Abstract
In the past decade, noticeable progress has been achieved regarding fluorescence imaging in the second near-infrared (NIR-II) window. Fluorescence imaging in the NIR-II window demonstrates superiorities of deep tissue penetration and high spatial and temporal resolution, which are beneficial for profiling physiological processes. Meanwhile, molecular imaging has emerged as an efficient tool to decipher biological activities on the molecular and cellular level. Extending molecular imaging into the NIR-II window would enhance the imaging performance, providing more detailed and accurate information of the biological system. In this progress report, selected achievements made in NIR-II molecular imaging are summarized. The organization of this report is based on strategies underlying rational designs of NIR-II imaging probes and their applications in molecular imaging are highlighted. This progress report may provide guidance and reference for further development of functional NIR-II probes designed for high-performance molecular imaging.
Keywords: molecular imaging, NIR-II window
Graphical Abstract
Molecular imaging enables visualization and understanding of physiological activities on the cellular and molecular level. The extension of the molecular imaging window into second near-infrared (NIR-II) window can better fulfill requirements of clinical applications. In this progress report, recent advances of NIR-II molecular imaging were summarized, providing insights for future development of NIR-II molecular imaging.
1. Introduction
Emerging as an intuitional and efficient way to monitor and understand physiological processes, fluorescence imaging demonstrates several advances of high resolution, real-time manner and multiple channels incorporation[1]. Recently a new fluorescence window, which is defined as the second near-infrared (NIR-II) window, extends the imaging wavelength to longer range of 1000–1700 nm compared with traditional imaging wavelength range of 400 – 900 nm (visible to near-infrared I window)[2]. The reduced light absorption, suppressed photon scattering and negligible autofluorescence have benefited NIR-II fluorescence imaging for improved penetration depth and high-spatial resolution, appearing to be a promising candidate for functional biomedical imaging[2–3]. To better understand the underlying mechanism of physiological processes, especially for disease-related systems, people begin to spare more efforts on deciphering relevant interactions on the molecular and cellular level. To achieve this, molecular imaging, which enables observation of cellular function and molecular process without perturbation, has received more and more attention[4]. In consideration of unique advantages of NIR-II fluorescence imaging, more and more attention has been switched to it for molecular imaging of particular targets and pathways. Obviously, the property of probes (e.g. brightness, biocompatibility, pharmacokinetics, binding affinity and specificity) is the key factor affecting the molecular imaging performance. In this perspective, through molecular engineering of fluorophore structures, our group obtained several NIR-II fluorophores with active functional epitopes suitable for conjugation of targeting moieties. With functional fluorophopres in hand, efficient molecular imaging has been successfully realized[5] [6]. Towards better molecular imaging performance, apart from fluorophores, imaging setup also matters much. Conventionally, fluorescence molecular imaging was performed under the two-dimensional (2D) wide-field mode, which relies on illumination of the entire view of a fluorescence microscope and simultaneous fluorescence detection with a camera, lacking the spatial information of imaging substances. With continuous efforts on the construction of the NIR-II confocal setup, our group has introduced a new imaging dimension along the z axis into conventional two dimensions of imaging along x and y axis, successfully realizing three-dimensional (3D) reconstruction of imaging tissues in the NIR-II window to reveal more detailed structural information and more distinct spatial distribution of imaging targets[7] [8]. Herein, we introduced a few progresses that have been recently made in NIR-II fluorescence molecular imaging.
2. Bright NIR-II fluorophores and their applications in molecular imaging
NIR-II fluorophores with high brightness emit fluorescence effectively under irradiation, enabling many exciting advances of high resolution and deep tissue penetration[9]. Enhancement of brightness could greatly improve signal-to-background ratio (SBR) and avoid from losing dynamic bioinformation for in vivo imaging. In the meantime, applied dose of fluorophores for bio-imaging can be controlled at a relatively low level, avoiding the potential toxicity. With regard to tumor specific molecular imaging, high brightness offers advantage on high tumor-to-normal tissue ratio (T/NT), which could delineate the tumor margin accurately and trace small tumor nodules. To date, a handful of NIR-II fluorophores with sufficient brightness for molecular imaging have been developed, including donor-acceptor-donor (D-A-D) architecture fluorophores[10], polymethine cyanine architecture fluorophores[11], fluorophore-protein/polymer complexes[7, 11d, 12] and inorganic material-based fluorophores[9a, 9c, 13]. Modifying fluorophores with functional groups (azide, amine, carboxyl, etc) and conjugating them with specific biomarkers will endow the fluorophores with binding affinity to pinpoint targeted regions. Here we aim to cover recently reported NIR-II fluorophores desirable for functional molecular imaging. The strategies of maintaining the brightness of each type of fluorophores are highlighted.
2.1. Small-molecule organic NIR-II fluorophores
To extend emission wavelength into the NIR-II window, the large conjugated structure has been implemented into organic fluorophores, in which strong electron-donating groups were built around strong electron acceptors through π-bridging moieties to lower the bandgap between LUMO and HOMO (lowest unoccupied molecular orbital/highest occupied molecular orbital). D-A-D and polymethine architecture fluorophores are typical small-molecule organic fluorophores emitting in the NIR-II window.
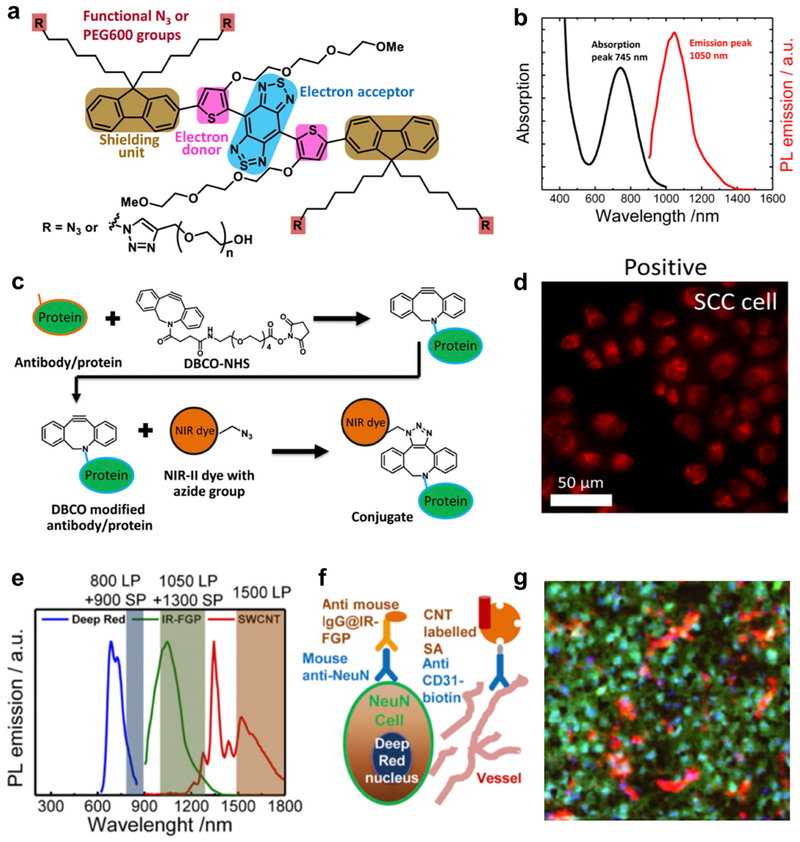
