Abstract
Transcriptional enhancers in the cell nuclei typically interact with the target promoters in cis over long stretches of chromatin, but the mechanism of this communication remains unknown. Previously we have developed a defined in vitro system for quantitative analysis of the rate of distant enhancer-promoter communication (EPC) and have shown that the chromatin fibers maintain efficient distant EPC in cis. Here we investigate the roles of linker histone H1 and HMGN5 protein in EPC. A considerable negative effect of histone H1 on EPC depending on its C- and N-tails was shown. Protein HMGN5 that affects chromatin compaction and is associated with active chromatin counteracts EPC inhibition by H1. The data suggest that the efficiency of the interaction between the enhancer and the promoter depends on the structure and dynamics of the chromatin fiber localized between them and can be regulated by proteins associated with chromatin.
Keywords: chromatin, enhancer-promotor communication, transcription in vitro, H1, HMGN5
INTRODUCTION
Communication between DNA regulatory regions and their targets in cis and in trans is involved in regulation of various cellular processes, including gene expression, chromosome translocation and splicing [1, 2]. At least in the case of transcriptional regulation, the activating DNA regions (enhancers) physically interact with their target promoters; these interactions are mediated by specifically DNA-bound proteins and accompanied by formation of chromatin loops [3–5]. Enhancers can efficiently activate transcription in cis over a wide range of distances (from hundreds to hundreds of thousands of base pairs [6]. To ensure the formation of loops of such different sizes, chromatin structure must be flexible and dynamic [7]. In order to participate in such regulatory interactions, chromatin structure has to be activated and decompacted to the level of the chromatin fiber [8, 9].
Although activated chromatin fibers are highly mobile structures [10, 11], the impact of this mobility on the efficiency of enhancer-promoter communication (EPC) in cis has not been established. DNA sequences separated by more than 1 kb do not communicate efficiently on linear DNA in vitro [12, 13]. Chromatin can facilitate short-range (within 200-bp distance) interactions between DNA elements in vitro [14, 15], Furthermore, chromatin structure can greatly facilitate intramolecular ligation of distantly spaced DNA ends [16] and EPC over a distance of at least 4 kb in vitro [17, 18]. Presence or absence of histone H1 and HMGN5 protein can affect compactness of chromatin in vitro [19], and correlates with inactive/active states of chromatin in vivo, respectively [20–23].
Based on the evidence that linker histone H1 and HMGN5 change chromatin structure and dynamics, we assumed that these proteins can change EPC in chromatin. Earlier we have developed an experimental approach that allows quantitative analysis of the rate of EPC in chromatin in vitro [24]. We have demonstrated that the chromatin fiber is an efficient communication device and that the histone N-terminal tails strongly facilitate EPC in chromatin [25]. In the current work, we have employed this experimental system for quantitative analysis of the roles of linker histone H1 and HMGN5 protein in EPC in chromatin.
METHODS
Proteins.
All transcriptional machinery proteins were purified as described [24]. Histone H1 was isolated as described [26] and further purified by two chromatography steps: using Pharmacia MonoS ion-exchange and Phenomenex Jupiter C4-bonded HPLC columns. All H1 subtypes were pooled together, except for H1°. HMGN5 was purified as described [19]. Recombinant H1 and its mutants were purified as described [27].
Chromatin assembly.
H1/H5-depleted chicken erythrocyte donor chromatin was prepared as described [28]. Tailless donor chromatin was prepared by digesting the long H1/H5-depleted chromatin with trypsin as described [29].
In vitro reconstitution of chromatin on supercoiled DNA templates was conducted at different mass ratios of donor chromatin DNA to template DNA (0.75 :1; 1 :1; 1.25 :1) using continuous dialysis from 1 M to 10 mM NaCl [28].
Analysis of chromatin templates in native gel.
Three nucleosome template for analysis of H1 binding with chromatin were assembled on 5’-radioactively labeled ApoI/EcoRI-fragment of pYP07 plasmid described before [25]. This fragment contains 601 sequences allowing precise nucleosome positioning and located on the distances of 30 bp between each over. The templates were incubated for 30 minutes at 37°C at a 1:1 molar ratio of H1 (or its mutant) to histone octamer in the transcription buffer followed by analysis by electrophoresis in 0.7% Agarose gel containing 0.5x TBE buffer during 4 hours at 4°C. Agarose gel was dried, exposed for phosphor imaging screen and scanned (BioRad, USA).
Analysis of H1 binding by micrococcal nuclease (MNase) digestion.
H1 binding to intact and tailless chromatin was analyzed by micrococcal nuclease digestion. Chromatin templates (150 ng) were digested by MNase (New England Biolabs) at 3.5 units/ml during 1 min at 37 °C in MNase buffer. The reaction was stopped by MNase stop solution (300 mM NaAc (ph=5.3), 20 mM EDTA (pH=8.0)). DNA was purified and radioactively labeled by T4 PNK (New England Biolabs). Labeled DNA was purified and analyzed by 8% PAG (39:1). The gel was dried, exposed and scanned (BioRad, USA).
Analysis of nucleosome positions and NPSs occupancy.
The expected positioning of nucleosomes within the array was verified using a different version of the restriction digestion sensitivity assay followed by primer extension. Chromatin was assembled on linearized pYP05 plasmid and digested with an excess of restriction enzymes AluI or ScaI. Purified DNA was subjected to primer extension with Taq DNA polymerase (New England Biolabs) using a radioactively end-labeled primer, which anneals immediately upstream of the promoter [30].
Transcription.
Conditions for in vitro transcription were optimized for maximal utilization of the chromatin templates. Transcription was conducted as described [24]. In all experiments identical molar amounts of the templates (DNA) were used.
