Abstract
Introduction:
The rapid development of clinical next generation sequencing (NGS) and the contemporaneous availability of molecular targeted therapies ignited and fueled the field of precision oncology. More recently there has been an explosion of immunotherapeutic agents, specifically the checkpoint inhibitors: PD-1, PD-L1 and CTLA-4 antibodies. These new classes of agents have produced durable responses in a variety of tumor subtypes.
Areas Covered:
In this review, the authors explore the role of NGS in identifying targets for molecular therapy. The authors also expand on the future uses of NGS in oncology including: prediction of checkpoint inhibitor response, quantification of tumor mutational burden, neoantigen calling using bioinformatics tools, and finally the personalization of cell transfer technologies and cancer vaccines.
Expert Commentary:
The near future will witness an increased understanding of the immune system and genomics in cancer. High throughput sequencing technology will expand in parallel with an ever-expanding array of novel therapies. Improved computational power coupled with bioinformatics algorithms will combine the fields of genomics and immunology. The emerging fields that stand to benefit from rapid translation of NGS technology include cancer vaccines and adoptive cell therapy, which will further refine precision oncology.
Keywords: Immunotherapy, Immuno-oncology, Targeted therapy, Precision Oncology, Personalized Medicine, Next Generation Sequencing
1. Introduction
“In the midst of chaos, there is also opportunity”
― Sun Tzu, Art of War
As the sun begins to set on a century of cytotoxic chemotherapy, we reflect on the successes: Hodgkin lymphoma, non-Hodgkin lymphoma, germ cell tumors, pediatric acute lymphoblastic leukemia, among many others [1]. The benefits and development of cytotoxic chemotherapy have reached a therapeutic plateau in many cancers. With the completion of sequencing of the human genome in 2001 [2], the pendulum of enthusiasm swung toward genomic profiling both for diagnosis and novel treatments of tumors [3, 4].
Over the ensuing half-decade, the cost of genome sequencing has decreased due to the rapid growth of commercially available clinical next-generation sequencing (NGS) tests [5]. NGS allows for the sequencing of millions of DNA or RNA snippets in a single run, thereby substantially reducing the cost while increasing the speed [6]. This technology has percolated from the research realm into clinical practice, enabling the detection of new targets for both diagnostic and prognostic purposes [7]. While NGS has advanced oncology research, clinical applicability still lags behind [8].
The advent of immune checkpoint inhibitors, specifically antibodies targeting program death-1 (PD-1), program death ligand-1(PD-L1) and cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) antibodies, have changed the landscape of oncology care. These new classes of agents have produced consistent results across multiple tumor types. Ipilimumab, an anti-CTLA-4 antibody, was the first checkpoint inhibitor approved in 2011 for metastatic. melanoma with initial response rates of over 10% [9]. Subsequently, the PD-1 antibodies pembrolizumab and nivolumab were evaluated in melanoma with corresponding improvements in overall survival [10, 11]. Immune checkpoint inhibitors targeting PD-1 and PD-L1 are approved by the United States Food and Drug Administration (FDA) for the treatment of non-small cell lung cancer (NSCLC), Hodgkin’s lymphoma, renal cell cancer, urothelial cancer, hepatocellular cancer, gastric cancer, and microsatellite instability-high (MSI-H) cancers. However, the response rates in diseases other than MSI-H cancers, Hodgkin’s lymphoma, and melanomas have not been over 20% even in the best selected patients [12, 13, 14]. More recently, better results have been achieved with combination of ipilimumab and nivolumab in metastatic melanoma with response rates as high as 85% in patients with high PD-L1 expression, but at the high cost of grade 3 and 4 immune related adverse events [15]. Although there are dramatic responders, the non-responders and patients who progress from these agents outnumber the responders in most diseases. In addition, these drugs carry financial toxicity for not only the drugs but associated hospital admissions for management of immune related adverse events. Companion diagnostics in the form of PD-L1 staining have been variable and remain a challenge as an effective biomarker [16].We need better biomarkers to predict clinical response to these agents. This is of paramount importance not only for the drugs currently in clinical use, but also for new immunotherapies in development both as single agents and in combination [17].
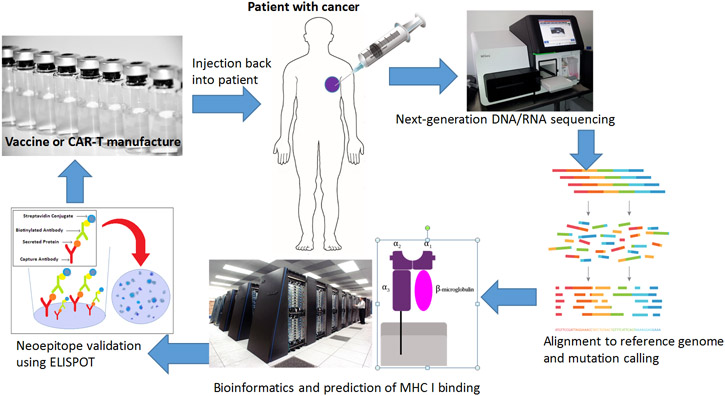
In this review, we examine the use of NGS as a diagnostic and prognostic tool to predict individual response to immunotherapy as well as to aid in the development of new immunotherapies. We also explore how NGS technology merges the fields of targeted therapy and immunotherapy (Figure 1).
Figure 1:
The merging of the fields of immunotherapy and targeted therapy
2. Next-Generation Sequencing (NGS) guided targeted therapy for Precision Oncology
NGS is a broad term used to describe several competing DNA sequencing technologies. The basis of NGS is to sequence short fragments of the genome, which are then aligned together to a reference genome. The end result is very deep and detailed coverage of a selected panel of genes that can be delivered quickly and inexpensively [18]. This technology has made a strong foothold in cancer research, refining diagnoses, and even providing prognostic information to patients [19, 20].
