Abstract
Myocardial hypertrophy is the result of sustained perturbations to the mechanical and/or neurohormonal homeostasis of cardiac cells and is driven by integrated, multiscale biophysical and biochemical processes that are currently not well defined. In this brief review, we highlight recent computational and experimental models of cardiac hypertrophy that span mechanisms from the molecular level to the tissue level. Specifically, we focus on: (i) molecular-level models of the structural dynamics of sarcomere proteins in hypertrophic hearts, (ii) cellular-level models of excitation-contraction coupling and mechanosensitive signaling in disease-state myocytes, and (iii) organ-level models of myocardial growth kinematics and predictors thereof. Finally, we discuss how spanning these scales and combining multiple experimental/computational models will provide new information about the processes governing hypertrophy and potential methods to prevent or reverse them.
Keywords: Multiscale Modeling, Cardiac Myofilament, Sarcomeric Point Mutations, Hypertrophic Cardiomyopathy
Introduction
Ventricular hypertrophy develops in response to environmental demands that alter the mechanical and biochemical loading on the myocardium, and it often increases the risk of heart failure [1]. The progression of remodeling is mediated by a combination of tissue-, cellular-, and molecular-level regulatory processes involving the mechanobiology of cardiomyocytes, the surrounding extracellular matrix (ECM), and cardiac fibroblasts. Thus, understanding the multiscale mechanisms involved in cardiac hypertrophy is a major challenge with the inherent need for a multiscale approach.
Perturbations in the mechanical homeostasis of cardiomyocytes that trigger a hypertrophic signaling response range from chronic systemic hypertension, aortic stenosis, ischemia, and genetic cardiac disorders. In particular, hypertrophic and dilated cardiomyopathy (HCM and DCM, respectively) are typically associated with genetic mutations related to the contractility of the myocardium. HCM and DCM manifest as anatomical and histological changes as well as a wide range of clinical manifestations. For instance, the overall cardiac mass in both HCM and DCM is increased due to ventricular growth, whereas the left ventricular chamber volume is diminished substantially in HCM and increased in DCM. HCM hearts typically display diastolic dysfunction due to thicker and stiffer ventricular walls, while DCM hearts typically have systolic dysfunction due to weak dilated ventricular walls. Histopathologically, the alignment of myocytes which characterizes the normal myocardium becomes distorted by the hypertrophic growth, which can produce enlarged and misshapen myocytes as well as disorientation of adjacent cells. These findings, collectively termed myocyte disarray, can be focal and juxtaposed beside normal-appearing myocardium or can be widespread throughout the ventricle. Cardiac fibroblast activation and remodeling of the associated ECM further contribute to the distortion in myocardial cell architecture
Thus, major efforts have been made to understand the multiscale mechanisms by which HCM and DCM develop with the ultimate goal of advancing preventative or restorative therapeutic discovery. In the following sections, we highlight computational models and experimental studies that have revealed important biophysical and biomechanical underpinnings of cardiac hypertrophy, and we speculate on the types of combinatorial modeling approaches that will be most informative in future studies.
Structural dynamics of the sarcomere in HCM hearts
Molecular and Brownian dynamics simulations of sarcomere proteins
Molecular dynamics (MD) simulations can be a powerful tool for exploring the effects of contractile protein mutations as altered atomic interactions cascade through the molecule and affect overall contractile mechanisms. By studying HCM mutations through the computational lens of MD, it is possible to answer fundamental questions regarding nanosecond-scale alterations to protein electrostatics and atomic interactions which can be combined with higher-order models and experiments to predict long term phenotypes.
A number of myosin binding protein C (cMyBPC) mutations have been associated with HCM development. An MD study of HCM-associated cMyBPC mutation Y235S demonstrated that the mutation critically alters intermolecular contacts and the electrostatic surface region within the C1 domain which is important for myosin interactions [7]*. These results were combined with mechanical myofilament experiments to suggest that the mutation leads to increased crossbridge attachment rates and overall increased calcium sensitivity, leading to hypercontractile behavior of the muscle as a whole. Another study which investigated C1 domain mutation E258K and C2 domain mutation E441K, both as single mutations and a double mutation, demonstrated that these mutations increase hydrogen binding in the C1-m-C2 complex of the molecule, leading to a more rigid overall structure and slower unbinding of the complex compared to wildtype simulations [8]. Overall electrostatic potential and phosphorylation site changes due to these mutations, and exaggerated in the double mutation, were also predicted to affect the actin binding mechanism of cMyBPC, which may result in declined diastolic function [9]. An earlier study of the C1 domain in isolation investigating mutations R177H, A216T and E258K demonstrates the diverse changes to hydrogen bonding and electrostatic surfaces that can occur due to different mutations associated with the same hypertrophic end stage phenotype [10].
Troponin I (TnI) mutations have also been associated with HCM. An MD study of inhibitory peptide mutation R146G and N-teminus mutation R21C found that these mutations strengthen interactions between the TnI switch peptide and Troponin C (TnC) by hindering the ability for PKA to phosphorylate Ser-23/Ser-24 in TnI [11]. TnI I-T arm mutation P83S has also been shown to stabilize the Ca2+ binding region of TnC and improve flexibility of TnC as a whole, which may lead to increased Ca2+ in mechanical experiments of mutated myofilaments [12]. A more recent study on N-terminal TnC mutations A8V, L29Q, and A31S used umbrella sampling to demonstrate that all three mutations cause lower free energy of hydrophobic patch opening of TnC, which precedes TnC-TnI interaction, leading to faster thin filament activation and higher Ca2+ sensitivity [13]**.