All templates were linearized, and the closed initiation complexes (RPC) were formed. Single round transcription assays were carried out in 50-μl aliquots in the transcription buffer (TB) containing 50 mM Tris-OAc (pH 8.0), 100 mM KOAc, 8 mM Mg(OAc)2, 27 mM NH4OAc, 0.7% PEG-8000, and 0.2 mM DTT at 1 nM DNA or chromatin concentrations and 10 nM core RNA polymerase, 300 nM σ54, 120 nM NtrC, and 400 nM NtrB transcription factors. Сначала все компоненты смешивали вместе в транскрипционном буфере (общий объем 40 мкл), инкубировали в течение 15 мин при 37°С для формирования закрытого комплекса. Далее 5 мкл 40 мМ АТР, разведенного в транскрипционном буфере, добавляли в реакцию до конечной концентрации 4 мМ. Реакцию проводили в течение 2 мин при 37°С для формирования открытого инициаторного комплекса (RPo). together in TB buffer and total volume of 40 μl; then the reaction mixture was incubated for 15 min at 37°C to form the closed initiation complex (RPC). Next, 5 μl of 40 mM ATP in 1x TB were added to the reaction to 4 mM final concentration, and the reaction was incubated at 37°C for 2 minutes (or for different time intervals) to form the open initiation complex (RPO), which is competent to begin elongation. Then a mixture of all four ribonucleotide-triphosphates (4 mM each) in 1x TB with 2.5 μCi of [α−32P]-GTP (3000 Ci/mmol) and 2 mg/ml heparin was added to the reaction to start transcript elongation reaction and to limit it to a single round. The reaction was continued at 37°C for 15 minutes before it was stopped by rapid addition of phenol:chloroform mixture (1:1). Labeled RNA was purified and analyzed by denaturing PAGE. The gel was dried, exposed and scanned as described above. The data were analyzed using the OptiQuant software.
Histone H1 (12 nM) was incubated with RPC at 37°C for 30 min. After that HMGN5 (24 nM) was added to the mixture and the reaction mixture was incubated on ice for 30 min before RPO formation.
Computational modeling.
Numerical simulations of long-distance enhancer-promoter communication along saturated (13×177-bp) nucleosome arrays were carried out along the lines described recently [25, 30]. The probabilities of communication were estimated from the distances (80–125 nm) between the centers of RNAP and NtrC protein assemblies attached to the binding sites on DNA. The predicted communication enhancement is the ratio of the contact probability determined for a nucleosome-bound DNA compared to that found for a nucleosome-free chain of the same length. New refinements in the methodology, that take detailed account of the wedge-like shape of the nucleosome, the electronic and spatial features of the regulatory proteins, and the precise pathways of the associated DNA, will be described elsewhere. DNA linkers are also now subject to incremental, as opposed to, random moves, and excluded volume is detected with software from a rigid-body simulator (OpenDE).
RESULTS
The experimental approach for analysis of EPC on saturated nucleosomal arrays
To obtain saturated arrays of regularly spaced, precisely positioned nucleosomes capable of spontaneous formation of chromatin fibers and to avoid nucleosome formation on the enhancer and promoter, we employed high-affinity histone-binding 601 DNA sequences that precisely position nucleosomes in vitro [31] (fig. 1а). Previously it was shown that DNA supercoiling strongly facilitates EPC [13]. To exclude effects of DNA supercoiling on EPC in chromatin, only linear nucleosomal arrays were studied. Using these nucleosomal arrays and the transcriptional assay, the rate of enhancer-promoter communication (EPC) was quantitatively measured as described by [25, 30].
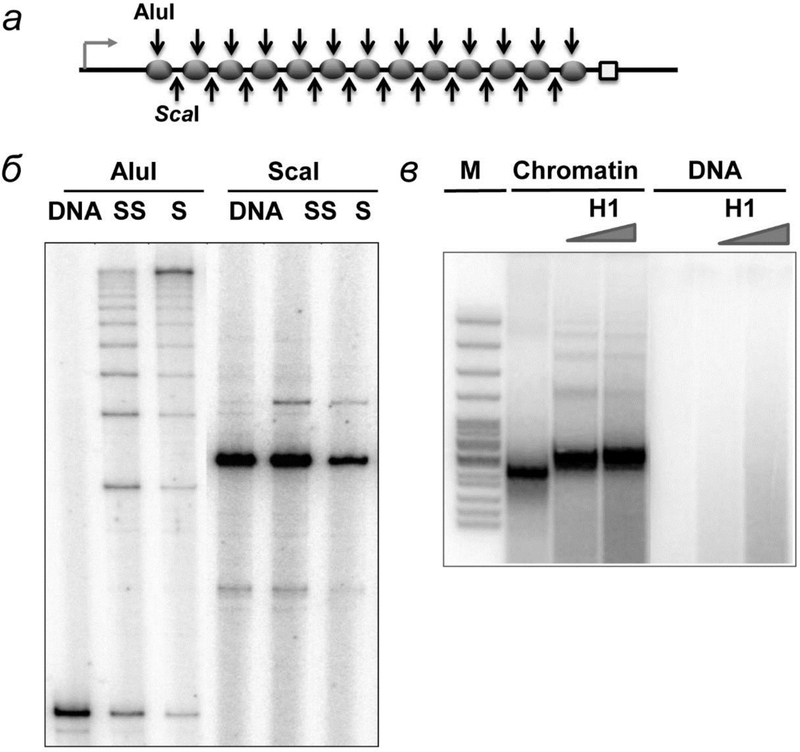
Fig. 1.
Analysis of chromatin templates assembly and H1 binding to them. а – Chromatin templates obtained using different ratios of donor chromatin and DNA templates were treated by restriction endonucleases AluI and ScaI, which cut DNA inside nucleosomal and linker DNA, respectively. б – Digested DNA was purified and amplified using one radioactively labeled primer and analyzed by denaturing PAGE. в – H1 binding to 13-nucleosome array was evaluated by micrococcal endonuclease digestion. H1 protects additional 20 bp from digestion.
The pYP05 plasmid contains thirteen strong 147-bp 601 nucleosome-positioning sequences (NPSs) between the enhancer and promoter (fig. 1а). Each of the NPSs lies within a 177-bp long repeating element (601177×13). The length of the spacer DNA between the NPSs is thus 30 bp. The 601177 nucleosomal arrays can form a chromatin fiber without linker histones [32, 33]. Chromatin reconstitution was conducted on negatively supercoiled pYP05 plasmid at different mass ratios of donor chromatin DNA to plasmid DNA (1 : 1, 1 : 0.75 и 1 : 0.5) by transfer of histone octamer from donor -H1 chromatin using dialysis from 1 M NaCl [30].
