Abstract
P-glycoprotein (Pgp) is a biomedically important member of the ABC transporter superfamily that mediates multidrug resistance in various cancer types. Substrate binding and transport in Pgp are modulated by the presence of cholesterol in the membrane. Structural information on cholesterol binding sites and mechanistic details of its redistribution are, however, largely unknown. In this study, a set of 40 independent molecular dynamics (MD) simulations of Pgp embedded in cholesterol-rich lipid bilayers are reported, totaling 8 μs, enabling extensive sampling of cholesterol-protein interactions in Pgp. Clustering analyses of the ensemble of cholesterol molecules (~5,740) sampled around Pgp in these simulations reveal specific and asymmetric cholesterol-binding regions formed by the transmembrane (TM) helices TM1-6 and TM8. Notably, not all the putative cholesterol binding sites identified by MD can be predicted by the primary sequence based cholesterol-recognition amino acid consensus (CRAC) or inverted CRAC (CARC) motifs, an observation that we attribute to inadequacy of these motifs to account for binding sites formed by remote amino acids in the sequence that can still be spatially adjacent to each other. Binding of cholesterol to Pgp occurs more frequently through its rough β-face formed by the two protruding methyl groups, whereas the opposite smooth α-face prefers packing alongside the membrane lipids. One full and two partial cholesterol flipping events between the two leaflets of the bilayer mediated by the presence of Pgp are also captured in these simulations. All flipping events are observed in a region formed by helices TM1, TM2, and TM11, featuring two full and two partial CRAC/CARC motifs, with Tyr49 and Tyr126 identified as key residues interacting with cholesterol during this event. Our study is the first to report direct observation of unconventional cholesterol translocation on the surface of Pgp, providing a secondary transport model for the known flippase activity of the transporter.
Introduction
ATP-binding cassette (ABC) transporters are a ubiquitous superfamily of membrane proteins [1] responsible for ATP-fueled translocation of a wide range of chemical species across the cellular membrane, including sugars [2], amino acids [3], lipids [4], and essential metals [5]. P-glycoprotein (Pgp), the most studied, biomedically significant member of the superfamily, is popularly known as a “hydrophobic vacuum cleaner”, as it exports a wide variety of lipophillic substrates, including drugs, and mediates multidrug resistance (MDR) in many cancer types [6-11]. The protein contains a large hydrophobic transmembrane (TM) cavity with a volume of ~6,000 Å3, that allows the protein to express poly-specificity in substrate binding [12-14]. Most drug molecules, e.g., chemotherapeutic agents, possess a chemical profile similar to Pgp substrates [15]. Export of these drug molecules by Pgp reduce their concentration, and thereby their pharmacological effect inside the cell, leading to the development of MDR.
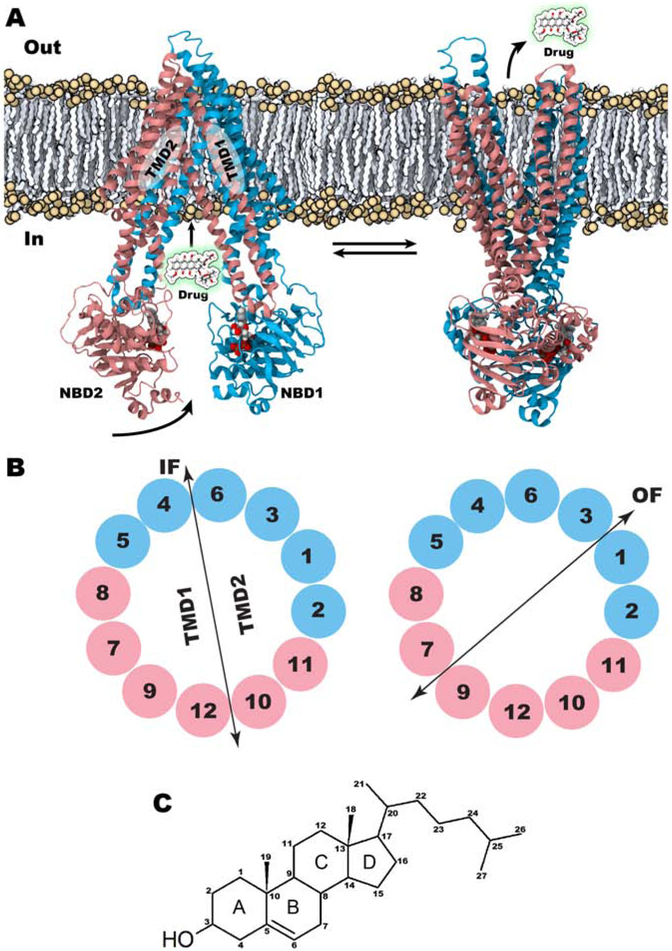
Pgp displays a unique architecture where a single polypeptide chain folds into two pseudo-symmetric bundles, comprised of two TM domains (TMDs) each with six helices, attached to two nucleotide-binding domains (NBDs), where ATP binding and hydrolysis take place (Fig. 1A) [13, 16, 17]. The transport process follows the widely accepted alternating-access mechanism [18, 19], where the transporter undergoes transition between inward-facing (IF) and outward-facing (OF) states, with the central substrate-binding cavity only accessible to either inside or outside of the cell at any given time [16, 20-24]. The conformational transition of the protein between the two states is brought about by the reorganization of the TM helices between the two TMD bundles (Fig. 1B). Nucleotide binding and hydrolysis at the NBDs energize this large-scale conformational transition, from the IF state to the OF state, thereby driving the vectorial transport of the substrate molecule from the cytoplasm to the extracellular space [25]. Subsequently, the transporter resets to the IF state, allowing for another round of transport [26, 27].
Figure 1:
Structural details of Pgp. (A) IF (left, PDB ID. 4M1M) and OF (right, model from reference [25]) states of Pgp are shown with their approximate membrane positioning. Arrows indicate vectorial transport of substrates through Pgp; doxorubicin (drug) is shown as an example. Horizontal arrows indicate the back and forth transition between the IF and OF states during the transport cycle. (B) Swapping of TM helices between the two TM bundles, TMD1 and TMD2 in IF (left) and OF (right) states is shown. TM helices are colored based on the protein sequence with the first half of the protein shown in cyan and the second half in pink. TMDs are defined based on the bundle formation. (C) Chemical structure of cholesterol along with its ring system (A-B-C-D) and atom numbering.
Lipid molecules have been shown to play a significant role in modulating the structure and function of membrane proteins like Pgp [28-35]. Cholesterol, an amphiphilic sterol molecule present in mammalian membranes [36, 37] (Fig. 1C), in particular, shows a complex relationship with the transporter. Pgp has been reported to be predominantly localized to cholesterol-rich membrane microdomains [38, 39]. The basal and drug-stimulated ATPase activity as well as the drug-binding and transport capacity of Pgp have also been shown to be modulated by cholesterol [40-45].
Various drugs show altered affinities to Pgp at higher cholesterol levels [46, 47]. In addition, cholesterol can actively participate in the transport cycle, and it has been reported to compete with daunorubicin, a known Pgp substrate, for its binding to the transporter [48]. In addition to its drug transporting capacity, Pgp can also function as a lipid flippase, redistributing and shuttling cholesterol and phospholipids from the inner to the outer leaflet of the cell membrane [45, 49]. Previous experimental studies with fluorescent measurements and bioinformatics-based analysis reported potential cholesterol binding motifs in Pgp [50]. However, no structural information is available for the prospective cholesterol binding sites on the surface of the protein or the redistribution mechanism of this sterol between the membrane leaflets by the transporter [45].
Here we present an extensive set of molecular dynamics (MD) simulations investigating cholesterol-protein interactions in Pgp embedded in cholesterol-rich lipid bilayers.
These simulations provide microscopic details on cholesterol-binding characteristics, including an asymmetric accumulation of cholesterol on the first TMD bundle of Pgp, TM1/2 interface as a high-density cholesterol binding region, and preferential binding between smooth and rough faces of cholesterol to Pgp. Notably, not all the putative cholesterol binding sites identified by MD can be predicted by the cholesterol-recognition amino acid consensus (CRAC) motifs, an observation that we attribute to inadequacy of these motifs to account for binding sites formed by remote amino acids in the sequence that can still be spatially adjacent to each other. Furthermore, we directly observe cholesterol flipping along the surface of the transporter, providing a novel alternative mechanism for Pgp-mediated lipid redistribution between the two leaflets of the membrane through a passive pathway provided by the protein along its surface.