An ambitious study modelled a full-length Tropomyosin (Tin) dimer in complex with 15 actin monomer to explore HCM-associated Tin mutations E62Q, A63V, K70T, V95A, D175N, E180G, L185R, and E192K [14]*. 20-ns simulations found that these mutations weaken Tm-actin interactions in the Tin Blocked (inactivated) state, potentially favoring the transition of Tm into the Closed and Open states required for activation of the thin filament [14]. A 30 ns study of Tm mutation E62Q strengthens the argument for reduced Tm-actin affinity in the Blocked state by finding a loss of negative charges on the actin-binding surface of Tm and a movement of Tm away from the actin thin filament [15]. These studies confirm predictions of an electrostatic Tm-actin interaction study by Orzechowski et al which found that interactions are weakened by Tm mutations E62Q, R160H, D175N, E180G, L185R, E192K, and others [16]. In addition to electrostatic differences, a study on persistence length of Tm with mutations D175N and E180G shows greater flexibility of the molecule near residue 180 in the mutated cases, which is a critical region for actin binding and may allow for enhanced myosin binding access to the actin filament [17].
The MD studies outlined above provide important insights into short timescale, local effects of HCM-associated mutations on contractile protein function. However, the computational expense of such simulations make it difficult to recreate larger protein systems and experimentally-relevant timescales. Therefore, it is important to contextualize MD findings using experimental work as well as larger-scale modeling frameworks in order to resolve the impact a single residue can have on whole cell function.
Mechanistic models of healthy and disease-state sarcomeres
Mechanistic models of the sarcomere have been developed over the past 50 years aiming to understand the thin filament physiology and the pathogenesis of thin filament-linked HCM cardiomyopathies. Some of these models have proven useful in answering/interpreting a variety of specific muscle contraction questions, but are often lacking sufficient detail to explain the complex disorders in sarcomeric cardiomyopathies and the associated impaired energetic metabolic activities during contraction. A commonly used model of thin filament activation and cross-bridge cycling is given in [5]. This model allows for a quick and efficient of solving a set of ODEs describing the cross-bridge cycling, but it lacks details of the spatial nearest-neighbour interactions of the myofilament. A more spatially resolved ODE-based model which was then developed to explicitly account for nearest-neighbour interactions of tropomyosin molecules can be found in [18]. Other models use Markov chain Monte Carlo (MCMC) processes to refine spatial detail, but these simulations can become computationally expensive too due to small time steps and the large number of repetitions/realizations needed to acquire statistically significant and smooth curves [3, 19]. In general, these low-level mechanistic models have limited predictive capability and rely on parameter optimization tools to fit mutation data.
The need for higher-resolution mechanistic insight was recently illustrated based on evidence from both patients and animal models [20]. Although the classical focus on hypotheses related to hyper-hypo contractile and force Ca2+ sensitivity has proven useful in many cases, there are significant limitations to this approach. Therefore, novel sarcomeric techniques coupled with a more modern understanding of the nature of sarcomeric cardiomyopathies are indeed required. Improved mechanistic models at the myofilament level can be coupled with robust data from animal models and intact in vitro systems to result in improved cardiac translational medicine power [21].
The direct mechanistic links between myofilament mutations, abnormalities in myocellular energetics and the early activation of myofilament signaling cascades remain unclear. The integration of high-level mechanistic models of the sarcomere and HCM-associated protein mutations remains a challenging problem in cardiac mechanics.
Revealing pathologic myofilament structure associated with cardiomyopathies
In addition to mechanistic spatiotemporal models of the sarcomere, structural models of the disease-state sarcomere have been developed by coupling experimental measurements with computational models to inform how HCM- and DCM-associated mutations affect sarcomere ultrastructure. Myosin mutations that are associated with hypertrophic and dilated cardiomyopathies are particularly prime candidates for studying the structural basis of the relationship between genotype, sarcomere ultrastructure, and myocardial hypertrophy. Advances in X-ray diffraction have enabled the characterization of the cardiac myosin filament structure in hearts with dilated or hypertrophic cardiomyopathy. Similar to skeletal muscle, the cardiac myosin filament in healthy hearts has a distinct relaxed state in which myosin motors are helically arranged on the thick filament backbone with ~ 14.3 nm periodicity [22]. The molecular interactions that stabilize this ‘sequestered’ state of the myosin motors, or the ‘OFF’ state of the thick filament, are not well understood in cardiac muscle but may involve myosin head-tail interactions [23], myosin-titin interactions [24], or interactions between myosin and MyBP-C [25, 26]. Consequently, recent focus has been on how cardiomyopathy-associated mutations in myosin might disrupt the diastolic structure of the thick filament, dysregulate contractility, and promote pathological remodeling.
Anderson et al. combined molecular structural models of myosin harboring HCM-causing mutations and X-ray diffraction analysis of myofilament structure, and demonstrated that a mutations that cause HCM indeed destabilize the resting conformation of myosin [27]. Moreover, work by the same group has led to the observation that a number of mutations associated with HCM are clustered in the so-called “myosin mesa”. A region of the myosin motor domain that is highly conserved in cardiac myosin isoforms across species [28]. A potential underlying mechanism of the destabilized resting conformation of myosin may be a mutation-induced disruption of myosin–MyBP-C interactions for mutations in the myosin mesa [29]**.
Conversely, Yuan and colleagues used X-ray diffraction techniques on permeabilized cardiac muscle from transgenic mouse hearts to find that a DCM-associated myosin mutation in the myosin regulatory light chain had little effect on the radial position of myosin motors or interfilament lattice spacing in the absence of calcium, but reduced the number of actin-interacting heads at submaximal calcium [30]. It may be that a reduction in the force-generating capacity of the sarcomere drives changes in myofibril structure and lattice geometry, exacerbating contractile dysfunction and the progression of ventricular dilation.
Taken together, structural models of the myofilaments harboring myosin-based mutations from both HCM and DCM hearts point towards a similar pathomechanism involving a perturbation in the structure of the thick filament. Future work using X-ray diffraction techniques and molecular modeling further will elucidating the structural basis of cardiomyopathies and may inform new therapeutic interventions that target the structure of the thick filament.