The extent of chromatin assembly and the quality of nucleosome positioning were evaluated by restriction endonucleases digestion method. It was demonstrated that increase of the donor chromatin:DNA ratio results in progressively better protection of the NPS DNA from cutting by enzymes [25, 30] (fig. 1б). Even at the highest ratio of donor chromatin:DNA the majority of nucleosomes are formed predominantly on the high-affinity NPSs, but not randomly along the pYP05 template and thus not on the enhancer and/or promoter sequences [30]. Less than 20% of the non-NPS plasmid sequences (including the promoter and enhancer regions) are covered by nucleosomes (fig. 1б). In summary, nearly all NPSs were covered by nucleosomes on saturated arrays, leaving the remaining (non-NPS) plasmid DNA largely nucleosome-free.
Linker histone H1 strongly inhibits EPC in chromatin
Nucleosome arrays with a repeat length of 177 bp spontaneously form chromatin fibers even in the absence of linker histones although linker histones can bind to the arrays without strongly affecting the structures of the arrays [32, 33]. Linker histone H1 binds to the entrance-exit regions of nucleosomal DNA [27, 35] and therefore is expected to decrease the rate of spontaneous uncoiling of nucleosomal DNA from the octamer [36, 37]. It has been proposed that the uncoiling of nucleosomal DNA from the octamer could facilitate EPC in chromatin [18]; if so, H1 would decrease the efficiency of EPC in chromatin. Previously it has been shown that histone H1 can strongly inhibit transcription in vitro [38, 39]. However, previous studies have focused on the effect of H1 on DNA-protein interactions and chromatin structure, while the role of H1 in distant communication has not been addressed.
To evaluate the effect of linker histone H1 on the rate of EPC in chromatin, transcription complexes were pre-formed on the 601177×13 nucleosomal arrays, followed by incubation in the presence of histone H1 (or its truncated variants) and subsequent transcription. Binding to the tail-containing and tailless arrays is saturated at approximately 1:1 nucleosome:H1 molar stoichiometry (fig. 1в), suggesting that one molecule of H1 binds to each nucleosome core.
The rate of EPC was measured using plasmid pYP05 containing NtrC-dependent enhancer, which activates glnAp2 promotor [30], and 13 nucleosomes located between them (fig. 2а). Experimental approach for EPC measuring is outlined on рис. 2б. Enhancer is activated by protein complex NtrC phosphorylated by NtrB kinase [40]. Phosphorylated NtrC interacts with holoenzyme Eσ54 and stimulates transition of inactive closed initiation complex to active open initiation complex [41, 42]. DNA forms a loop during EPC [43].
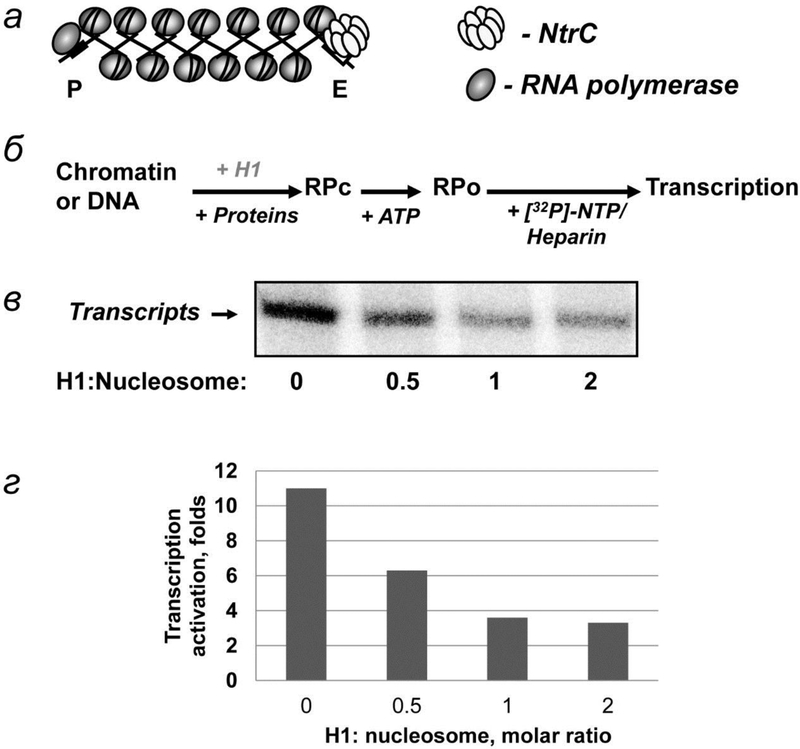
Fig. 2.
H1 effect on EPC in chromatin. а – Schematic diagram of the 13-nucleosome array. The structure of the fiber is depicted using the two-start model. The positions of the RNA polymerase and enhancer-binding complex (NtrС) are indicated. б – The experimental approach for analysis of the rate of EPC in chromatin. Closed initiation complexes (RPC) were formed and converted to open (RPO) complexes after addition of ATP, followed by addition of NTPs and heparin to limit transcription to a single round and to remove the nucleosomal barrier to transcription. в – Analysis of the labeled transcripts by denaturing PAGE at different H1 to nucleosome ratios. г – Quantitative analysis of the specific transcripts shown in 2в.
Transcriptional analysis (fig. 2в) indicates that binding of H1 to native and tailless arrays at a 1:1 nucleosome:H1 molar ratio results in a strong inhibition of EPC (fig. 2в, г). Binding of H1 to the fibers does not cause a change in the structure of the fiber [33]. Therefore the negative effect of H1 on EPC could be explained by: (a) a decrease of the rate of uncoiling of nucleosomal DNA from the octamer [18], as H1 binds and locks DNA entering and exiting the nucleosome (fig. 3а, б), and/or (b) an increase in the rigidity of the fiber. The rigidity of the chromatin fiber is determined in part by the flexibility of the linker DNA, which is likely to be decreased after H1 binding (fig. 3а).
