, 79, 685–702. [DOI] [PubMed] [Google Scholar]
- (51).Huang Y; Yue Z; Tsai C-C; Henderson JA; Shen J Predicting Catalytic Proton Donors and Nucleophiles in Enzymes: How Adding Dynamics Helps Elucidate the Structure-Function Relationships. J. Phys. Chem. Lett 2018, 9, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Goh GB; Hulbert BS; Zhou H; Brooks CL III Constant pH Molecular Dynamics of Proteins in Explicit Solvent with Proton Tautomerism. Proteins 2014, 82, 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Liu R; Yue Z; Tsai C-C; Shen J Assessing Lysine and Cysteine Reactivities for Designing Targeted Covalent Kinase Inhibitors. J. Am. Chem. Soc 2019, 141, 6553–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.