Methods
System preparation
From the available crystal structures of Pgp in the Protein Data Bank (PDB), a recent structure of Pgp (PDB ID: 4M1M) was downloaded. Except chain A, which is used for model construction, all protein chains and non-protein atoms were removed. The Membrane Builder module of CHARMM-GUI [51, 52] was used to build the system by placing the protein in a patch of POPC:cholesterol (70:30) bilayer of 100×100 Å2 dimension and solvated with water. The system was neutralized with Na+ and Cl− ions to achieve a net ionic concentration of 150 mM. The final system contained ~240,000 atoms with dimensions of 100×100×170 Å3 before any simulations. The prepared system was minimized using steepest descent algorithm for 5,000 steps. To prepare the nucleotide-bound system, Mg2+ and ATP were docked into their respective binding sites in the two NBDs following the protocol described by Wen et al. [53], which is based on the nucleotide binding characteristics observed in the high-resolution crystal structure of another ABC transporter, HlyB (PDB ID: 1XEF) [54]. Independent membrane building steps for apo and Mg2+/ATP-bound systems ensured randomization of the initial placement of lipids and cholesterols in the two systems (Fig. S1).
MD Simulation protocol
Simulations of Pgp in its apo and Mg2+/ATP-bound forms were performed in POPC:cholesterol (70:30) bilayers (Table 1). The simulations were performed at 310K, maintained constant using Langevin dynamics [55] with a damping coefficient of γ = 0.5 ps−1, and at 1 bar constant pressure maintained using the Nosé-Hoover Langevin piston method [56, 57]. The particle mesh Ewald (PME) method [58] was used for the calculation of long-range electrostatic forces. Non-bonded interactions were calculated with switching and cutoff distances of 10 Å and 12 Å, respectively, and a 2 fs timestep was used for integration.
Table 1:
Details of simulation systems.
| System | Protein | Membrane (POPC%:CHL%) |
Simulation copies |
# of cholesterol flipping events |
|---|---|---|---|---|
| Set 1 | apo | 70:30 | 20×200 ns | 1 |
| Set 2 | Mg2+/ATP | 70:30 | 20×200 ns | 2 |
To remove steric clashes between the protein and the surrounding lipids, the system was energy minimized and equilibrated following a step-wise protocol with restraints applied on different regions of the system and gradually removed in 6 steps. To restrain the protein, the following force constants were used: 10/5/2.5/1/0.5/0.1 kcal/mol/Å2 for backbone and 5/2.5/1/0.5/0.1/0 kcal/mol/Å2 for side chains during the 6-step equilibration protocol. The force constants used to restrain the position of lipid head groups along the z-axis in the bilayer during these 6 steps were 5/5/2/1/0.2/0 kcal/mol/Å2, respectively. Phosphorous and oxygen atoms were selected for POPC and cholesterol, respectively, to apply these bilayer restraints. Additionally, dihedral restraints were applied on the POPC lipids using extrabonds function of NAMD2 to maintain chirality of the lipid head group and isomerization around the double bond, with force constants of 500/200/100/100/50/0 kcal/mol/rad2 in 6 steps. Following the standard CHARMM-GUI protocol, the 6 equilibrium steps were run for 50, 50, 50, 200, 200 and 200 ps, respectively. Additional distance restraints were added between Mg2+/ATP and their interacting water molecules, walker-sequence residues, signature motif, and A-loop residues present in NBDs to help stabilize the docked Mg2+/ATP molecules at their respective binding sites [53]. The target distances of Mg2+/ATP molecules from their respective interacting residues in the NBDs were obtained from the crystal structure of dimerized NBDs of another ABC transporter, HlyB (PDB ID: 1XEF) [54]. For the production run, all simulations were performed for 200 ns using NAMD2 [59] with the CHARMM36 force-field representing all protein, lipid, and nucleic acids, and TIP3P model for water [60-62].
Analysis
The generated MD trajectories were analyzed using custom-written TCL scripts running in VMD [63] and plots were generated with Gnuplot [64] and R. In order to identify potential cholesterol binding sites, clustering analysis was performed using measure cluster command in VMD. Trajectories obtained from the last 50 ns of the simulations of both apo and Mg2+/ATP-bound systems were used in clustering calculations. All cholesterol molecules within 4 Å of any protein atoms were extracted and clustered based on the root-mean squared deviation (RMSD) values calculated using the heavy atoms of the cholesterol molecule after superimposing the protein. Two cholesterol molecules were considered to be in the same cluster if their RMSD was ≤ 3 Å. Three independent clustering calculations were performed with 10, 20, and 30 set as the maximum number of output clusters. Furthermore, the 200-ns trajectories were divided into four equal parts at 50 ns intervals and used in the clustering analysis to investigate the convergence of the obtained clusters over the course of the simulation.
The amino acid sequence of Pgp was scanned for Cholesterol Recognition Amino acid Consensus (CRAC) motifs and also for the inverted CRAC (CARC) motifs, using ScanProSite tool [65], as available in ExPASy Bioinformatics Resource Portal. A CRAC motif contains (L/V)-X1–5-(Y)-X1–5-(K/R) residues in which X can be any amino acid [50, 66]. CARC motif, on the other hand, contains the reverse sequence (K/R)-X1–5-(Y/F)-X1–5-(L/V), which is distinct from CRAC at the central aromatic amino acid which can be either Tyr or Phe. We also investigated the precise location of these motifs in the TM region of Pgp and reported their accessibility from the bilayer.
Preferential binding between the smooth α-face and rough β-face of cholesterol to Pgp was calculated based on their distance to the closest protein atom. Cholesterol molecules with their atoms located within 5 Å of any protein atom were selected for this analysis. Distances were calculated between the nearest protein atom and the center of mass of the carbon atoms C18 and C19 of the two methyl groups in cholesterol, representing the rough β-face, and between the nearest protein atom and the center of mass of carbon atoms C10 and C13 present at the interface of the A-B and C-D ring systems in cholesterol, representing the smooth α-face of the molecule (Fig. 1C). These distances were compared to the distances of the α- and β-faces to the closest POPC or cholesterol atom to identify sandwiched cholesterol molecules, where both smooth and rough faces of cholesterol lie closer to the protein than to the bilayer.
