Dear Editor,
Fatty acid synthase (FAS) is the key enzyme for de novo fatty acid synthesis and exists widely from bacteria to mammalian cells (Schweizer and Hofmann, 2004). It is involved in membrane organization, metabolism regulation, energy storage, and many other processes (Maier et al., 2010). The ataxia telangiectasia-mutated and Rad3-related kinase (ATR) is a master regulator of DNA damage response (DDR) and is highly conserved. ATR is activated by DNA double-strand breaks and various types of DNA replication stresses (Cortez et al., 2001). Recent researches have reported that FAS seems closely related to genome stability (Wu et al., 2016). It has been proved that palmitic acid, the product of FAS, can inhibit ATR phosphorylation and kinase activity in mouse fibroblast (Zeng et al., 2008). These studies suggest that there is a crosstalk between fatty acid metabolism and DDR pathway, but the mechanism remains unknown.
Here, we found that FAS was stably co-purified with Rad3/Rad26 (ATR/ATRIP homolog in Schizosaccharomyces pombe, ATRIP (ATR-interacting protein)) from whole cell extraction of S. pombe (Figure 1A). We prepared electron microscope specimen of co-purified elution stained with 0.75% uranyl formate (UF). Typical micrographs directly show the FAS–Rad3/Rad26 assemblies and suggest there is an interaction between FAS and Rad3/Rad26 (Figure 1B). Their interaction was also verified by in vivo co-IP assay. Fas1 (β subunit of FAS) could be reproducibly co-precipitated with IgG-Rad3 beads (double protein A tagged) from cell lysate (Figure 1C).
Figure 1.
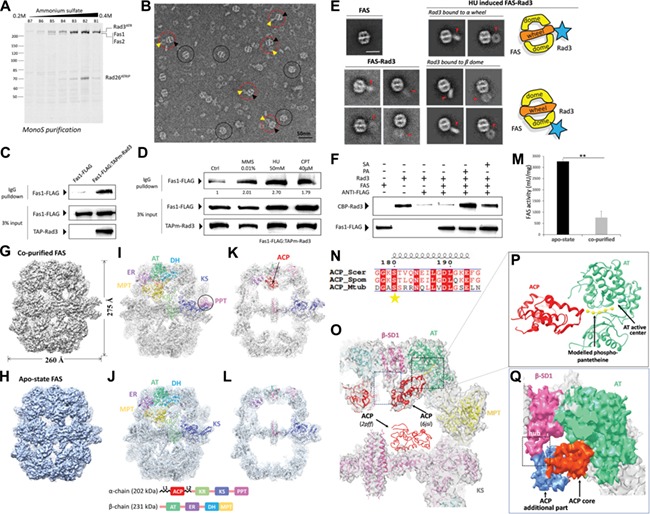
The interaction between S. pombe FAS and Rad3/Rad26 and structure/activity comparisons between co-purified and apo-state FAS. (A) Ruby-stained SDS–PAGE gel of Mono S chromatography fractions. (B) Negative stain EM micrograph of co-purified FAS–Rad3/Rad26. Black circles indicate FAS. Red circles indicate FAS–Rad3/Rad26 complex. Yellow and black arrows indicate Rad3/Rad26 and FAS, respectively in the complex. Scale bar, 50 nm. (C) In vivo co-IP assays in the Fas1-FLAG/TAPm-Rad3 double tagging strain and the Fas1-FLAG single tagging strain (negative control). TAPm-Rad3 has affinity with IgG resin and Fas1-FLAG was co-precipitated by TAPm-Rad3. Western blot analysis using indicated antibodies detecting FAS and Rad3. (D) Western blot of in vivo co-IP assays in cells treated with MMS, HU, and CPT. (E) 2D averages of apo-state FAS (left top panel), non-induced co-purified FAS–Rad3/Rad26 (left bottom panel) and HU-induced co-purified FAS–Rad3/Rad26 (right panel). Red arrows indicate Rad3/Rad26 density. Diagrams representing the typical complex configurations are shown beside the averages. Scale bar, 30 nm. (F) In vitro pull-down assays with affinity of ANTI-FLAG resin to Fas1-FLAG, to test if TAPm-Rad3 could be pulled down by Fas1-FLAG with/without PA or SA. PA, palmitic acid; SA, stearic acid. (G and H) Cryo-EM maps of co-purified FAS (G) and apo-state FAS (H). The resolutions are 4.7 Å and 5.1 Å, respectively. (I and J) The EM map and structural models of co-purified (I, PDB ID code 6JSI) and apo-state (J, PDB ID code 6JSH) FAS. The eight catalytic domains are shown with distinct colors. Domain organization of α and β subunits are shown at the bottom. PPT domain is highlighted by the circle in co-purified FAS structure. (K and L) A slice through the middle of co-purified FAS (K) and apo-state FAS (L) cryo-EM structure in side view. The ribbon of ACP domains is colored red. (M) The specific activity of apo-state and co-purified FAS. **P < 0.01 (two tails student’s t-test). Independent experiments were repeated for three times. (N) Alignment of FAS ACP recognition helix sequences