Abstract
Intrinsically disordered proteins (IDPs) are important functional proteins, and their deregulation are linked to numerous human diseases including cancers. Understanding how disease-associated mutations or drug molecules can perturb the sequence-disordered ensemble-function-disease relationship of IDPs remains challenging, because it requires detailed characterization of the heterogeneous structural ensembles of IDPs. In this work, we combine the latest atomistic force field a99SB-disp, enhanced sampling technique replica exchange with solute tempering, and GPU-accelerated molecular dynamics simulations to investigate how four cancer-associated mutations, K24N, N29K/N30D, D49Y, and W53G, and binding of an anti-cancer molecule, epigallocatechin gallate (EGCG), modulate the disordered ensemble of the transactivation domain (TAD) of tumor suppressor p53. Through extensive sampling, in excess of 1.0 μs per replica, well-converged structural ensembles of wild-type and mutant p53-TAD as well as WT p53-TAD in the presence of EGCG were generated. The results reveal that mutants could induce local structural changes and affect secondary structural properties. Interestingly, both EGCG binding and N29K/N30D could also induce long-range structural reorganizations and lead to more compact structures that could shield key binding sites of p53-TAD regulators. Further analysis reveals that the effects of EGCG binding are mainly achieved through nonspecific interactions. These observations are generally consistent with on-going NMR studies and binding assays. Our studies suggest that induced conformational collapse of IDPs may be a general mechanism for shielding functional sites, thus inhibiting recognition of their targets. The current study also demonstrates that atomistic simulations provide a viable approach for studying the sequence-disordered ensemble-function-disease relationships of IDPs and developing new drug design strategies targeting regulatory IDPs.
Keywords: intrinsically disordered proteins, molecular dynamics simulations, induced conformational collapse
1. Introduction
As key components of cellular signaling and regulatory networks, intrinsically disordered proteins (IDPs) do not have stable structures under physiological conditions and deviate from the traditional protein structure-function paradigm1–10. Sequence analysis have shown that IDPs are highly prevalent in biology11, suggesting that intrinsic disorder provides major advantages in supporting related functions. Many IDPs have been shown to interact with multiple targets, often working as signaling hubs in protein interaction networks5, 7–8, 11–13. Mutation of IDPs or altered IDP abundance are frequently associated with human diseases, including cancers, neurodegenerative diseases, cardiovascular disorders, and diabetes11, 13–16. Nearly one fourth of disease mutations could be mapped to disordered regions15, 17, and many of them may alter the residual structure level of IDPs15. Therefore, there is a strong need to understand the molecular mechanisms of how IDPs carry out various biological functions and contribute to various diseases. A persisting bottleneck, however, is the challenge of detailed characterization of the structural and dynamic properties of disordered protein states. The highly dynamic conformations of IDPs do not lend themselves to traditional experimental characterizations, which are geared toward determining well-defined structures of folded proteins18–24. Several approaches have also been described to generate models of the disordered ensembles using experimental data from nuclear magnetic resonance (NMR) spectroscopy, small angle X-ray scattering, and others.25–30 However, a key challenge is that experimentally measured ensemble averages are generally insufficient to define the heterogeneous ensemble by themselves. The underdetermined nature renders the resulting ensembles prone to inevitable biases in the model generation protocol.20 Instead, physics-based molecular dynamics (MD) simulations have a crucial and unique role to play in understanding IDPs. It can generate structural ensembles with atomistic details, given highly accurate force fields and adequate sampling of relevant conformational space. When properly validated using experimental data, the simulated ensembles could provide the molecular details required for establishing the physical basis on how intrinsic conformational disorder mediates protein functions and how such functional mechanisms fail in human diseases.
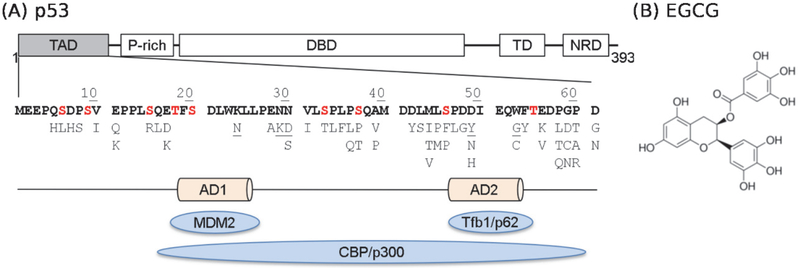
In this work, we focus on the intrinsically disordered N-terminal transactivation domain (TAD) of tumor suppressor p53, one of the most frequently mutated proteins in cancers31–32. The stability and activity of p53 are tightly regulated by interactions with other proteins, such as the E3 ubiquitin ligase MDM2 and transcriptional coactivator CBP/p30033. These interactions are mediated by p53-TAD, which contains two functional sites, AD1 and AD2, for specific recognition of target proteins (see Figure 1). In unstressed cells, the level of p53 remains low, since it binds tightly to the E3 ubiquitin ligase MDM2 through TAD and becomes polyubiquitinated and degraded34–35. Under genotoxic stress, p53 becomes phosphorylated at multiple sites in TAD (Figure 1), which reduces binding affinity to MDM2 and enhanced binding affinity to CBP, thus stabilizing and activating p5336–40. The underlying mechanisms of how phosphorylation and cancer-associated mutations modulate p53 interactions with these key regulators remains unclear. Importantly, many of these phosphorylation and mutation sites are not located at the known binding interfaces (AD1 and AD2) identified in known structures of p53-TAD complexes41–44. Therefore, modulation of binding affinities by phosphorylation at many sites cannot be explained by disruption of inter-molecular interaction itself. Instead, the unbound structural ensemble of p53-TAD is likely poised to respond to various cellular signals, including phosphorylation and mutations, which can shield key binding sites and affect the entropic and enthalpic costs of binding. As such, the unbound state of p53-TAD can provide a central conduit that integrates various cellular signals to regulate the activity of p53. Indeed, it has been shown that modulating the helicity of p53-TAD alone can have important consequences on binding and potentially activation in cells.45 Along this line, detailed characterization of the unbound ensemble is critical to understanding the sequence-disordered ensemble-function-disease relationship of p53.
