The increasing availability of DNA sequence data and access to sophisticated bioinformatic algorithms mean that an unbiased bioinformatics-based assessment of the predicted impact of a genomic variant is rapidly available. The key point of this Viewpoint article is that such bioinformatic assessments are not equivalent to an expert diagnostic interpretation and may be misleading in both research and clinical care.
Predication algorithms in genomic medicine
Recently published examples involving monogenic diabetes demonstrate how pathogenicity prediction algorithms can be very inaccurate for predicting which genetic variants are likely causal of dominant monogenic disease (1–4). Here, we highlight the potential pitfalls of variant classification and how they can be avoided.
A recent study used a bioinformatic algorithm to identify 88 “likely pathogenic” monogenic diabetes variants in 80 individuals (8.6%) from a cohort of 1019 individuals with type 1 diabetes for 50 or more years (4). Application of the widely used American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) standards and guidelines (5) classifies only nine of these 88 variants as likely pathogenic or pathogenic variants that would be reported by our clinical diagnostic laboratory as likely causative of the patients’ diabetes. This is not an isolated occurrence; other published research studies with an overreliance on in silico prediction tools have reported high levels (~90%) of false positive “likely pathogenic” monogenic diabetes variants (1–3). We have seen clinical diagnostic reports from laboratories in eight countries across Europe, Asia, the Middle East, and the United States that have similarly reported such variants as incorrectly likely causative of a patient’s diabetes.
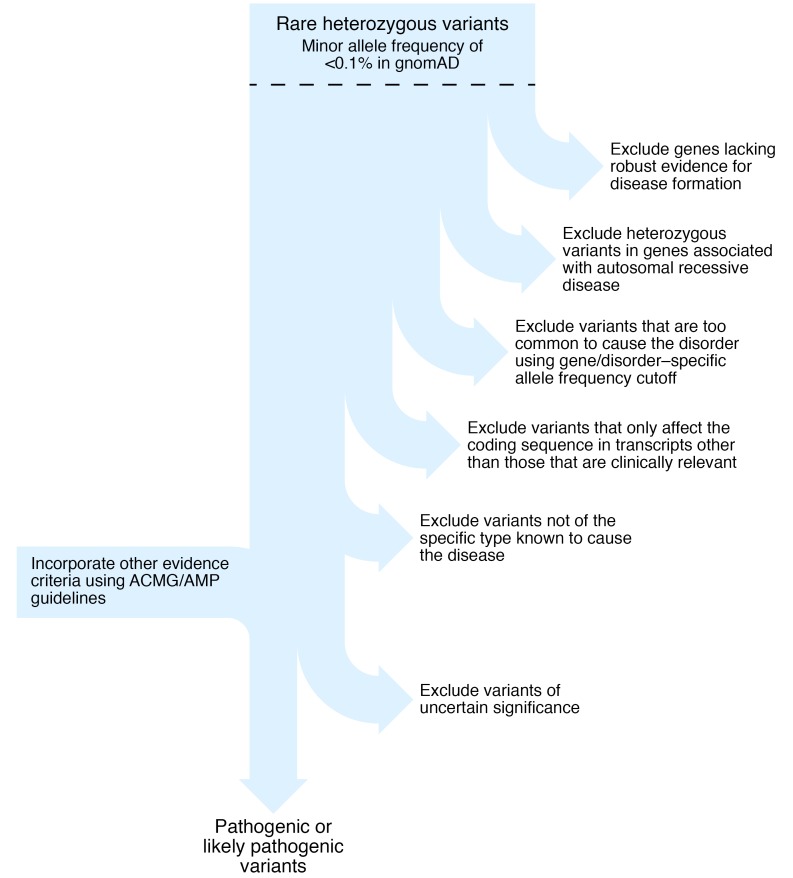
Such great discrepancies occur not because the bioinformatic algorithm is wrong, or even based on incorrect scientific principles, but because variant interpretation in the clinical setting requires other information in addition to the predicted effect upon protein function. This additional information includes knowledge regarding the gene-disease validity, mode of inheritance, appropriate allele frequency cutoff thresholds, most clinically relevant transcript, and specificity of disease-causing variant type for each gene. Each of these is discussed in turn below and illustrated in Figure 1.