Results
Locations of identified CRAC and CARC motifs
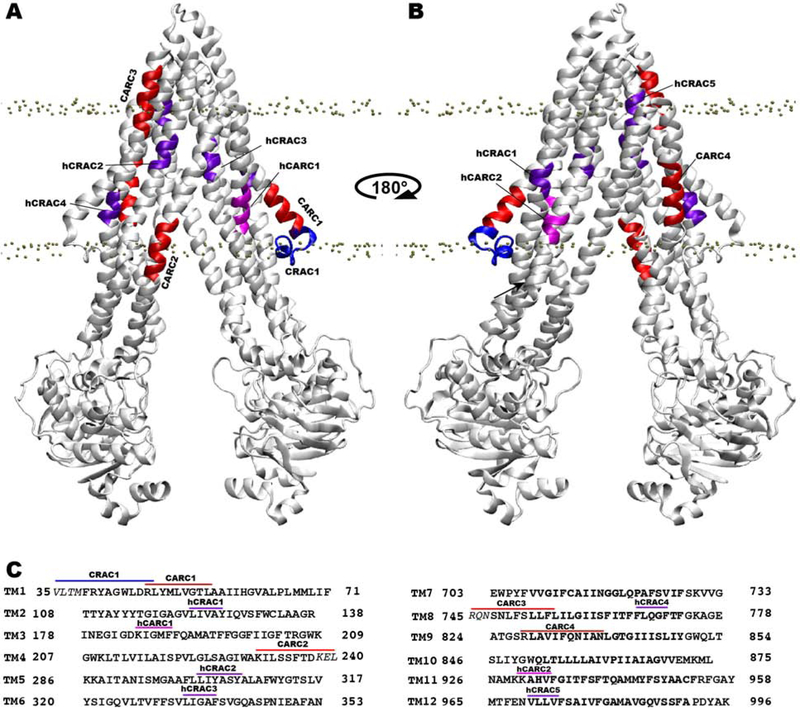
Scanning for the CRAC and CARC motifs in the Pgp sequence resulted in a total of 9 CRAC and 20 CARC motifs. Of these, only one CRAC and four CARC motifs are fully or partially located within the TM core (TM region present within the membrane) of Pgp (Fig. 2), and the others are present either in the intracellular/extracellular regions of the TM helices or within the NBDs, therefore, not expected to be able to contribute to binding of cholesterol. The only TM CRAC motif (V35-R47; labeled CRAC1) is located within the first elbow helix, whereas the CARC motif (R47-L55; labeled CARC1) immediately following it is located fully within the TM core (Fig. 2). Another CARC motif (R828-L839, labeled CARC4) is located fully within the TM core in TM9, whereas the two other CARC motifs, K230-L240 (TM4; labeled CARC2) and R745-L756 (TM8; labeled CARC3), are only partially located within the TM core. In addition to the CRAC/CARC motifs, we also report a set of half CRAC/CARC (hCRAC/hCARC) motifs in the TM core, defined to contain at least one half of the CRAC/CARC motifs including the aromatic Phe/Tyr residue present at the center (Fig. 2C). Scanning for hCRAC/hCARC motifs was motivated by the identification of Y126 from TM2, which is not part of a CRAC/CARC motif, as a cholesterol-interacting residue from the cholesterol clustering analysis performed in this study (see below). A total of seven hCRAC/hCARC motifs, including 5 hCRAC and 2 hCARC motifs, were identified within the TM core (Fig. 2). Proximity of these partial motifs in a three dimensional fold may allow them to behave similar to a full motif, especially when the missing part in a half motif is donated by the adjacent one. Eight out of 12 motifs are located within the intracellular half, 2 (hCRAC2 and hCRAC3) in the middle, and 2 (CARC3 and hCRAC5) in the extracellular half of the TM core of Pgp.
Figure 2:
Locations of identified CRAC and CARC motifs within the TM core of Pgp. (A) Front and (B) back views of Pgp viewed from the membrane. CRAC, hCRAC, CARC, and hCARC motifs are colored blue, purple, red, and magenta, respectively, and are labeled with consecutive numbering based on their appearance in the protein sequence. Phosphorous atoms of POPC are shown in gray spheres to denote the location of the bilayer. (C) Amino acid sequences of all TM helices with labeled motifs using the same color scheme as in A and B. Amino acid residues lying outside the TM core are shown in italicized text.
Calculated membrane accessibility of full and half motifs showed that CARC1, hCARC1, CARC2, CARC3, and CARC4 are highly accessible from the bilayer and thus available to bind cholesterol molecules present in the bilayer, whereas other motifs are generally buried inside the protein in the IF conformation, and may become accessible during IF to OF transition of Pgp (Fig. S2).
Asymmetric accumulation of cholesterol on the surface of Pgp
Both apo and Mg2+/ATP-bound simulations captured binding of a large number of cholesterol molecules to the surface of Pgp, indicating a close relationship between the protein and the lipid environment.
Our results reported here are based on the top 10, 20 and 30 clusters obtained from the clustering analysis of bound cholesterol molecules as detailed in Methods, in that other binding regions, which were sampled only rarely by cholesterol are not included in the clusters discussed.
Overall, the results show that the clusters were spread almost equally among the extracellular and intracellular leaflets (Table S1), although the majority of the highly populated (top 10) clusters are located in the extracellular leaflet. Most cholesterol clusters (8 out of 10, 16 out of 20, and 24 out of 30 clusters) are located on one side of Pgp (first half) formed by the first six TM helices (Fig. 3, Fig. S3 and Fig. S4), pointing to an asymmetric cholesterol binding preference. This asymmetric accumulation somewhat correlates with the 7 cholesterol-recognition motifs present in the first half compared to 5 in the second half of the protein, and also with their accessibility profiles from the membrane (Fig. 2 and Fig. S2). Six out of the top 30 clusters are located at the portal formed by TM4/6 helices in comparison to no cluster present at the symmetrically positioned TM10/12 portal (Fig. S4), which may be due to the presence of a CARC motif (CARC2) in TM4 (Fig. 2). Further analysis of these clusters showed accumulation of cholesterols at specific TM regions of Pgp formed by the recognition motifs. Clusters identified in the first half of Pgp are generally located around CARC1 (TM1), hCRAC1 (TM2), and CARC2 (at the portal in TM4) motifs, whereas other cholesterol motifs present in the first half of Pgp are buried within the protein and not accessible from the bilayer in the IF conformation of Pgp studied here (Fig. S4, Fig. 2 and Fig. S2). Similarly, some of the clusters in the second half of the protein are located around CARC3 (TM8) and CARC4 (TM9) motifs, whereas other cholesterol motifs in this half are inaccessible from the bilayer in the IF conformation.
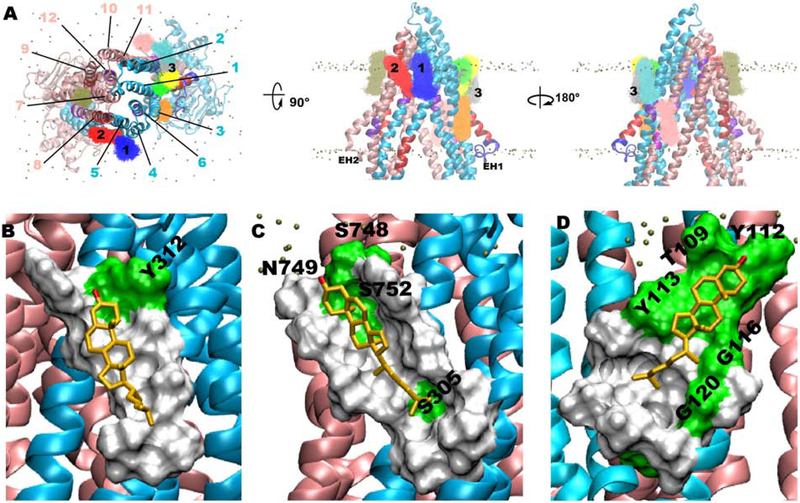
Figure 3:
Clustering analysis of cholesterol molecules located within 4 Å of Pgp using last 50 ns of both apo and Mg2+/ATP-bound simulation data (see Fig. S5 for comparison of cholesterol clusters between Mg2+/ATP-bound and apo systems). Top 10 clusters viewed from (A) top-down view from the extracellular side (left), and rotated side views from the membrane (middle and right) (see Fig. S3 and Fig. S4 for an overlay of the top 20 and 30 clusters, respectively). TM helices are numbered using black lines and first 3 clusters are numbered on top of the corresponding clusters. Representative binding modes of cholesterol from clusters 1 (B), 2 (C), and 3 (D) are shown in gold stick representation along with molecular surface formed by the protein residues within 5 Å. Hydrophobic and polar residues of the protein are shown in white and green color surface, respectively, with only polar residues labeled. Number of members present in each of these clusters is given in Table S1.
Representative conformations from the first 3 cholesterol clusters, collectively accounted for ~50% of the cholesterol population in the top 10 clusters (Fig. 3 and Table S1), were selected and their locations along with their interacting residues further analyzed. Cholesterol molecules in cluster-1 bind at the interface of TM4/5 in the extracellular side of the membrane, with the hydroxyl group interacting with Y312 of TM5 helix (Fig. 3B). Rest of the molecule binds to the hydrophobic region formed by residues V213, A216, I217, P219, V220, L308, W311 and Y312 of TM4/5 located adjacent to the hCRAC2 motif (Fig. 2 and Fig. 3A,B). The long axis of cholesterol in this cluster assumes an angle of ~22° to the membrane normal. Cholesterols present in cluster-2 bind at the TM5/8 interface in the extracellular half. The orientation of the molecules is similar to cluster-1 with an angle more tilted away from the membrane normal (~33°) (Fig. 3C). The polar region formed by S748, N749, and S752 interacts with the hydroxyl group of cholesterols present in this cluster and residues I217, V220, L221, L301, A304, S305, A307, L308, W311, L750, L753, F755 and L756 form the complementing hydrophobic surface for binding of the rest of the molecule. This cluster is sandwiched between hCRAC2 and CARC3 motifs (Fig. 2 and Fig. 3A,C). Cluster-3 binds at the amphiphilic (polar/hydrophobic) region formed by TM1/2 interface in the extracellular half, assuming angle of ~44° to the membrane normal (Fig. 3D). The polar part is majorly formed by residues T109 and aromatic Y112 and Y113 of TM2 and the hydrophobic part is formed by residues I59, V62, A63, L66, I117, A119, V121, and V124.