Cardiomyocyte dysfunction: EC coupling, mechanosensing, and remodeling
Mathematical EC coupling models in HCM
Cardiac cells can be considered as a coupled electromechanical network in which the action potential (AP) triggers an increase in intracellular Ca2+ concentration and contraction of the myofilaments that allows the heart to pump blood in a process called excitation-contraction (EC) coupling. In principle, the contractile force production by cardiac muscles depends on the relative sliding of the thick-thin filaments within sarcomeres. This sliding motion and the developed force are regulated by several mechanochemical-enzymatic mechanisms, including ATP hydrolysis, crossbridge cycling of myosin heads, and troponin-tropomyosin-actin interactions. The contraction-relaxation interplay of cardiac muscle is controlled sterically by the tropomyosin protein, which covers myosin binding interaction sites on the actin subunits to inhibit crossbridge formation during relaxation. Therefore, tropomyosin is believed to play a critical role not only in controlling the strength and duration of contraction, but also in promoting the following activation events and cooperativity. The dynamic motions of tropomyosin as governed by a multi-well energy landscape on thin filaments is influenced by both Ca2+ binding to troponin-C and myosin binding to actin [4, 31–34], but the effects of hypertrophy-causing mutations in these proteins on this energetic landscape have not yet been examined.
There are many different mathematical myofilament models that have been developed by several groups trying to understand mechanisms and responses of length-dependent cardiac muscle activation, thin filament regulation, myosin (thick) filament function, and ventricular mechanics [35]. The vast majority of these models couple a mechanistic myofilament activation model to a crossbridge cycling model and include length-dependent active force and passive tissue properties. Some of these myofilament models include phenomenological representations of crossbridge cycling [5], while others account for cooperative activation/nearest-neighbor interaction between troponin/tropomyosin units [3, 18]. Although several approximations were used in these models, the overall model structure attempts to agree well with the underlying biophysics. In addition, other simplified filament scale contraction models [35] were integrated into larger EC tissue scale models and have also been used as the basis of a multiscale electromechanical model of left ventricle simulation [36]. Although simulated responses using these models have been compared with a wide range of experimental measures, their applicability to study myocardial diseases are limited. For example, simplified contraction models can not be used to describe the link between sarcomeric point mutations and the HCM phenotype. Instead, multiscale atomistic-to-tissue type models are required, for reasons we outline below.
Sarcomeric point mutations alter cardiac muscle contractility and can lead to HCM. The majority of these mutations are distributed on residues located on the Tm-actin interface, which can modify the Tm-actin interaction and affect Tm positioning and mobility on the surface of actin filaments. These mutations and post-translational modifications influence not only Tm dynamics, but they also affect myofilament Ca2+ sensitivity and alter cooperative interactions between actin, Tm, troponin-complex and myosin [37, 38]. Therefore, the exact molecular-to-filament mechanism by which these alterations provide the trigger for disease progression and remodeling is still poorly understood. Because mutations can perturb the interaction energy landscape between Tm and actin, their effects cannot be explained by a filament-scale model that is either based on the mean-field approximation theory or relies purely on Markov type simulations.
Accurate modeling of the myofilament biophysics problem requires a multi-scale approach to modeling thin filament activation that can link atomistic molecular simulations of protein-protein interactions in the thin filament regulatory unit to sarcomere level activation dynamics, and to the tissue/whole organ level (see Figure 1). Nevertheless, several attempts using Markov Chain Monte Carlo (MCMC) type of myofilament models with parametric optimization were used to investigate and predict the effects of point mutations on sarcomere contractility [3, 39]. None of these models have directly addressed the thin filament activation process based on the detailed atomisitic dynamics of the Tm-coil on the surface of actin filaments. However, they are still attractive and flexible methods where the cooperativity due to the nearest-neighbour interactions among the thin filament RUs can be included explicitly in the system [3, 18].
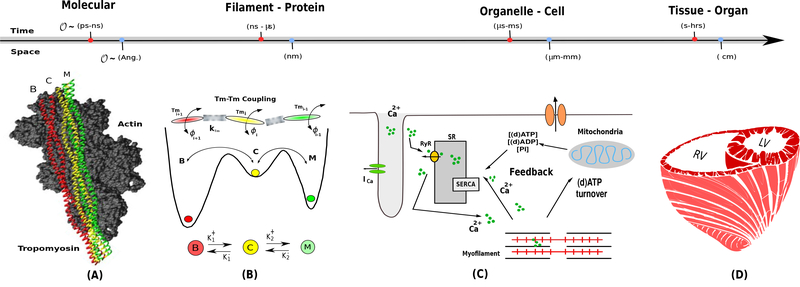
Figure 1:
Schematic illustrating the scope of cardiac multi-scale mathematical modeling techniques, spanning spatial and temporal scales. For example, pannel A shows tha molecular and Brownian dynamics simulations can be used to study thin filament action at the molecular scale [2]. Panel B shows that Markov Chain Monte Carlo (MCMC) technique can possibly integrate the molecular scale results to study the sarcomere protein interactions at the filament scale [3, 4]. Panel C shows that an organelle-cellular scale model can be then used to calculate contraction force [5]. Finally, this contraction force can be then integrated in an tissue-organ-scale FEM model to study growth and remodeling in hypertrophic cardiomyopathies [6].
Although myocardial contraction research has reached a point where detailed separate information about the molecular, cellular, and tissue-level mechanisms that drive and regulate contraction is available, the integration of this information in a single big-data modeling approach remains a challenging problem. The Brownian ratchet theory coupled with the Langevin dynamic principle could offer a solution to this problem using a coarse-grained mechanistic approach to bridging the molecular-to-filament scales [2, 34, 40]. The essence of this approach lies in computing the interacting Tm-actin energy landscape from atomistic Brownian dynamics simulations and integrating into a sarcomere level activation dynamics [2]. Moreover, we also agree with the recent opinion by K. Campbell et al [41], which points out that it is time for myofilament researchers to emphasizing on leveraging the available knowledge to a potential applied approach based on multiscale computer modeling and simulation to improve HCM cardiac patient care.