Figure 1. Flowchart to illustrate key steps in the interpretation of genetic variants to identify autosomal dominant (likely) pathogenic variants for clinical diagnostic or research reporting.
gnomAD, Genome Aggregation Database.
How should expert disease-related knowledge guide the interpretation and reporting of genetic variants that might cause autosomal dominant monogenic disease?
Do not analyze genes without robust evidence to support the gene-disease association (6), for example the BLK, KLF11, and PAX4 genes, where current evidence is limited (7).
Reputable clinical diagnostic laboratories do not include these genes in their monogenic diabetes testing.
Do not report a heterozygous variant in autosomal recessive disorders caused by biallelic variants (homozygous or compound heterozygous).
For example, biallelic pathogenic WFS1 variants cause Wolfram syndrome (8), a very rare disorder with an estimated prevalence of 1 in 500,000 (heterozygous carrier frequency ~0.3%). WFS1 is an extremely polymorphic gene with rare (allele frequency < 0.1%) missense variants present in more than 2% of the population. Although there are reports of autosomal dominant diabetes caused by heterozygous WFS1 variants, these are extremely rare: one family with dominantly inherited nonsyndromic diabetes (9) and five patients with neonatal- or infancy-onset diabetes, deafness, and cataracts (10). The finding of a rare heterozygous variant is therefore highly unlikely to be causative of monogenic diabetes and should not be reported.
Use appropriate gene-specific allele frequency cutoffs in control population data to exclude variants that are too common to be highly penetrant disease-causing variants.
For example, Ming-Qiang et al. found that PAX4 variants were the second most common cause of monogenic diabetes in their Chinese cohort (3), but the missense variants they reported are present in more than 1% of the East Asian population cohort (approximately 15,000 individuals) in the publicly available gnomAD database (https://gnomad.broadinstitute.org) (11). A tool is available (http://cardiodb.org/alleleFrequencyApp) that allows the user to input inheritance mode, disease prevalence, penetrance, genetic heterogeneity (how many cases can be attributed to the gene), and allelic heterogeneity (how many cases can be attributed to a single variant) and calculate a maximum credible allele frequency (12). Using HNF1A monogenic diabetes as an example with monoallelic inheritance, disease prevalence of 1 in 10,000, allelic heterogeneity 0.16, genetic heterogeneity 0.35, and penetrance 0.95, the maximum tolerated pathogenic allele count is 2 in the gnomAD database (n = 141,456). In a study of diabetic subjects from South India, Mohan et al. reported HNF1A variants as the most common subtype of monogenic diabetes in their study (1). However, six of the 11 patients had variants that are too common in the European ancestry population in gnomAD (allele counts of 4 or more in approximately 60,000 Europeans) to be highly penetrant variants causative of monogenic diabetes. It is essential to check the variant frequency in large variant data sets to avoid this type of misclassification.
Check that the most clinically relevant transcript is used.
For example, there are multiple isoforms of the transcription factor HNF4A. For interpretation of monogenic diabetes variants, the messenger RNA transcript that encodes the pancreatic isoform, rather than the liver isoform, should be used because there is a pancreatic specific promoter and exon 1 (NM_175914.4). Using this transcript, the HNF4A variant p.A417T Yu et al. reported (4) is noncoding (c.1063+120) and likely benign.
Identify the specific subtypes of heterozygous mutations that result in the specific change of function required to cause monogenic disease.
Heterozygous pathogenic variants in the GCK, HNF1A, HNF1B, and HNF4A genes cause diabetes by reducing the level of functional protein, described as haploinsufficiency (7). For other monogenic diabetes subtypes there is a different disease mechanism, and heterozygous predicted loss-of-function (frameshift, nonsense, or essential splice site) variants are not causative. The KCNJ11 and ABCC8 genes encode the subunits of the β cell potassium channel. Activating variants prevent the channel from closing in response to raised blood glucose, and this prevents insulin release in patients with diabetes (13). Recessive loss-of-function KCNJ11 and ABCC8 variants cause the opposite phenotype of hyperinsulinism (14). This means that a heterozygous loss of function variant in one of these genes confers carrier status for congenital hyperinsulinism but does not cause monogenic diabetes. Other examples include the CEL gene, where only variants within the first or fourth repeats of the VNTR region are pathogenic (15); RFX6, where there is only evidence to implicate protein truncating variants (16); and the sole heterozygous PDX1 pathogenic variant, p.P63fs, known to cause monogenic diabetes through a dominant negative effect (17).