Figure 1.
(A) Domain structure of p53, sequence of p53-TAD and its key interaction partners (blue ovals). Phosphorylation sites are colored in red and known cancer mutants are listed below the sequence. Mutants studied in this work are underlined. (B) Chemical structure of EGCG.
Recent advances in developing more accurate protein force fields and enhanced sampling techniques have paved the way for using atomistic simulations to study IDPs of biological and biomedical importance23, 46. We have critically examined the ability of several latest force fields for describing both local and long-range structural features of wild-type (WT) p53-TAD47. In comparison with a wide array of experimental observables, including NMR chemical shifts, paramagnetic relaxation enhancement (PRE) effects and single-molecule and time resolved FRET measurements, we found that the force field a99SB-disp48 in particular could accurately describe virtually all key structural features of WT p53-TAD, including overall dimension, secondary structural properties and transient long-range contact formation. a99SB-disp has also been shown to reliably model many other folded proteins and disordered proteins and peptides48, thus providing a solid base for us to integrate atomistic simulation and biophysical experiment to further investigate how cancer-associated mutations and ligand binding may modulate the disordered ensemble of p53-TAD to perturb its interactions with key regulators and biological activities.
Here, we combined the highly accurate atomistic force field a99SB-disp48, enhanced sampling technique replica exchange with solute tempering (REST2)49–50, and GPU-accelerated MD simulations to investigate how four cancer-associated mutations, K24N, N29K/N30D, D49Y, and W53G, and binding of an anti-cancer drug, epigallocatechin gallate (EGCG) (Fig. 1), may modulate the disordered ensemble of p53-TAD. Combination of enhanced sampling and GPU acceleration proves effective in overcoming the computational demand of sufficient sampling of the disordered conformational space. The effects of cancer-associated mutations and EGCG binding were examined at multiple levels, including the overall dimension, secondary structures and conformational distributions, and compared with on-going NMR and binding studies.
2. Methods
2.1. Simulation details
The 61-residue p53-TAD domain (MEEPQ SDPSV EPPLS QETFS DLWKL LPENN VLSPL PSQAM DDLML SPDDI EQWFT EDPGP D) and its four cancer-associated mutants, K24N, N29K/N30D, D49Y, and W53G, were studied in this work. Each peptide was capped with an acetyl group at the N-terminus and N-methyl amide at the C-terminus. The a99SB-disp force field48 was used in all simulations. To study the effects of an anti-cancer drug EGCG, another system was constructed with one WT p53-TAD and 10 EGCG molecules simulated together, where EGCG was modelled using the general AMBER force field51. For each WT and mutant p53-TAD, we performed two independent REST2 simulations starting from contrasting structures, either highly helical or fully extended extracted from our previous simulations of p53-TAD52. This allows us to critically evaluate simulation convergence, because well converged ensembles should be independent of the starting conformation. In the case of EGCG molecules interacting with WT p53-TAD, two independent REST2 simulations were also performed, but the starting structures were centroids of top 16 most populated clusters derived from previous clustering analysis of WT p53-TAD47, with 10 EGCG molecules randomly inserted into the simulation box. The protein was solvated using ~24,000 water molecules in a truncated octahedron box, whose volume was ~710 nm3. Counter ions (14 Na+ ions in all systems, except for 15 Na+ in K24N and 13 Na+ in D49Y) was included to neutralize the system.