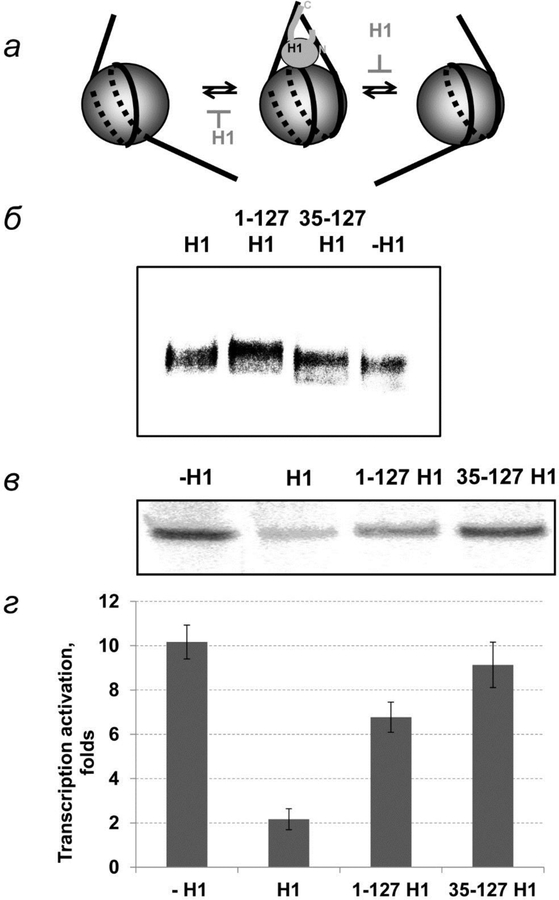
Fig. 3.
The role of histone H1 domains in EPC in chromatin. а – Schematic diagrams of histone H1 binding to a nucleosome. H1 binds DNA at DNA entrance and exit regions from nucleosome making it less flexible. б – Binding of H1 and its truncated forms with chromatin. в – Histone H1 and its truncated form lacking C-terminal domain or both terminal domains were added to chromatin template after transcription machinery proteins. The rate of EPC was measured as described on fig. 2б. Analysis of labeled transcripts by denaturing PAGE. г – Quantitative analysis of the specific transcripts shown on fig. 3в.
To discriminate between these possibilities and determine the functional domains of H1, the role of H1 with the positively charged C-tail deleted (1–127 H1) and that of tailless H1 (35–127 H1) in EPC were studied. Both truncated versions of H1 bind to the chromatin fiber на (fig. 3б). High mobility of H1-containg templates can be explained by their higher rigidity [44].
Transcriptional analysis shows that deletion of the H1 C-terminal tail results in a 3-fold decrease of H1 inhibition of EPC (fig. 3в, г). Thus the C-terminal tail of H1 plays a dominant role in H1-dependent inhibition of EPC. The C-terminal tail could affect the rate of EPC through both mechanisms described above. Further support for the first (DNA uncoiling) mechanism is provided by the observation that H1 considerably inhibits EPC on tailless chromatin (fig. 4). Since tailless chromatin is characterized by minimal internucleosomal interactions [36, 45] (fig. 4а), H1 most likely inhibits coiling and uncoiling of nucleosomal DNA from the octamer or decreases the flexibility of linker DNA. Deletion of the N-terminal tail of histone H1 results in a further ~30% decrease of H1 inhibition of EPC; tailless H1 minimally affects EPC in chromatin (fig. 3в, г). The negative effect of the N-terminal tail of H1 on EPC is unlikely to be explained by internucleosomal interactions because the tail is too short to reach a neighboring nucleosome. Although tailless H1 interacts with DNA exiting and entering the nucleosome [27], these interactions are not sufficient to inhibit EPC by either of the mechanisms proposed above.
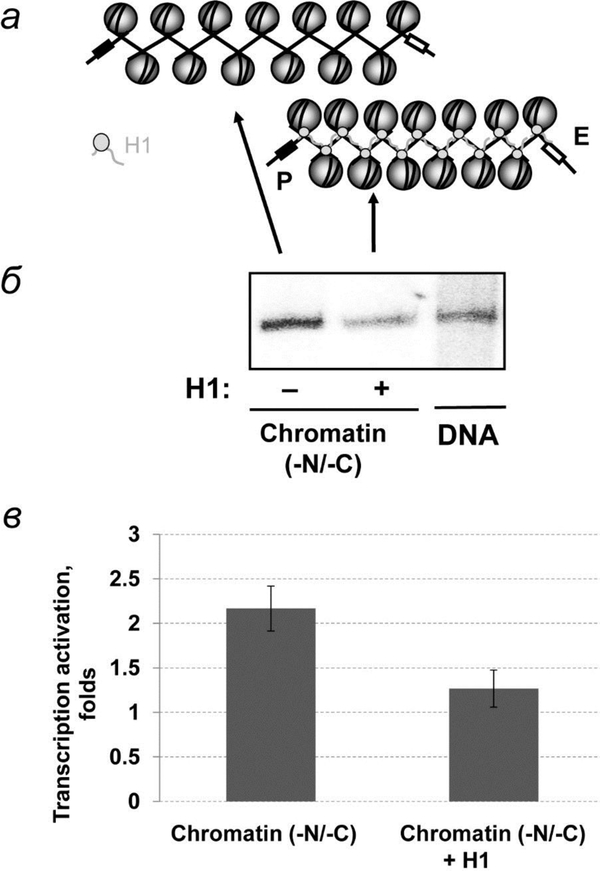
Fig. 4.
Binding of linker histone H1 to tailless chromatin fibers prevents efficient EPC. а – Schematic diagrams of histone H1 binding with tailless chromatin (-N/-C). б – Analysis of labeled transcripts with and without H1 by denaturing PAGE as described on fig. 2б. в – Quantitative analysis of the specific transcripts shown on fig. 4б.