The first D-A-D structure NIR-II fluorophore, named CH-1055, allowed targeted molecular imaging of tumors in vivo after conjugating with anti-epidermal growth factor receptor (EGFR) affibody[10a]. Through high specific targeting of a xenograft human squamous cell carcinoma tumor, NIR-II molecular imaging improved tumor-to-normal tissue (T/NT) ratio to ~5, higher than traditional tumor imaging utilizing enhanced permeability and retention (EPR) effect. However, one major issue concerning the large conjugated D-A-D fluorophore for molecular imaging is the significant nonradiative energy transfer and quenching effect when interacting with water molecules in the physiological environment[10d, 14]. Enhancing the brightness of the fluorophores will further improve the T/NT ratio for pinpointing tumors, enabling early detection of small tumors as well as tracing therapeutic response. To deal with the quenching effect, the shielding unit was introduced to the D-A-D structure, which forms the new shielding unit-donor-acceptor-donor-shielding unit (S-D-A-D-S) structure, followed by systematic optimization of components for corresponding units to achieve biocompatible NIR-II fluorophores with high QY. A representative fluorophore (IR-FGP) adopted benzo[1,2-c:4,5-c′]bis([1,2,5] thiadiazole) (BBTD) as the acceptor unit, tert(ethylene glycol) (TEG)-substituted thiophene as the donor unit and dialkyl chain-substituted fluorene as the shielding unit (Figure 1a and b)[6c]. More specifically, TEG substituted thiophene can increase the dihedral angle between BBTD and TEG-thiophene and distort the conjugated backbone, affording reduced molecular interaction for bright emission. Futhermore, dialkyl chains stretching out of the conjugated backbone plane could efficiently prevent paralleled intermolecular interactions[6c]. For conjugation of targeting moieties, azide groups were incorporated at the termini of alkyl chains on the shielding unit (Figure 1a). Through mild copper-free click chemistry[15], Erbitux (Erb), an antibody specifically binding (EGFR)[16], was covalently conjugated onto IR-FGP without affecting the protein’s activity (Figure 1c). The Erb@IR-FGP conjugate efficiently differentiated the expression level of EGFR between squamous cell carcinoma (SCC, EGFR positive)[17] and Uppsala 87 malignant glioma (U87MG, EGFR negative) cells[9b](Figure 1d). Furthermore, another NIR-II fluorophore single walled carbon nanotube (SWCNTs), whose emission does not overlap with that of IR-FGP (Figure 1e), was conjugated with a protein called streptavidin (SA) to obtain a new molecular imaging probe. With above probes in hand, molecular imaging of the SCC tumor in multiple colors was conducted with IR-FGP highlighting the EGFR, SWCNT highlighting vessels and a commercial fluorophore (Deep Red) highlighting the nucleus (Figure 1e), paving a new way to study different components simultaneously. To broaden the molecular imaging application in the NIR-II window, a secondary antibody (immunoglobulin G, IgG) was conjugated with IR-FGP to demonstrate the multiplex staining of mouse brains with the assistance of SWCNT and Deep Red. After overlapping these three colors, the distribution of each imaging component in the mouse brain was clearly profiled (Figure 1f and g).[18]
Figure 1. NIR-II fluorophore IR-FGP with high QY for the molecular imaging application.
a) Molecular structure of NIR-II fluorophore IR-FGP. The fluorophore is modified with active functional groups -N3 for subsequent bio-conjugation. b) Emission and adsorption spectra of IR-FGP. c) Scheme of conjugation of IR-FGP with proteins under mild copper-free click chemistry. d) In vitro staining of EGFR positive SCC cell line using conjugate Erb@FGP. e) PL emission spectra of Deep Red, IR-FGP, and NIR-IIb SWCNT including the long-pass/short- pass filter ranges used for three-color imaging. Three ranges without overlapping were chosen for the multi-color molecular imaging application. f) Scheme regarding how three-color molecular imaging is performed. Deep Red was used to stain nucleus. IR-FGP was chosen to stain the neuron cells. CNT was applied to stain vessels. g) Multicolor brain tissue imaging with high resolution by NIR-II confocal microscopy. Reproduced with permission[6c]. Copyright 2017, National Academy of Sciences.
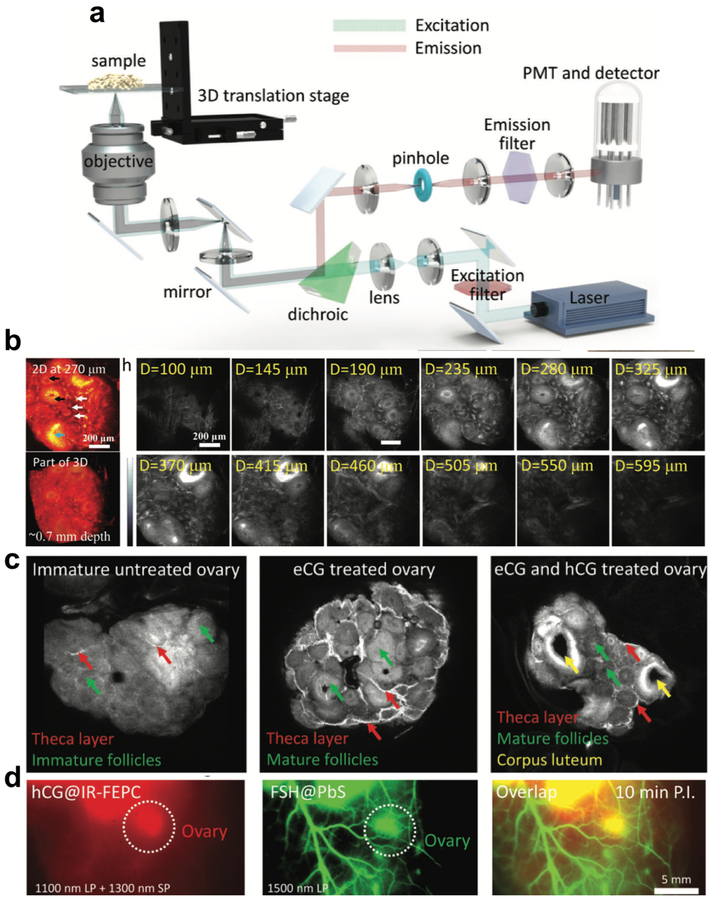
Maintaining the molecular brightness by S-D-A-D-S structure has also enabled the possibility to profile dynamic molecular process in a 3D manner. Besides conventional 2D wide -field imaging, our group also succeed in introducing another dimension along z axis into the NIR-II imaging using our home-built confocal setup (Figure 2a)[6a, 19]. 3D NIR-II molecular imaging could provide more structural information and more distinct spatial distribution of interested components, appearing to be a more efficient way to profile imaging targets. Unlike 2D wide-field imaging, confocal imaging only collects signal from a small probe volume at the focus, allowing for 3D imaging in a layer-by-layer manner [20]. To perform deep 3D imaging within the limit of safe laser intensity, confocal imaging is more demanding on fluorophore brightness. By using bright fluorophore, sufficiently high scanning speed in a approporiate time frame compatible with biological samples can be achieved. In other words, one of the main reasons that imaging in the NIR-II window is limited to 2D fluorescence imaging without the ability of gleaning 3D deep tissues is the lack of bright biocompatible fluorophores. Taking advantage of the high QY of S-D-A-D-S structure-based NIR-II fluorophores, we introduced carboxylic groups at the termini of shielding units to obtain a fluorophore candidate (IR-FEPC) suitable for 3D molecular imaging of biological samples. Under the mild conjugation condition using EDC coupling method[21], human chorionic gonadotropin (hCG) was conjugated with IR-FEPC to form a probe specific to luteinizing hormone (LH) receptors in three stages of ovary[22]. This probe together with our developed NIR-II confocal technique facilitated a layer-by-layer imaging of ovaries along the z axis, reaching a ~ 900 μm imaging depth (Figure 2b). After reconstruction of all scanned images, high-quality 3D structure of ovaries was obtained, clearly distinguishing theca, granulosa, and luteal cell enriched sub-regions during three different ovarian stages (Figure 2c). Furthermore, to better understand the process of ovarian follicle maturation[23], two related proteins hCG and follicle-stimulating hormone (FSH) were bioconjugated to IR-FEPC and lead sulfide (PbS) nanocrystal, (two high QY NIR-II fluorophores without emission overlapping) respectively. With these two probes, the first demonstration of two-color NIR-II molecular imaging was achieved, which can realize both 2D wide-field and 3D confocal imaging, paving a new way to study physiological processes (Figure 2d).