from S. cerevisiae, S. pombe, and M. tuberculosis. Strictly conserved and similar residues are surrounded by red and colored red, respectively. Catalytic residues are marked by stars. (O) Superimposition of ACP domain in co-purified FAS map is compared with that in the crystal structure (PDB ID code 2PFF). Modeled phosphopantetheinyl arm was fitted in the extra density observed around the active site of ACP domain. (P) Model of interaction between ACP and AT domain is shown in ribbon. The model was separated from O. (Q) The core (orange–red) and additional part (cornflower blue) of ACP domain are packed against AT (green) and SD1 (pink) domains, respectively in surface representation.
As Rad3/Rad26 participates in DDR, we examined the effect of genotoxic stress on the interaction between FAS and Rad3. Cells were treated with methyl methanesulfonate (MMS), hydroxyurea (HU), and camptothecin (CPT) for 2 h, respectively after growing to log phase, and the interaction between FAS and Rad3 was enhanced in different levels upon treatment with these DNA damaging agents (Figure 1D). HU made the most significant effect with 2.70 times compared with control. MMS and CPT were less effective inducers with 2.01 and 1.79 times compared with control, respectively. From HU-induced cell lysis, we efficiently separated FAS–Rad3/Rad26 assembly. We also purified apo-state FAS from Fas1-FLAG tagged cell lysis (Supplementary Figure S1). Negative stain electron micrographs of these samples were taken and 2D analysis was carried out. Comparing to 2D images of non-induced FAS–Rad3/Rad26, those of HU-induced FAS–Rad3/Rad26 reveal more stable interaction (Figure 1E). Rad3/Rad26 interacting with FAS harbors both compact and loose conformations. At the same time, Rad3/Rad26 is loosely coupled to FAS along the top of dome and proximity of the wheel. In vitro pull-down assays were carried out with apo-state FAS and Rad3/Rad26 and revealed that FAS hardly interacted with Rad3. Interestingly, saturated fatty acid (palmitic acid or stearic acid) could promote FAS–Rad3/Rad26 interaction and the ratio of assembly is significantly increased (Figure 1F).
We then determined the cryo-EM structures of both co-purified and apo-state FAS and the resolution reached 4.7 Å and 5.1 Å, respectively (Supplementary Figures S2–S5). Similar to Saccharomyces cerevisiae FAS, the structure of S. pombe FAS is barrel-shaped and comprised of a central wheel and two domes on each side. Inside the dome is a cavity. The overall structure takes 260 Å width and 275 Å height (Figure 1G and H). We used the crystal structure of S. cerevisiae FAS (PDB: 2PFF) to fit the EM map as a rigid body and found substantial differences. Then, the crystal structure was segmented and manually adjusted followed with model refinement and validation (Supplementary Figure S6 and Table S1). The refined model matched well with the cryo-EM map (Figure 1I and J; Supplementary Figure S7). The structure of co-purified FAS shows discernible acyl carrier protein (ACP) domains and phosphopantetheine transferase (PPT) domains, which are responsible for substrate shuttling among catalytic sites and ACP activation, respectively. However, there was little density corresponding to ACP and PPT domains in apo-state FAS (Figure 1I and K vs. Figure 1J and L; Supplementary Figure S8). This observation suggests that Rad3/Rad26 could probably stabilize the conformation of these FAS domains. To investigate the influence of FAS activity, the enzymatic activity of co-purified and apo-state FAS were measured. Surprisingly, the catalytic activity of co-purified FAS is sharply decreased with only 23.3% of the activity of apo-state FAS (760 vs. 3266 milliunits/mg) (Figure 1M).