One way to utilize NGS is to identify a driver mutation that can predict response to targeted therapy. This is best illustrated in the treatment of NSCLC, which has recently become the poster child for precision oncology. While multiple diagnostic modalities exist to detect actionable mutations in NSCLC, Rozenblum and colleagues asked if NGS could improve diagnostic yield [21]. They report that NGS was able to detect actionable aberrations in 15% of patients after previous negative or inconclusive standard testing for mutations in these genes. Overall, they were able to identify actionable alterations in half of patients in genes such as EGFR, RET, ALK, MET, and ERBB2. NGS testing resulted in a therapy change for 42% of patients and led to an overall response rate of 65%. This is consistent with other investigators who have reported NSCLC to have about 50% actionable driver mutations [22].
NGS is a major step forward in DNA sequencing technology, but is not without limitations. The technology is dependent on alignment with a reference genome, and DNA fragments can be aligned to the wrong chromosome or be unmapped completely. This results in the inability to read insertions and deletions properly. Additionally, single nucleotide polymorphisms that occur with low frequency are challenging to detect. This becomes especially problematic when a somatic mutation occurs at a locus near a germline polymorphism in an area that already has high rates of polymorphisms [23]. Another limitation is tissue acquisition. Even though NGS can be performed with as little as 50ng of DNA [24], occasionally a sample may be inadequate. This can occur for a variety of reasons, most commonly insufficient quantities of DNA due to low cellularity and small samples [25]. Samples that return with inconclusive results are often marked by the performing laboratory. Clinicians have the option of obtaining a fresh biopsy with more tumor tissue or sending a liquid biopsy. This remains true when clinical suspicion for a genomic aberration remains high, but NGS testing does not detect the aberrant gene.
Liquid biopsy in the context of NGS refers to the sequencing of cell-free DNA – fragments of DNA shed by the tumor and freely floating in the blood or circulating tumor cells. Even more than NGS performed on tissue, liquid biopsies are dependent on pre-analytic variables such as serum or blood source and time to centrifugation [26]. Additionally, because cell-free DNA exists as fragments, difficulties arise during alignment to a reference genome. This becomes especially problematic when attempting to identify oncogenic fusions which most readily manifest as RNA products. Unfortunately, performing NGS on cell free RNA is technically challenging and beyond the ability of most clinical labs [27]. Despite these limitations, liquid biopsies provide another NGS option for clinicians when additional tumor tissue is unavailable or a patient is unwilling to undergo additional biopsy.
3. Basket studies, NCI-MATCH and other programs
Many molecularly targeted therapies are initially evaluated in a defined histologic subtype. However, the pathways that are targeted with these therapies are present across many different tumor types. One of the first “basket studies,” involved the use of vemurafenib (BRAF inhibitor) in various non-melanoma cancers harboring BRAFV600E mutations. This study resulted in response rates above 40% in specific arms [28] and led to the approval of vemurafenib in Erdheim Chester disease and Langerhans Cell Histiocytosis [29]. Subsequent studies such as combination BRAF+ MEK inhibitor studies have further validated this concept and have demonstrated efficacy in other cancers like BRAFV600E mutant non-small cell lung cancer [30] and anaplastic thyroid cancer [31]. However, this level of success was not the case in all tumors. Single agent vemurafenib was not sufficient in BRAFV600E mutant colorectal cancer to produce clinical responses. Rather, a combination with EGFR inhibition was needed to overcome the innate resistance pathways active in BRAFV600E mutant colorectal cancer [32]. Hence, tissue of origin remains important in the field of precision oncology.
There are several over-arching basket trials such as the Novartis Signature® study, Genentech’s MY Pathway trial, in addition to national programs such as Investigation of Serial Studies to Predict Your Therapeutic Response through Imaging and Molecular Analysis 2 (I-SPY2), NCI cooperative group’s Targeted Therapy Directed by Genetic Testing in Treating Patients With Advanced Refractory Solid Tumors, Lymphomas, or Multiple Myeloma (NCI-MATCH))and American Society of Clinical Oncology (ASCO)’s Targeted Agent and Profiling Utilization Registry (TAPUR) Study. These trials should further contribute to our understanding of achieving optimal efficacy with genomically targeted therapy. Some of these studies, such as the NCI-MATCH and MY Pathway have added immunotherapy arms as well, based on MSI-H cancers or PD-L1 gene amplification.
4. Immunotherapy
4.1. Immune Checkpoint inhibitors
The immune system is regulated by a series of inhibitory and stimulatory receptors, including CTLA-4 and PD-1. Tumor cells evade immune surveillance and destruction by inhibiting the immune system using these receptors. For example, tumors can hyper-express PD-L1, which is a ligand for PD-1 expressed on immune cells, specifically T-cells. The interaction between PD-L1 and PD-1 will dampen the T-cell response and enable the tumor to hide from immune response[33]. Blocking the PD-1/PD-L1 axis allows for the T-cell to recognize and destroy tumor cells. Immune modulation can also occur upstream of the interaction of the immune system and the tumor cells. CTLA-4 was one of the first receptors to be targeted for cancer therapy. CTLA-4 is found on the T-cells and is a mechanism involved in the activation and proliferation of the immune response. Drugs such as ipilimumab, nivolumab, pembrolizumab, avelumab, atezolizumab and durvalumab are able to reverse this inhibition and allow the immune system to continue uninhibited in tumor killing [34].
4.2. Vaccines
Tumor induced inhibition of the immune system implies that tumors are immunogenic. Since tumors arise from our own cells, they have a high degree of self-antigens, those derived from proteins existing in normal human cells. With increased mutational burden, tumors produce more non-self-antigens or tumor associated antigens. These tumor associated antigens can thereby be identified and targeted by the host’s immune system [35]. Many vaccines have been developed over the years to target these tumor associated antigens. The major obstacles in vaccine development have been tumor heterogeneity, as one single antigen is not expressed in every tumor cell. Other mechanisms of evasion include loss of major histocompatibility complex (MHC) [36] or beta-2 microglobulin [37]. At worst, cancer cells become so dedifferentiated that they stop expressing any tumor associated antigens, making a vaccine futile [38].
4.3. Adoptive Cellular therapy
Rather than showing the immune system fragments of tumor using a vaccine, educated T-cells that already “see” the tumor can be used as a therapeutic modality. These tumor infiltrating lymphocytes (TILs) can be isolated from the tumor, grown ex-vivo, and reinjected to produce dramatic clinical response. In the pre-immune checkpoint era, the therapeutic use of TIL’s have demonstrated response rates as high as 34% in melanoma [39].
