The rapid accrual of knowledge in genomic medicine has prompted the reanalysis of pre-existing data.1,2 We clinically reanalyzed data from two patient series that had undergone diagnostic proband-only exome sequencing.
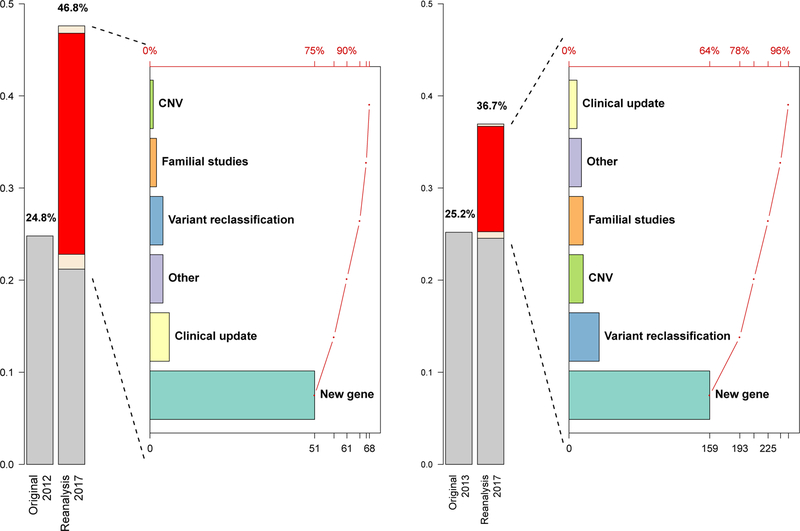
The exome sequences of the first series of 250 patients were obtained between 2011 and 2012.3 Cumulative reanalyses of extant data after the release of the initial clinical report increased the molecular diagnostic yield of cases from 24.8% to 46.8% (comprehensive manual reanalysis performed based on knowledge source from 12/2017) (Figure 1). Although clinical laboratories often use a “manual” approach (see Supplementary Appendix) for routine exome-sequence analysis, this approach may not be sustainable at scale, particularly for iterative re-analyses accruing historical cases. We designed a semi-automated reanalysis process, which prioritizes variants based on genetic and/or functional evidence and genes based on semantic phenotypic similarity scores (Supplemental Appendix). This approach, when used to analyze the first series, achieved a diagnostic sensitivity of 92.9% (with the manual reanalysis as the reference standard), while sustaining a manageable workload under a model of annual reanalysis over a five-year period (Supplemental Appendix). This semi-automated reanalysis approach was then applied on a second cohort of 2000 consecutive cases originally analyzed in 2012–2013,4 augmenting the molecular diagnostic rate from 25.2% to 36.7% (Figure 1). We did not carry out manual reanalysis of the exome sequences of the second, much larger cohort, given the burden of the task. Potential factors contributing to the difference in the overall diagnostic rates between the two series (46.8% vs. 36.7%) include different percentages of patients with Mendelian disorders and a difference in sensitivity between comprehensive manual and semi-automated analysis.
Figure 1. Systematic reanalysis increases diagnostic rates in two patient series referred to clinical exome sequencing.
The left (Cohort #1, N=250) and the right (Cohort #2, N=2000) panels depict the molecular diagnostic rate changes from the original rates, evidenced by respective publications3,4, and the reanalysis rates. The yield alteration originates from patients whose diagnostic molecular findings changed from zero to one or more (new diagnosis, highlighted in red in the reanalysis bar), from one to two or more (multilocus pathogenic variation; the white segment below the red segment in the reanalysis bar), and from one to zero (overturned molecular diagnosis, see Supplemental Appendix). The white segment above the red segment represents the proportion of patients who received a partial diagnosis from reanalysis (n=2 for Cohort #1, n=5 for Cohort #2), which is not counted towards the diagnostic rate. The Pareto charts to the right of the bar charts illustrate the breakdown of the interpretive reasons contributing to each new molecular diagnosis, with cumulative counts from each category shown in the bottom axis and the cumulative percentages shown in the top axis.