Figure 2. 3D confocal molecular imaging in the NIR-II window.
a) A simplified optical diagram of the stage-scanning NIR-II fluorescence confocal microscopy. The stage can move along the z-axis, affording the z axis sectioning ability. b) Confocal imaging of ovary in the NIR-II window at different imaging depths. Owing to the high QY of IR-FEPC, the confocal imaging can reach as deep as ~ 900 μm. c) 3D confocal imaging of the ovary after removal from the body showed strong signals in the theca layer of the follicles, mature granulosa cells of the follicles of eCG pretreated mice, and corpora lutea of the eCG/hCG pretreated mice. 3D reconstruction of scanned images can provide more detailed structural information. d) In vivo two-color imaging of the adult female ovary at 10 and 150 min post injection time points. Reproduced with permission[6a]. Copyright 2018, John Wiley and Sons.
Despite of the high QY of S-D-A-D-S structure NIR-II contrast agents, the brightness of this category of fluorophores are restricted by low absorption extinction coefficients. On the contrast, polymethine cyanine-based fluorophores, which consist of two heterocyclic terminal groups connected by the polymethine linker, afford high absorption extinction coefficients (ε > 105M−1 cm−1)[11a, 11c, 11f]. Moreover, cyanine fluorophores can not only achieve NIR-II emission, but also NIR-II region excitation[11d, 11e]. The long absorption/emission wavelength enables molecular imaging with deep penetration, high SBR and T/NT ratio. For example, clickable polymethine fluorophores 5H5 was conjugated to an integrin αVβ3 targeting peptide c(RGD)fk by copper-catalyzed azide-alkyne cycloaddition for U87MG glioma tumor imaging[11b]. Highest T/NT ratio was obtained at 8 h post injection, achieving 6.8 under 1064 nm excitation. Fluorophores with high extinction coefficients demonstrate to be a direction of future development of NIR-II molecular imaging.
2.2. Organic NIR-II fluorophores encapsulated in an amphiphilic matrix
Large conjugated backbone is necessary to tune the excitation of small organic fluorophores to long wavelength window. However, large conjugated planar backbones tend to interact strongly with surrounding molecules in aqueous solution, for which excited states possess large tendency to be attacked and quenched. Encapsulating fluorophores into an amphiphilic matrix could build a hydrophobic environment around fluorophore molecules, which would efficiently minimize nonradiative energy transfer and suppress the quenching effect. As a result, the fluorophore brightness is largely maintained after being transferred into biological environment. To date, a handful of bright protein-fluorophore or polymer-fluorophore complexes have been reported based on above mechanism[7, 11d, 24].
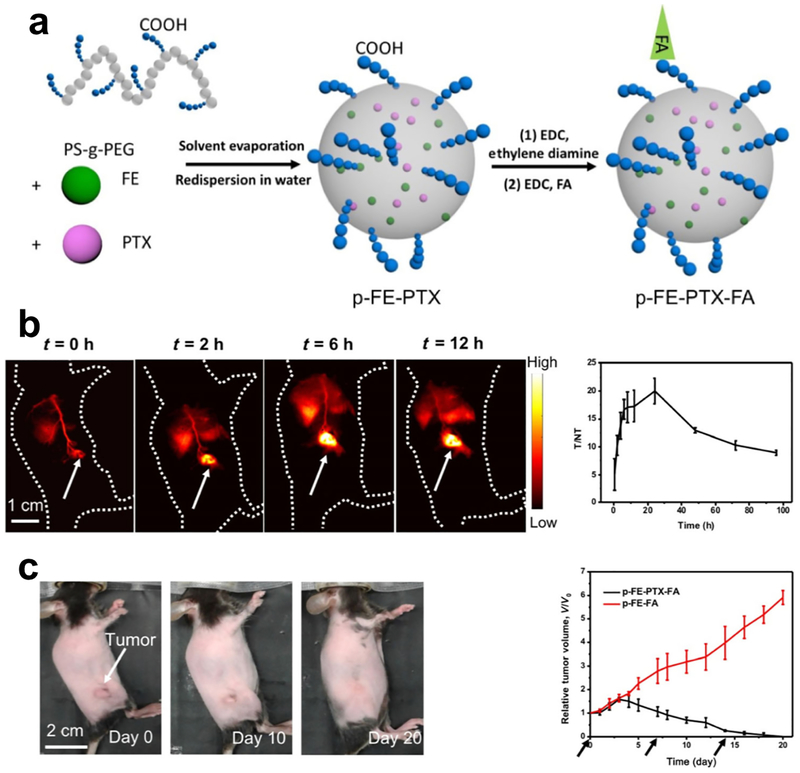
For example, an evaporation induced assembly method was developed to acquire a NIR-II fluorophore complex with high QY for molecular imaging. The hydrophobic fluorophore (FE) demonstrating high QY in organic solvents was synthesized. Then, an polymer (poly(styrene-co-chloromethyl styrene)-graft-poly(ethylene glycol) (PS-g-PEG) with the amphiphilic property, was rationally synthesized for the encapsulation of FE. PS-g-PEG and FE were mixed homogenously in toluene and the encapsulation process was induced by the gradual evaporation of toluene, forming a biocompatible NIR-II fluorophore with minimized quenching effect (p-FE, Figure 3a)[12, 19a]. The hydrophobic part in PS-g-PEG (PS chains) mimics the environment of organic solvents to prevent FE from interacting with water molecules, while the hydrophilic part (PEG chains) maintains the stability of p-FE in aqueous environment. Through above molecular assembly strategy, QY of p-FE can be as high as ~ 16.5%, demonstrating to be the highest QY among reported NIR-II organic fluorophores[3a, 3b, 10b, 14, 25]. For subsequent non-invasive molecular imaging of the mouse breast tumor (4T1) in vivo, carboxylic groups were introduced at the termini of PEG chains for the conjugation of the targeting moiety folic acid to form a NIR-II probe (p-FE-FA) specific to folate receptor proteins (FRP)[26]. Before in vivo applications, molecular imaging of FRP by the NIR-II probe was firstly tested in vitro by paralleled staining of 4T1 (FRP positive)[27] and human embryonic kidney 293 (HEK293, FRP negative) cell lines[28]. Due to the high brightness of p-FE, the injected dose can be controlled at a relatively low level to avoid potential toxicity and the off-peak emission beyond 1300 nm can be utilized for superior imaging quality. After intravenous injection of p-FE-FA into mice inoculated with 4T1 tumors, p-FE-FA targeted the tumor and selectively accumulated within the tumor overtime as the conjugate circulated (Figure 3b). Fluorescence detection above 1300 nm when irradiated under an 808 nm laser afforded a large Stokes shift, enabling molecular imaging of the 4T1 tumor at a high signal-to-background ratio. Molecular imaging of 4T1 tumor reached a high T/NT ratio of 20.0 ± 2.3, highlighting the tumor to be easily differentiated from surrounding normal tissues (Figure 3b). Meanwhile, NIR-II molecular imaging-guided chemotherapy was successfully realized when anti-cancer drugs were encapsulated within the amphiphilic polymer PS-g-PEG along with the NIR-II fluorophore FE. The efficiency of chemotherapy was monitored overtime through collecting fluorescence emitting from FE in the NIR-II window. As indicated by the fluorescence images, 4T1 tumors shrank overtime and were completed eradicated within 20 days of the first injection (Figure 3c), indicating the great potential of molecular imaging in guiding therapy process.