A newer approach involves generating T-cells with a unique receptor targeting a molecule expressed on cancer cells. Chimeric antigen receptor T-cells (CAR-Ts) have recently received FDA approval for treatment of CD19 positive B-cell ALL and aggressive B-cell lymphomas [40]. Part of the success behind the use of CAR-T cells in leukemia and lymphoma is the ubiquitous expression of a specific antigen, specifically CD19. Creating CAR-Ts for solid tumors is more challenging as these are complicated by intra-tumor heterogeneity and lack of a shared cell surface antigen. For example, CAR-T cells targeting HER-2 and CEA have yielded minimal benefit [41].
4.4. Immunotherapy after targeted therapy
One of the pressing issues in oncology is that of sequencing targeted strategies and immunotherapy. Unfortunately, there is a paucity of data to guide clinicians. In melanoma, for example, expert opinion suggest that when possible, immunotherapy should be used ahead of BRAF inhibition, but that the reverse strategy is also effective [42]. For instance when there is a bulky tumor with a BRAFV600E mutation, targeted therapy could be used to “debulk” tumor followed by immunotherapy to consolidate the response. Conversely, others report that PD-1 checkpoint blockade is not highly effective in EGFR and ALK mutant lung cancer, especially for those with a T790M mutation [43, 44].
4.5. Combinations of immunotherapy and targeted therapy
A current trend in clinical research has been the combination of immunotherapy with other available cancer treatments. This is based on the premise that immunotherapy in combination with other appropriate therapies will enhance efficacy. Examples of combination trials include immunotherapy with another immunotherapy [15], immunotherapy with a targeted agent [45], and immunotherapy with a chemotherapy [46]. Combinations of an immunotherapy with either another immunotherapy or a chemotherapeutic can be applied across a broad population, but the combination with a targeted agent is almost invariably biomarker driven.
There is good rationale for combining these two modalities. For example, Kammerer-Jacquet et al demonstrated that in renal cell carcinoma a strong correlation exists between high c-MET and PD-L1 expression [47]. Similarly, Callahan and colleagues demonstrated that RAF inhibition causes enhanced activity of CTLA-4 blockade in preclinical models [48]. This highlights the importance of identifying mutations in the combinatorial use of immunotherapy.
The role of NGS in development of such a drug medley becomes important. It is possible to design a single trial where a single gene (eg. BRAF V600E) is tested. However, as the number of targeted agents increases, it becomes inefficient to screen patients individually for each study. Instead, a design such as NCI-MATCH becomes necessary to test all of the possible immunotherapy and targeted therapy combinations. Thus, NGS becomes an invaluable tool for the development and evaluation of immunotherapy combinations.
Toxicity remains a significant concern with the combination of immunotherapy and targeted therapy. Ribas et al reported unacceptable hepatotoxicity when ipilimumab (anti-CTLA-4) was combined with vemurafenib in BRAF V600E mutant melanoma [49]. Similarly, Ahn et all describe inadmissible toxicity when EGFR inhibitors are combined with PD-1 inhibitors, including pulmonary and hepatotoxicity [50]. There are several trials ongoing with sequential combinations and different dosing schemas of targeted and immunotherapy that may answer these questions in the near future [51].
5. Can NGS predict response to immunotherapy?
The true question is not whether NGS can help determine immunotherapy combinations, but if it can predict response or resistance to immunotherapy. PD-L1 staining using IHC emerged as the first clinically approved biomarker to predict response to immunotherapy, specifically in NSCLC [16]. However, standardization and interpretation of what PD-L1 positivity is remains an issue given discrepancies in antibodies used for staining. Furthermore, PD-L1 negative patients can still respond to immune checkpoint inhibitors, meaning that it is not entirely predictive[52].
5.1. PD-L1 gene amplification:
Roemer et al examined the prognostic significance of PD-L1 and PD-L2 expression in Hodgkin lymphoma [53]. They found that patients with advanced stage Hodgkin’s had amplification of locus 9p24.1 which contains PD-L1 and PD-L2. This finding, using fluorescent in situ hybridization assay (FISH), provided rationale for efficacy of PD-1 inhibition in Hodgkin’s lymphoma [54]. One major flaw in PD-L1 testing whether by FISH or more commonly immunohistochemistry (IHC) is that there is no standardized expression level that exists across tumor types [55]. From a practical standpoint, testing using multiple modalities (IHC, FISH, genomic sequencing) is cumbersome and can sometimes require sending specimens to different labs with significant delays in therapy. PD-L1 amplification could be detected using NGS which can be incorporated in a multi-gene panel testing. Incorporating this into one set of testing could alter future immunotherapy use in studies as well as in routine practice.
5.2. Tumor mutational burden
Tumor mutational burden (TMB) has emerged as a promising new predictive tool in patient selection for immunotherapy. Several investigators have reported a favorable response to immunotherapy in patients who exhibit a high mutational burden[56, 57, 58]. Roszik et al developed a novel way to predict total mutational load using a small NGS panel and found that this method could strongly predict outcomes to immunotherapy in both melanoma and NSCLC[59]. Alternatively, Hanna et al defined a new inflammatory phenotype in head and neck squamous cell carcinoma that does have improved survival with immunotherapy[60]. Unfortunately, this inflammatory phenotype did not correlate with tumor mutational burden. This inflamed phenotype had high CD8+ T cell infiltrates and high PD-1/TIM3 co-expression[60]. It is likely that tumor mutational burden plays a role in predicting response to immunotherapy, but the strength of correlation might be variable among different tumor types.
Defining thresholds for determining the significance of tumor mutational burden remains a dilemma. Chalmers et al sequenced 100,000 genomes and evaluated their TMB. They found that melanoma, lung adenocarcinoma, and renal clear cell carcinoma had 14.4, 6.3, and 2.7 mutations per megabase respectively [61, 62]. A trend emerges from this data and others that the higher mutational burden patients have, the more likely they are to respond to immunotherapy. When we reach the highest degree of TMB we enter the realm of microsatellite unstable tumors. Some investigators have defined high TMB as greater than 8.8 mutations/MB[63] to greater than 13.8 mutations/MB[64]. It is clear that TMB varies significantly with a patient’s age and type of malignancy, with pediatric patients having almost 4 times fewer mutations/MB on average than elderly patients[65]. Though multiple studies across a number of tumor types (e.g., melanoma, NSCLC, bladder cancer) suggest an association between higher TMB and immunotherapy response, recent data suggests that this may not always be the case as evidenced by an absence of such a correlation between TMB and response in renal cell carcinoma.