This cluster binds to the hCRAC1 recognition motif present in TM2 (Fig. 2 and Fig. 3A,D). Other locations where cholesterol accumulates are elbow helices (EH) present between TMDs and NBDs. These EHs lie perpendicular to the membrane normal and are located at the level of the head group region of the cytoplasmic leaflet. A representative conformation from a cluster located at EH1 (cluster-14), where the only fully TM CRAC motif is present (CRAC1 in TM1), binds almost perpendicular to the membrane normal by assuming an angle of ~78° to the membrane normal (Fig. S3). The hydroxyl group of cholesterols within this cluster interacts with R355, a charged residue of TM6 located at the level of the head group region of the cytoplasmic leaflet.
Some clusters within the top 30 clusters calculated are not located close to any recognition motifs indicating that CRAC/CARC motifs may not be prerequisite for binding cholesterol molecules but rather the 3-dimensional structure of the protein may allow engagement of residues from different parts of the structure, forming the binding sites for cholesterol. Additionally, regions were identified in the TMD showing depletion of cholesterol on the surface of Pgp. These were TM10 and extracellular part of TM11 (Fig. S4) where no cluster was obtained from our simulation data. This also matches with sequence-based CRAC/CARC motif prediction that shows TM10 and extracellular part of TM11 are void of these motifs (Fig. 2). Comparison of clustering analyses performed separately on apo and Mg2+/ATP-bound systems showed similar clusters obtained for both these systems, except for two clusters obtained between TM7 and TM8 in the case of apo simulations (Fig. S5). The occupancy maps obtained by using the last 50 ns of the simulations show results similar to those obtained from the clustering analysis (Fig. S6). The top 10 clusters identified over different time segments of the trajectory indicate convergence of the identified clusters (Fig. S7).
Cholesterol majorly binds to Pgp though its rough β-face
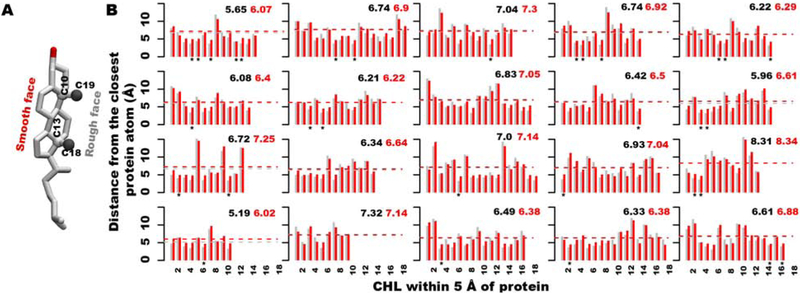
Cholesterol has an asymmetric ring system comprising a smooth α-face and a rough β-face due to the presence of two methyl groups at atoms C10 and C13 of the tetracyclic cyclopentanophenanthrene ring system (Fig. 4A).
Figure 4:
Binding preference between rough and smooth faces of cholesterol molecules to Pgp in its Mg2+/ATP-bound form (see Fig. S8 for apo system). (A) 3D structure of cholesterol with descriptions of the atoms used in distance calculations. C18/C19 atoms and C10/C13 atoms were selected to represent the rough and smooth faces of cholesterol, respectively, and are shown in gray and white ball representations. (B) Calculated center-of-mass distance profiles between rough (C18 and C19 atoms) and smooth (C10 and C13 atoms) faces of cholesterol and the closest atom of Pgp are shown in gray and red color bars, respectively. X-axis lists the number of cholesterols present within 5Å around the protein and selected for this analysis. Mean distance values are provided as black and red dashed lines for the rough and smooth faces, respectively. Asterisks in the plots denote the sandwiched cholesterols between protein atoms.
The center of mass distances between the selected atoms representing the α- and β-faces of cholesterol and the closest protein atom were calculated to distinguish between binding of cholesterol’s rough and smooth faces to Pgp.
Based on the calculated distance profiles using the final snapshots from 40 copies of apo and Mg2+/ATP-bound simulations, cholesterol molecules are found to bind to Pgp more likely via their rough β-face (Fig. 4B and Fig. S8). Out of 686 selected cholesterol molecules located within 5 Å of the protein in all the simulations, 386 molecules (56.3%) bind facing their rough face and 229 molecules (33.4%) bind facing their smooth face, whereas 71 cholesterols (10.3%) are sandwiched between the protein atoms, i.e., both the rough and smooth faces lie closer to the protein atoms compared to the neighboring membrane atoms.
Unconventional cholesterol flipping along the surface of Pgp
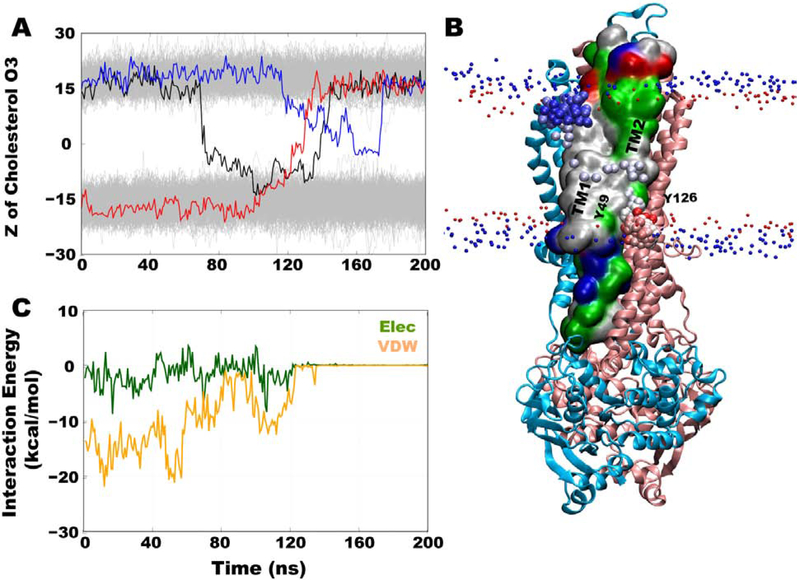
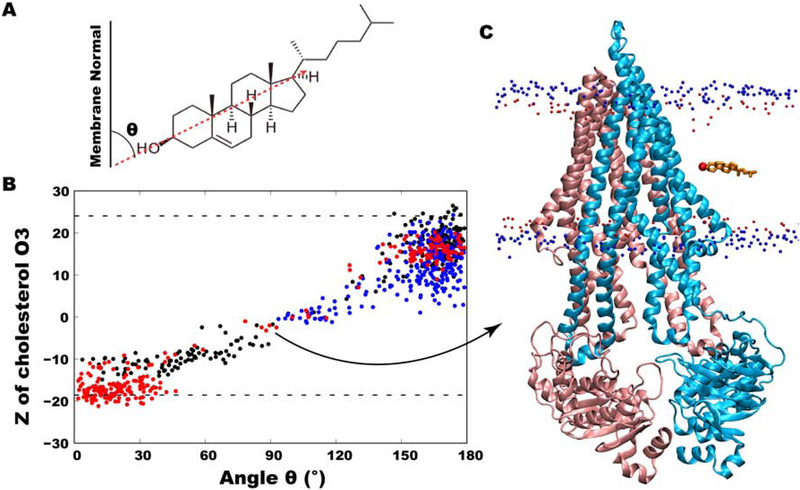
In order to monitor cholesterol positioning within the leaflets, the z-coordinates of the hydroxyl group oxygen atoms of ~5,740 cholesterol molecules in all 40 simulations were tracked as a function of the simulation time. Three cholesterol flipping events were observed along the surface of Pgp (Fig. 5A). Although these events are still rare in our data sets and only observed in a small fraction of the simulations (likely related to the short timescale of individual simulations compared to diffusion rate of lipids), their analysis can provide useful information about the cholesterol behavior and its dynamics at the protein interface. Out of these three events, one successful flipping occurred from the intracellular to the extracellular leaflet, with cholesterol stably merging with the lipids present in the latter leaflet (Fig. 5A, red trace). This event occurred in ~40 ns and was initiated at TM1, where the only full CRAC motif (V35-R47) and a CARC motif (R47-L55) are present (Fig. 5B and Fig. 2). In the second flipping event, cholesterol initially flipped from the extracellular to the intracellular leaflet, sampling the hydrophobic half of the intracellular leaflet for ~55 ns before flipping back to the original leaflet (Fig. 5A, black trace). In the third flipping event, the cholesterol molecule started to flip from the extracellular leaflet but did not complete the process, instead maintaining a ~90° angle to the membrane normal for a short duration before returning to the original leaflet (Fig. 5A, blue trace). The z-coordinates of the rest of the cholesterol hydroxyl groups remained stable at their positions throughout the simulation; indicating generally a low cholesterol flipping probability. All three cholesterol molecules that underwent partial or complete flipping achieved a 90° angle to the membrane normal at the middle of the membrane (Fig. 6A,B).