Figure 3. A NIR-II theranostic platform for imaging guided chemotherapy.
a) Scheme of fabrication of the NIR-II theranostic platform. b) The NIR-II theranostic platform gradually lit up the 4T1 tumors inoculated on the mice. The T/NT ratio reach the peak value of 20 ± 2.3 at 24 h post injection. c) The chemotherapy effect of the NIR-II theranostic platform. 4T1 tumors shrank overtime after intravenous injection of p-FE-PTX-FA. Reproduced with permission [12]. Copyright 2018, Springer Nature.
2.3. Inorganic material-based NIR-II fluorophores
Inorganic material-based NIR-II fluorophores, including quantum dots (QDs)[9a, 9c, 13c] and rare-earth nanoparticles (RENPs) [13a, 29], exhibit excellent performance in fluorescence imaging. While organic fluorophores benefit from more favorable pharmacokinetics, advantages of high QY, superior photostability and tunable narrow emission make inorganic material-based fluorophores ideal candidates for molecular imaging with deep penetration, ensured spatial and temporal resolution, and color multiplexity. Introducing a core-shell structure to separate the emitting center apart from water molecules is a universal strategy to retain brightness of inorganic particle fluorophores after phase transfer. Further surface modification of the particles not only increase water solubility but also introduce functional groups. For example, rare-earth down conversion nanoparticle (DCNP), NaGdF4:5 % Nd@NaGdF4 with the emission peak at ~1060 nm was applied for metastatic tumor imaging[30]. Direct surface conjugation of DCNPs with complementary DNA was realized through ligand exchange strategy. Then follicle-stimulating hormone (FSHβ) peptide specific to the epithelium ovarian cancer was anchored onto the nanoparticle by EDC/NHS reaction. Superior T/NT ratio of ~12.5 was achieved successfully and metastases with size less than 1 mm can be clearly visualized, making the particle desirable for precise tumor diagnosis and imaging-guided surgery.
3. NIR-II fluorophores with improved pharmacokinetics and biocompatibility and their applications in molecular imaging
Molecular imaging is regarded as a promising tool for tumor diagnosis and imaging-guided surgery. NIR-II molecular imaging offers the prospect of non-invasive tumor detection with several advantages of deep penetration, real-time manner, superior temporal and spatial resolution. However, in terms of clinical applications, pharmacokinetics and biocompatibility are major concerns which need to be fully taken into consideration. Majority of NIR-II fluorophores can not be easily excreted out of body post imaging, raising potential safety concerns in the long term[6a, 10d, 14, 24, 31]. To alleviate toxicity concern and facilitate FDA-approval of NIR-II molecular imaging, systematical pharmacokinetics and biocompatibility study should be done before NIR-II probes are utilized. Blood half-life time and biodistribution are important criteria to investigate the pharmacokinetics while different fluorophores vary largely on circulation time and tissue accumulation. For instance, aforementioned p-FE exhibit ~16 h blood half-life time[7], which was 8–16 times longer than previously reported organic molecules emitting in NIR-II window including CH1055[3b], IR-E1[10b] and IR-FEP[6c]. In vivo toxicity study, including body weight measurement, serum biochemistry test, pathology analysis by H&E staining, should be investigated to evaluate the biocompatibility of NIR-II probes. Only NIR-II probes with superior biocompatibility can be applied for molecular imaging.
3.1. Renal-excretable NIR-II fluorophores
Long retention of NIR-II fluorophores within body not only raises considerable safety concern but also influence the performance of molecular imaging. Accumulation of fluorescence probes in the organs of the reticuloendothelial system (RES) will increase background signals to interfere with the targeted navigation. In this regard, NIR-II fluorophores with superior excretion feature demonstrate to be favorable candidates for in vivo molecular imaging. So far, a handful of explorations have been done on developeing molecular NIR-II fluorophores with fast excretion kinetics and their superiorities in molecular imaging have been demonstrated.
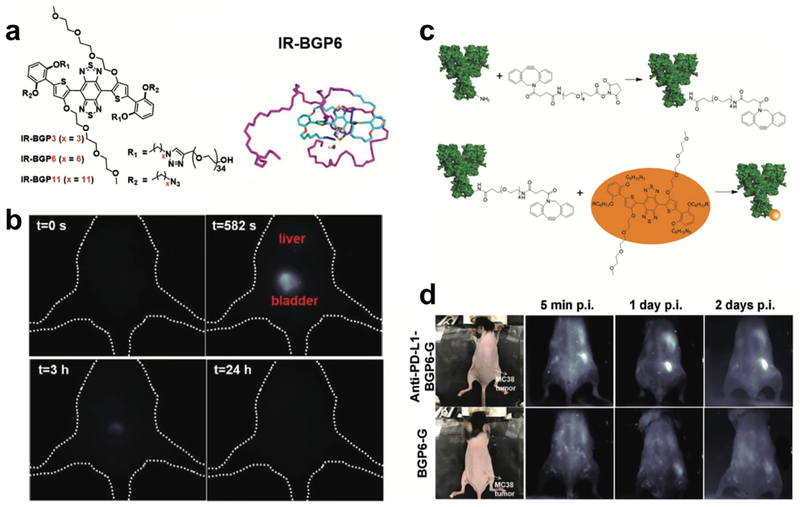
Taking advantage of previous experience on manipulating the molecular structure of S-D-A-D-S backbone-based fluorophores, our group synthesized several NIR-II fluorophores with similar building units (Figure 4a)[6b]. Through chemical engineering of components of shielding units and donors, a bright NIR-II fluorophore (IR-BGP6) with high QY of ~ 1.5% was obtained, which reveals fast renal excretion kinetics and low tissue accumulation (Figure 4b)[6b]. Molecular dynamic simulations decipher factors affecting the excretion behavior and brightness of the NIR-II fluorophore, providing insights for future development of renal-excreted NIR-II fluorophores. Then we conjugated IR-BGP6 with programmed cell death ligand-1 monoclonal antibody (PD-L1 mAb) under mild copper free click chemistry to obtain a probe (anti-PD-L1-BGP6) specific to PD-L1 (Figure 4c). Sensitivity and specificity of this probe to PD-L1 were firstly validated by in vitro staining of various cell lines to differentiate PD-L1 expression level. After that, this probe was injected into mice inoculated with murine colon adenocarcinoma (MC38) tumors to perform in vivo NIR-II molecular imaging of immune-checkpoint PD-L1. Through collection of fluorescence emission above 1200 nm, the location of MC38 tumors can be clearly pinpointed, and the tumor can be easily distinguished from surrounding normal tissues, leading to a high tumor-to-normal tissue (T/NT) ratio of ~ 9.5. Meanwhile, without the attachment of PD-L1 mAb, IR-BGP6 only gave a low T/NT ratio of 2.6, clearly reflecting the molecular imaging process (Figure 4d). In striking contrast, T/NT ratio does not reveal much difference in the case of non-renal excreted NIR-II fluorophore IR-FEP with and without the attachment of PD-L1 mAb, further reflecting the superiority of renal- excreted NIR-II fluorophores in the molecular imaging. Surprisingly, even after the attachment of big size mAb, anti-PD-L1-BPG6 still demonstrated relatively fast excretion behavior, which could be ascribed to the gradual digestion of mAb in vivo to release IR-BGP6 for renal excretion, making this probe promising to be translated into the clinical application in the future.
Figure 4. Molecular imaging of immune checkpoint PD-L1 based on the renal-excreted NIR-II fluorophores.
a) Molecular structure and molecular dynamics simulation of renal-excreted NIR-II fluorophore IR-BGP6. b) Renal excretion behavior of IR-BGP6. Most IR-BGP6 went to bladder after intravenous injection. c) Scheme of conjugation of PD-L1 mAb onto IR-BGP6. d) Non-invasive molecular imaging of PD-L1 using anti-PD-L1-BGP6 probe. Without attachment of PD-L1 mAb negligible IR-BGP6 accumulated within the MC38 tumors. Reproduced with permission[6b]. Copyright 2018, John Wiley and Sons.