5.3. Microsatellite instability
Patients with mismatch repair deficiency (MMR-d) lack the ability to detect and correct mistakes made by DNA polymerase in certain repetitive sequences of DNA called microsatellites[66]. This results in microsatellite instability, and since some of these microsatellites are located inside genes, this affects gene structure and transcription. The result is a high mutational burden[66]. These “frame-shifted” peptides (neoantigens) are highly immunogenic and are the basis for immunotherapy success in MMR-d colorectal cancer. Specifically, the Lynch syndrome associated genes MSH2, MSH6, MLH1, and PMS2 are currently exploited as a method to detect high microsatellite instability (MSI-H) tumors[66]. The recent US FDA approval of pembrolizumab for all tumors in pediatric and adult patients with MSI-H cancer is a landmark event in precision oncology and immunotherapy[67]. Vuzman et al used a large NGS panel to investigate tumor mutational burden (TMB) as a predictor of response to immunotherapy[68]. They found that TMB is predictive of response to immunotherapy only when the mutation burden is equivalent to that found in patients with MMR-d.
5.4. Neoantigens
Neoantigens are the critical step in immune surveillance. These peptides are transported by antigen presenting cells to lymph nodes. These antigens are then presented on major histocompatibility complex class I (MHC) to CD8+ T-cells, which become “activated” and “hunt” the tumor [69]. A series of co-stimulatory and inhibitory molecules on the T-cell play an intricate role in this process. Agents such as ipilimumab serve to enhance activation of the immune system by targeting these checkpoint inhibitor molecules that downregulate the immune system. Ultimately, tumor cells are killed by T-cells, which releases more neoantigens and feeds the process further. Tumors with higher mutation burden often have higher numbers of neoantigens[70].
Mutational burden may only predict response in the minority of patients who have extremely high load and fall into MMR-d. Some investigators suggest that mutational burden cannot explain the response to immunotherapy and have proposed more robust techniques. Snyder et al examined response to ipilimumab in melanoma patients and found that while a correlation existed between response and TMB, this alone was not sufficient to predict response [71]. Using NGS and a novel algorithm they were able to identify peptide sequences common to responders, but absent in non-responders. Importantly, the presence of these neoantigen peptides was not dependent on TMB and was sufficient to predict response even in the absence of high TMB. Similarly, Brown and colleagues used NGS derived RNA-seq data from The Cancer Genome Atlas (TCGA) to predict immunogenicity [72]. They found that detection of predicted immunogenic mutations can identify patients who respond to immunotherapy. From a clinical perspective this is critical and demonstrates the power of NGS as a predictor when discussing the use of an expensive drug with potential deadly side effects.
Some investigators are using neoantigens not to predict response to checkpoint inhibitors, but rather to create custom therapies. Rajasagi and co-workers performed NGS on patients with chronic lymphocytic leukemia (CLL) and algorithmically predicted neoantigens [73]. Patients underwent hematopoietic stem cell transfer and responders were further evaluated for T-cells to these specific predicted neoantigens. The authors concluded that an NGS based neoantigen prediction strategy could be used to create personalized cancer immunotherapy. Similarly, Robbins et al developed a novel method using NGS based whole-exome sequencing to find neoantigens in melanoma [74]. They were able to validate their findings in patients who received tumor-infiltrating lymphocytes (TILs) and had objective responses. The advantage of their technique is that it skips the need to create cDNA libraries and can speed the development of TIL therapy as well as to create custom cancer vaccines.
5.5. Hyper-progression
Recently, reports started to surface that immunotherapy may not only be ineffective, but in some cases may also cause accelerated growth of the disease. [75, 76, 77]. This is extremely worrisome and highlights the need to identify patients who will benefit from immunotherapy as well as those for whom immunotherapy should be avoided. Kato et al attempted to characterize the hyper-progressor phenotype using NGS. They found that patients with MDM2/4 amplification as well as EGFR aberrations were more prone to hyper-progression [78]. As more data becomes available, NGS will likely emerge as the best tool to predict this aggressive phenotype and prevent immunotherapy treatment in these patients.
5.6. Bioinformatics
There is mounting evidence that NGS will play a role in further development of immunotherapy, both as a predictive tool for response to checkpoint inhibitors and ultimately to develop powerful new personalized immunotherapies such as TIL. Hundal et al have created a tool to predict neoantigens using NGS. Their approach focuses on epitope to MHC binding affinity[79]. These epitopes are validated as recognized by T-cells that have been activated by checkpoint blockade. The group has since created an entire suite of software that uses not just DNA, but RNA-seq data as well as HLA typing for accurate prediction of neoantigen epitopes.
5.7. Personalized vaccines
The failure of cancer vaccines as described previously is at least partially due to a lack of correct antigens in the vaccine. With the help of NGS and computational techniques, a complete antigen map can be personally created for each individual patient and tumor. One can even go so far as to create multiple maps for a patient based on several metastatic sites[35]. Several groups have shown clinical efficacy with this approach. Sahin et al describe a method where they used NGS to predict neoantigens and manufactured a vaccine based on these neoantigens [80]. They treated five melanoma patients, two of whom had a documented objective response that was shown to be related to the vaccine-targeted epitopes. Similarly, Ott et al created a personal melanoma vaccine based on NGS and computational neoantigen prediction [69]. The vaccine consisted of 20 neoantigens. Six patients with high-risk stage III and IV melanoma, treated with curative surgery, received the vaccine. At the end of 25 months, only 2 had recurred. Taken together, these small studies demonstrate the potential for further development of personalized cancer vaccines.