GROMACS 201853–54 patched with PLUMED 2.3.055–57 was used to carry out all REST2 simulations. Initial conformations were energy minimized to remove any steric clashes. The system was then equilibrated at 298 K for 1 ns under constant temperature (NVT) conditions, followed by 1 ns constant temperature and constant pressure (NPT) simulation. All production runs were carried out at 298 K under NVT conditions. Short-range nonbonded interactions were truncated at 1.2 nm, and long-range electrostatic interactions were calculated using the Particle Mesh Ewald (PME) method58. LINCS algorithm59 was applied to constrain lengths of all bonds involving hydrogen atoms. The MD time step was 2 fs. In the subsequent REST2 simulations, 16 replicas were used with the effective temperatures of protein spaced exponentially between 298 K and 500 K. The higher effective temperature of protein facilitates its conformational transitions, which can be achieved by reducing potential energies. Specifically, we scaled the solute-solute and solute-solvent interactions by λ and , respectively, with λ ranging from 1.0 to 0.6. Exchange between replicas was attempted every 2 ps, and the average acceptance ratio was approximately 0.25. Trajectories were saved every 2 ps. All simulations lasted for at least 1 μs/replica. The total aggregated REST2 MD time was 192 μs, making this one of the most extensive explicit solvent simulations of p53-TAD.
2.2. Analysis
The simulated ensembles were derived only from unbiased replicas of λ = 1.0, with the first 300 ns trajectories of each simulation discarded to remove the initial equilibration phase. All analyses were conducted using the GROMACS toolset53–54 unless otherwise specified.
3. Results and Discussion
3.1. Assessment of simulation convergence
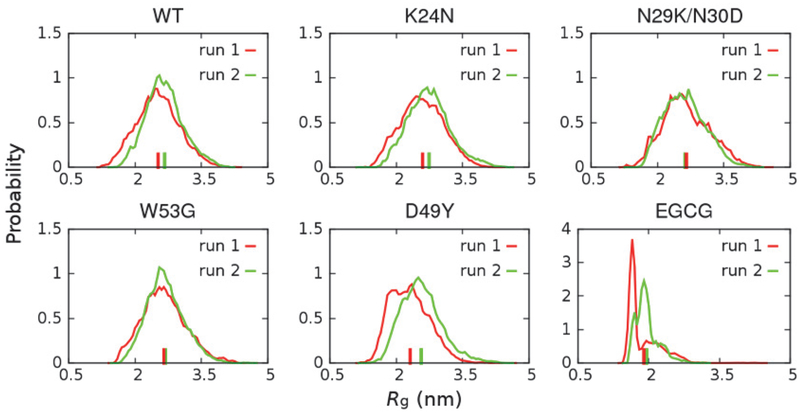
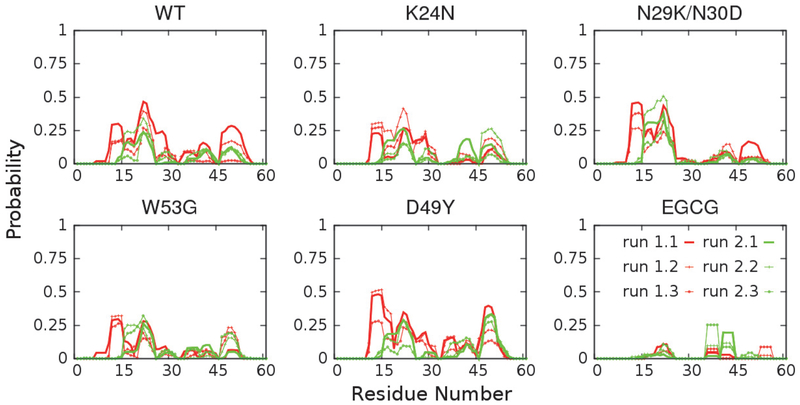
Achieving a sufficient level of convergence is critical for examining how the disordered ensembles respond to mutations and/or ligand binding. In this work, the simulation convergence was examined by comparing the structural ensembles obtained from two independent runs (see Methods for details). As illustrated in Figure 2 and Figure 3, key structural features of p53-TAD, including the chain dimension, measured by radius of gyration Rg, and secondary structural properties, are quite consistent between two independent runs, suggesting that these simulations are reasonably well converged. We note that substantially differences persist between results from control and folding simulations, which reflects the highly challenging nature of generating extremely well converged disordered ensembles. This is an important limitation that will prevent one from reliably detecting and resolve small perturbations of mutations and/or drug binding on IDPs. Nonetheless, it has been shown that a99SB-disp could accurately describe the key structural features of many biologically important IDPs48 and particularly p53-TAD47. These simulated ensembles thus should provide solid insights on how ligand binding and mutations may perturb the properties of unbound p53-TAD and modulate its interactions with key regulators.
Figure 2.
Distributions of Rg of p53-TAD at 298 K calculated from two independent REST2 runs. The corresponding ensemble averaged values are indicated using vertical bars on the x-axis.
Figure 3.