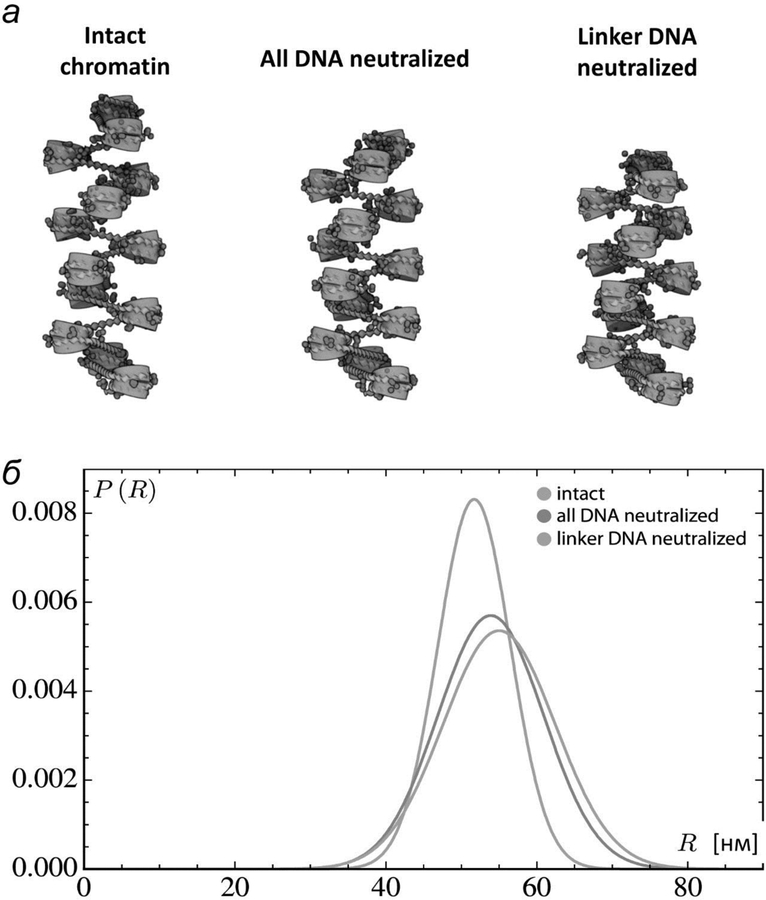
While the structure of H1-nucleosome complex is resolved with low resolution [22, 46], it remains unclear how H1 effect EPC in the absence of high resolution structures. The precise positioning of the globular DNA-binding domain of H1 remains an open question [47], and structural understanding of how the intrinsically disordered linker histone termini interact with DNA is even less certain [48]. Moreover, the positioning of the globular domain may depend upon the H1 variant [48] and the stem-like structures captured in electron microscopic images of trinucleosomal arrays [27, 49], may not be relevant to the longer arrays with differently spaced nucleosomes that we consider. Here, as a crude approximation of H1-DNA interactions, we examine how neutralization of DNA affects the overall configuration and deformability of fibers with the same 13 evenly spaced, fully intact nucleosomes treated above (fig. 5а). Whereas complete neutralization of the DNA has a relatively small effect on the simulated structures, the neutralization of linker DNA alone compacts and stiffens the simulated fibers appreciably (fig. 5а). The distribution of distances between terminal base pairs shifts toward lower values and narrows more substantially when only the linker DNA is neutralized (fig. 5б), suggesting that H1 binding could stiffen and slightly change the overall geometry of the fiber and make the fiber less flexible (fig. 5). Unraveling the precise forces responsible for the reduction in EPC introduced by the presence of H1 requires further investigation.
Fig. 5.
Computational simulation of the chromatin fiber: distant internucleosomal interaction are increased after DNA charge neutralization. а – Models of chromosomal fragments based on the average configurations of the central residues of simulated structures with 13 intact nucleosomes on fully charged, fully neutralized, and linker-neutralized DNA The central residues are expected to be representative of the configurational states adopted by a long fiber with evenly spaced nucleosomes. б – Distribution of distances between the first and last base pairs on simulated chromatin constructs with 13 intact nucleosomes, spaced at 177-bp increments (30-bp linkers), on fully charged, fully neutralized, and linker-neutralized DNA.
Taken together, the data suggest that H1 blocks EPC by binding with linker DNA through its C-terminal (primarily) and N-terminal tails. This binding could inhibit coiling-uncoiling of nucleosomal DNA from the histone octamer and (or) reduce flexibility of the chromatin fiber. Removal of the histone tails prevents formation of a stable chromatin fiber and strongly reduces the inhibiting effect of H1 on EPC.
HMGN-like HMGN5 protein counteracts the negative effect of histone H1 on EPC
To further evaluate the role of linker histone H1 in EPC in chromatin and to evaluate a candidate protein that could positively affect EPC in H1-containing chromatin, we have analyzed the effect of the nucleosome-binding protein HMGN5 protein on EPC. The HMGN5 binds to H1-containing nucleosomal arrays and induces partial unfolding of the 30-nm chromatin fiber formed with 60-bp internucleosomal DNA linkers [19]. The negatively charged C-terminal region of HMGN5 interacts with the positively charged H1 C-terminal “tail” [19], thereby interfering with the H1-DNA interactions mediated by the H1 C-terminal tail. Given these findings, it could be expected that HMGN5 would prevent H1-mediated inhibition of EPC in chromatin. In the case of arrays containing a shorter (30-bp) internucleosomal linker DNA (our experimental system), H1 binding to nucleosomal arrays does not strongly affect the structure of the fiber [32, 33]; therefore, HMGN5 is expected to affect H1-chromatin interactions, but not the overall structure of the fiber.
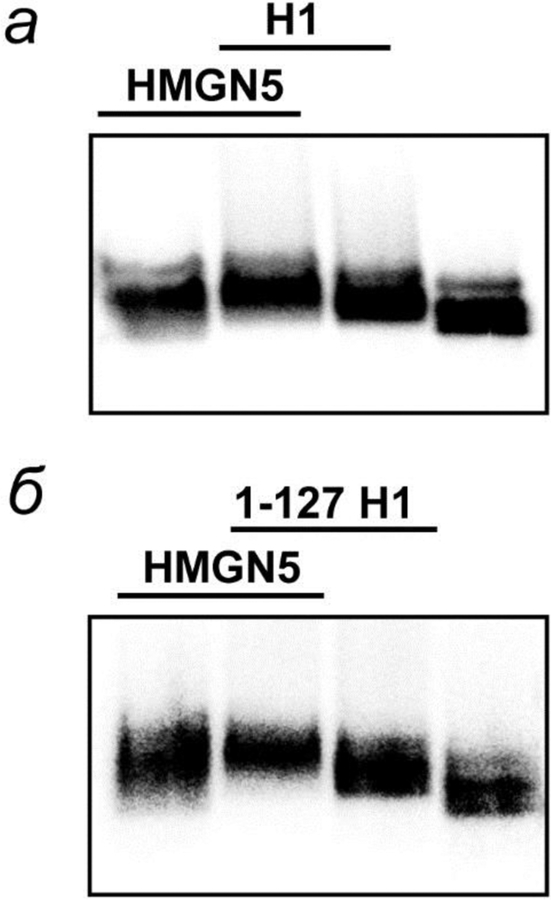
To evaluate the effect of HMGN5 on the rate of EPC in H1-containing chromatin and 1–127 H1 containing chromatin, transcription complexes were pre-formed on the 601177×13 nucleosomal arrays, incubated in the presence of histone H1, 1–127 H1 and/or HMGN5, and transcribed (fig. 6). The mobility of H1-containing arrays in native gel is affected in the presence of HMGN5 and is different from the mobility of the nucleosome array with HMGN5, indicating that H1 remains bound to the array in the presence of HMGN5 (fig. 6). Thus, all versions of H1 and HMGN5 bind to the chromatin in a non-exclusive way.