Experimental cellular models of sarcomerogenesis and hypertrophy
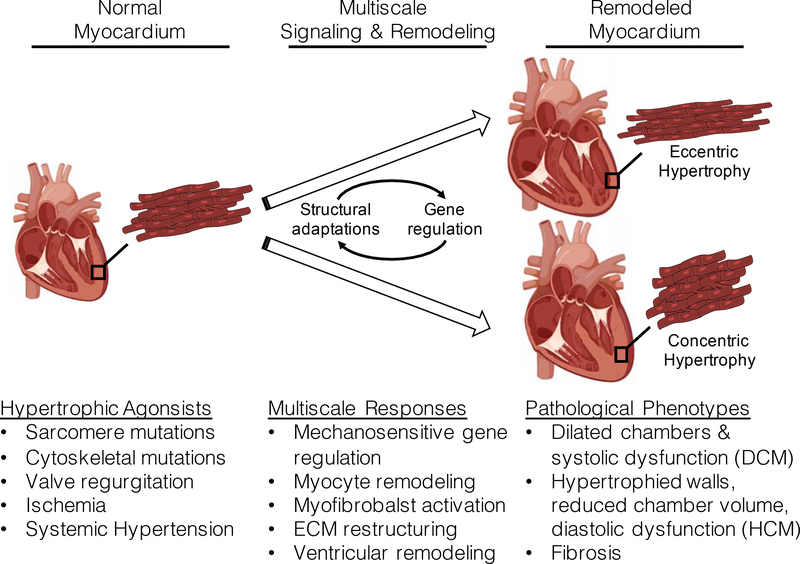
Cardiomyocyte growth and remodeling is typically characterized by increased protein synthesis and sarcomere formation. However, hypertrophy can either be concentric (reduced length-width ratio) or eccentric (increased length-width ratio). Concentric hypertrophy at the myocyte level is the result of sarcomeres added in parallel, and manifests as a thickening of the ventricular walls with diastolic dysfunction. As such, this type of cellular hypertrophy is typically seen in HCM hearts. Conversely, eccentric hypotrophy at the cellular level is the result of sarcomeres added in series and normally is associated with dilated ventricles and systolic dysfunction, and is typically associated with DCM. While each type of hypertrophy has been well documented, the molecular determinants of each type remain largely unknown (see Figure 2).
Figure 2:
Various hypertrophic agonists acting on normal myocardium trigger multiscale regulatory pathways that ultimately produce either eccentric or concentric hypertrophy. The precise molecular mechanisms involved in translating the mechanical stimuli into changes in gene expression that lead to either eccentric or concentric hypertrophy and remodeling remain unresolved. This illustration is made using biorender application (www.biorender.com).
In-vitro and in-vivo models of mechanically induced hypertrophy have been helpful in identifying structural and genetic responses to different loads. Neonatal rodent cardiomyocytes that are cultured under static load applied longitudinally (i.e., along the myofibrillar axis) respond with serial addition of sarcomeres and eccentric hypertrophy [42, 43], while myocytes under transverse load respond with parallel sarcomere formation and concentric hypertrophy [44]. Differential gene regulation in response to transverse and longitudinal load suggests that the structural adaptations are driven by unique mechanosensitive gene expression [45]. Furthermore, concentric and eccentric myocyte hypertrophy is typically associated with HCM and DCM, respectively [46], and defining the molecular regulators of concentric versus eccentric hypertrophy in each case remains an important problem in cardiopathology.
Because the sarcomere has a putative role in the mechanosensing and mechanotransduction of the heart [for a more comprehensive review on this subject, see Reference [47]], the vast number of mutations associated with HCM and DCM in genes that encode sarcomere proteins strongly implicates sarcomere mechanics in determining the type of cardiac hypertrophy. Functional experimental assays using human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) are useful for elucidating mechanisms of hypertrophy in healthy hearts and in hearts containing patient-specific, pathogenic sarcomere protein mutations. For example, Leonard et al. highlighted the mechanosensitivity of hiPSC-CMs by demonstrating that engineered heart tissues (EHTs) made from hiPSC-CMs that were cultured in high-afterload conditions exhibited greater cellular growth and expression of hypertrophic biomarkers [48]. Also using EHTs with hiPSC-CMs, Hinson and colleagues were able to characterize the effects of truncating mutations associated with DCM in the giant sarcomere protein titin on sarcomerogenesis, cardiac contractility, and hypertrophic biomarker signaling [49]. The authors demonstrated that DCM-associated titin-truncating mutations blunt sarcomerogenesis, which may be an underlying cause of DCM in patients with titin truncations [49]. Titin also may play a role in stress-dependent sarcomerogenesis [50]**. Experimental models using EHTs such as these may help lead to a precision medicine model for preventing or reversing HCM and DCM phenotypes.
Models of mechanical and biochemical regulatory signaling networks
Cardiomyocytes undergo tremendous mechanical and geometrical dynamics during systole-diastole cycles yet maintain the ability to acutely sense abnormalities in the mechanical microenvironment. Prolonged exposure to abnormal mechanics generally causes myocyte remodeling and hypertrophy that often translates to organ-level dysfunction. Precise molecular links that provide intracellular mechanosensing and translate mechanical signals into genetic responses that promote myocyte remodeling and heart failure are far from being completely understood, but correlations between mechanical stimuli, hypertrophic agonists, and molecular signaling have been identified through experimental and computational models of hypertrophy pathways (see Reference [51] for a comprehensive review of mechanosensitive gene regulation in the heart). Key signaling pathways in cardiac hypertrophy have been identified in recent in-vitro and in-vivo experimental models. Among these, major pathways that have emerged involve calcium-dependent interactions between the transcription factor Nuclear Factor of Activated T-cells (NFAT), calcineurin, and the mitogen-activated protein kinase 1 (MAPK1). These regulators of hypertrophy also regulate other signaling pathways, such as activation of transcription factors like GATA4 or factors involved in regulating the actin cytoskeleton [52]. Interestingly, a critical relationship was recently identified between O-linked β-N-acetylglucosamine (O-GlcNAc), a carbohydrate involved in posttranslational modifications of serine and threonine residues, and NFAT signaling [53]. Facundo et al. demonstrated that NFAT activation associated with pressure overload-induced hypertrophy in mice requires O-GlcNAc signaling, suggesting a key metabolic-based regulatory mechanism of hypertrophy [53]. Future investigations of mitochondrial function in response to altered myocyte mechanics may be quite informative in the context of metabolic signaling in cardiac hypertrophy. More generally, the cross-talk between the various signaling pathways and other downstream effectors of cardiac hypertrophy quickly creates a complex regulatory landscape that will require a highly integrated modeling approach to elucidate. Recent efforts to synthesize the extensiveness of biochemical signaling in cardiomyocytes in response to mechanical stretch have generated a systems-level predictive network of mechanosensitive gene regulation pathways [54, 55]. Tan and colleagues developed a systems-level computational model of mechanosensitive signaling networks in cardiomyocytes that accurately predicts stretch-induced changes in gene expression and cellular growth [55]. The model uses a system of logic-based differential equations coupled with experimental observations from published literature to construct a signaling cascade initiated by cardiomyocyte stretch and was validated against in-vitro assessments of stretch-induced gene regulation and cell growth in isolated neonatal rat cardiomyocytes. Other experimental efforts have focused on the transcriptome of hearts in hypertrophic heart failure [56, 57], but combining these experimental observations with a predictive genetic model in future studies will provide a more comprehensive understanding of these pathomechanisms.