Figure 5:
Tracking cholesterol flipping events along the surface of Pgp. (A) z-coordinates of hydroxyl group O3 atoms of cholesterol in all simulations. Three cholesterol molecules that flipped during the simulations are shown in colored lines with the rest of the cholesterol molecules shown in gray. The only complete flipping event from intracellular to extracellular leaflet is shown in red line, whereas two partial/incomplete flipping events from extracellular to intracellular leaflets are shown in black and blue lines. (B) Molecular representation of Pgp in blue (TM1-6) and pink (TM 7-12) cartoon representation with positions of O3 atom of completely flipped cholesterol shown in red (beginning) to blue (end) beads coloring during the simulation. TM1 and TM2 helices are shown in surface representation and colored based on residue type (white: hydrophobic, green: hydrophilic, red: acidic, blue: basic). Two Tyr (Y49 of TM1 and Y126 of TM2) residues are labeled to denote the location of CARC1 and hCRAC1 motifs. Phosphorus atoms of POPC and O3 atoms of other cholesterol molecules are shown in small blue and red spheres, respectively, to demarcate the locations of the head groups. (C) Interaction energy profiles between the completely flipped cholesterol molecule and Pgp is shown as a function of simulation time.
Figure 6:
Cholesterol orientation in the membrane, defined as the angle between the long axis of cholesterol and membrane normal. (A) A schematic showing the vector used in measuring the angle. (B) z-coordinates of the hydroxyl group O3 atom of cholesterol molecules involved in complete or partial flipping as a function of their angles with respect to membrane normal. The completely flipped cholesterol molecule is shown in red dots. (C) Molecular representation of Pgp shown in cartoon representation (blue and pink colors) with the completely flipped cholesterol molecule present at its 90 degree angle during flipping. Phosphorus atoms of POPC and O3 atoms of other cholesterol molecules are shown in small blue and red spheres in the background.
The interaction energies calculated between the fully flipped cholesterol molecule and Pgp are dominated by van der Waals interactions, with small electrostatic contributions via its hydroxyl group (Fig. 5C). As the molecule makes a ~90° angle to the membrane normal, only the hydroxyl group interacts with the protein, with the rest of the molecule buried inside the membrane core (Fig. 6C). At this point, all Pgp-cholesterol interactions were lost and the remainder of the flipping occurred without any interactions with the protein, suggesting that only intracellular half of the TM region of the protein is involved in this event. For the rest of the simulation time, i.e., before and after the flipping event, this cholesterol molecule sampled conformations parallel to the membrane normal (Fig. 6B).
Pgp-cholesterol interactions during flipping
All three flipping events sampled in our simulations occurred in the same region, located at the intracellular leaflet, formed by TM1, TM2, and TM11 helices.
Analysis of the amino acid composition of these helices revealed that along with the presence of a full CRAC and a full CARC motifs in TM1, one partial CRAC and one partial CARC motifs are also present in the same region, making it highly conducive for cholesterol binding (Fig. 2).
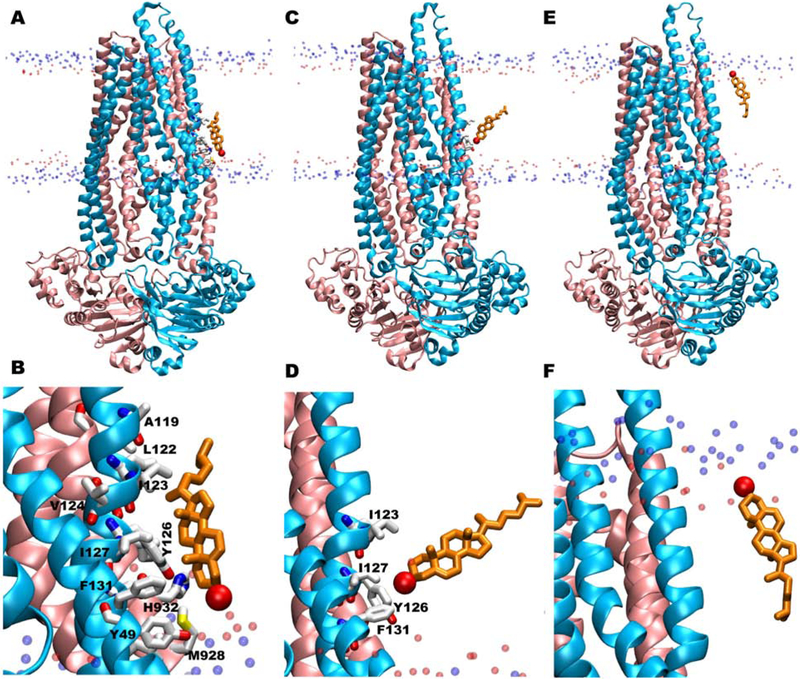
The fully flipped cholesterol initially interacts with Y49 of TM1 and H932 of TM11, with both side chains pointing towards the membrane (Fig. 7A,B). F131 of TM2 and M928 of TM11 also take part in forming a non-polar groove for cholesterol. Another Tyr residue from a partial CRAC motif (Y126 in TM2), located slightly above the membrane head groups in the intracellular leaflet, favorably interacts with the cholesterol molecule to facilitate upward movement at the start of the flipping event (Fig. 7C,D). Nonpolar aliphatic residues, I123 and I127 in TM2, also interact with cholesterol at this time. All interactions between Pgp and cholesterol are lost after cholesterol reaches ~90° angle with respect to the membrane normal (Fig. 7E,F). Two cholesterol molecules from the extracellular leaflet, that show incomplete/partial flipping, were seen to visit the same region formed by Y49, Y126, M928, and H932, before returning to the original leaflet, indicating this region is important not only for cholesterol binding but also for initiating flipping events.
Figure 7:
Snapshots of the complete flipping of cholesterol along the surface of Pgp observed in our simulations. (A, C, E) Different orientations and locations of the fully flipped cholesterol molecule within the bilayer during its flipping. (B, D, F) Corresponding zoomed-in views of the binding orientations in A, C, and E showing protein residues within 5 Å of the cholesterol molecule in white licorice representation. Most of the residues are from TM1 and TM2 helices where both CARC1 and hCRAC1 motifs are located. The flipped cholesterol molecule is shown in orange licorice representation with its oxygen atom in red. Phosphorus (blue) and oxygen (red) atoms of POPC and cholesterol molecules in the membrane are shown in the background.
Discussion
Plasma membranes of higher organisms like mammals are rich in cholesterol, where it plays an important role not only in modulating the physical properties of the membrane but also the functions of both integral and peripheral membrane proteins [37, 41, 67]. Despite many studies performed over the last decade investigating the relationship between Pgp, a widely studied integral membrane ABC-transporter, and cholesterol, very limited microscopic detail has been presented on how this protein interacts with cholesterol and mediates its redistribution within the lipid bilayer [42]. The present study, with simulations of multiple copies of Pgp in cholesterol-rich lipid bilayers in both apo and Mg2+/ATP-bound states, enabled the direct observations of cholesterol binding sites on the surface of the protein and even unconventional flipping events between the membrane leaflets.