The ACP domain from S. cerevisiae and S. pombe are highly conserved (Supplementary Figure S9). Several catalytic residues are also shared between yeast and Mycobacterium tuberculosis FAS (Figure 1N). In the co-purified S. pombe FAS, the ACP domains abut to the acetyl transferase (AT) domains (Figure 1O). Whereas, the ACP domains are located at the dimer interface of KS domains in the crystal structure of S. cerevisiae FAS (Lomakin et al., 2007). The active site Ser180 of ACP domain stretches toward the substrate binding cleft of AT domain, where active sites of AT lay deeply (Figure 1P; Supplementary Figure S7D). The distance between ACP Ser180 and AT active sites is nearly 18 Å, which matches with the length of phosphopantetheine arm (Leibundgut et al., 2007). The additional part of ACP, which is apart from ACP core and composed with 4 α-helices, packs against the structural domain1 in β subunit (β-SD1) (Figure 1Q). β-SD1 acts as a hub, anchoring ACP additional part to stabilize the interaction between ACP core and AT domain. ACP shuttles to AT domain at priming step during the fatty acid synthesis, which is the first step after ACP activation by PPT domain (Schuster et al., 1995; Fichtlscherer et al., 2000). The observation that ACP is immobilized at AT domain suggests that Rad3/Rad26 binding probably inhibits the fatty acid synthesis at the priming step, which offers structural insight into the inhibition of FAS by Rad3/Rad26.
Our work illuminated that FAS could play a role in DDR through interaction with ATR–ATRIP complex (Wu et al., 2016). We characterized the interaction between FAS and Rad3/Rad26 in vivo and in vitro. Our structural and functional analysis consistently suggest that the fatty acid synthesis may be inhibited at the priming step by Rad3/Rad26 binding. This work disclosed the close relevance between FAS and DDR and paved the way for future mechanistic studies.
[Atomic coordinates and structure factors for the reported structures have been deposited in the Protein Data bank under accession numbers 6JSH and 6JSI. The electron density map for the cryo-EM structure has been deposited in the EMDataBank under accession numbers EMD-9881 and EMD-9882. Supplementary material is available at Journal of Molecular Cell Biology online. EM data were collected at the Center for Bio-imaging, Institute of Biophysics, Chinese Academy of Sciences, and Center for Integrative Imaging of Hefei National Laboratory for Physical Sciences at the Microscale, respectively. We thank Gang Ji, Xiaojun Huang, and Peiping Tang for technical help and support with electron microscopy. We are grateful to Zhihui Zhang for IBP cryo data collection. This work was supported by grants from the National Natural Science Foundation of China (31870734 and 31890783) to G.C. and the National Basic Research Program (2018YFC1003400) to X.W.]
Supplementary Material
References
- Cortez D., Guntuku S., Qin J., et al. (2001). ATR and ATRIP: partners in checkpoint signaling. Science 294, 1713–1716. [DOI] [PubMed] [Google Scholar]
- Fichtlscherer F., Wellein C., Mittag M., et al. (2000). A novel function of yeast fatty acid synthase. Subunit alpha is capable of self-pantetheinylation. Eur. J. Biochem. 267, 2666–2671. [DOI] [PubMed] [Google Scholar]
- Leibundgut M., Jenni S., Frick C., et al. (2007). Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science 316, 288–290. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Xiong Y., and Steitz T.A. (2007). The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 129, 319–332. [DOI] [PubMed] [Google Scholar]
- Maier T., Leibundgut M., Boehringer D., et al. (2010). Structure and function of eukaryotic fatty acid synthases. Q. Rev. Biophys. 43, 373–422. [DOI] [PubMed] [Google Scholar]
- Schuster H., Rautenstrauss B., Mittag M., et al. (1995). Substrate and product binding sites of yeast fatty acid synthase. Stoichiometry and binding kinetics of wild-type and in vitro mutated enzymes. Eur. J. Biochem. 228, 417–424. [PubMed] [Google Scholar]
- Schweizer E., and Hofmann J. (2004). Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68, 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dong Z., Wang C.J., et al. (2016). FASN regulates cellular response to genotoxic treatments by increasing PARP-1 expression and DNA repair activity via NF-κB and SP1. Proc. Natl Acad. Sci. USA 113, E6965–E6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Wu G.Z., Goh K.J., et al. (2008). Saturated fatty acids modulate cell response to DNA damage: implication for their role in tumorigenesis. PLoS One 3, e2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