Cellular- and tissue-level models of the kinematics of myocardial growth and remodeling
Strain and tension-based predictive growth models
Understanding the mechanical stimuli that produce a particular hypertrophic phenotype (eccentric versus concentric) is a major challenge. As discussed, cardiac growth and remodeling at the cellular and tissue level is often the result of altered mechanical load, and in-vitro experiments have shown that myocytes tend to respond with growth in the same direction as the applied stretch. However, it is difficult to apply these experiments to understand the pathogenesis of inherited genetic-based cardiomyopathies as it is unknown whether intrinsic mechanical abnormalities (e.g., altered sarcomere contractility) trigger a similar mechanosignaling cascade as extrinsic mechanical loading. As such, multiscale models that combine phenomenological stress- and strain-based growth with genetic sarcomere mutations would provide a powerful tool for predicting pathological remodeling and designing new preventative or restorative treatments.
To date, several models have been developed based on growth laws derived from pressure or volume overloading (see Reference [58] for a comparison of different growth models). Goktepe et al. have employed mathematical models of the kinematics of concentric and eccentric growth based on pressure and volume overload from the myofibril level to the ventricular level [59]. Similarly, Kerckoffs and colleagues developed a nonlinear finite element model of the right and left ventricles superimposed with realistic fiber alignment [6]. The proposed strain-dependent growth law stated that the maximum strain in the fiber direction stimulates cardiomyocyte lengthening and maximum cross-fiber strain stimulates cardiomyocyte thickening, and it was able to predict growth due to pressure and volume overload [6], postnatal hemodynamic loading [60], and asymmetric hypertrophy in response to ventricular pacing [61].
More recently, Davis and colleagues introduced a new paradigm for myocardial growth based on a ‘tension index’ generated by computational models of cardiac twitches for various disease conditions [62]. The authors demonstrated a strong correlation between the change in the impulse of a cardiac twitch (i.e., the tensiontime integral) for HCM or DCM hearts compared to healthy hearts and the type and severity of myocardial remodeling—a large positive impulse relative to healthy hearts correlated with concentric hypertrophy and the HCM phenotype while a large negative impulse relative to healthy hearts correlated with eccentric hypertrophy and the DCM phenotype [62]. An interesting question that stems from this work is whether the altered contractility investigated in the work by Davis and colleagues causes abnormal fiber and cross-fiber strain, or if altered contractility has unique mechanosignaling pathways associated with the resulting growth. Future work that combines a stress- and strain-based growth laws at the myocyte and ventricle levels will be informative in deciphering the mechanical signals initiated by HCM- and DCM-associated mutations.
Finite elements models for variations in ventricular HCM morphology
The vast majority of the current cardiac finite element modeling (FEM) uses phenomenological models of myofibril contraction. This simplification in the dynamics of contractile apparatus adds limitations on using such FEM approaches to simulate, for example patient-specific cardiac mechanics problems. Moreover, the mathematical formalization of these models makes it difficult to scale up from atomistic-to-organ level, which adds extra challenge to simulate mechanical responses of hearts under diseased state, e.g., mutations-linked HCM. However, a left ventricular FEM study [63] has recently been proposed to overcome this limitation, where the model was developed based on integrating a filament-level model of contraction [64].
Another limitation of classical cardiac FEM approaches is, the use of semi-empirical methods to describe the growth and remodeling of heart cells triggered by cardiac disease. Most of the current implementations of cardiac growth and remodeling algorithms within FEM have focused on geometrical representations (parallel/serial filling approach) of the sarcomeres [65]. Unfortunately, these models are too simplistic for clinical use because they don’t account for the patient specificity disease factors. Nevertheless, more advanced algorithms have recently been proposed to better integrate continuum growth and remodeling models into morphic finite element simulations that can change and evolve in time [6, 66, 67]. More specifically, these models couple element-level mechanics to long term changes in tissue-level structure and function [68]**.
Modern cardiac FEM can be a useful approach to study the mechanics of variations in ventricular morphology due to HCM. For instance, FEM should have the capability to integrate experimental and clinical data, so it can help in both therapeutic and diagnostic phases. In other words, FEM could help clinicians draw better quantitative understanding of cardiac disease state, which in turn will inspire rational therapies.
Conclusion
In this review, we presented current experimental and computational models of cardiac mechanics spanning multiple spatial and temporal scales that are aimed at understanding the pathomechanisms of cardiac hypertrophy. Models of cardiac hypertrophy agonists that span mechanisms from the molecular level to the tissue level were also given. In particular, we focused on atomistic-level models of the structural dynamics of sarcomere proteins in hypertrophic hearts, cellular-level models of excitation-contraction coupling and mechanosensitive signaling in disease-state myocytes, and organ-level models of myocardial growth and remodelling. Because of the complex combination of molecular mechanobiology, genetic signaling, cardiomyocyte and ECM remodeling, as well as tissue mechanics involved in myocardial hypertrophy, a multiscale approach is required to elucidate genotype-to-phenotype mechanisms. We finally discuss how spanning these scales will provide new information about the processes governing hypertrophy and potential methods to prevent cardiomyopathies or generate novel therapeutics.