The conceptual cholesterol recognition motifs, CARC and CRAC, primarily defined based on the primary sequence of a membrane protein, have been used in studies focusing on predicting cholesterol interaction with membrane protein [66]. Given its limited scope, specially with regard to spatial arrangements of residues to form a binding site, which cannot be necessarily predicted merely based on the primary sequence, we should not expect a thorough survey of putative cholesterol binding sites by this metric. Compared to previous studies focusing on the cholesterol recognition motifs in Pgp [50], we have identified additional CRAC/CARC motifs, including partial recognition motifs, along with their relative membrane locations (Fig. 2). This includes a complete CARC motif in TM1 and an incomplete CRAC motif in TM2, missing only the terminal positively charged residue (K/R). Although the missing charged residues in the incomplete CRAC motifs may decrease the interactions with the hydroxyl group of cholesterol, neighboring partial recognition motifs present in the 3-dimensional structure may be able to compensate for these missing interactions. Scanning other ABC transporters for CRAC/CARC motifs with missing terminal residues may also allow for the identification of partial recognition motifs in these proteins that can successfully interact with and bind cholesterol.
A coarse-grained MD simulation study had previously reported cholesterol binding at the two drug-entry portals, formed by TM4/6 helices on one side and TM10/12 helices on the other [68]. In the present study, clustering analyses performed on all cholesterol molecules within 4 Å of the protein showed that they tend to accumulate substantially more on one side of the TMD, formed by TM1-6 of TMD1 and TM8 of TMD2 (Fig. 3), with only few cholesterol molecules observed in the vicinity of the TM10/12 helices on the opposite side (Fig. S4).
The clustering also showed asymmetric accumulation of cholesterol molecules between the two membrane leaflets, with most of the highly populated top 10 clusters generated found to be located in the extracellular leaflet. The difference might have resulted from the conical shape of the TMD of Pgp, where the intracellular half is significantly wider than its extracellular half, resulting in less-dense cholesterol clusters that are not ranked in top 10. Overall, the results indicate a preference for a non-uniform distribution of cholesterol on the surface of Pgp, that may in turn differentially modulate the dynamics of different regions of the transporter, promoting or inhibiting specific conformational changes in the protein. Previous MD simulations reported that removal of the methyl groups forming the rough β-face of cholesterol lowers its membrane ordering and condensing properties, and disturbs the optimal orientation of the molecule [69]. An experimental study combining calorimetry and fluorescence measurements also confirmed the importance of the rough and smooth faces of cholesterol in sterol orientation in the bilayer that in turn improves cholesterol-phospholipid interactions [70].
In this study, we observe preferential binding of cholesterol to Pgp through its rough β-face (Fig. 4 and Fig. S8). As the smooth α-face of the cholesterol ring is shown to be favored in packing nearby lipid tails via stronger van der Waals interactions [69, 70], the rough β-face might be more available to interact with the surface of the transporter.
Out of a large dataset containing ~5,740 copies of cholesterol, one cholesterol molecule showed complete flipping and translation from the intracellular leaflet to the extracellular one (Fig. 5 and Fig. 6). Two other cholesterol molecules from the extracellular leaflet showed incomplete or partial flipping, forming ~90° angles with the membrane normal at the interface of the two leaflets.
During the complete flipping event, which was analyzed in more detail, the initial time is spent in achieving the correct binding interactions and orientation of cholesterol with respect to the recognition CRAC/CARC motifs present in TM1 and TM2 (Fig. 5 and Fig. 7). These motifs present in the intracellular part of TM1 and TM2 helices may in fact contribute to the flippase activity of Pgp [49].
Computational studies with free energy calculations have reported that cholesterol prefers to tilt and rotate first, embedding its polar head group into the hydrophobic core of the bilayer, which appears to be the most energetically unfavorable and likely the rate-limiting step, before translating to the opposite leaflet [71, 72]. It is important to note that these studies were performed with pure membrane bilayers with no proteins. Interestingly, the same trend is observed in our study in the presence of a transporter, where cholesterol first interacts with the binding-motifs, engaging its hydroxy group with the protein surface thereby reducing the energy barrier, before flipping to the opposite leaflet (Fig. 7). Previous coarse-grained MD simulations of asymmetric lipid bilayers without protein, have also reported multiple flip-flop events for cholesterol with a mean time of 350 - 2,000 ns [73, 74]. In our study, the fully flipped cholesterol showed a shuttling time of only ~40 ns between the two leaflets. Pgp may in fact provide a surface with lower hydrophobicity for binding the polar head group of cholesterol compared to the purely hydrophobic core of the bilayer. This may reduce the energy barrier arising from cholesterol-lipid interactions in pure membranes, resulting in faster shuttling time compared to pure lipid bilayers [71].
Our results on spontaneous cholesterol flipping along the surface of Pgp calls into question the conventional understanding about the flippase activities of ABC exporters for cholesterol. This activity has been proposed to occur through the poly-specific translocation chamber of Pgp, present at the center of the TMD. In fact, this conventional mechanism still appears to be the only way cholesterol transport can be unidirectional (from inside to outside) and coupled to ATP binding and hydrolysis (active transport). Here we present an alternate mechanism of lipid flipping activity of the transporter on the surface of the protein, a passive mechanism where the conical shape of Pgp in its IF state, forming a broader base at the intracellular leaflet and a narrower apex at the extracellular leaflet, may play a role in the surface flipping of these molecules (Fig. 5). In this way, for a successful flipping to take place, the molecule has to cross only one-half of the protein, before diffusing into the opposite membrane leaflet, reducing the energetic cost involved in the lipid redistribution between the two membrane leaflets i.e. this mode of transport will be passive. For this alternate mechanism, the role of structural changes of the protein in the mechanism can be simply bringing about a conformation that is most conducive to this mode of cholesterol transport. The protein surface-mediated cholesterol flipping mechanism, in which the energy barrier is lowered by providing polar and hydrogen bonding surfaces within the membrane, may also represent a mechanism relevant to other membrane proteins exposing polar residues within the membrane.
Conclusion
Here we have shown the different cholesterol-binding regions in Pgp and the preferential binding between its smooth α-face and rough β-face to the transporter, using simulation data providing extensive sampling of cholesterol molecules around Pgp. Locations of full and partial cholesterol-recognition motifs with respect to the lipid bilayer are also reported, though these motifs do not appear to sufficiently account for all cholesterol binding sites/modes in the protein. Our unique and direct observation of cholesterol flipping on the surface of Pgp provide a new passive mechanism by which Pgp might contribute to the redistribution of lipids in membrane. As a single successful flipping event was observed between the two leaflets, further studies will be necessary for deriving a general mechanism of passive transport in membrane proteins. The work provides a snapshot into understanding the intimate relationship between Pgp and cholesterol, an essential membrane molecule known to closely regulate the function of membrane proteins.
Our simulations show the importance of studying membrane proteins in a membrane bilayer that mimics the biological environments as much as possible in order to make biologically relevant conclusions.
Supplementary Material
Highlights.
Extensive set of molecular simulations probe cholesterol binding to P-glycoprotein.
Cholesterol binding hotspots are identified for both apo and ATP-bound states.
CARC/CRAC motifs are found to be inadequate descriptors for cholesterol binding.