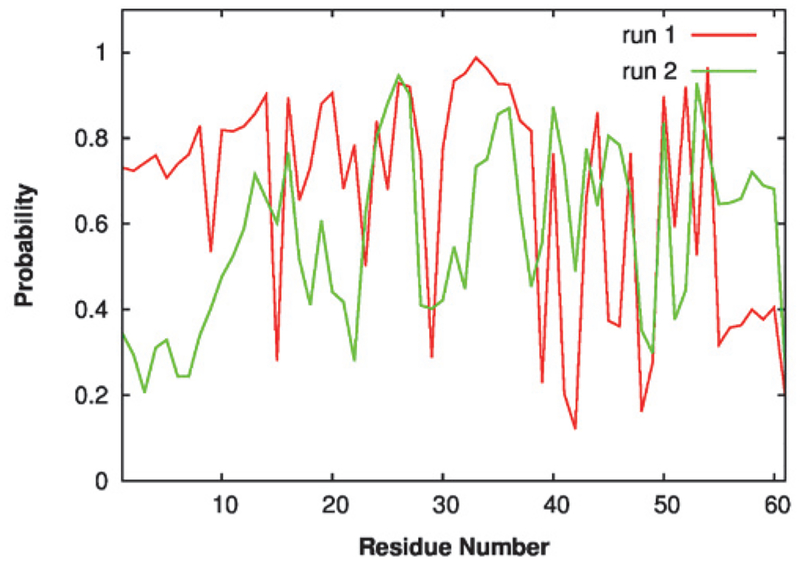
Residual helicity profiles of p53-TAD at 298 K calculated from two independent REST2 runs. Each run was equally divided into three portions. Secondary structure of each snapshot was calculated using the DSSP60 program, and probabilities of forming α-helix were reported here.
3.2. Modulation of p53-TAD secondary structural properties
IDPs could interact with different cellular targets under different conditions, thus often working as signaling hubs in protein interaction networks5, 7–8, 11–13. The secondary structural properties of IDPs are considered an important determinant of signaling fidelity8. For example, missense disease mutations in IDPs alter residual secondary structure with higher probabilities than neutral evolutionary substitutes15, which implies that changing IDP secondary structural properties may have detrimental impacts on their functions. Transiently formed partial helices have also been observed in unbound p53-TAD61–62, and they are critical in mediating interactions with p53 regulators like MDM2 and CBP33. Therefore, we would like to examine how ligand binding and cancer-associated mutations may modulate p53-TAD secondary structural properties. As shown in Figure 3, p53-TAD in all cases contains residual helices in regions similar to those observed for the WT protein. This is generally consistent with ongoing NMR characterizations, where secondary chemical shift analysis has confirmed that mutations do not lead to significant changes in the secondary structural propensity (unpublished data, Prakash and Zolkiewski labs, Kansas State University). However, some subtle and local effects can also be observed. For example, N29K/N30D almost completely abolishes residual helicity at residues 25–35, while D49Y increases helical probability at residues 45–55. Since these regions often undergo disorder-to-order transitions upon binding to target proteins, such changes in helical propensities of unbound p53-TAD may lead to changes in folding conformational entropy cost in the coupled binding and folding, thus affecting the binding affinities.
3.3. Modulation of p53-TAD overall chain dimension
The activity of p53 is tightly regulated by TAD’s interaction with MDM2 (the degradation pathway) and transcriptional coactivator CBP (activation pathway), forming ternary complexes that mediate p53 turnover37. Multi-site phosphorylation of TAD under prolonged genotoxic stress stabilizes p53 and activates its tumor suppressor function by weakening binding to MDM2 and at the same time enhancing binding to CBP36–40. Considering the cooperative nature of p53 regulation and existence of multiple binding and phosphorylation sites in p53-TAD, changes in overall chain dimension may directly affect the availability of binding sites, thus perturbing the balance between degradation and activation pathways. As summarized in Figure 2, cancer-associated mutations do not appear to dramatically change the overall dimension of p53-TAD, while EGCG binding leads to significant compaction of peptide chain. Such effect of EGCG binding is consistent with NMR studies, where enhanced R2 relaxation rate have been observed across the entire sequence of p53-TAD in the presence of EGCG (unpublished data, Wang lab, Rensselaer Polytechnic Institute). Surface plasmon resonance competition assay has further confirmed that EGCG binding indeed could disrupt the interaction between p53 and its regulator, the E3 ubiquitin ligase MDM2, thus stabilizing p53 (unpublished data, Wang lab, Rensselaer Polytechnic Institute).