Acknowledgement
This work was supported in part by National Institutes of Health awards No.R01 HL105242, R01 HL137100, U01 HL122199, U01 HL126273 to A.D.M.
Conflict of Interest
Y.A., J.P. and K.M. declare no conflict of interest. A.D.M. is a co-founder of and has an equity interest in Insilicomed and Vektor Medical. He serves on the scientific advisory board of Insilicomed and as scientific advisor to both companies. Some of his research grants, including those acknowledged here, have been identified for conflict of interest management based on the overall scope of the project and its potential benefit to these companies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
* * of outstanding interest
- [1].Hill JA, Olson EN, Cardiac plasticity, N Engl J Med 358 (2008) 1370–1380. [DOI] [PubMed] [Google Scholar]
- [2].Aboelkassem Y, McCabe KJ, Huber G, Regnier M, McCammon JA, McCulloch AD, A stochastic coarse-grained multiscale model of thin filament activation using brownian and langevin dynamics, Submitted to Biophys J :in press (2019) 10.1016/j.bpj.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aboelkassem Y, Bonilla JA,. K. J. McCabe, S. Campbell, Contributions of ca2+-independent thin filament activation to cardiac muscle function, Biophys J 109 (2015) 2101–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Aboelkassem Y, Trayanova N, Tropomyosin dynamics during cardiac muscle contraction as governed by a multi-well energy landscape, Prog Biophys Mol Biol 144 (2019) 102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rice J, Wang F, Bers D, Tombe PD, Approximate model of cooperative activation and crossbridge cycling in cardiac muscle using ordinary differential equations, Biophysical Journal 95(5) (2008) 2368–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kerckhoffs RC, Omens JH, McCulloch AD, A single strain-based growth law predicts concentric and eccentric cardiac growth during pressure and volume overload, Mech. Res. Commun. 42 (2012) 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Doh C, Li J, Mamidi R, Stelzer J, The hcm-causing y235s cmybpc mutation accelerates contractile function by altering c1 domain structure, Biochim Biophys Acta Mol Basis Dis 1865 (3) (2019) 661–677.[*, This article uses Molecular Dynamics (MD) studies as well as mechanical experiments to determine the effects of HCM–associated myosin binding protein C (cMyBPC) mutation Y235S on contractile function. Experimentally, studies on mouse myocardium virally transfected with the mutation determined increased Ca2+ sensitivity of force and overall hypercontractile behaviour.].
- [8].Krishnamoorthy N, Gajendrarao P, Olivotto I, Yacoub M, Impact of disease-causing mutations on inter-domain interactions in cmybp-c: a steered molecular dynamics study, J Biomol Struct Dyn 35 (9) (2017) 1916–1922. [DOI] [PubMed] [Google Scholar]
- [9].Gajendrarao P, Krishnamoorthy N, Selvaraj S, Girolami F, Cecchi F, Olivotto I, Yacoub M, An investigation of the molecular mechanism of double cmybp-c mutation in a patient with end-stage hypertrophic cardiomyopathy, J Cardiovasc Transl Res 8 (4) (2015) 232–43. [DOI] [PubMed] [Google Scholar]
- [10].Gajendrarao P, Krishnamoorthy N, Kassem HS, Moharem-Elgamal S, Cecchi F, Olivotto I, Yacoub MH, Molecular modeling of disease causing mutations in domain c1 of cmybp-c, PLoS One 8 (3) (2013) e59206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cheng Y, Rao V, yue Tu A, Lindert S, Wang D, Oxenford L, McCulloch AD, McCammon JA, Regnier M, Troponin i mutations r146g and r21c alter cardiac troponin function, contractile properties, and modulation by protein kinase a (pka)-mediated phosphorylation, J Biol Chem 290 (46) (2015) 27749–27766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cheng Y, Lindert S, Oxenford L, Tu A, AD AM, Regnier M, Effects of cardiac troponin i mutation p83s on contractile properties and the modulation by pka-mediated phosphorylation, J Phys Chem B 120 (33) (2016) 8238–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bowman J, Lindert S, Molecular dynamics and umbrella sampling simulations elucidate differences in troponin c isoform and mutant hydrophobic patch exposure, J Phys Chem B 122 (32) (2018) 7874–7883.[**, This study uses MD simulations to investigate several HCM (A8V and A31S) and DCM (Y5H, Q50R, and E59D/D75Y) mutations to troponin C (TnC). In order to capture the likelihood of long-timescale TnC transitions while running 1 microsecond MD simulations, umbrella sampling was employed to determine free energy landscapes for each mutated TnC molecule.].
- [14].Zheng W, Hitchcock-DeGregori S, Barua B, Investigating the effects of tropomyosin mutations on its flexibility and interactions with filamentous actin using molecular dynamics simulation, J Muscle Res Cell Motil 37 (4–5) (2016) 131–147.[*, The authors conducted extensive molecular dynamics simulations for the wild–type and mutant Tpm in complex with F-actin. The results indicate that mutants exhibited distinct changes (i.e., increase or decrease) in the overall and local flexibility of the Tpm coiled-coil, with each mutation causing both local and long-range modifications of the Tpm flexibility.].