5.8. Chimeric antigen receptor T-cell therapies
Chimeric antigen receptor T-cell (CAR-T) therapies have overcome major technical hurdles including transport to the tumor, tumor derived inhibitory cytokines, immunosuppressive T-cells, and regulatory mechanisms [41]. The major challenge in solid tumors is finding appropriate neoantigens for the chimeric T-cells to target. One way to do so is to clone genes from a T-cell receptor isolated at the tumor site and then use NGS to select the most potent neoantigens in order to genetically engineer a CAR-T using the selected clones [81]. In effect, this process creates a truly personal CAR-T therapy based on not only a patient’s own T-cell, but tumor antigens as well. Recent US FDA approval of CAR-T cell therapies in hematological malignancies have created great enthusiasm in this area for both liquid and solid tumors. However, the therapy is expensive, carries significant toxicity such as cytokine release syndrome, and does result in treatment failure by a variety of mechanism such as downregulation of CD19. Further personalization of CAR-T’s using NGS would allow higher efficacy and better patient selection.
6. Expert commentary
As our understanding of the immune system, genomics, and their interplay in cancer grows, and as our high throughput sequencing ability increases, the two are poised to merge. This will start with universal genomic testing at diagnosis[82].
Two recent FDA approvals of NGS based testing as well as the anticipated availability of clinical trials and targeted therapies will fuel this area in the near future. Immunotherapy has seemingly made in-roads into almost all cancer types. However, response rates are generally in the sub-20 percent range, and there is a danger of treating checkpoint inhibitors as a kind of blanket therapy that may be offered to everyone with a hope that benefit will result.
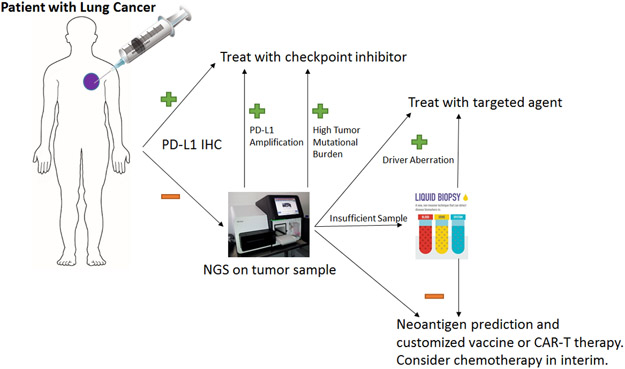
By combining the disciplines of immunotherapy and next-generation sequencing, we can begin to predict who will benefit from checkpoint inhibitors. However, this is just the beginning of a much more powerful fusion as shown in the Figure 2 for non-small cell lung cancer. The modern paradigm will merge all therapeutic modalities including chemotherapy, radiation, immunotherapy, and genomically targeted therapy.
Figure 2:
Algorithm for a Non-small cell Lung cancer patient in the clinic
This dream of personalized immunotherapy, while within reach, is still on the horizon. One of the major challenges that is being solved near-term is computational. The necessary computing power is now becoming a reality and the algorithms to find neoantigens are being perfected. With time, NGS will determine neoantigens individual to a patient’s tumor that can be used to make a custom cancer vaccine or other personalized immunotherapy. This will hopefully raise the response rate to immunotherapies and produce some very dramatic responses. Still, these computing innovations need further validation to demonstrate that they can reliably find the right peptides to target with immunotherapy. Even when the identification of neoantigens becomes dependable, personalized immunotherapies need to be perfected. The most likely technology to benefit from neoantigens is cell transfer therapy such as TIL and CAR-T. These therapies can be directly engineered based on specific epitopes and the appropriate T-cells can be transferred. Still, cell transfer therapy promises to be extraordinarily expensive and with devastating potential side effects such as cytokine release syndrome, making it inappropriate for a large number of patients. Such hurdles as cost, development time, and toxicity need to be resolved before NGS based adoptive cell therapy can be applied on a truly wide scale.
The other technology that stands to benefit from NGS is cancer vaccines. These have the advantage of standing on a century of vaccine knowledge in infectious disease. They have the potential to be relatively inexpensive and with potentially minimal toxicity. There has been minimal therapeutic success, likely because the epitopes around which the vaccine was developed cannot overcome the heterogeneity of a tumor. With the knowledge gained from NGS about individual tumors, vaccines may finally have a clinical breakthrough. Still, the vaccine alone may not create a robust enough immune response to produce clinically meaningful results. Whatever personalized vaccine is developed in the future will likely rely on co-stimulation with an immune checkpoint inhibitor.
7. Five-year view
We speculate that in the coming years NGS will be firmly embedded as a tool in oncology, with more and more genes being added to cancer panels. The computational ability and bioinformatics algorithms will continue to increase in speed and precision with the goal of quickly and reliably identifying neoantigens. The added knowledge gained from neoantigen prediction will likely be applied in cell transfer therapy first, as this is the technology with momentum currently. With the recent FDA approval of CAR-T and mounting successes in leukemia and lymphoma, the interest and funding will flow toward this personalized immunotherapy. As clinicians, companies, and patients become more comfortable with this burgeoning technology, personalized cancer therapy will become even more personal with the help of NGS.
Key issues.
Next-generation sequencing (NGS) is able to quickly, reliably, and affordably detect actionable alterations for targeted therapy in cancers.
Neoantigens are highly immunogenic protein fragments and are the critical step in immune surveillance.
NGS and novel computational algorithms are being used to identify neoantigens.
NGS has the advantage of speed and cost when compared to older cDNA based neoantigen identification.
The identification of neoantigens is used to construct cell transfer therapy such as tumor infiltrating lymphocytes (TILs).
Chimeric antigen receptor T-cell therapy (CAR-T) is proving extremely successful in leukemias and lymphomas, but has not yet made in-roads in solid tumors because of a lack of consistent antigen target.
NGS based identification of neoantigens may make personalized CAR-Ts or cancer vaccines possible for individual solid tumor patients.
The combination of NGS and immunotherapy promises to bring truly personalized cancer care to an increasing number of cancer patients.
Acknowledgments
Funding
This paper was supported by the National Institutes of Health Cancer Center Support Grant CA016672.