3.2. Renal-excretable NIR-II molecular imaging probes
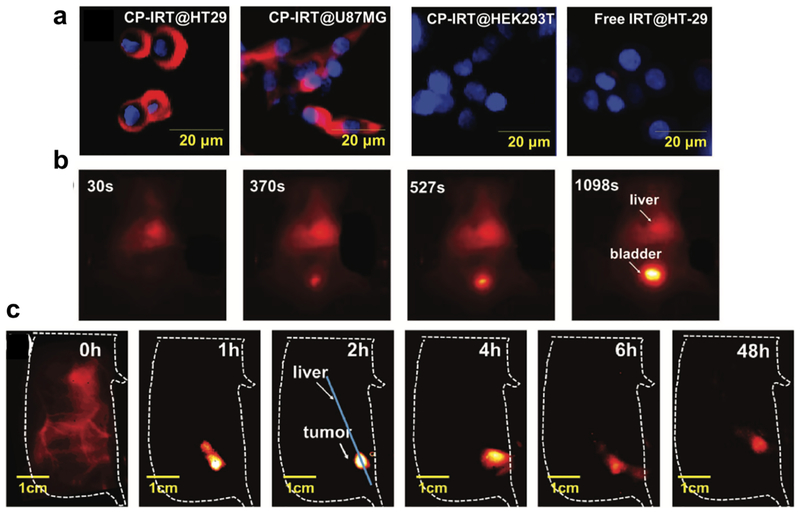
As a conventional strategy to realize molecular imaging, big size antibody (~ 150 kDa) with high specificity and sensitivity towards targets are covalently attached to small molecular fluorophores[32]. For NIR-II fluorophores with renal excretion, the attachment of antibody would sacrifice their rapid excretion behaviors, which is not optimal for in vivo molecular imaging. Compared with commonly used antibody targeted imaging, peptides can afford targeting probes with smaller size, higher penetration, and faster excretion[33]. In view of this, our group screened a pool of peptides and achieved to select a peptide (named CP) showing high binding affinity and specificity towards a biomarker of tumor stem cell (CD133). We then utilized the small size peptide CP for the covalent conjugation of renal-excreted NIR-II fluorophores (IRT)[5]. After conjugation, the size of peptide-based NIR-II probe (CP-IRT) is still below the renal cutoff, affording fast renal excretion. First, the targeting ability of CP-IRT towards CD133 marker was validated by microarray detection comparison of HT-29 and U87 cell lysate (CD133 positive) with HEK293T cell lysate (CD133 negative), which was further confirmed by in vitro staining of HT29, U87MG and HEK293T cells (Figure 5a). To move forward, CP-IRT probe was injected into mice inoculated with HT-29 tumors to evaluate its in vivo molecular imaging ability. Seen from the results, the tumor can be selectively highlighted as the probe circulated. Peak T/NT ratio reached ~ 8.3 at 2 h post injection, demonstrating a fast in vivo tumor binding kinetics (Figure 5c). Moreover, besides the tumor, the CP-IRT probe also accumulated within bladder and was then excreted through urine (~ 87% excretion within 6 h, Figure 5b). This is the first demonstration of NIR-II molecular imaging probe which can be fast excreted outside body, holding the great potential for future clinical translation.
Figure 5. A renal-excreted probe for molecular imaging of CD133 marker.
a) In vitro profile of CD133 expression level using CP-IRT probe. The CD133 expression level was high on cancer cell lines, i.e. HT29 and U87MG. The CD133 expression level was low on normal cell lines, i.e. HEK293. b) Most of CP-IRT probe accumulated within bladder after intravenous injection. c) The in vivo molecular imaging of CD133 on HT-29 tumors using CP-IRT. CP-IRT can selectively home to the HT-29 tumors without too much non-specific accumulation within other normal tissues. Reproduced with permission [5]. Copyright 2018, John Wiley and Sons.
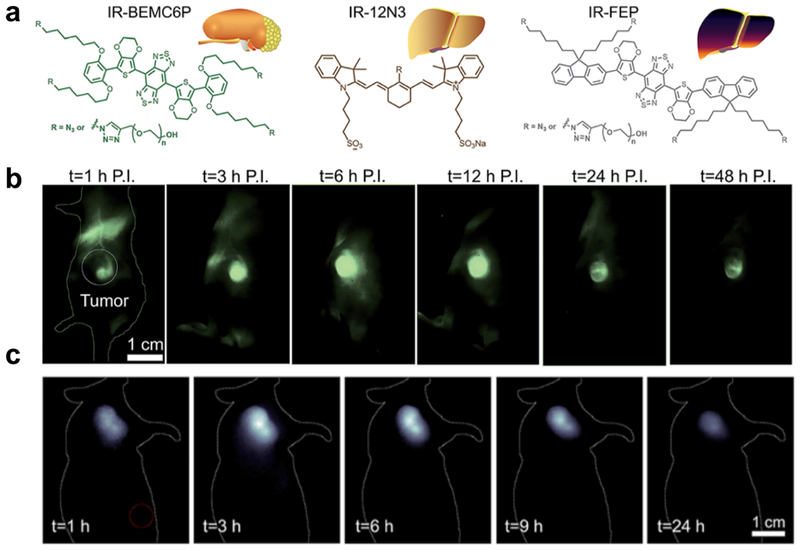
With the progress on the development of molecular imaging probes with renal excretion, potential clinical applications of these probes were further explored. Chen et al[27] compared fluorophores IR-BEMC6P with fast renal excretion, IR-12N3 with fast hepatobiliary clearance, and IR-FEP/IR-FGP with long liver retention in parallel. Several properties, including small size, near-neutral functional groups, fast dissociation with proteins, and low uptake by macrophages, were identified to be beneficial for the fast excretion kinetics (Figure 6a)[34]. Tumor targeting RGD peptide was then conjugated to IR-BEMC6P under mild conditions to perform in vivo molecular imaging of U87MG tumors, reaching a high T/NT ratio of ~ 9 at 6 h post injection to benefit the differentiation of tumors and normal tissues (Figure 6b). Encouraged by this, octreotate (TATE) peptide[35], which targets somatiostatin receptors (SSTRs) in gastroenteropancreatic neuroendocrine tumors, was also conjugated with IR-BEMC6P. Similar to above probe CP-IRT, IR-BEMC6P@TATE demonstrated superior molecular imaging ability without the loss of the renal excretion ability. Benefiting from the reduced normal tissue exposure of IR-BEMC6P@TATE, peak T/NT ratio for mice inoculated with AR42J tumor could reach as high as ~ 10 (Figure 6c), comparable to conventionally used PET imaging. With these advances, a tumor removal surgery was performed under the guidance of NIR-II imaging and white light. NIR-II molecular imaging guided micro-surgery was additionally performed utilizing IR-BEMC6P@FSH, successfully realizing the cutting of single follicles which are hundreds of micrometer in size.
Figure 6. A super-contrast NIR-II fluorophore for molecular imaging guided microsurgery.
a) The chemical structure of renal excretion and liver-uptake NIR-II fluorophores. b) Molecular imaging of the U87 tumor-bearing mouse intravenously injected with IR-BEMC6P@RGD in the NIR-II window. c) NIR-II molecular imaging of the AR42J tumor mice after tail vein injection of IR-BEMC6P@TATE. Reproduced with permission [34]. Copyright 2019, Royal Society of Chemistry.