Exclude variants of uncertain significance.
These are variants lacking evidence for classification as pathogenic or likely pathogenic (5). Examples include novel missense variants in constrained genes where the amino acid substitution is predicted to have a deleterious effect upon protein function. Any individual has about 100 variants of this type and should be treated as “uncertain until proven guilty” (18).
Why are errors in the interpretation of genetic variants common in both academic and diagnostic reports?
Misinterpretation of genetic variants is common, both in the published literature (19) and in reports from diagnostic laboratories. Next-generation sequencing technology has facilitated both the ease and scale of genetic testing, but obtaining genotype data is far more straightforward than interpreting it correctly.
Academic studies often use bespoke criteria for defining pathogenicity rather than applying guidelines developed to improve the quality and consistency of variant interpretation (5). Studies may base their variant classifications solely on in silico prediction of pathogenicity using tools such as REVEL or SIFT, PolyPhen, and MutationTaster that provide only supporting evidence within the ACMG/AMP guidelines framework recommended for diagnostic reporting (5).
The availability of large variant data sets such as the gnomAD (11) has shown that many previously reported pathogenic variants are too common to be highly penetrant disease-causing variants (20). In some cases the evidence supporting gene-disease associations is no longer valid, but publications refuting these genes are rare and may consider only a single putative mutation (21, 22). The utility of population variant databases will increase as more exome and genome sequence data are aggregated from a wider range of populations (11).
An overemphasis on bioinformatic tools for predicting pathogenicity has resulted in false positive assertions. Although curated databases of pathogenic/likely pathogenic variants are widely used, the level of curating varies, and there are often insufficient data available for users to assess the provenance of individual variant pathogenicity assertions.
The prior probability of a monogenic etiology is an important consideration for variant classification. For monogenic diabetes the prior probability will be low because only a small proportion of patients with diabetes have a monogenic subtype (3.6% of patients diagnosed at age ≤ 30 years; ref. 23). Sensitivity/specificity estimates for pathogenicity prediction tools like REVEL and PolyPhen are calculated from a set of pathogenic and benign variants in genes with a higher likelihood of a monogenic etiology (24).
How can the accuracy of variant interpretation and reporting be improved?
A number of initiatives are addressing various aspects of variant interpretation. For example the NIH-funded ClinGen resource (https://clinicalgenome.org/) includes curating of gene-disease validity evidence, the ClinVar variant repository, and expert groups developing gene- or disease-specific criteria for variant classification. Sharing genetic variant data on a global scale is an essential requirement (25).
The ACMG/AMP variant classification guidelines have been adopted in many countries, and we recommend that all academic studies use these guidelines for variant classification (5). Genetic testing for clinical diagnosis should be performed in an accredited laboratory that participates in external quality assessment schemes that include variant classification.
Conclusion
We have entered a new era in which the generation of massive quantities of accurate genetic data from an individual is no longer difficult, but the new challenge is how to correctly interpret this data. This Viewpoint emphasizes how disease-specific expertise is required when interpreting genetic data and how failure to use this information will result in errors. Misdiagnosis not only affects the individual patients for whom testing is being performed but also can be amplified through predictive testing of relatives and use of incorrect variant classifications in databases and publications for interpretation of the same variant in other patients. Accurate genetic diagnosis is needed to predict disease prognosis and guide clinical management.
Acknowledgments
SE and ATH are Wellcome Senior Investigators (098395/Z/12/Z). ATH is a Senior Investigator at the National Institute for Health Research and is supported by the National Institute for Health Research Exeter Clinical Research Facility. KAP has a postdoctoral fellowship funded by the Wellcome Trust (110082/Z/15/Z).
Version 1. 12/09/2019
Electronic publication
Version 2. 01/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):14–16. https://doi.org/10.1172/JCI133516.
Contributor Information
Sian Ellard, Email: sian.ellard@nhs.net.
Kevin Colclough, Email: kevin.colclough@nhs.net.
Andrew T. Hattersley, Email: A.T.Hattersley@exeter.ac.uk.