Footnotes
Declaration of Interest
V Subbiah receives research funding for clinical trials from Novartis, Bayer, GSK, Nanocarrier, Vegenics, Northwest Biotherapeutics, Berghealth, Incyte, Fujifilm, Pharmamar, D3, Pfizer, Multivir, Amgen, Abbvie, Bluprint medicines, LOXO and Roche/ Genentech. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer Disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose
References
Reference annotations
* Of interest
** Of considerable interest
- 1.DeVita VT, Chu E. A History of Cancer Chemotherapy. Cancer Res. 2008. 2008/November/01/;68(21):8643–8653. doi: 10.1158/0008-5472.CAN-07-6611. en. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001. 2001/February/15/;409(6822):860–921. doi: 10.1038/35057062. en. [DOI] [PubMed] [Google Scholar]
- 3.Cross D, Burmester JK. The Promise of Molecular Profiling for Cancer Identification and Treatment. Clin Med Res. 2004. 2004 August;2(3):147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petty RD, Nicolson MC, Kerr KM, et al. Gene Expression Profiling in Non-Small Cell Lung Cancer: From Molecular Mechanisms to Clinical Application. Clin Cancer Res. 2004. 2004/May/15/;10(10):3237–3248. doi: 10.1158/1078-0432.CCR-03-0503. en. [DOI] [PubMed] [Google Scholar]
- 5.The Cost of Sequencing a Human Genome: National Human Genome Research Institute; 2016. [updated July 6, 2016;4/2/18]. Available from: https://www.genome.gov/27565109/the-cost-of-sequencing-a-human-genome/
- 6.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends in Genetics. 2008. 2008/March/01/;24(3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Groisberg R, Hong DS, Holla V, et al. Clinical genomic profiling to identify actionable alterations for investigational therapies in patients with diverse sarcomas. Oncotarget. 2017. June 13;8(24):39254–39267. doi: 10.18632/oncotarget.16845. PubMed PMID: 28424409; PubMed Central PMCID: PMCPMC5503611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisberg R, Roszik J, Conley A, et al. The Role of Next-Generation Sequencing in Sarcomas: Evolution From Light Microscope to Molecular Microscope. Curr Oncol Rep. 2017. 2017/December/01/;19(12):78. doi: 10.1007/s11912-017-0641-2. en. [DOI] [PubMed] [Google Scholar]
- 9.Hodi FS, O’Day SJ, McDermott DF, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. New England Journal of Medicine. 2010. 2010/August/19/;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robert C, Long GV, Brady B, et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. New England Journal of Medicine. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. PubMed PMID: 25399552. [DOI] [PubMed] [Google Scholar]
- 11.Robert C, Schachter J, Long GV, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. New England Journal of Medicine. 2015. 2015/June/25/;372(26):2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 12.Ferris RL, Blumenschein G Jr., Fayette J, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. New England Journal of Medicine. 2016. 2016/November/10/;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim SH, Sun J-M, Lee S-H, et al. Pembrolizumab for the treatment of non-small cell lung cancer. Expert Opinion on Biological Therapy. 2016. 2016/March/03/;16(3):397–406. doi: 10.1517/14712598.2016.1145652. [DOI] [PubMed] [Google Scholar]
- 14.Bellmunt J, de Wit R, Vaughn DJ, et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. New England Journal of Medicine. 2017. 2017/March/16/;376(11):1015–1026. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. *.Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Overall Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. New England Journal of Medicine. 2017. 2017/October/05/;377(14):1345–1356. doi: 10.1056/NEJMoa1709684.First evidence of combination immunotherapy having clinical efficacy.
- 16.Diggs LP, Hsueh EC. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomarker Research. 2017. 2017/March/15/;5:12. doi: 10.1186/s40364-017-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J, Shalabi A, Hubbard-Lucey VM. Comprehensive analysis of the clinical immuno-oncology landscape. Annals of Oncology. 2017. 2017/December/07/. doi: 10.1093/annonc/mdx755. en. [DOI] [PubMed] [Google Scholar]
- 18.Behjati S, Tarpey PS. What is next generation sequencing? Arch Dis Child Educ Pract Ed. 2013. 2013 December;986:236–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shyr D, Liu Q. Next generation sequencing in cancer research and clinical application. Biol Proced Online. 2013. 2013/February/13/;15:4. doi: 10.1186/1480-9222-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamps R, Brandão RD, van den Bosch BJ, et al. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. International Journal of Molecular Sciences. 2017. 2017/January/31/;18(2). doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. *.Rozenblum AB, Ilouze M, Dudnik E, et al. Clinical Impact of Hybrid Capture-Based Next-Generation Sequencing on Changes in Treatment Decisions in Lung Cancer. J Thorac Oncol. 2017. February;12(2):258–268. doi: 10.1016/j.jtho.2016.10.021. PubMed PMID: 27865871.Demonstrates the power of NGS to detect actionable mutations that make a clinical impact.
- 22.Gupta A, Connelly C, Frampton G, et al. P2.03b-068 The Druggable Mutation Landscape of Lung Adenocarcinoma. Journal of Thoracic Oncology. 2017. 12(1):S977 [Google Scholar]
- 23.Daber R, Sukhadia S, Morrissette JJ. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet. 2013. December;206(12):441–8. doi: 10.1016/j.cancergen.2013.11.005. PubMed PMID: 24528889. [DOI] [PubMed] [Google Scholar]
- 24.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013. November;31(11):1023–31. doi: 10.1038/nbt.2696. PubMed PMID: 24142049; PubMed Central PMCID: PMCPMC5710001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goswami RS, Luthra R, Singh RR, et al. Identification of Factors Affecting the Success of Next-Generation Sequencing Testing in Solid Tumors. Am J Clin Pathol. 2016. February;145(2):222–37. doi: 10.1093/ajcp/aqv023. PubMed PMID: 27124905. [DOI] [PubMed] [Google Scholar]
- 26.Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res. 2016. October;5(5):505–510. doi: 10.21037/tlcr.2016.10.08. PubMed PMID: 27826531; PubMed Central PMCID: PMCPMC5099511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguado C, Gimenez-Capitan A, Karachaliou N, et al. Fusion gene and splice variant analyses in liquid biopsies of lung cancer patients. Transl Lung Cancer Res. 2016. October;5(5):525–531. doi: 10.21037/tlcr.2016.09.02. PubMed PMID: 27826534; PubMed Central PMCID: PMCPMC5099519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. **.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. New England Journal of Medicine. 2015. 2015/August/20/;373(8):726–736. doi: 10.1056/NEJMoa1502309.Demonstration of basket studies and clinical benefit to treating based on driver mutation.