4. Summary and future outlook
As a promising strategy for cancer diagnosis and imaging-guided surgery and therapy, molecular imaging has been widely adopted. The last decade has witnessed the rapid development of NIR-II fluorescence imaging. Owing to advances of reduced photon scattering and suppressed autofluorescence, imaging in the NIR-II window demonstrates high resolution and deep penetration, which fulfill requirements to obtain high signal-to-background ratios for molecular imaging to visually locate targets on the cellular and molecular level. As a result, more and more people are performing NIR-II molecular imaging. To achieve this, numerous efforts are being spared on the development of probes with qualities of high binding affinity and specificity, high brightness, excellent biocompatibility and favorable pharmacokinetics. Beside above endeavors, some efforts are also being made on the optimization of optical instruments and parameters for enhancing imaging performance. Even with the significant progress made on the molecular imaging in the NIR-II window, there is still room for further improvement of this field:
Develop NIR-II probes with high brightness and photostability. The quantum efficiency QE, which is directly related to brightness of probes, is determined by two key factors: absorption extinction coefficient and QY. As for D-A-D architecture fluorophores, depite of the impressive QY, the low adsorption extinction coefficients largely limits the brightness, resulting in the potential safety concerns by increasing the irradiation power and hindering the monitoring of dynamic physiological processes caused by long applied exposure time. On the other hand, although cyanine fluorophores show high absorption coefficients, red-shift of the fluorophore by lengthening the polymethine chain will compromise QY and decrease the photostability. Therefore, developing photo-stable NIR-II molecular probes with enhanced extinction coefficient and QY are still urgently needed. Molecular simulation can be used to assist the design of NIR-II fluorophores for high quantum efficiency. Meanwhile, more attentions should also be put into the applied conjugation method and affinity moieties that are going to be attached, making sure these two aspects will not affect the brightness, adsorption and emission wavelength of NIR-II fluorophores a lot.
Optimize the binding moiety of NIR-II probes. As another major constitution of molecular imaging probes, the binding moiety dominates the sensitivity and specificity of probes, largely affecting the molecular imaging performance. As of today, antibodies with large molecular weight (~ 150 kDa) and big size (~ 3.5 nm) appear to be the most widely used binding moieties for the construction of molecular imaging probes. Although antibodies possess high binding selectivity and affinity, the long blood half-life time and big size beyond the size-cutoff of renal excretion would lower the excretion kinetics of probes. As the field of bioengineering progresses, small-size and low-molecular weight moieties (e.g. peptide, aptamer and affibody) possessing comparable binding specificity and affinity towards targets with those of conventionally used antibodies have been developed. These moieties seem to be promising alternatives of antibodies to be conjugated with NIR-II fluorophores for the formation of molecular imaging probes with the size below the cutoff of renal excretion, achieving low tissue accumulation. Moreover, compared with conventionally used antibodies, these small size moieties demonstrate better stability and can be more easily manipulated for the conjugation with fluorophores even under relatively harsh conditions, providing more choice for selecting conjugation methods to obtain an optimal probe.
Increase clinical translatability and applicability of NIR-II probes. For the clinical translation of NIR-II probes, water solubility and biocompatibility are prerequisites. To achieve superior biocompatibility, researchers spare many efforts to modify organic NIR-II fluorophores with biocompatible moieties (e.g. PEG, zwitterion and polysaccharide) to increase their solubility in physiological environment. While organic NIR-II fluorophores show more favorable pharmacokinetics properties and mitigated toxicity, this type of fluorophores still suffers from broad emission spectrum, low fluorescence intensity and difficulty in tuning emission wavelength. Inorganic nanomaterials benefit from high photostability, narrow fluorescence spectrum, and ability to emit in in the long end of NIR-II window (i.e., the NIR-IIb window within the range of 1500 to 1700 nm), leading to better imaging performance. Nowadays, inorganic nanoparticles including QDs, RENPs, and carbon tubes still compromise the majority of long wavelength emitting probes. However, all current inorganic NIR-II probes struggle from slow excretion kinetics and potential toxicity. One possible prospective to solve this problem is optimizing the surface modification strategy and increase the compatibility of the particles in aqueous environment. Exploring surface functionalization strategy will also prevent nanoparticles from being recognized and repelled by the immune system to promote the particle stability in biological system for the enhancement of their biocompatibility.
NIR-II molecular imaging based on the bioorthogonal chemistry. Bioorthogonal chemistry refers to any chemical reaction that can occur inside of living systems without interfering with native biochemical processes. Conventional visible and NIR fluorophores have been applied to combine with the bioorthgonal chemistry to study biomolecules in living systems without cellular toxicity. The in-situ fluorescence imaging approach would decipher the intrinsic status of biomolecules, including spatial distribution, content and activity, which demonstrates to be suitable for understanding the underlying mechanism of physiological activities. As of today, no NIR-II fluorophores have been utilized for bioorthogonal chemistry-based molecular imaging. Superiorities of NIR-II imaging would extend the in vivo imaging application of bioorthogonal chemistry, further approaching to disclose the original internal status of monitored targets. A number of chemical ligation strategies have been developed to fulfill the requirements of biorthogonality, e.g. 1,3-dipolar cycloaddition between azides and cyclooctynes, oxime/hydrazine formation from aldehydes and ketones and tetrazine ligation. One straightforward method to introduce the concept of bioorthogonal chemistry into NIR-II molecular imaging is to modify NIR-II fluorophores with active functional groups (e.g. cyclooctynes, ketones, tetrazine) without affecting too much of their optical properties.
Acknowledgements
H.W. and H.D. contributed equally to this work. This study was also supported by National Institutes of Health DP1-NS-105737 and the Calbrain program.
Biographies
Hao Wan is a postdoctor researcher in Stanford University (Dai lab). He received his Ph.D. from a joint Ph.D. program between Dalian Institute of Chemical Physics (DICP), Chinese Academy of Sciences (CAS) and East China University of Science and Technology. And his B. Eng. degree was obtained from East China University of Science and Technology. His current research is centered on bio-imaging/detection.

Haotian Du received her bachelor’s degree in chemistry from Tsinghua University in China in 2018. She is currently a Ph.D. student under the guidance of Prof. Hongjie Dai in the Department of Chemistry at Stanford University. Her current research focus is developing novel fluorescence imaging agents in second near-infrared window for understanding of physiological process in living organisms.