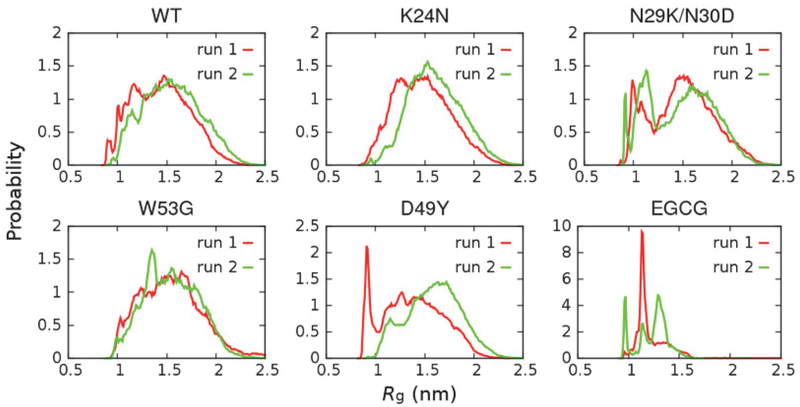
We further examined the local compactness of around AD1, which is a key recognition site, such as for with high binding affinity to MDM2 and moderate binding affinity to CBP domains37 (see Figure 1). As shown in Figure 4, the results reveal that N29K/N30D can induce significant compaction around AD1, with a greatly enhanced probability adopting highly compact structures with Rg ≤ 1.25 nm. This is consistent with NMR results showing that N29K/N30D is the only mutant among the four examined here that leads to significant increase in R2 relaxation rates (unpublished data, Prakash lab, Kansas State University). Importantly, preliminary binding assay has also revealed that the N29K/N30D double mutation abolishes binding of p53-TAD to MDM2 and CBP domains (unpublished data, Zolkiewski lab, Kansas State University). The conformational consequence of N29K/N30D appears to be similar to those caused by EGCG binding (Figure 4), and both lead to impairment of binding. These results support our hypothesis that increasing chain collapse may shield the functional sites of p53-TAD and perturb its interaction network.
Figure 4.
Probability distributions of Rg of p53-TAD subdomain AD1 (residues 10 – 40) calculated from two independent simulations.
3.4. Mutations and ligand binding dramatically shift p53-TAD conformational equilibria
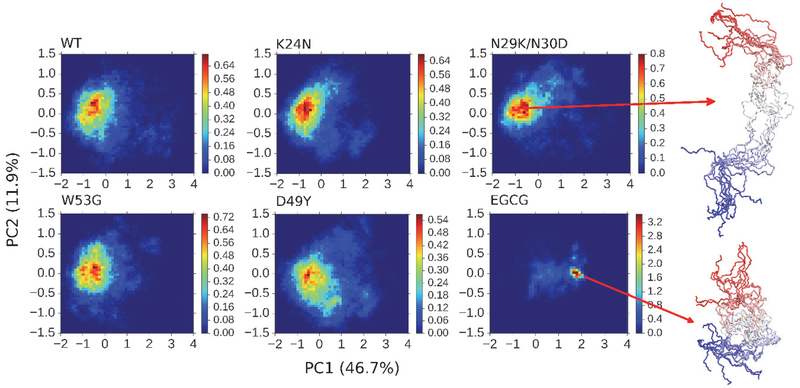
The above analyses of peptide chain dimension and secondary structural properties suggest that these cancer-associated mutations and ligand binding could perturb the structural ensemble of unbound p53-TAD. To directly visualize the conformational space available to p53-TAD and changes induced by mutations and ligand binding, we have performed principal component analysis (PCA). For this, we combined conformational ensembles obtained from all simulations, and performed featurization on peptide backbone heavy atoms using the DRID algorithm, distribution of reciprocal of interatomic distances63, as implemented in MSMBuilder 3.6.164. PCA analysis in DRID space (Figure 5) reveals that WT p53-TAD and all four mutants could visit a very large conformational space, as expected for a highly dynamic and disordered peptide. On the other hand, the conformational space available to p53-TAD becomes highly restricted in the presence of EGCG molecules (Figure 5). As illustrated by the representative snapshots, the conformational ensemble of p53-TAD becomes dominated by compact conformers in the presence of EGCG. This is consistent with the previous observation that EGCG could lead to significant compaction of peptide chain (Figure 2). Interestingly, D49Y appears to slightly increase the conformational heterogeneity of p53-TAD, leading to a broader distribution (Figure 5).
Figure 5.
Projection of simulated structural ensembles of p53-TAD onto the first two principal components. The heat maps indicate probability distributions derived from simulation statistics. Values in the parenthesis are percentages of variance in each direction. Ten representative snapshots are shown for each of the two selected states, with the color changing from red at the N-terminus to blue at the C-terminus.
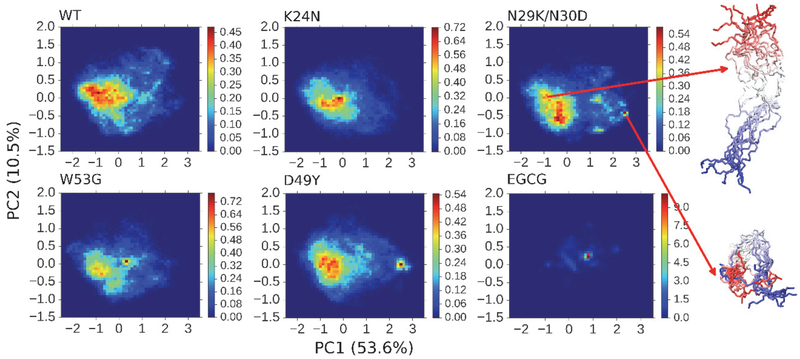
We have further examined the impacts of four cancer-associated mutations and EGCG binding on the conformational space of residues 10 – 40, which includes AD1 subdomain of p53-TAD. As shown in Figure 6, PCA analysis of AD1 conformations in DRID space suggests that besides EGCG binding, the double mutation N29K/N30D could also significantly perturb the conformational equilibria of AD1 subdomain, which is consistent with above observation that both EGCG binding and N29K/N30D could induce more compact AD1 structures (Figure 4). The apparent changes induced by D49Y cannot be faithfully evaluated, since AD1 conformations in this system are not well converged (e.g., Figure 4).