In line with the rapid pace of improvement in knowledge of genetic causes of disease, the vast majority of new molecular diagnoses resulted from newly discovered disease genes (75% and 64% from Cohorts #1 and #2, respectively; Supplemental Appendix, Figure S1). Additionally, several newly implicated genes initially associated with childhood-onset neurologic disorders, including PURA, DDX3X, and TANGO2, rank highly amongst the most frequently mutated in our clinical cohorts (Fig. S2 in the Supplementary Appendix). New molecular diagnoses also resulted from upgraded variant classifications in known disease genes (5.9% and 14% from Cohorts #1 and #2, respectively; Supplemental Appendix, Table S2). Other contributors include the emergence of additional clinically observed phenotypes, the identification of pathogenic copy-number variants, and the positive identification of a genetic cause that had been previously missed (Supplemental Appendix). The number of patients with multiple molecular diagnoses increased from 25 to 48 (in the combined series) after reanalysis, illustrating the contribution of continued genomic analyses to the deconvolution of clinically blended phenotypes (Supplemental Appendix, Table S5).5
Education is necessary for physicians and patients to understand the concept of an ‘evolving’ molecular interpretation. The communication and counseling processes are complicated pragmatically by the temporal separation between the initial and the updated reports and the dilemma of whom to send the report to. We sent a survey to 55 healthcare providers of the 64 patients in the first series for whom the reanalysis had yielded new molecular diagnoses. We received a response from 23 (42%) providers regarding follow-up data for 42 (66%) patients. Of these 23 providers, 10 reported a preference that all physicians involved in the patient’s care be informed of the results of the reanalysis, and 13 reported a preference that only the physician who ordered the reanalysis or who had ordered the original analysis be informed. The respondents reported that about three quarters of their patients (30) had received genetic counseling for the updated findings, and of these 30, about half (17) had their clinical management changed as a consequence of the new results (Supplemental Appendix, Table S7). The 12 patients who did not receive genetic counseling about their new diagnoses were reported to have relocated, died, or received their new diagnosis but did not keep their follow-up appointment. These findings suggest that periodic reanalysis may benefit patients and their families and physicians, and that they should be counselled about this at the time that clinical exome sequencing is ordered. However, an important caveat to our conclusions is that our study was not designed to measure clinical benefit of the intervention.
Supplementary Material
Acknowledgments
Funding
Research in the laboratories of JRL and RAG is partially supported by the NIH including a National Institutes of Neurological Disorders and Stroke (NINDS, R35 NS105078) and National Human Genome Research Institute (NHGRI, U54 HG003273) grants, and a jointly funded National Human Genome Research Institute (NHGRI)/National Heart, Lung, and Blood Institute (NHLBI) grant to the Baylor-Hopkins Center for Mendelian Genomics (UM1 HG006542). JEP is supported by NHGRI K08 HG008986.
Disclosures
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), which performs clinical exome sequencing and chromosomal microarray genomics assay services. The following authors, PL, LM, FX, AB, PW, HD, BY, WB, RX, XW, VRS, CS, CME, and YY, are employees of BCM and derive support through a professional services agreement with BG. TC, XW, and YY own stocks of AiLife Diagnostics. YY is a member of the Scientific Advisory Board of Veritas Genetics China. JRL serves on the Scientific Advisory Board of the BG. JRL has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and the Regeneron Genetics Center, has stock options in Lasergen, Inc. and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting.
Contributor Information
Pengfei Liu, Baylor College of Medicine, Houston, TX.
Linyan Meng, Baylor College of Medicine, Houston, TX.
Elizabeth A. Normand, Baylor College of Medicine, Houston, TX.
Fan Xia, Baylor College of Medicine, Houston, TX.