Fig. 6.
Binding of HMGN5 protein to linear 3-nucleosome arrays containing intact histone H1 and 1–127 H1. The saturated arrays were assembled using end-labeled DNA and analyzed by native agarose gel after incubation with H1 (a), 1–127 H1(б) and/or HMGN5.
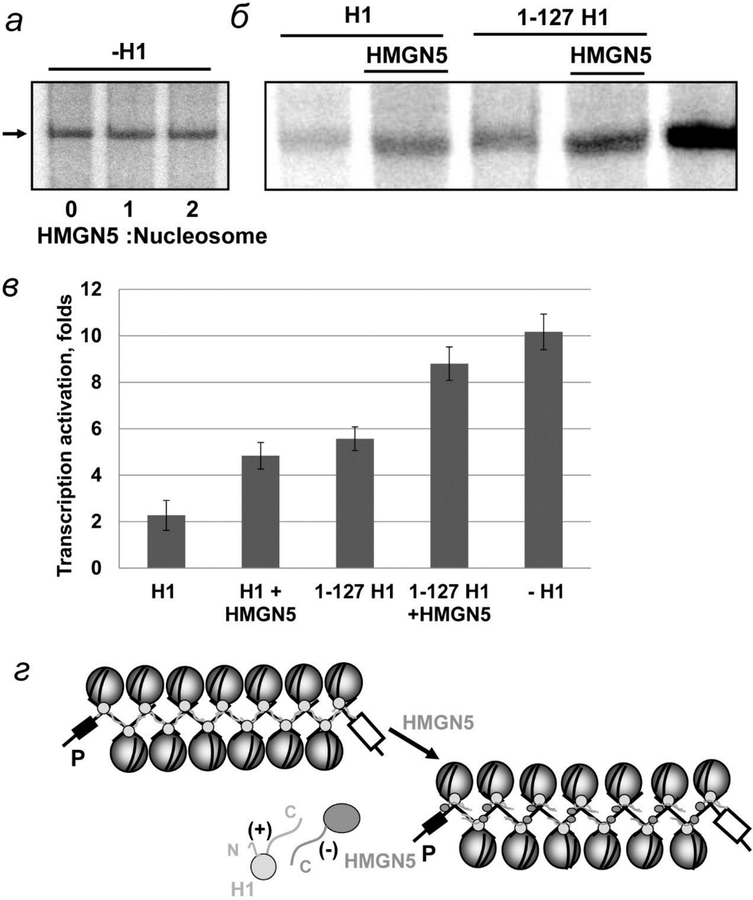
As expected, HMGN5 positively affects transcription of H1-containing chromatin and does not affect transcription of H1-depleted chromatin (fig. 7а–в). The maximal effect is observed at 2:1 molar ratio of HMGN5 to core nucleosomes (piс. 7в). In the presence of HMGN5 the rate of EPC is increased 2-fold, as compared with H1-containing chromatin fiber, indicating that HMGN5 reverses the negative effect of H1 and its -C-tail mutant on EPC. A lower (~1.5-fold) positive effect of HMGN5 on EPC with the array containing 1–127 H1 could be explained by the interaction between HMGN5 and N-tail of H1, which is expected to be less strong than the HMGN5-C-tail interaction due to the lower positive charge of the N-tail. HMGN5 does not affect EPC on the array containing the globular (35–127) domain of H1, indicating that the majority of H1-HMGN5 interactions occur through the H1 tails.
Fig. 7.
The HMG-like HMGN5 protein relieves the negative effect of linker histone H1 on EPC in chromatin. а, б – Transcription of linear DNA and saturated 13-nucleosome arrays carried out in the absence and presence of linker histone H1 or 1–127 H1 and HMGN5 protein. Analysis of labeled transcripts by denaturing PAGE as on fig. 2б. в – Quantitative analysis of the specific transcripts shown on fig. 7б. г – Schematic diagrams of histone H1 and HMGN5 interaction with chromatin.
In summary, HMGN5 protein partially reverses the negative effect of H1 on distant EPC in chromatin, most likely interfering with the H1-DNA interactions mediated by the positively charged H1 “tails” (fig. 7г).
DISCUSSION
In this work, we have employed an in vitro experimental system [30], that allows quantitative analysis of distant enhancer-promoter communication (EPC) in cis on physiologically relevant, saturated arrays of precisely positioned nucleosomes that spontaneously form chromatin fibers (fig. 1). Using this system we have shown that: 1) EPC in chromatin occurs primarily in cis, through looping of the intervening chromatin that separates the enhancer and promoter (fig. 2–4). 2) Binding of linker histone H1 to the arrays results in strong inhibition of EPC on intact and tailless chromatin (fig. 3, 5). Inhibition of EPC by H1 is mediated primarily by the C-terminus of H1 that most likely interacts with linker DNA and either reduces the flexibility of the chromatin fiber or inhibits coiling/uncoiling of nucleosomal DNA from the octamer. 3) The HMGN5 protein counteracts the negative effect of histone H1 on EPC (fig. 7), most likely interfering with H1-DNA interactions through binding with the H1 “tails”.