A novel pathway for cholesterol translocation across the lipid bilayer is reported.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Dean M, Annilo T, Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates, Annu. Rev. Genomics Hum. Genet 6 (2005) 123–142. [DOI] [PubMed] [Google Scholar]
- [2].Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W, Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon thermococcus litoralis, EMBO J. 19 (2000) 5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hosie AH, Poole PS, Bacterial ABC transporters of amino acids, Res. Microbiol 152 (2001) 259–270. [DOI] [PubMed] [Google Scholar]
- [4].Tarling EJ, de Aguiar Vallim TQ, Edwards PA, Role of ABC transporters in lipid transport and human disease, Trends Endocr. Metab 24 (2013) 342–50. URL: http://www.ncbi.nlm.nih.gov/pubmed/23415156. doi: 10.1016/j.tem.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gerber S, Comellas-Bigler M, Goetz BA, Locher KP, Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter, Science 321 (2008) 246–250. [DOI] [PubMed] [Google Scholar]
- [6].Eckford PDW, Sharom FJ, ABC efflux pump-based resistance to chemotherapy drugs, Chem. Rev 109 (2009) 2989–3011. [DOI] [PubMed] [Google Scholar]
- [7].Loo TW, Clarke DM, A salt bridge in intracellular loop 2 is essential for folding of human P-glycoprotein, Biochemistry 52 (2013) 3194–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Stege A, Priebsch A, Nieth C, Lage H, Stable and complete overcoming of MDR1/P-glycoprotein-mediated multidrug resistance in human gastric carcinoma cells by rna interference, Cancer Gene Ther. 11 (2004) 699 – 706. [DOI] [PubMed] [Google Scholar]
- [9].Ferté J, Analysis of the tangled relationships between P-glycoprotein-mediated multidrug resistance and the lipid phase of the cell membrane, Eur. J. Biochem 267 (2000) 277–294. URL: 10.1046/j.1432-1327.2000.01046.x. doi: 10.1046/j.1432-1327.2000.01046.x. [DOI] [PubMed] [Google Scholar]
- [10].Mechetner EB, Roninson IB, Efficient inhibition of P-glycoprotein-mediated multidrug resistance with a monoclonal antibody, Proc. Natl. Acad. Sci. USA 89 (1992) 5824–5828. URL: http://www.pnas.org/content/89/13/5824.abstract arXiv:http://www.pnas.org/content/89/13/5824.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Szakàcs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM, Targeting multidrug resistance in cancer, Nat. Rev. Drug Disc 5 (2006) 219–234. [DOI] [PubMed] [Google Scholar]
- [12].Li J, Jaimes KF, Aller SG, Refined structures of mouse P-glycoprotein, Prot. Sci 23 (2014) 34–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G, Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding, Science 323 (2009) 1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Omote H, Al-Shawi MK, Interaction of transported drugs with the lipid bilayer and P-glycoprotein through a solvation exchange mechanism, Biophys. J 90 (2006) 4046–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wasan KM, Brocks DR, Lee SD, Sachs-Barrable K, Thornton SJ, Impact of lipoproteins on the biological activity and disposition of hydrophobic drugs: implications for drug discovery, Nat. Rev. Drug Disc 7 (2008) 84–99. doi: 10.1038/nrd2353. [DOI] [PubMed] [Google Scholar]
- [16].Jin MS, Oldham ML, Zhang Q, Chen J, Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans, Nature 490 (2012) 566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rosenberg MF, Callaghan R, Modok S, Higgins CF, Ford RC, Three-dimensional structure of P-glycoprotein: the transmembrane regions adopt an asymmetric configuration in the nucleaotide-bound state, J. Biol. Chem 280 (2005) 2857–2862. URL: http://www.jbc.org/content/280/4/2857.abstract. doi: 10.1074/jbc.M410296200. arXiv:http://www.jbc.org/content/280/4/2857.full.pdf+html. [DOI] [PubMed] [Google Scholar]
- [18].Mitchell P, A general theory of membrane transport from studies of bacteria., Nature 180 (1957) 134–136. [DOI] [PubMed] [Google Scholar]
- [19].Jardetzky O, Simple allosteric model for membrane pumps, Nature 211 (1966) 969–970. [DOI] [PubMed] [Google Scholar]
- [20].Dawson RJ, Locher KP, Structure of a bacterial multidrug ABC transporter, Nature 443 (2006) 180–185. [DOI] [PubMed] [Google Scholar]
- [21].Oldham ML, Davidson AL, Chen J, Structural insights into ABC transporter mechanism, Curr. Opin. Struct. Biol 18 (2008) 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Locher KP, Structure and mechanism of ATP-binding cassette transporters, Phil. Trans. R. Soc. Lond. B 364 (2009) 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ward A, Reyes CL, Yu J, Roth CB, Chang G, Flexibility in the ABC transporter MsbA: Alternating access with a twist, Proc. Natl. Acad. Sci. USA 104 (2007) 19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hohl M, Briand C, Grütter MG, Seeger MA, Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation, Nat. Struct. Mol. Biol 19 (2012) 395–402. [DOI] [PubMed] [Google Scholar]
- [25].Verhalen B, Dastvan R, Thangapandian S, Peskova Y, Koteiche HA, Nakamoto RK, Tajkhorshid E, Mchaourab HS, Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein, Nature 543 (2017) 738–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM, P-glycoprotein: from genomics to mechanism, Oncogene 22 (2003) 7468–7485. [DOI] [PubMed] [Google Scholar]
- [27].Sauna ZE, Ambudkar SV, About a switch: how P-glycoprotein (ABCB1) harnesses the energy of ATP binding and hydrolysis to do mechanical work, Mol. Cancer Ther 6 (2007) 13–23. URL: http://mct.aacrjournals.org/content/6/1/13.abstract. doi: 10.1158/1535-7163.MCT-06-0155 arXiv:http://mct.aacrjournals.org/content/6/1/13.full.pdf+html. [DOI] [PubMed] [Google Scholar]
- [28].Hong WC, Amara SG, Membrane cholesterol modulates the outward facing conformation of the dopamine transporter and alters cocaine binding., J. Biol. Chem 285 (2010) 32616–32626. URL: 10.1074/jbc.M110.150565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].North P, Fleischer S, Alteration of synaptic membrane cholesterol/phospholipid ratio using a lipid transfer protein. effect on gamma-aminobutyric acid uptake., J. Biol. Chem 258 (1983) 1242–1253. [PubMed] [Google Scholar]
- [30].Scanlon SM, Williams DC, Schloss P, Membrane cholesterol modulates serotonin transporter activity., Biochemistry 40 (2001) 10507–10513. [DOI] [PubMed] [Google Scholar]
- [31].Schroeder C, Heider H, Möncke-Buchner E, Lin TI, The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein, Eur. Biophys. J 34 (2005) 52–66. [DOI] [PubMed] [Google Scholar]
- [32].Levitan I, Barrantes FJ (Eds.), Cholesterol regulation of ion channels and receptors, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2012. URL: http://doi.wiley.com/10.1002/9781118342312. doi: 10.1002/9781118342312. [DOI] [Google Scholar]
- [33].Oates J, Watts A, Uncovering the intimate relationship between lipids, cholesterol and GPCR activation., Curr. Opin. Struct. Biol 21 (2011) 802–7. URL: http://www.ncbi.nlm.nih.gov/pubmed/22036833. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- [34].Rosenhouse-Dantsker A, Noskov S, Durdagi S, Logothetis DE, Levitan I, Identification of novel cholesterol-binding regions in Kir2 channels., J. Biol. Chem 288 (2013) 31154–31164. URL: http://www.ncbi.nlm.nih.gov/pubmed/24019518. doi: 10.1074/jbc.M113.496117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jones KT, Zhen J, Reith ME, Importance of cholesterol in dopamine transporter function, J. Neurochem 123 (2012) 700–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Goluszko P, Nowicki B, Membrane cholesterol: a crucial molecule affecting interactions of microbial pathogens with mammalian cells, Infect. Immun 73 (2005) 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Maxfield FR, van Meer G, Cholesterol, the central lipid of mammalian cells, Curr. Opin. Cell Biol 22 (2010) 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Luker GD, Pica CM, Kumar AS, Covey DF, Piwnica-Worms D, Effects of cholesterol and enantiomeric cholesterol on P-glycoprotein localization and function in low-density membrane domains, Biochemistry 39 (2000) 7651–7661. URL: http://pubs.acs.org/doi/abs/10.1021/bi9928593. doi: 10.1021/bi9928593. arXiv:http://pubs.acs.org/doi/pdf/10.1021/bi9928593. [DOI] [PubMed] [Google Scholar]