- [15].Farman G, Rynkiewicz M, Orzechowski M, Lehman W, Moore J, Hcm and dcm cardiomyopathy-linked -tropomyosin mutations influence off-state stability and crossbridge interaction on thin filaments, Arch Biochem Biophys 647 (2018) 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Orzechowski M, Fischer S, Moore JR, Lehman W, Farman GP, Energy landscapes reveal the myopathic effects of tropomyosin mutations, Arch. Biochem. Biophys 564 (2014) 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li X, Suphamungmee W, Janco M, Geeves M, Marston S, et al SF, The flexibility of two tropomyosin mutants, d175n and e180g, that cause hypertrophic cardiomyopathy, Biochem. Biophys. Res. Commun 424 (2012) 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Campbell SG, Lionetti FV, Campbell KB, McCulloch AD, Coupling of adjacent tropomyosins enhances crossbridge-mediated cooperative activation in a markov model of the cardiac thin filament, Biophys J 98 (2010) 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tanner BCW, Daniel TL, Regnier M, Filament compliance influences cooperative activation of thin filaments and the dynamics of force production in skeletal muscle, PLoS Comp Biol 8 (2012) e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Coats C, Pavlou M, Watkinson O, Protonotarios A, Moss L, Hyland R, Rantell K, Pantazis A, Tome M, McKenna W, Frenneaux M, Omar R, Elliott P, Effect of trimetazidine dihydrochloride therapy on exercise capacity in patients with nonobstructive hypertrophic cardiomyopathy: a randomized clinical trial, JAMA Cardiol. 4 (2019) 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deranek AE, Klass MM, Tardiff JC, Moving beyond simple answers to complex disorders in sarcomeric cardiomyopathies: the role of integrated systems, Eur. J. Physio 471 (2019) 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Massimo R, Marco C, Francesca P, Theyencheri PJD,N, JM SG, Marco L, Vincenzo L, Gabriella P, Myosin filament activation in the heart is tuned to the mechanical task, Proc Natl Acad Sci 114 (2017) 3240–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].AL-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP, Atomic model of the human cardiac muscle myosin filament, Proc Natl Acad Sci 110 (2012) 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Muhle-Goll C, Habeck M, Cazorla O, Nilges M, Labeit S, Granzier H, Structural and functional studies of titin’s fn3 modules reveal conserved surface patterns and binding to myosin s1 - a possible role in the frank-starling mechanism of the heart, jmb 313 (2001) 431–447. [DOI] [PubMed] [Google Scholar]
- [25].Kensler RW, Craig R, Moss RL, Phosphorylation of cardiac myosin binding protein c releases myosin heads from the surface of cardiac thick filaments, Proc Natl Acad Sci 114 (2017) E1355–E1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang X, Kampourakis T, Yan Z, Sevrieva I, Irving M, Sun Y, Distinct contributions of the thin and thick filaments to length-dependent activation in heart muscle, Elife 6 (2017) e24081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Anderson R, Trivedi D, Sarkar S, Henze M, Ma W, Gong H, Rogers C, Gorham J, Wong F, Morck M, Seidman J, Ruppel K, Irving T, Cooke R, Green E, Spudich J, Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers., Proc Natl Acad Sci 115 (2018) E8143–E8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Spudich J, The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy, Biochem Soc Trans 43 (2015) 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nag S, Trivedi D, Sarkar S, Adhikari A, Sunitha M, Sutton S, Ruppel K, Spudich J, The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations, Nat Struct Mol Biol. 24 (2017) 525–533. [**The authors not only report insightful structural features of HCM—related myosin mutations and the potential importance of the ‘myosin mesa’ region, but they also elucidate molecular mechanisms regulating the resting state of the thick filament.].
- [30].Yuan C, Kazmierczak K, Liang J, Zhou Z, Yadav S, Gomes A, Irving T, Szczesna-Cordary D, Sarcomeric perturbations of myosin motors lead to dilated cardiomyopathy in genetically modified myl2 mice, Proc Natl Acad Sci 115 (2018) E2338–E2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Orzechowski M, Moore JR, Fischer S, Lehman W, Tropomyosin movement on f-actin during muscle activation explained by energy landscapes, Arch. Biochem. Biophys 545 (2014) 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Moore JR, Campbell SG, Lehman WJ, Structural determinaants of muscle thin filament cooperativity, Arch. Biochem. Biophys 594 (2016) 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Aboelkassem Y, Trayanova N, Tropomyosin dynamics during cardiac thin filament activation as governed by a multi-well energy landscape, Biophys J 110 (2016) 524a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aboelkassem Y, McCabe KJ, Huber G, Sundnes J, McCulloch AD, Turning the azimuthal motions of adjacent tropomyosins into a coupled n-body problem in a brownian model of cardiac thin filament activation, Biophys J 114 (2018) 502a–503a. [Google Scholar]
- [35].Niederer SA, Campbell KS, Campbell SG, A short history of the development of mathematical models of cardiac mechanics, J Mol Cell Cardiol 127 (2019) 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Krishnamurthy A, Villongco CT, Chuang J, Frank LR, Nigam V, Belezzuoli E, Stark P, Krummen DE, Narayan S, Omens JH, McCulloch AD, Kerckhoffs RC, Patient-specific models of cardiac biomechanics, J. Comput. Phys 244 (2013) 4–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Redwood C, Robinson P, Alpha-tropomyosin mutations in inherited cardiomyopathies, J. Muscle Res. Cell Motil 34 (2013) 285–294. [DOI] [PubMed] [Google Scholar]
- [38].Farman GP, Rynkiewicz MJ, Orzechowski M, Lehman W, Moore JR, Hcm and dcm cardiomyopathy-linked alphatropomyosin mutations influence off-state stability and crossbridge interaction on thin filament, Arch. Biochem. Biophys 647 (2018) 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sewanan LR, Moore JR, Lehman WJ, Campbell SG, Predicting effects of tropomyosin mutations on cardiac muscle contraction through myofilament modeling, Front. Physiol. 7 (2016) 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Aboelkassem Y, Trayanova N, Tropomyosin fluctuations over a multi-well energy landscape: A brownian ratchet model of cardiac muscle contraction, Biophys J 112 (2017) 259a–260a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Campbell KB, Yengo CM, Lee L-C, Kotter J, Sorrell VL, Guglin M, Wenk JF, Closing the therapeutic loop, Arch. Biochem. Biophys 663 (2019) 129–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mansour H, de Tombe P, Samarel A, Russell B, Restoration of resting sarcomere length after uniaxial static strain is regulated by protein kinase cepsilon and focal adhesion kinase, Circ Res. 94 (2004) 642–9. [DOI] [PubMed] [Google Scholar]