Figure 6.
Projection of simulated structural ensembles of p53-TAD subdomain AD1 (residues 10 – 40) onto the first two principal components. The heat maps indicate probability distributions derived from simulation statistics. Values in the parenthesis are percentages of variance in each direction. Ten representative snapshots are shown for each of the two selected states, with the color changing from red at the N-terminus to blue at the C-terminus.
3.5. Nonspecific binding of EGCG to p53-TAD
To further understand how EGCG induces collapse of p53-TAD conformations, we computed the contact probabilities between EGCG molecules and each residue of the peptide. A contact is considered formed when the minimum heavy-atom distance between any EGCG molecule and the residue in p53-TAD is less than or equal to 0.42 nm. As shown in Figure 7, although the contact profiles are not well converged at the level of each individual residue, it’s obvious that most residues in the peptide sequence could form contacts with EGCG molecules with significant probabilities. This seems to suggest that EGCG binds to p53-TAD through highly nonspecific interactions. The EGCG molecule is rich in hydroxyl groups and aromatic rings, which could then form various hydrogen bonds and hydrophobic contacts with the peptide. We have also observed similar nonspecific interactions of small drug molecules that could induced local compaction of Aβ42, another IDP that is associated with Alzheimer’s diseases, to suppress oligomerization and potentially aggregation65. Taken together, these results suggest that nonspecific interaction may likely be a common and effective mode of action for small molecules targeting IDPs.
Figure 7.
Probability of forming contacts between each residue of p53-TAD and any EGCG molecule calculated from two independent simulations.
4. Conclusions
IDPs are important functional proteins in cellular signaling and regulation, and their mutations are often associated with human diseases. However, it remains challenging to determine the underlying molecular mechanisms of IDP function and design effective therapeutic strategies to target these IDPs. In this work, we combined the state-of-the-art atomistic force field a99SB-disp with enhanced sampling and GPU accelerated MD simulation to investigate how cancer-associated mutations and ligand binding may modulate the disordered ensemble of unbound p53-TAD. Through microsecond-timescale REST2 enhanced sampling, all simulated ensembles are reasonably converged at levels of overall dimension and secondary structural propensities. The impacts of K24N and W53G on the unbound state of p53-TAD do not seem to be obvious according to this study. D49Y, on the other hand, has local effect on the intrinsic helical propensities of p53-TAD. Similar local effects have been observed in N29K/N30D as well, but this double mutation could also induce long-range effects, like the collapse of p53-TAD subdomain AD1. The impacts of EGCG binding seems to be the most dramatic, which bind nonspecifically to p53-TAD and lead to a highly restricted compact conformational ensemble. These observations are generally consistent with ongoing experimental studies, which have also confirmed the ability of N29K/N30D and EGCG binding to affect p53-TAD binding to key regulators. Structural insights derived from these simulations will lay the foundation for establishing the sequence-disordered ensemble-function-disease relationship of p53-TAD and shed light on new drug design strategies targeting p53 and other regulatory IDPs.
Our studies suggest that induced compaction may to be a general mechanism for shielding functional sites of IDPs to achieve inhibitory effects. This is consistent with previous studies suggesting a key role of entropic modulation in IDP-drug interactions65–66. Furthermore, nonspecific interaction is likely a common and effective mode of action for small molecules targeting IDPs, which is in contrast to traditional drug design strategy relying on specific interactions between drug molecules and target proteins. Our study also demonstrates a viable approach that integrates physics-based atomistic simulation and biophysical experiments to unravel how the disordered ensemble of IDPs may be modulated by various cellular signals including mutations, post-translational modifications and ligand binding. We note that generating reliable structural ensembles of IDPs remains a formidable task, due to the sampling requirement and more importantly persisting limitations in atomistic force fields. It is thus imperative to examine the properties of the simulated ensembles across of a series of mutations and/or with and without the presence of drug molecules, such that systematic biases may be cancelled and reliable features identified.
5. Acknowledgements
We thank Om Prakash, Michal Zolkiewski and Chunyu Wang for collaboration and extremely helpful discussions, whose labs have also provided unpublished experimental data for the validation and interpretation of simulation results. This work is supported by the National Institutes of Health (GM114300). The computing was performed on the Pikes cluster housed in the Massachusetts Green High-Performance Computing Center (MGHPCC).