- 29.Diamond EL, Subbiah V, Lockhart AC, et al. Vemurafenib for BRAF V600–Mutant Erdheim-Chester Disease and Langerhans Cell Histiocytosis: Analysis of Data From the Histology-Independent, Phase 2, Open-label VE-BASKET Study. JAMA Oncol. 2017. 2017/November/29/. doi: 10.1001/jamaoncol.2017.5029. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF V600E -mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. The Lancet Oncology. 2017;18(10):1307–1316. [DOI] [PubMed] [Google Scholar]
- 31.Subbiah V, Kreitman RJ, Wainberg ZA, et al. Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600–Mutant Anaplastic Thyroid Cancer. Journal of Clinical Oncology. 2017. 2017/October/26/:JCO.2017.73.678. doi: 10.1200/JCO.2017.73.6785. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong DS, Morris VK, El Osta B, et al. Phase IB Study of Vemurafenib in Combination with Irinotecan and Cetuximab in Patients with Metastatic Colorectal Cancer with BRAFV600E Mutation. Cancer Discov. 2016;6(12):1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. *.Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012. March 28;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. PubMed PMID: 22461641; PubMed Central PMCID: PMCPMC3568523.Historical importance as pre-clinical rationale for development of PD-1 inhibitors.
- 34.Sharpe AH. Introduction to checkpoint inhibitors and cancer immunotherapy. Immunol Rev. 2017. 2017/March/01/;276(1):5–8. doi: 10.1111/imr.12531. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Sharma PK, Peter Goedegebuure S, et al. Personalized cancer vaccines: Targeting the cancer mutanome. Vaccine. 2017. 2017/February/15/;35(7):1094–1100. doi: 10.1016/j.vaccine.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angell TE, Lechner MG, Jang JK, et al. MHC class I loss is a frequent mechanism of immune escape in papillary thyroid cancer that is reversed by interferon and selumetinib treatment in vitro. Clin Cancer Res. 2014. December 1;20(23):6034–44. doi: 10.1158/1078-0432.CCR-14-0879. PubMed PMID: 25294906; PubMed Central PMCID: PMCPMC4252612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.del Campo AB, Kyte JA, Carretero J, et al. Immune escape of cancer cells with beta2-microglobulin loss over the course of metastatic melanoma. Int J Cancer. 2014. January 1;134(1):102–13. doi: 10.1002/ijc.28338. PubMed PMID: 23784959. [DOI] [PubMed] [Google Scholar]
- 38.Bodey B, Bodey B, Siegel SE, et al. Failure of cancer vaccines: the significant limitations of this approach to immunotherapy. Anticancer Res. 2000. 2000 August- Julundefined;204:2665–2676. eng.Overview of oncologic vaccine therapy and why it has failed so far.
- 39.Rosenberg SA, Yannelli JR, Yang JC, et al. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J Natl Cancer Inst. 1994. 1994/August/03/;86(15):1159–1166. eng.Seminal paper on TIL therapy.
- 40.Neelapu SS, Locke FL, Bartlett NL, et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. New England Journal of Medicine. 2017. 2017/December/10/;0(0):null. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newick K, Moon E, Albelda SM. Chimeric antigen receptor T-cell therapy for solid tumors. Molecular Therapy - Oncolytics. 2016. 2016/January/01/;3(Supplement C):16006. doi: 10.1038/mto.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atkins MB, Larkin J. Immunotherapy Combined or Sequenced With Targeted Therapy in the Treatment of Solid Tumors: Current Perspectives. J Natl Cancer Inst. 2016. 2016/June/01/;108(6). doi: 10.1093/jnci/djv414. [DOI] [PubMed] [Google Scholar]
- 43.Gainor JF, Shaw AT, Sequist LV, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res. 2016. 2016/September/15/;22(18):4585–4593. doi: 10.1158/1078-0432.CCR-15-3101. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haratani K, Hayashi H, Tanaka T, et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann Oncol. 2017. 2017/July/01/;28(7):1532–1539. doi: 10.1093/annonc/mdx183. eng. [DOI] [PubMed] [Google Scholar]
- 45.Hu-Lieskovan S, Robert L, Homet Moreno B, et al. Combining Targeted Therapy With Immunotherapy in BRAF-Mutant Melanoma: Promise and Challenges. Journal of Clinical Oncology. 2014. 2014/July/20/;32(21):2248–2254. doi: 10.1200/JCO.2013.52.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langer CJ, Gadgeel SM, Borghaei H, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 2016. 2016;17(11):1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kammerer-Jacquet S-F, Medane S, Bensalah K, et al. Correlation of c-MET Expression with PD-L1 Expression in Metastatic Clear Cell Renal Cell Carcinoma Treated by Sunitinib First-Line Therapy. Targ Oncol. 2017. 2017/August/01/;12(4):487–494. doi: 10.1007/s11523-017-0498-1. en. [DOI] [PubMed] [Google Scholar]
- 48.Callahan MK, Masters G, Pratilas CA, et al. Paradoxical activation of T cells via augmented ERK signaling mediated by a RAF inhibitor. Cancer Immunol Res. 2014. 2014January;2(1):70–79. CIR-13-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribas A, Hodi FS, Callahan M, et al. Hepatotoxicity with Combination of Vemurafenib and Ipilimumab. New England Journal of Medicine. 2013. 2013/April/04/;368(14):1365–1366. doi: 10.1056/NEJMc1302338. [DOI] [PubMed] [Google Scholar]
- 50.Ahn M-J, Sun J-M, Lee S-H, et al. EGFR TKI combination with immunotherapy in non-small cell lung cancer. Expert Opinion on Drug Safety. 2017. 2017/April/03/;16(4):465–469. doi: 10.1080/14740338.2017.1300656. [DOI] [PubMed] [Google Scholar]
- 51.Karachaliou N, Gonzalez-Cao M, Sosa A, et al. The combination of checkpoint immunotherapy and targeted therapy in cancer. Ann Transl Med. 2017. October;5(19):388. doi: 10.21037/atm.2017.06.47. PubMed PMID: 29114546; PubMed Central PMCID: PMCPMC5653508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015. April;14(4):847–56. doi: 10.1158/1535-7163.MCT-14-0983. PubMed PMID: 25695955. [DOI] [PubMed] [Google Scholar]