- [39].Corsetto PA, Colombo I, Kopecka J, Rizzo AM, Rigant C, ω-3 long chain polyunsaturated fatty acids as sensitizing agents and multidrug resistance revertants in cancer therapy, Int. J. Mol. Sci 18 (2017). doi: 10.3390/ijms18122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Belli S, Elsener PM, Wunderli-Allenspach H, Krämer SD, Cholesterol-mediated activation of P-glycoprotein: Distinct effects on basal and drug-induced ATPase activities, J. Pharmaceut. Sci 98 (2009) 1905–1918. URL: 10.1002/jps.21558. doi: 10.1002/jps.21558. [DOI] [PubMed] [Google Scholar]
- [41].Meyer dos Santos S, Meyer Dos Santos S, Weber C-C, Franke C, Müller WE, Eckert GP, Cholesterol: Coupling between membrane microenvironment and ABC transporter activity., Biochem. Biophys. Res. Commun 354 (2007) 216–21. URL: http://www.ncbi.nlm.nih.gov/pubmed/17223079. doi: 10.1016/j.bbrc.2006.12.202. [DOI] [PubMed] [Google Scholar]
- [42].Eckford PDW, Sharom FJ, Interaction of the P-glycoprotein multidrug efflux pump with cholesterol: effects on ATPase activity, drug binding and transport., Biochemistry 47 (2008) 13686–98. URL: http://www.ncbi.nlm.nih.gov/pubmed/19049391. doi: 10.1021/bi801409r. [DOI] [PubMed] [Google Scholar]
- [43].Fenyvesi F, Fenyvesi E, Szente L, Goda K, Bacsó Z, Bácskay I, Váradi J, Kiss T, Molnár E, Janáky T, Szabó G, Vecsernyés M, P-glycoprotein inhibition by membrane cholesterol modulation., Eur. J. Pharm. Sci 34 (2008) 236–42. URL: http://www.ncbi.nlm.nih.gov/pubmeFd/18539442. doi: 10.1016/j.ejps.2008.04.005. [DOI] [PubMed] [Google Scholar]
- [44].Aurade RM, Jayalakshmi SK, Udikeri SS, Sreeramulu K, Modulation of P-glycoprotein ATPase of Helicoverpa armigera by cholesterol: effects on ATPase activity and interaction of insecticides., Arch. Insect Biochem. Physiol 79 (2012) 47–60. URL: http://www.ncbi.nlm.nih.gov/pubmed/23589220. doi: 10.1002/arch.21004. [DOI] [PubMed] [Google Scholar]
- [45].Rothnie A, Theron D, Soceneantu L, Martin C, Traikia M, Berridge G, Higgins CF, Devaux PF, Callaghan R, The importance of cholesterol in maintenance of P-glycoprotein activity and its membrane perturbing influence, Eur. Biophys. J 30 (2001) 430–442. doi: 10.1007/s002490100156. [DOI] [PubMed] [Google Scholar]
- [46].Sharom FJ, Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: Its role in modulating protein function., Front. Oncol 4 (2014) 1–19. URL: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3939933/. doi: 10.3389/fonc.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Subramanian N, Schumann-Gillett A, Mark AE, O’Mara ML, Understanding the accumulation of P-glycoprotein substrates within cells: The effect of cholesterol on membrane partitioning, Biochim. Biophys. Acta, Biomembr 1858 (2016) 776–782. doi: 10.1016/j.bbamem.2015.12.025. [DOI] [PubMed] [Google Scholar]
- [48].Wang E, Casciano CN, Clement RP, Johnson WW, Cholesterol interaction with the daunorubicin binding site of P-glycoprotein, Biochem. Biophys. Res. Commun 276 (2000) 909–916. [DOI] [PubMed] [Google Scholar]
- [49].Garrigues A, Escargueil AE, Orlowski S, The multidrug transporter, P-glycoprotein, actively mediates cholesterol redistribution in the cell membrane, Proc. Natl. Acad. Sci. USA 99 (2002) 10347–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Clay AT, Lu P, Sharom FJ, Interaction of the P-glycoprotein multidrug transporter with sterols, Biochemistry 54 (2015) 6586–6597. doi: 10.1021/acs.biochem.5b00904. [DOI] [PubMed] [Google Scholar]
- [51].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: A web-based graphical user interface for CHARMM, J. Comp. Chem 29 (2008) 1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- [52].Jo S, Lim JB, Klauda JB, Im W, CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes, Biophys. J 97 (2009) 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wen P-C, Verhalen B, Wilkens S, Mchaourab HS, Tajkhorshid E, On the origin of large flexibility of p-glycoprotein in the inward-facing state, J. Biol. Chem 288 (2013) 19211–19220. doi: 10.1074/jbc.M113.450114. arXiv:http://www.jbc.org/content/288/26/19211.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L, H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB, EMBO J. 24 (2005) 1901–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ryckaert J-P, Ciccotti G, Berendsen HJC, Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes, J. Comp. Phys 23 (1977) 327–341. [Google Scholar]
- [56].Martyna GJ, Tobias DJ, Klein ML, Constant pressure molecular dynamics algorithms, J. Chem. Phys 101 (1994) 4177–4189. [Google Scholar]
- [57].Feller SE, Zhang Y, Pastor RW, Constant pressure molecular dynamics simulation: The Langevin piston method, J. Chem. Phys 103 (1995) 4613–4621. [Google Scholar]
- [58].Darden T, York D, Pedersen L, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, J. Chem. Phys 98 (1993) 10089–10092. [Google Scholar]
- [59].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K, Scalable molecular dynamics with NAMD, J. Comp. Chem 26 (2005) 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Huang J, MacKerell AD, Charmm36 all-atom additive protein force field: Validation based on comparison to nmr data, J. Comp. Chem 34 (2013) 2135–2145. URL: 10.1002/jcc.23354. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Feller SE, MacKerell AD Jr., An improved empirical potential energy function for molecular simulations of phospholipids, J. Phys. Chem. B 104 (2000) 7510–7515. [Google Scholar]
- [62].Foloppe N, MacKerell AD Jr., All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data, J. Comp. Chem 21 (2000) 86–104. [Google Scholar]
- [63].Humphrey W, Dalke A, Schulten K, VMD – Visual Molecular Dynamics, J. Mol. Graphics 14 (1996) 33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [64].Williams T, K. C., Gnuplot: An interactive plotting program., 1993.
- [65].de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N, Scanprosite: detection of prosite signature matches and prorule-associated functional and structural residues in proteins, Nucleic Acids Res. 34 (2006) W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fantini J, Barrantes F, How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains, Front. Physiol 4 (2013) 31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wen P-C, Mahinthichaichan P, Trebesch N, Jiang T, Zhao Z, Shinn E, Wang Y, Shekhar M, Kapoor K, Chan CK, Tajkhorshid E, Microscopic view of lipids and their diverse biological functions, Curr. Opin. Struct. Biol 51 (2018) 177 – 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Domicevica L, Kolds H, Biggin PC, Multiscale molecular dynamics simulations of lipid interactions with P-glycoprotein in a complex membrane, J. Mol. Graph. Model 77 (2017) 250 – 258. URL: http://www.sciencedirect.com/science/article/pii/S1093326317302474. doi: 10.1016/j.jmgm.2017.09.002. [DOI] [PubMed] [Google Scholar]
- [69].Smondyrev AM, Berkowitz ML, Molecular dynamics simulation of the structure of dimyristoylphosphatidylcholine bilayers with cholesterol, ergosterol, and lanosterol, Biophys. J 80 (2001) 1649–1658. doi: 10.1016/S0006-3495(01)76137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Róg T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M, What happens if cholesterol is made smoother: Importance of methyl substituents in cholesterol ring structure on phosphatidylcholine-sterol interaction, Biophys. J 92 (2007) 3346–3357. doi: 10.1529/biophysj.106.095497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jo S, Rui H, Lim JB, Klauda JB, Im W, Cholesterol flip-flop: Insights from free energy simulation studies, J. Phys. Chem. B 114 (2010) 13342–13348. doi: 10.1021/jp108166k. [DOI] [PubMed] [Google Scholar]
- [72].Parisio G, Sperotto MM, Ferrarini A, Flip-flop of steroids in phospholipid bilayers: Effects of the chemical structure on transbilayer diffusion, J. Am. Chem. Soc 134 (2012) 12198–12208. doi: 10.1021/ja304007t. [DOI] [PubMed] [Google Scholar]
- [73].Yesylevskyy SO, Demchenko AP, How cholesterol is distributed between monolayers in asymmetric lipid membranes, Eur. Biophys. J 41 (2012) 1043–1054. doi: 10.1007/s00249-012-0863-z. [DOI] [PubMed] [Google Scholar]
- [74].Yesylevskyy SO, Demchenko AP, Cholesterol Behavior in Asymmetric Lipid Bilayers: Insights from Molecular Dynamics Simulations, Springer New York, New York, NY, 2015, pp. 291–306. doi: 10.1007/978-1-4939-1752-5_20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.