6. References
- 1.Wright PE; Dyson HJ, J Mol Biol 1999, 293 (2), 321–331. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK; Brown CJ; Lawson JD; Iakoucheva LM; Obradovic Z, Biochemistry (Mosc.) 2002, 41 (21), 6573–6582. [DOI] [PubMed] [Google Scholar]
- 3.Tompa P, Trends Biochem. Sci 2002, 27 (10), 527–533. [DOI] [PubMed] [Google Scholar]
- 4.Dyson HJ; Wright PE, Nat. Rev. Mol. Cell Biol 2005, 6 (3), 197–208. [DOI] [PubMed] [Google Scholar]
- 5.Uversky VN; Oldfield CJ; Dunker AK, J. Mol. Recognit 2005, 18 (5), 343–384. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK; Silman I; Uversky VN; Sussman JL, Curr. Opin. Struct. Biol 2008, 18 (6), 756–764. [DOI] [PubMed] [Google Scholar]
- 7.Smock RG; Gierasch LM, Science 2009, 324 (5924), 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright PE; Dyson HJ, Nat. Rev. Mol. Cell Biol 2015, 16 (1), 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Click TH; Ganguly D; Chen J, Int J Mol Sci 2010, 11 (12), 5292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teilum K; Olsen JG; Kragelund BB, Cellular and molecular life sciences : CMLS 2009, 66 (14), 2231–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iakoucheva LM; Brown CJ; Lawson JD; Obradovic Z; Dunker AK, J. Mol. Biol 2002, 323 (3), 573–584. [DOI] [PubMed] [Google Scholar]
- 12.Mitrea DM; Kriwacki RW, FEBS Lett. 2013, 587 (8), 1081–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu MM; van der Lee R; de Groot NS; Gsponer J, Curr Opin Struct Biol 2011, 21, 432–40. [DOI] [PubMed] [Google Scholar]
- 14.Lu HC; Chung SS; Fornili A; Fraternali F, Frontiers Mol Biosci 2015, 2, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vacic V; Markwick PR; Oldfield CJ; Zhao X; Haynes C; Uversky VN; Iakoucheva LM, PLoS Comput Biol 2012, 8 (10), e1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uversky VN; Dave V; Iakoucheva LM; Malaney P; Metallo SJ; Pathak RR; Joerger AC, Chemical reviews 2014, 114 (13), 6844–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vacic V; Iakoucheva LM, Mol. BioSyst 2012, 8 (1), 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brucale M; Schuler B; Samori B, Chemical reviews 2014, 114 (6), 3281–3317. [DOI] [PubMed] [Google Scholar]
- 19.Kruschel D; Zagrovic B, Mol. BioSyst 2009, 5 (12), 1606–1616. [DOI] [PubMed] [Google Scholar]
- 20.Fisher CK; Stultz CM, Curr. Opin. Struct. Biol 2011, 21 (3), 426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuler B; Eaton WA, Curr. Opin. Struct. Biol 2008, 18 (1), 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eliezer D, Curr. Opin. Struct. Biol 2009, 19 (1), 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JH, Archives of biochemistry and biophysics 2012, 524 (2), 123–131. [DOI] [PubMed] [Google Scholar]
- 24.Ganguly D; Chen J, J Mol Biol 2009, 390 (3), 467–477. [DOI] [PubMed] [Google Scholar]
- 25.Choy WY; Forman-Kay JD, J. Mol. Biol 2001, 308 (5), 1011–32. [DOI] [PubMed] [Google Scholar]
- 26.Varadi M; Kosol S; Lebrun P; Valentini E; et al. , Nucleic Acids Res. 2014, 42, D326–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindorff-Larsen K; Kristjansdottir S; Teilum K; Fieber W; Dobson CM; Poulsen FM; Vendruscolo M, J. Am. Chem. Soc 2004, 126 (10), 3291–3299. [DOI] [PubMed] [Google Scholar]
- 28.Jensen MR; Salmon L; Nodet G; Blackledge M, J. Am. Chem. Soc 2010, 132, 1270–2. [DOI] [PubMed] [Google Scholar]
- 29.Salmon L; Nodet G; Ozenne V; Yin GW; Jensen MR; Zweckstetter M; Blackledge M, J. Am. Chem. Soc 2010, 132 (24), 8407–8418. [DOI] [PubMed] [Google Scholar]
- 30.Vise P; Baral B; Stancik A; Lowry DF; Daughdrill GW, Proteins-Structure Function and Bioinformatics 2007, 67 (3), 526–530. [DOI] [PubMed] [Google Scholar]
- 31.Muller PAJ; Vousden KH, Cancer Cell 2014, 25 (3), 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandoth C; McLellan MD; Vandin F; Ye K; Niu BF; Lu C; Xie MC; Zhang QY; McMichael JF; Wyczalkowski MA; Leiserson MDM; et al. , Nature 2013, 502 (7471), 333.-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris SL; Levine AJ, Oncogene 2005, 24 (17), 2899–2908. [DOI] [PubMed] [Google Scholar]