- 53.Roemer MGM, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol. 2016. 2016/August/10/;34(23):2690–2697. doi: 10.1200/JCO.2016.66.4482. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 Blockade with Nivolumab in Relapsed or Refractory Hodgkin’s Lymphoma. New England Journal of Medicine. 2015. 2015/January/22/;372(4):311–319. doi: 10.1056/NEJMoa1411087. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udall M, Rizzo M, Kenny J, et al. PD-L1 diagnostic tests: a systematic literature review of scoring algorithms and test-validation metrics. Diagn Pathol. 2018. February 9;13(1):12. doi: 10.1186/s13000-018-0689-9. PubMed PMID: 29426340; PubMed Central PMCID: PMCPMC5807740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonta I, Isac JF, Meiri E, et al. Correlation between tumor mutation burden and response to immunotherapy. Journal of Clinical Oncology. 2017. 2017/May/20/;35(15_suppl):e14579–e14579. doi: 10.1200/JCO.2017.35.15_suppl.e14579. [DOI] [Google Scholar]
- 57.Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Molecular Cancer Therapeutics. 2017. 2017/January/01/:molcanther.0386.2017. doi: 10.1158/1535-7163.MCT-17-0386. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yaghmour G, Pandey M, Ireland C, et al. Role of Genomic Instability in Immunotherapy with Checkpoint Inhibitors. Anticancer Res. 2016. 2016/August/01/;36(8):4033–4038. en. [PubMed] [Google Scholar]
- 59.Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Medicine. 2016. 2016/October/25/;14:168. doi: 10.1186/s12916-016-0705-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanna GJ, Liu H, Jones RE, et al. Defining an inflamed tumor immunophenotype in recurrent, metastatic squamous cell carcinoma of the head and neck. Oral Oncology. 2017. 2017/April/01/;67(Supplement C):61–69. doi: 10.1016/j.oraloncology.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013. 2013/August/14/;500(7463):nature12477. doi: 10.1038/nature12477. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. New England Journal of Medicine. 2015. 2015/November/05/;373(19):1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morse CB, Elvin JA, Gay LM, et al. Elevated tumor mutational burden and prolonged clinical response to anti-PD-L1 antibody in platinum-resistant recurrent ovarian cancer. Gynecol Oncol Rep. 2017. 2017/June/27/;21:78–80. doi: 10.1016/j.gore.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017. June;23(6):703–713. doi: 10.1038/nm.4333. PubMed PMID: 28481359; PubMed Central PMCID: PMCPMC5461196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Medicine. 2017. 2017/April/19/;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Westdorp H, Fennemann FL, Weren RDA, et al. Opportunities for immunotherapy in microsatellite instable colorectal cancer. Cancer Immunol Immunother. 2016. 2016/October/01/;65(10):1249–1259. doi: 10.1007/s00262-016-1832-7. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. New England Journal of Medicine. 2015. 2015/June/25/;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vuzman D, Powers W, Huang X, et al. Tumor mutation burden derived from large NGS panel as biomarker for immunotherapy response. Journal of Clinical Oncology. 2017. 2017/May/20/;35(15_suppl):e23077–e23077. doi: 10.1200/JCO.2017.35.15_suppl.e23077. [DOI] [Google Scholar]
- 69.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017. 2017;547(7662):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chee J, Robinson BWS, Holt RA, et al. Immunotherapy for Lung Malignancies: From Gene Sequencing to Novel Therapies. Chest. 2017. 2017/April/01/;151(4):891–897. doi: 10.1016/j.chest.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 71.Snyder A, Makarov V, Merghoub T, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma [research-article]. http://dxdoiorg/101056/NEJMoa1406498. 2014. 2014/December/03/. En. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. *.Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014. 2014/May/01/;24(5):743–750. doi: 10.1101/gr.165985.113. en.Basis for clinical application of neo-antigen prediction.
- 73.Rajasagi M, Shukla SA, Fritsch EF, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014. 2014/July/17/;124(3):453–462. doi: 10.1182/blood-2014-04-567933. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robbins PF, Lu Y-C, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nature Medicine. 2013. 2013;19(6):747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Champiat S, Dercle L, Ammari S, et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clin Cancer Res. 2017;23(8):1920–1928. [DOI] [PubMed] [Google Scholar]
- 76.Sharon E Can an Immune Checkpoint Inhibitor (Sometimes) Make Things Worse? Clin Cancer Res. 2017;23(8):1879–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saâda-Bouzid E, Defaucheux C, Karabajakian A, et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2017. 2017/July/01/;28(7):1605–1611. doi: 10.1093/annonc/mdx178. eng. [DOI] [PubMed] [Google Scholar]
- 78.Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res. 2017. 2017/August/01/;23(15):4242–4250. doi: 10.1158/1078-0432.CCR-16-3133. en. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. *.Hundal J, Carreno BM, Petti AA, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Medicine. 2016. 2016/January/29/;8. doi: 10.1186/s13073-016-0264-5.Bioinformatics tools for neoantigen identification.
- 80.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222. [DOI] [PubMed] [Google Scholar]
- 81.Bethune MT, Joglekar AV. Personalized T cell-mediated cancer immunotherapy: progress and challenges. Current Opinion in Biotechnology. 2017. 2017/December/01/;48(Supplement C):142–152. doi: 10.1016/j.copbio.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 82.Subbiah V, Kurzrock R. Universal Genomic Testing Needed to Win the War Against Cancer: Genomics IS the Diagnosis. JAMA Oncol. 2016. 2016/June/01/;2(6):719. doi: 10.1001/jamaoncol.2016.0078. en. [DOI] [PubMed] [Google Scholar]