References
- 1.Mohan V, et al. Comprehensive genomic analysis identifies pathogenic variants in maturity-onset diabetes of the young (MODY) patients in South India. BMC Med Genet. 2018;19(1):22. doi: 10.1186/s12881-018-0528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pezzilli S, et al. Insights from molecular characterization of adult patients of families with multigenerational diabetes. Diabetes. 2018;67(1):137–145. doi: 10.2337/db17-0867. [DOI] [PubMed] [Google Scholar]
- 3.Ming-Qiang Z, et al. Maturity onset diabetes of the young (MODY) in Chinese children: genes and clinical phenotypes. J Pediatr Endocrinol Metab. 2019;32(7):759–765. doi: 10.1515/jpem-2018-0446. [DOI] [PubMed] [Google Scholar]
- 4.Yu MG, et al. Residual β cell function and monogenic variants in long-duration type 1 diabetes patients. J Clin Invest. 2019;129(8):3252–3263. doi: 10.1172/JCI127397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strande NT, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017;100(6):895–906. doi: 10.1016/j.ajhg.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald TJ, Ellard S. Maturity onset diabetes of the young: identification and diagnosis. Ann Clin Biochem. 2013;50(Pt 5):403–415. doi: 10.1177/0004563213483458. [DOI] [PubMed] [Google Scholar]
- 8. National Library of Medicine. Wolfram syndrome. Genetics Home Reference website. https://ghr.nlm.nih.gov/condition/wolfram-syndrome Accessed November 12, 2019.
- 9.Bonnycastle LL, et al. Autosomal dominant diabetes arising from a Wolfram syndrome 1 mutation. Diabetes. 2013;62(11):3943–3950. doi: 10.2337/db13-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Franco E, et al. Dominant ER stress-inducing WFS1 mutations underlie a genetic syndrome of neonatal/infancy-onset diabetes, congenital sensorineural deafness, and congenital cataracts. Diabetes. 2017;66(7):2044–2053. doi: 10.2337/db16-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. doi: 10.1101/531210. Posted on bioRxiv August 13, 2019. [DOI]
- 12.Whiffin N, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med. 2017;19(10):1151–1158. doi: 10.1038/gim.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloyn AL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 14.Thomas PM, et al. Mutations in the sulfonylurea receptor gene in familial persistent hyperinsulinemic hypoglycemia of infancy. Science. 1995;268(5209):426–429. doi: 10.1126/science.7716548. [DOI] [PubMed] [Google Scholar]
- 15.Johansson BB, et al. The role of the carboxyl ester lipase (CEL) gene in pancreatic disease. Pancreatology. 2018;18(1):12–19. doi: 10.1016/j.pan.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Patel KA, et al. Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun. 2017;8(1):888. doi: 10.1038/s41467-017-00895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoffers DA, Stanojevic V, Habener JF. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J Clin Invest. 1998;102(1):232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weck KE. Interpretation of genomic sequencing: variants should be considered uncertain until proven guilty. Genet Med. 2018;20(3):291–293. doi: 10.1038/gim.2017.269. [DOI] [PubMed] [Google Scholar]
- 19.Bell CJ, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3(65):65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnefond A, et al. Reassessment of the putative role of BLK-p.A71T loss-of-function mutation in MODY and type 2 diabetes. Diabetologia. 2013;56(3):492–496. doi: 10.1007/s00125-012-2794-8. [DOI] [PubMed] [Google Scholar]
- 22.Laver TW, Weedon MN, Caswell R, Hussain K, Ellard S, Flanagan SE. Analysis of large-scale sequencing cohorts does not support the role of variants in UCP2 as a cause of hyperinsulinaemic hypoglycaemia. Hum Mutat. 2017;38(10):1442–1444. doi: 10.1002/humu.23289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shields BM, et al. Population-based assessment of a biomarker-based screening pathway to aid diagnosis of monogenic diabetes in young-onset patients. Diabetes Care. 2017;40(8):1017–1025. doi: 10.2337/dc17-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis NM, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright CF, et al. Genomic variant sharing: a position statement. Wellcome Open Res. 2019;4:22. doi: 10.12688/wellcomeopenres.15090.1. [DOI] [PMC free article] [PubMed] [Google Scholar]