- 34.Grossman SR; Deato ME; Brignone C; Chan HM; Kung AL; Tagami H; Nakatani Y; Livingston DM, Science 2003, 300 (5617), 342–4. [DOI] [PubMed] [Google Scholar]
- 35.Brooks CL; Gu W, Mol. Cell 2006, 21 (3), 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruse JP; Gu W, Cell 2009, 137 (4), 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreon JC; Lee CW; Arai M; Martinez-Yamout MA; Dyson HJ; Wright PE, Proc. Natl. Acad. Sci. U. S. A 2009, 106 (16), 6591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teufel DP; Bycroft M; Fersht AR, Oncogene 2009, 28 (20), 2112–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CW; Ferreon JC; Ferreon ACM; Arai M; Wright PE, Proc. Natl. Acad. Sci. U. S. A 2010, 107 (45), 19290–19295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine AJ; Hu W; Feng Z, Cell Death Differ. 2006, 13 (6), 1027–1036. [DOI] [PubMed] [Google Scholar]
- 41.Kussie PH; Gorina S; Marechal V; Elenbaas B; Moreau J; Levine AJ; Pavletich NP, Science 1996, 274 (5289), 948–953. [DOI] [PubMed] [Google Scholar]
- 42.Lee CW; Martinez-Yamout MA; Dyson HJ; Wright PE, Biochemistry (Mosc.) 2010, 49 (46), 9964–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CW; Arai M; Martinez-Yamout MA; Dyson HJ; Wright PE, Biochemistry (Mosc.) 2009, 48 (10), 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Lello P; Jenkins LMM; Jones TN; et al. , Mol. Cell 2006, 22 (6), 731–740. [DOI] [PubMed] [Google Scholar]
- 45.Borcherds W; Theillet FX; Katzer A; et al. , Nat. Chem. Biol 2014, 10 (12), 1000–2. [DOI] [PubMed] [Google Scholar]
- 46.Chen J; Kriwacki RW, J. Mol. Biol 2018, 430 (16), 2275–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X; Chen J, J Chem Theory Comput 2019, 15 (8), 4708–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robustelli P; Piana S; Shaw DE, Proc. Natl. Acad. Sci. U. S. A 2018, 115 (21), E4758–E4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P; Kim B; Friesner RA; Berne BJ, Proc. Natl. Acad. Sci. U. S. A 2005, 102 (39), 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang LL; Friesner RA; Berne BJ, J Phys Chem B 2011, 115 (30), 9431–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang JM; Wolf RM; Caldwell JW; Kollman PA; Case DA, J Comp Chem 2004, 25 (9), 1157–1174. [DOI] [PubMed] [Google Scholar]
- 52.Ganguly D; Chen JH, PLoS computational biology 2015, 11 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abraham MJ; Murtola T; Schulz R; Páll S; Smith JC; Hess B; Lindahl E, SoftwareX 2015, 1, 19–25. [Google Scholar]
- 54.Pall S; Abraham MJ; Kutzner C; Hess B; Lindahl E, Lect Notes Comput Sc 2015, 8759, 3–27. [Google Scholar]
- 55.Tribello GA; Bonomi M; Branduardi D; Camilloni C; Bussi G, Comput Phys Commun 2014, 185 (2), 604–613. [Google Scholar]
- 56.Terakawa T; Kameda T; Takada S, J Comp Chem 2011, 32 (7), 1228–1234. [DOI] [PubMed] [Google Scholar]
- 57.Bussi G, Mol Phys 2014, 112 (3–4), 379–384. [Google Scholar]
- 58.Darden T; York D; Pedersen L, J. Chem. Phys 1993, 98 (12), 10089–10092. [Google Scholar]
- 59.Hess B; Bekker H; Berendsen HJC; Fraaije JGEM, J. Comput. Chem 1997, 18 (12), 1463–1472. [Google Scholar]
- 60.Kabsch W; Sander C, Biopolymers 1983, 22 (12), 2577–2637. [DOI] [PubMed] [Google Scholar]
- 61.Lee H; Mok KH; Muhandiram R; Park KH; Suk JE; Kim DH; Chang J; Sung YC; Choi KY; Han KH, J. Biol. Chem 2000, 275 (38), 29426–29432. [DOI] [PubMed] [Google Scholar]
- 62.Zhan YA; Wu HW; Powell AT; Daughdrill GW; Ytreberg FM, Proteins 2013, 81 (10), 1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou T; Caflisch A, J Chem Theory Comput 2012, 8 (8), 2930–2937. [DOI] [PubMed] [Google Scholar]
- 64.Beauchamp KA; Bowman GR; Lane TJ; Maibaum L; Haque IS; Pande VS, J Chem Theory Comput 2011, 7 (10), 3412–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang C, S. S, Fejzo J, Eyles S, and Chen J, J. Chem. Theory Comput. 2019, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heller GT; Bonomi M; Vendruscolo M, J. Mol. Biol 2018, 430 (16), 2288–2292. [DOI] [PubMed] [Google Scholar]