- [43].B Russell YKAS, MW Curtis, Mechanical stress-induced sarcomere assembly for cardiac muscle growth in length and width, J Mol Cell Cardiol. 48 (2010) 817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yang H, Schmidt LP, Wang Z, Yang X, Shao Y, Borg TK, Markwald R, Runyan R, Gao BZ, Dynamic myofibrillar remodeling in live cardiomyocytes under static stretch, Sci Rep 6 (2016) 20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gopalan S, Flaim C, Bhatia S, Hoshijima M, Knoell R, Chien K, Omens J, McCulloch A, Anisotropic stretch- induced hypertrophy in neonatal ventricular myocytes micropatterned on deformable elastomers, Biotechnol Bioeng 81 (2003) 578–87. [DOI] [PubMed] [Google Scholar]
- [46].Marian A, Genetic determinants of cardiac hypertrophy, Curr Opin Cardiol 23 (2008) 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lyon R, Zanella F, Omens J, Sheikh F, Mechanotransduction in cardiac hypertrophy and failure, Circ Res 116 (2015) 1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Leonard A, B. A, P. JD, B. KM, B. S, R. M, M. CE, S. NJ, fterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues, J Mol Cell Cardiol 118 (2018) 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A, Homsy J, Hubner N, Church G, Cook SA, Linke WA, Chen CS, Seidman JG, Seidman CE, Titin mutations in ips cells define sarcomere insufficiency as a cause of dilated cardiomyopathy, Science 349 (2015) 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chopra A, Kutys M, Zhang K, Polacheck W, Sheng C, Luu R, Eyckmans J, Hinson J, Seidman J, Seidman C, Chen C, Force generation via β-cardiac myosin, titin, and a-actinin drives cardiac sarcomere assembly from cell-matrix adhesions, Dev Cell 44 (2018) 87–96.e5, [**, The authors demonstrate the powerful utility of combining CRISPR-edited hiPSC-CM models of disease with high-resolution time-lapse microscopy, and reveal how DCM-causing titin truncations disrupt the mechanisms involved in sarcomere assembly. Using this type of integrative approach coupled with computational models of the subcellular organizational dynamics in dysfunctional myocytes will be extremely valuable in future work towards understanding pathomechanisms of cardiac remodeling.
- [51].Saucerman J, Tan P, Buchholz K, AD AM, Omens J, echanical regulation of gene expression in cardiac myocytes and fibroblasts, Nat Rev Cardiol 16 (2019) 361–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lauriol J, Keith K, Jaffre F, Couvillon A, Saci A, Goonasekera S, McCarthy J, Kessinger C, Wang J, Ke Q, Kang P, Molkentin J, Carpenter C, Kontaridis M , Rhoa signaling in cardiomyocytes protects against stress-induced heart failure but facilitates cardiac fibrosis, Sci Signal 348 (2014) ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Facundo H, Brainard R, Watson L, Ngoh G, Hamid T, Prabhu S, Jones S, O-glcnac signaling is essential for nfat- mediated transcriptional reprogramming during cardiomyocyte hypertrophy, Am J Physiol Heart Circ Physiol. 302 (2012) H2122–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ryall K, Holland D, Delaney K, Kraeutler M, Parker A, Saucerman J, Network reconstruction and systems analysis of cardiac myocyte hypertrophy signaling, J Biol Chem. 287 (2012) 42259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tan PM, Buchholz KS, Omens JH, McCulloch AD, Saucerman JJ, Predictive model identifies key network regulators of cardiomyocyte mechano-signaling, PLoS Comput Biol 13 (2017) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lee J, Gao C, Peng G, Greer C, Ren S, Wang Y, Xiao X, Analysis of transcriptome complexity through rna sequencing in normal and failing murine hearts, Circ Res. 109 (2011) 1332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang K, Yamada K, Patel A, Topkara V, George I, Cheema F, Ewald G, Mann D, Nerbonne J, Deep rna sequencing reveals dynamic regulation of myocardial noncoding rnas in failing human heart and remodeling with mechanical circulatory support, Circulation 129 (2014) 1009–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Witzenburg C, Holmes J, A comparison of phenomenologic growth laws for myocardial hypertrophy, Journal of Elasticity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Goktepe S, Abilez O, Parker K, Kuhl E, A multiscale model for eccentric and concentric cardiac growth through sarcomerogenesis, Journal of Theoretical Biology. [DOI] [PubMed] [Google Scholar]
- [60].Kerckhoffs R, Computational modeling of cardiac growth in the post-natal rat with a strain-based growth law, Journal of Biomechanics 45 (2012) 865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kerckhoffs R, nad JO, McCulloch AD AD, Mechanical discoordination increases continuously after the onset of left bundle branch block despite constant electrical dyssynchrony in a computational model of cardiac electromechanics and growth, Europace 42 (2012) 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Davis J, Davis L, Correll R, Makarewich CA, Schwanekamp JA, Moussavi-Harami F, Wang D, York AJ, Wu H, Houser S, Seidman C, Seidman J, Regnier M, Metzger J, Wu J, Molkentin J, A tension-based model distinguishes hypertrophic versus dilated cardiomyopathy, Cell 165 (2016) 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhang X, Liu Z, Campbell K, Wenk J, Evaluation of a novel finite element model of active contraction in the heart, Front. Physiol 9 (2018) 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Campbell KB, Dynamic coupling of regulated binding sites and cycling myosin heads in striated muscle, J. Gen. Physiol 143 (2014) 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gerdes JCAM, Clark LC, Regression of cardiac hypertrophy after closing an aortocaval fistula in rats, Am. J. Physiol 268 (1995) H2345–H2351. [DOI] [PubMed] [Google Scholar]
- [66].Lee L, Genet M, Acevedo-Bolton G, Ordovas K, Guccione J, Kuhl E, A computational model that predicts reverse growth in response to mechanical unloading, biomechanics model, Mechanobiol. 14 (2015) 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee L, Sundnes J, Genet M, Wenk J, Wall S, An integrated electromechanical growth heart model for simulating cardiac therapies, Biomechanics Model. Mechanobiol. 15 (2016) 791–803. [DOI] [PubMed] [Google Scholar]
- [68].Costabal FS, Choy J, Sack K, Guccione J, Kassab G, Kuhl E, Multiscale characterization of heart failure, Acta Biomaterialia 86 (2019) 66–76. [**,The authors provide a better quantitative understanding of the correlations between sarcomerenumber, myocyte length, and ventricular dilation in a multiscale model of heart failure].