Chromatin could affect the rate of EPC by compacting DNA and/or providing an additional dynamic component facilitating communication. Increasing evidence suggests that the dynamic component is highly important during EPC in chromatin. [50]. Our previous data suggested that the strong effect of chromatin structure on the rate of EPC cannot be explained only by DNA compaction [18]. The new data identify several factors that affect the dynamic properties of chromatin independently of their effect on chromatin compactness and vice versa: (1) Binding of histone H1 to the intact and tailless arrays dramatically decreases the rate of EPC in chromatin (fig. 1–3). Since H1 minimally affects the structure of the 30 nm fiber formed on 177-bp arrays [32, 33, 36], it must affect the dynamic properties of the fiber that are essential for efficient EPC. 2) Most likely, the HMGN5 protein has only a minimal effect on the structure of the fiber; nevertheless, it can fully counteract the inhibitory effect of histone H1 on EPC (fig. 7). Taken together, the data suggest that chromatin dynamics and a competitive interplay between nucleosome binding proteins play a critical role in determining the rate of distant EPC.
Binding of linker histone H1 strongly inhibits EPC in chromatin (fig. 2–4). This inhibition most likely occurs through H1-DNA interactions because the disruption of some of these interactions, either by removal of histone tails (fig. 4) or by HMGN5 protein (fig. 7), results in at least partial relief of H1 inhibition. Since binding of H1 does not affect the structure of the chromatin fiber [32], an effect on chromatin flexibility and coiling-uncoiling of nucleosomal DNA from the octamer is more likely. Chromatin-bound H1 decreases the flexibility of simulated fibers [44, 51]. Such a decrease in flexibility is consistent with the observed lower efficiency of both short-range and long-range EPC in the presence of H1.
EPC over a distance of more than 1 kb on linear or relaxed histone-free DNA occurs very slowly and constitutes the rate-limiting step during transcription in vitro [13]. Distant EPC in chromatin occurs at a much higher rate that can be modulated by various factors identified in this study. Therefore EPC in chromatin could constitute a novel early rate-limiting step in gene regulation. In particular, it is possible that histone modifications that destabilize the chromatin fiber (e.g., H4K16 acetylation) [52], could decrease the efficiency of H1 binding [53] and allow efficient EPC. Another possibility is that modifications or replacement of linker histones could be essential for activation of a chromatin domain. Thus it has been shown that phosphorylation of linker histones prevents inhibition of ATP-dependent chromatin remodeling [54], and histone H2A deubiquitinase facilitates eviction of histone H1 during gene activation [55]. The HMGN5 protein is associated with active chromatin [19], facilitates EPC in chromatin in vitro (fig. 7) and could also destabilize the chromatin fiber in vivo.
In summary, our work suggests that the chromatin fiber is a highly dynamic, efficient communication device that can maintain a high level of DNA compaction during EPC. We have identified factors (such as core linker histones, and the HMGN5 protein) that strongly affect the rate of EPC in chromatin. More generally, our data suggest that EPC could constitute a novel regulated step during gene expression in eukaryotes.
Acknowledgments
We thank T. Richmond (ETH Zürich, Institute of Molecular Biology and Biophysics, Zürich, Switzerland) for the plasmid containing 601177×12 nucleosome positioning sequences and S. Dimitrov (Université Grenoble Alpes, CNRS UMR 5309, INSERM U1209, Institute for Advanced Biosciences (IAB), Site Santé – Allé e des Alpes, France) for intact and mutant histone H1.
This work was supported by NIH RO1GM119398 and R21CA220151 to Studitsky V.M., RO1GM34809 to W.K. Olson and by Russian Science Foundation Grant (14-24-00031). Polikanov Y.S. work was sponsored by Illinois State start-up funds, USA. Work of Y. V. Postnikov was sponsored by National Institutes of Health, National Cancer Institute, USA.
All procedures performed in this work comply with the ethical standards of the institutional committee on research ethics and the 1964 Helsinki Declaration and its subsequent changes or comparable standards of ethics.
Footnotes
The authors declare that they have no conflict of interest.
REFERENCES
- 1.de Laat W, Duboule D (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 502, 499–506. [DOI] [PubMed] [Google Scholar]
- 2.Gibcus JH, Dekker J (2013) The hierarchy of the 3D genome. Mol. Cell 49, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmston N, Lenhard B (2013) Chromatin and epigenetic features of long-range gene regulation. Nucl. Acids Res 41, 7185–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krivega I, Dean A (2012) Enhancer and promoter interactions-long distance calls. Curr. Opin. Genet. Dev 22, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nizovtseva EV, Todolli S, Olson WK, Studitsky VM (2017) Towards quantitative analysis of gene regulation by enhancers. Epigenomics. 9, 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W (2008) Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet 4, e1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringrose L, Chabanis S, Angrand PO, Woodroofe C, Stewart AF (1999) Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J 18, 6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghirlando R, Felsenfeld G (2008) Hydrodynamic studies on defined heterochromatin fragments support a 30-nm fiber having six nucleosomes per turn. J. Mol. Biol 376, 1417–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA (2004) Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell. 118, 555–566. [DOI] [PubMed] [Google Scholar]
- 10.Platani M, Goldberg I, Lamond AI, Swedlow JR (2002) Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell. Biol 4, 502–508. [DOI] [PubMed] [Google Scholar]
- 11.Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM (2004) Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119, 955–967. [DOI] [PubMed] [Google Scholar]
- 12.Bellomy GR, Record MT Jr. (1990) Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog. Nucl. Acid Res. Mol. Biol 39, 81–128. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Bondarenko V, Ninfa A, Studitsky VM (2001) DNA supercoiling allows enhancer action over a large distance. Proc. Natl. Acad. Sci. USA 98, 14883–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson JR, Benyajati C (1993) DNA-histone interactions are sufficient to position a single nucleosome juxtaposing Drosophila Adh adult enhancer and distal promoter. Nucl. Acids Res 21, 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schild C, Claret FX, Wahli W, Wolffe AP (1993) A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J 12, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein A, Dalal Y, Fleury TJ (2002) Circle ligation of in vitro assembled chromatin indicates a highly flexible structure. Nucl. Acids Res 30, 5103–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laybourn PJ, Kadonaga JT (1992) Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science. 257, 1682–1685. [DOI] [PubMed] [Google Scholar]