Xiaofei Song, Baylor College of Medicine, Houston, TX.
Andrew Ghazi, Baylor College of Medicine, Houston, TX.
Jill Rosenfeld, Baylor College of Medicine, Houston, TX.
Pilar L. Magoulas, Baylor College of Medicine, Houston, TX.
Alicia Braxton, Baylor College of Medicine, Houston, TX.
Patricia Ward, Baylor College of Medicine, Houston, TX.
Hongzheng Dai, Baylor College of Medicine, Houston, TX.
Bo Yuan, Baylor College of Medicine, Houston, TX.
Weimin Bi, Baylor College of Medicine, Houston, TX.
Rui Xiao, Baylor College of Medicine, Houston, TX.
Xia Wang, Baylor College of Medicine, Houston, TX.
Theodore Chiang, Baylor College of Medicine, Houston, TX.
Francesco Vetrini, Baylor Genetics, Houston, TX.
Weimin He, Baylor Genetics, Houston, TX.
Hanyin Cheng, Baylor Genetics, Houston, TX.
Jie Dong, Baylor Genetics, Houston, TX.
Charul Gijavanekar, Baylor Genetics, Houston, TX.
Paul J. Benke, Joe DiMaggio Children’s Hospital, Hollywood, Florida.
Jonathan A. Bernstein, Stanford University School of Medicine, Stanford, California.
Tanya Eble, Baylor College of Medicine, Houston, TX.
Yasemen Eroglu, Oregon Health and Science University, Portland, Oregon.
Deanna Erwin, Baylor College of Medicine, Houston, TX.
Luis Escobar, St. Vincent’s Peyton Manning Children’s Hospital, Indianapolis, Indiana.
James B. Gibson, Dell Children’s Medical Group, Austin, Texas.
Karen W. Gripp, A. I. DuPont Hospital for Children, Wilmington, Delaware.
Soledad Kleppe, Hospital Italiano de Bs.As. Buenos Aires, Argentina.
Mary Kay Koenig, UTHealth at McGovern Medical School, Houston, TX.
Andrea M. Lewis, Baylor College of Medicine, Houston, TX.
Marvin Natowicz, Cleveland Clinic, Cleveland, Ohio.
Pedro Mancias, UTHealth at McGovern Medical School, Houston, TX.
LaKeesha Minor, UTHealth at McGovern Medical School, Houston, TX.
Fernando Scaglia, Baylor College of Medicine, Houston, TX.
Christian P. Schaaf, Heidelberg University, Heidelberg, Germany.
Haley Streff, Baylor College of Medicine, Houston, TX.
Hilary Vernon, Johns Hopkins School of Medicine, Baltimore, Maryland.
Crescenda L Uhles, Children’s Health Hospital, Dallas, Texas, USA.
Elaine H. Zackai, The Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania.
Nan Wu, Peking Union Medical College Hospital, Beijing, China.
V. Reid Sutton, Baylor College of Medicine, Houston, TX.
Arthur L. Beaudet, Baylor College of Medicine, Houston, TX.
Donna Muzny, Baylor College of Medicine, Houston, TX.
Richard A. Gibbs, Baylor College of Medicine, Houston, TX.
Jennifer E. Posey, Baylor College of Medicine, Houston, TX.
Seema Lalani, Baylor College of Medicine, Houston, TX.
Chad Shaw, Baylor College of Medicine, Houston, TX.
Christine M. Eng, Baylor College of Medicine, Houston, TX.
James R. Lupski, Baylor College of Medicine, Houston, TX.
Yaping Yang, Baylor College of Medicine, Houston, TX.
References
- 1.Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med 2017;19:209–14. [DOI] [PubMed] [Google Scholar]
- 2.Eldomery MK, Coban-Akdemir Z, Harel T, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med 2017;9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013;369:1502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Posey JE, Harel T, Liu P, et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N Engl J Med 2017;376:21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.