Hongjie Dai is the J. G. Jackson and C. J. Wood Professor of Chemistry at Stanford University. His research interfaces with chemistry, physics, materials science, and biological and medical sciences. Thus far, his group has made advances to the basic science of carbon nanotubes and graphene and potential applications in the areas of nanoelectronics, nanobiotechnology, nanomedicine, energy storage, and catalysis. He has been elected to the American Academy of Arts and Sciences, the AAAS and the National Academy of Sciences

References
- [1].a) Frangioni JV, Current Opinion in Chemical Biology 2003, 7, 626; [DOI] [PubMed] [Google Scholar]; b) Pinaud F, Michalet X, Bentolila LA, Tsay JM, Doose S, Li JJ, Iyer G, Weiss S, Biomaterials 2006, 27, 1679; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Falati S, Gross P, Merrill-Skoloff G, Furie BC, Furie B, Nat. Med 2002, 8, 1175. [DOI] [PubMed] [Google Scholar]
- [2].Hong G, Antaris AL, Dai H, Nat. Biomed. Eng 2017, 1, 0010. [Google Scholar]
- [3].a) Hong G, Lee JC, Robinson JT, Raaz U, Xie L, Huang NF, Cooke JP, Dai H, Nat. Med 2012, 18, 1841; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Antaris AL, Chen H, Cheng K, Sun Y, Hong G, Qu C, Diao S, Deng Z, Hu X, Zhang B, Zhang X, Yaghi OK, Alamparambil ZR, Hong X, Cheng Z, Dai H, Nat. Mater 2015, 15, 235; [DOI] [PubMed] [Google Scholar]; c) Hong G, Robinson JT, Zhang Y, Diao S, Antaris AL, Wang Q, Dai H, Angew. Chem. Int. Ed 2012, 51, 9818; [DOI] [PubMed] [Google Scholar]; d) Hong G, Diao S, Chang J, Antaris AL, Chen C, Zhang B, Zhao S, Atochin DN, Huang PL, Andreasson KI, Kuo CJ, Dai H, Nat. Photonics 2014, 8, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Choy G, Choyke P, Libutti SK, Mol. Imaging 2003, 2, 15353500200303142; [DOI] [PubMed] [Google Scholar]; b) Weissleder R, Mahmood U, Radiology 2001, 219, 316; [DOI] [PubMed] [Google Scholar]; c) Weissleder R, Science 2006, 312, 1168; [DOI] [PubMed] [Google Scholar]; d) Massoud TF, Gambhir SS, Genes & Dev 2003, 17, 545. [DOI] [PubMed] [Google Scholar]
- [5].Wang W, Ma Z, Zhu S, Wan H, Yue J, Ma H, Ma R, Yang Q, Wang Z, Li Q, Qian Y, Yue C, Wang Y, Fan L, Zhong Y, Zhou Y, Gao H, Ruan J, Hu Z, Liang Y, Dai H, Adv. Mater 2018, 30, 1800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Zhu S, Herraiz S, Yue J, Zhang M, Wan H, Yang Q, Ma Z, Wang Y, He J, Antaris AL, Zhong Y, Diao S, Feng Y, Zhou Y, Yu K, Hong G, Liang Y, Hsueh AJ, Dai H, Adv. Mater 2018, 30, 1705799; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wan H, Ma H, Zhu S, Wang F, Tian Y, Ma R, Yang Q, Hu Z, Zhu T, Wang W, Ma Z, Zhang M, Zhong Y, Sun H, Liang Y, Dai H, Adv. Funct. Mater 2018, 28, 1804956; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhu S, Yang Q, Antaris AL, Yue J, Ma Z, Wang H, Huang W, Wan H, Wang J, Diao S, Zhang B, Li X, Zhong Y, Yu K, Hong G, Luo J, Liang Y, Dai H, Proc. Natl. Acad. Sci 2017, 114, 962; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhang M, Yue J, Cui R, Ma Z, Wan H, Wang F, Zhu S, Zhou Y, Kuang Y, Zhong Y, Pang DW, Dai H, Proc. Natl. Acad. Sci. U S A 2018, 115, 6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wan H, Yue J, Zhu S, Uno T, Zhang X, Yang Q, Yu K, Hong G, Wang J, Li L, Ma Z, Gao H, Zhong Y, Su J, Antaris AL, Xia Y, Luo J, Liang Y, Dai H, Nat. Commun 2018, 9, 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Zhu S, Herraiz S, Yue J, Zhang M, Wan H, Yang Q, Ma Z, Wang Y, He J, Antaris AL, Zhong Y, Diao S, Feng Y, Zhou Y, Yu K, Hong G, Liang Y, Hsueh AJ, Dai H, Adv Mater 2018, 30, e1705799; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ma Z, Zhang M, Yue J, Alcazar C, Zhong Y, Doyle TC, Dai H, Huang NF, Adv. Funct. Mater 2018, 28; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhong Y, Ma Z, Zhu S, Yue J, Zhang M, Antaris AL, Yuan J, Cui R, Wan H, Zhou Y, Wang W, Huang NF, Luo J, Hu Z, Dai H, Nat. Commun 2017, 8, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Li C, Zhang Y, Wang M, Zhang Y, Chen G, Li L, Wu D, Wang Q, Biomaterials 2014, 35, 393; [DOI] [PubMed] [Google Scholar]; b) Hong G, Zou Y, Antaris AL, Diao S, Wu D, Cheng K, Zhang X, Chen C, Liu B, He Y, Wu JZ, Yuan J, Zhang B, Tao Z, Fukunaga C, Dai H, Nat. Commun 2014, 5, 4206; [DOI] [PubMed] [Google Scholar]; c) Zhang Y, Hong G, Zhang Y, Chen G, Li F, Dai H, Wang Q, ACS Nano 2012, 6, 3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Antaris AL, Chen H, Cheng K, Sun Y, Hong G, Qu C, Diao S, Deng Z, Hu X, Zhang B, Nature materials 2016, 15, 235; [DOI] [PubMed] [Google Scholar]; b) Zhang X-D, Wang H, Antaris AL, Li L, Diao S, Ma R, Nguyen A, Hong G, Ma Z, Wang J, Zhu S, Castellano JM, Wyss-Coray T, Liang Y, Luo J, Dai H, Adv. Mater 2016, 28, 6872; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Feng Y, Zhu S, Antaris AL, Chen H, Xiao Y, Lu X, Jiang L, Diao S, Yu K, Wang Y, Chem. Sci 2017, 8, 3703; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yang Q, Hu Z, Zhu S, Ma R, Ma H, Ma Z, Wan H, Zhu T, Jiang Z, Liu W, Jiao L, Sun H, Liang Y, Dai H, J. Am. Chem. Soc 2018, 140, 1715; [DOI] [PubMed] [Google Scholar]; e) Yang Q, Ma Z, Wang H, Zhou B, Zhu S, Zhong Y, Wang J, Wan H, Antaris A, Ma R, Adv. Mater 2017, 29, 1605497. [DOI] [PubMed] [Google Scholar]
- [11].a) Carr JA, Franke D, Caram JR, Perkinson CF, Saif M, Askoxylakis V, Datta M, Fukumura D, Jain RK, Bawendi MG, Proc. Natl. Acad. Sci. U S A 2018, 115, 4465; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ding B, Xiao Y, Zhou H, Zhang X, Qu C, Xu F, Deng Z, Cheng Z, Hong X, J. Med. Chem 2019, 62, 2049; [DOI] [PubMed] [Google Scholar]; c) Cosco ED, Caram JR, Bruns OT, Franke D, Day RA, Farr EP, Bawendi MG, Sletten EM, Angew. Chem. Int. Ed 2017, 56, 13126; [DOI] [PubMed] [Google Scholar]; d) Li B, Lu L, Zhao M, Lei Z, Zhang F, Angew. Chem 2018, 57, 7483; [DOI] [PubMed] [Google Scholar]; e) Wang S, Fan Y, Li D, Sun C, Lei Z, Lu L, Wang T, Zhang F, Nat. Commun 2019, 10, 1058; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Zhu S, Hu Z, Tian R, Yung BC, Yang Q, Zhao S, Kiesewetter DO, Niu G, Sun H, Antaris AL, Adv. Mater 2018, 30, 1802546. [DOI] [PubMed] [Google Scholar]