- 18.Rubtsov MA, Polikanov YS, Bondarenko VA, Wang YH, Studitsky VM (2006) Chromatin structure can strongly facilitate enhancer action over a distance. Proc. Natl. Acad. Sci. USA 103, 17690–17695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rochman M, Postnikov Y, Correll S, Malicet C, Wincovitch S, Karpova TS, McNally JG, Wu X, Bubunenko NA, Grigoryev S, Bustin M (2009) The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chromatin compaction and modulates transcription. Mol. Cell 35, 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodcock CL, Skoultchi AI, Fan Y (2006) Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14, 17–25. [DOI] [PubMed] [Google Scholar]
- 21.Shimada M, Chen WY, Nakadai T, Onikubo T, Guermah M, Rhodes D, Roeder RG (2019) Gene-specific H1 eviction through a transcriptional activator→p300→NAP1→H1 pathway. Mol. Cell 74, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Saez I, Menoni H, Boopathi R, Shukla MS, Soueidan L, Noirclerc-Savoye M, Le Roy A, Skoufias DA, Bednar J, Hamiche A, Angelov D, Petosa C, Dimitrov S (2018) Structure of an H1-bound 6-nucleosome array reveals an untwisted two-start chromatin fiber conformation. Mol. Cell 72, 902–915. e7. [DOI] [PubMed] [Google Scholar]
- 23.Azad GK, Ito K, Sailaja BS, Biran A, Nissim-Rafinia M, Yamada Y, Brown DT, Takizawa T, Meshorer E (2018) PARP1-dependent eviction of the linker histone H1 mediates immediate early gene expression during neuronal activation. J. Cell Biol 217, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polikanov YS, Rubtsov MA, Studitsky VM (2007) Biochemical analysis of enhancer-promoter communication in chromatin. Methods. 41, 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulaeva OI, Zheng G, Polikanov YS, Colasanti AV, Clauvelin N, Mukhopadhyay S, Sengupta AM, Studitsky VM, Olson WK (2012) Internucleosomal interactions mediated by histone tails allow distant communication in chromatin. J. Biol. Chem 287, 20248–20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodwin GH, Johns EW (1973) Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur. J. Biochem 40, 215–219. [DOI] [PubMed] [Google Scholar]
- 27.Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, Everaers R, Angelov D, Bednar J, Dimitrov S (2010) Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. USA 107, 9620–9625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter W, Studitsky VM (2004) Construction, analysis, and transcription of model nucleosomal templates. Methods. 33, 18–24. [DOI] [PubMed] [Google Scholar]
- 29.Polach KJ, Lowary PT, Widom J (2000) Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol 298, 211–223. [DOI] [PubMed] [Google Scholar]
- 30.Polikanov YS, Studitsky VM (2009) Analysis of distant communication on defined chromatin templates in vitro. Methods Mol. Biol 543, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J (1999) Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol 288, 213–229. [DOI] [PubMed] [Google Scholar]
- 32.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 306, 1571–1573. [DOI] [PubMed] [Google Scholar]
- 33.Routh A, Sandin S, Rhodes D (2008) Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA 105, 8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol 327, 85–96. [DOI] [PubMed] [Google Scholar]
- 35.Caterino TL, Fang H, Hayes JJ (2011) Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol. Cell. Biol 31, 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carruthers LM, Hansen JC (2000) The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem 275, 37285–37290. [DOI] [PubMed] [Google Scholar]
- 37.Brockers K, Schneider R (2019) Histone H1, the forgotten histone. Epigenomics. 11, 363–366. [DOI] [PubMed] [Google Scholar]
- 38.Ding HF, Bustin M, Hanse U (1997) Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region in chromosomal protein HMG-14. Mol. Cell. Biol 17, 5843–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Neil TE, Meersseman G, Pennings S, Bradbury EM (1995) Deposition of histone H1 onto reconstituted nucleosome arrays inhibits both initiation and elongation of transcripts by T7 RNA polymerase. Nucl. Acids Res 23, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ninfa AJ, Magasanik B (1986) Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 83, 5909–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buck M, Cannon W (1992) Activator-independent formation of a closed complex between sigma 54- holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol. Microbiol 6, 1625–1630. [DOI] [PubMed] [Google Scholar]
- 42.Popham DL, Szeto D, Keener J, Kustu S (1989) Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 243, 629–635. [DOI] [PubMed] [Google Scholar]
- 43.Su W, Porter S, Kustu S, Echols H (1990) DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. USA 87, 5504–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langowski J, Heermann DW (2007) Computational modeling of the chromatin fiber. Semin. Cell. Dev. Biol 18, 659–667. [DOI] [PubMed] [Google Scholar]
- 45.Gordon F, Luger K, Hansen JC (2005) The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem 280, 33701–33706. [DOI] [PubMed] [Google Scholar]
- 46.Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, Syed SH, Lone IN, Tonchev O, Crucifix C, Menoni H, Papin C, Skoufias DA, Kurumizaka H, Lavery R, Hamiche A, Hayes JJ, Schultz P, Angelov D, Petosa C, Dimitrov S (2017) Structure and dynamics of a 197 bp nucleosome in complex with linker histone H1. Mol. Cell 66, 384–397. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou BR, Feng H, Kato H, Dai L, Yang Y, Zhou Y, Bai Y (2013) Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. USA 110, 19390–19395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang H, Clark DJ, Hayes JJ (2012) DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucl. Acids Res 40, 1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer S, Becker NB, Syed SH, Goutte-Gattat D, Shukla MS, Hayes JJ, Angelov D, Bednar J, Dimitrov S, Everaers R (2011) From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem. Nucl. Acids Res 39, 9139–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nizovtseva EV, Clauvelin N, Todolli S, Polikanov YS, Kulaeva OI, Wengrzynek S, Olson WK, Studitsky VM (2017) Nucleosome-free DNA regions differentially affect distant communication in chromatin. Nucl. Acids Res 6, 3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T (2009) Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA 106, 13317–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 311, 844–847. [DOI] [PubMed] [Google Scholar]
- 53.Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D (2008) 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol 381, 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL (2002) Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol 9, 263–267. [DOI] [PubMed] [Google Scholar]
- 55.Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG (2007) A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]