- [12].Ma Z, Wan H, Wang W, Zhang X, Uno T, Yang Q, Yue J, Gao H, Zhong Y, Tian Y, Sun Q, Liang Y, Dai H, Nano Res 2018, 12, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Zhong Y, Ma Z, Zhu S, Yue J, Zhang M, Antaris AL, Yuan J, Cui R, Wan H, Zhou Y, Nat. Commun 2017, 8, 737; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang M, Yue J, Cui R, Ma Z, Wan H, Wang F, Zhu S, Zhou Y, Kuang Y, Zhong Y, Proc. Natl. Acad. Sci. U S A 2018, 115, 6590; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Franke D, Harris DK, Chen O, Bruns OT, Carr JA, Wilson MW, Bawendi MG, Nat. Commun 2016, 7, 12749; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bruns OT, Bischof TS, Harris DK, Franke D, Shi Y, Riedemann L, Bartelt A, Jaworski FB, Carr JA, Rowlands CJ, Nat. Biomed. Eng 2017, 1, 0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang Q, Ma Z, Wang H, Zhou B, Zhu S, Zhong Y, Wang J, Wan H, Antaris A, Ma R, Zhang X, Yang J, Zhang X, Sun H, Liu W, Liang Y, Dai H, Adv. Mater 2017, 29, 1605497. [DOI] [PubMed] [Google Scholar]
- [15].a) Chang PV, Prescher JA, Sletten EM, Baskin JM, Miller IA, Agard NJ, Lo A, Bertozzi CR, Proc. Natl. Acad. Sci. U S A 2010,107, 1821; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR, Proc. Natl. Acad. Sci. U S A 2007, 104, 16793; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Koo H, Lee S, Na JH, Kim SH, Hahn SK, Choi K, Kwon IC, Jeong SY, Kim K, Angew. Chem 2012, 124, 12006. [DOI] [PubMed] [Google Scholar]
- [16].a) Buchsbaum DJ, Bonner JA, Grizzle WE, Stackhouse MA, Carpenter M, Hicklin DJ, Bohlen P, Raisch KP, Int. J. Radiat. Oncol. Bio. Phy 2002, 54, 1180; [DOI] [PubMed] [Google Scholar]; b) Lynch TJ, Patel T, Dreisbach L, McCleod M, Heim WJ, Robert H, Eugene P, Virginie P, Weber MR, Woytowitz D, J. Thorac. Oncol 2007, 2, S340. [Google Scholar]
- [17].Knowles JA, Heath CH, Saini R, et al. , Arch. Otolaryngol–Head & Neck Surg 2012, 138, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, Schnitzer MJ, Science 2015, 350, 1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Wan H, Yue J, Zhu S, Uno T, Zhang X, Yang Q, Yu K, Hong G, Wang J, Li L, Ma Z, Gao H, Zhong Y, Su J, Antaris AL, Xia Y, Luo J, Liang Y, Dai H, Nat. Commun 2018, 9, 1171; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang M, Yue J, Cui R, Ma Z, Wan H, Wang F, Zhu S, Zhou Y, Kuang Y, Zhong Y, Pang D-W, Dai H, Proc. Natl. Acad. Sci. U S A 2018, 115, 6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, Stelzer EHK, Nat. Methods 2007, 4, 311; [DOI] [PubMed] [Google Scholar]; b) Petroll WM, Jester JV, Cavanagh HD, Scanning 1996, 18, 45; [DOI] [PubMed] [Google Scholar]; c) Dinsmore AD, Weeks ER, Prasad V, Levitt AC, Weitz DA, Appl. Opt 2001, 40, 4152. [DOI] [PubMed] [Google Scholar]
- [21].Shi Kam NW, Jessop TC, Wender PA, Dai H, J. Am. Chem. Soc 2004, 126, 6850. [DOI] [PubMed] [Google Scholar]
- [22].a) Hsueh AJW, Dafau ML, Catt KJ, Biochemical and Biophysical Research Communications 1976, 72, 1145; [DOI] [PubMed] [Google Scholar]; b) Hsueh AJ, Dufau ML, Catt KJ, Proc. Natl. Acad. Sci. U S A 1977, 74, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].a) Kumar TR, Wang Y, Lu N, Matzuk MM, Nature Genetics 1997, 15, 201; [DOI] [PubMed] [Google Scholar]; b) Patiño R, Yoshizaki G, Thomas P, Kagawa H, Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol 2001, 129, 427. [DOI] [PubMed] [Google Scholar]
- [24].Antaris AL, Chen H, Diao S, Ma Z, Zhang Z, Zhu S, Wang J, Lozano AX, Fan Q, Chew L, Zhu M, Cheng K, Hong X, Dai H, Cheng Z, Nat. Commun 2017, 8, 15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].a) Tao Z, Hong G, Shinji C, Chen C, Diao S, Antaris AL, Zhang B, Zou Y, Dai H, Angew. Chem. Int. Ed 2013, 52, 13002; [DOI] [PubMed] [Google Scholar]; b) Feng Y, Zhu S, Antaris AL, Chen H, Xiao Y, Lu X, Jiang L, Diao S, Yu K, Wang Y, Herraiz S, Yue J, Hong X, Hong G, Cheng Z, Dai H, Hsueh AJ, Chem. Sci 2017, 8, 3703; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sun Y, Ding M, Zeng X, Xiao Y, Wu H, Zhou H, Ding B, Qu C, Hou W, Er-bu AGA, Zhang Y, Cheng Z, Hong X, Chem. Sci 2017, 8, 3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].a) Weitman SD, Lark RH, Coney LR, Fort DW, Frasca V, Zurawski VR, Kamen BA, Cancer Res 1992, 52, 3396; [PubMed] [Google Scholar]; b) Piedrahita JA, Oetama B, Bennett GD, van Waes J, Kamen BA, Richardson J, Lacey SW, Anderson RGW, Finnell RH, Nat. Genetics 1999, 23, 228. [DOI] [PubMed] [Google Scholar]
- [27].Reddy JA, Xu L-C, Parker N, Vetzel M, Leamon CP, J. Nucl. Med 2004, 45, 857. [PubMed] [Google Scholar]
- [28].Mansouri S, Cuie Y, Winnik F, Shi Q, Lavigne P, Benderdour M, Beaumont E, Fernandes JC, Biomaterials 2006, 27, 2060. [DOI] [PubMed] [Google Scholar]
- [29].a) Fan Y, Wang P, Lu Y, Wang R, Zhou L, Zheng X, Li X, Piper JA, Zhang F, Nat. Nanotechnol 2018, 13, 941; [DOI] [PubMed] [Google Scholar]; b) Naczynski D, Tan M, Zevon M, Wall B, Kohl J, Kulesa A, Chen S, Roth C, Riman R, Moghe P, Nat. Commun 2013, 4, 2199; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang R, Li X, Zhou L, Zhang F, Angew. Chem. Int. Ed 2014, 53, 12086. [DOI] [PubMed] [Google Scholar]
- [30].Wang P, Fan Y, Lu L, Liu L, Fan L, Zhao M, Xie Y, Xu C, Zhang F, Nat. Commun 2018, 9, 2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].a) Hong G, Lee JC, Robinson JT, Raaz U, Xie L, Huang NF, Cooke JP, Dai H, Nat. Med 2012, 18, 1841; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Robinson JT, Hong G, Liang Y, Zhang B, Yaghi OK, Dai H, J. Am. Chem. Soc 2012, 134, 10664; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ma Z, Zhang M, Yue J, Alcazar C, Zhong Y, Doyle TC, Dai H, Huang NF, Adv. Funct. Mater 2018, 28, 1803417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].a) England CG, Hernandez R, Eddine SBZ, Cai W, Mol. Pharm 2016, 13, 8; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pan Y, Volkmer J-P, Mach KE, Rouse RV, Liu J-J, Sahoo D, Chang TC, Metzner TJ, Kang L, van de Rijn M, Skinner EC, Gambhir SS, Weissman IL, Liao JC, Sci. Transl. Med 2014, 6, 260ra148; [DOI] [PubMed] [Google Scholar]; c) Ogawa M, Regino CAS, Choyke PL, Kobayashi H, Mol. Cancer Ther 2009, 8, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].a) Lee S, Xie J, Chen X, Biochemistry 2010, 49, 1364; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sturm MB, Joshi BP, Lu S, Piraka C, Khondee S, Elmunzer BJ, Kwon RS, Beer DG, Appelman HD, Turgeon DK, Wang TD, Sci. Transl. Med 2013, 5, 184ra61; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hsiao J-K, Wu H-C, Liu H-M, Yu A, Lin C-T, Nanomed.: Nanotechnol., Biology and Med 2015, 11, 1425. [Google Scholar]
- [34].Tian R, Ma H, Yang Q, Wan H, Zhu S, Chandra S, Sun H, Kiesewetter DO, Niu G, Liang Y, Chen X, Chem. Sci 2019, 10, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kam BLR, Teunissen JJM, Krenning EP, de Herder WW, Khan S, van Vliet EI, Kwekkeboom DJ, Eur. J. Nucl. Med. Mol. Imaging 2012, 39 Suppl 1, S103. [DOI] [PMC free article] [PubMed] [Google Scholar]