Abstract
Unconventional T cell subsets, including donor-unrestricted T cells (DURTs) and γδ T cells, are promising new players in the treatment and prevention of infectious diseases. In this issue of the JCI, Ogongo et al. used T cell receptor (TCR) sequencing to characterize unconventional T cell subsets in surgical lung resections and blood from Mycobacterium tuberculosis–infected (Mtb-infected) individuals with and without HIV coinfection. The study revealed highly localized expansions of γδ T cell clonotypes not previously associated with the immune response to Mtb and demonstrates the power of high-throughput analysis of the TCR repertoire directly from infected tissue. The findings contribute to our understanding of tuberculosis control and have implications for the development of both therapeutic and vaccination strategies.
The promise of donor-unrestricted T cells
Donor-unrestricted T cells (DURTs) comprise mucosa-associated invariant T cells (MAITs), invariant NK T cells (iNKTs), and germline-encoded mycolyl lipid–reactive T cells (GEMs) (1). Generally speaking, these specialized T cell subsets are characterized by the expression of (semi)invariant T cell receptors (TCRs) and restricted by monomorphic antigen-presenting molecules that bind and display nonpeptide antigens such as microbial metabolites and lipids (1, 2). Similarly, γδ T cells express a limited number of somatically rearranged TCRs and are activated through nonclassical antigen presentation pathways including a recently characterized inside-out presentation mechanism for phosphoantigens involving butyrophilin 3A1 (2, 3).
Given their limited TCRα chain usage, DURT TCRs are more comparable to the pattern recognition receptors of the innate immune system than the highly diverse TCR repertoire that recognizes extensively processed peptide antigens in the context of classical, polymorphic MHC class I and II molecules (2, 4, 5). The chemical nature of the DURT and γδ T cell nonproteinaceous antigens further renders antigen presentation to these cells less susceptible to many pathogen immune evasion strategies specifically developed to subvert the classical peptide antigen–processing machinery. In addition, DURTs differ from classical peptide-specific T cells by their unusually high frequency in both the blood and mucosal tissues and their poised, memory-like phenotype (2). Together, these characteristics make DURTs and γδ T cells attractive targets for both therapeutic and vaccination strategies (2, 5, 6). However, to effectively leverage these unique assets, a better understanding of their function in infectious disease settings is required.
The power of TCR sequencing
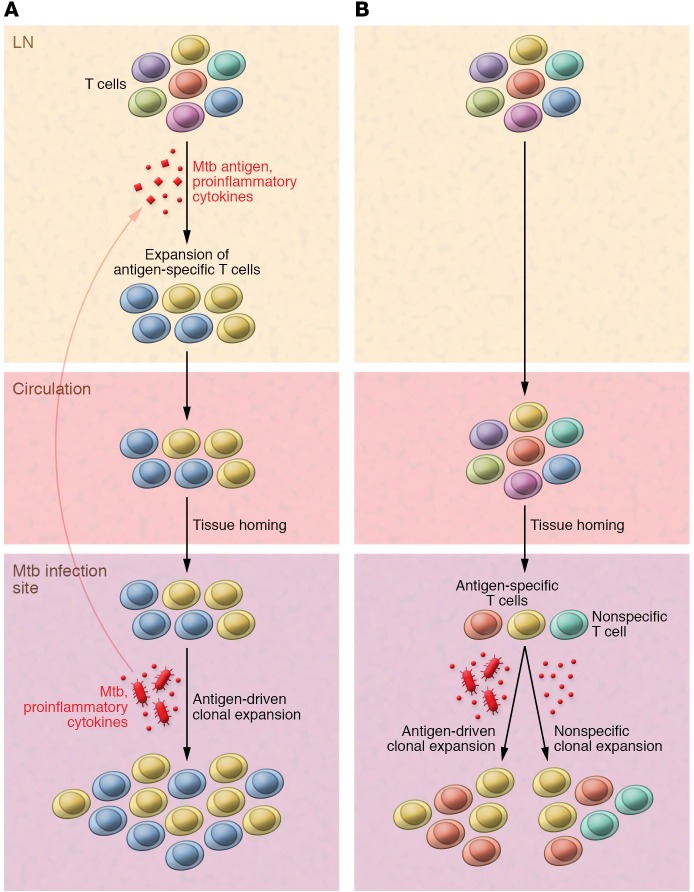
One hallmark of the adaptive immune response is the specific proliferation of T cells that express a pathogen-reactive TCR (7). Thus, tracking T cell subset frequency over the course of an immune response or comparing T cell clone frequency in the affected tissue with its frequency among the total circulating T cells provides a measure of T cell activation. Enrichment of a particular T cell clone or subset in the tissue relative to the blood indicates clonal expansion. If this expansion is antigen driven, it reflects the ability of those T cells to recognize and mount an immune response against a particular pathogen. In this scenario, exposure to cognate antigen in the lymph node activates pathogen-specific T cells, which then recirculate and migrate into the infected tissue. There, they may continue to expand locally due to activation-induced proliferation. As a result, a small number of highly antigen-reactive clones dominate the TCR repertoire at the site of infection (Figure 1A). Since the pool of specifically expanded T cell clones can be a source of long-lived memory T cells, this skewing of the TCR repertoire is an important factor when evaluating potential vaccine targets (7). Alternatively, a stochastic process driven by the limited number of tissue-resident T cell clones already present at the site of infection could explain increased clonality in infected tissue (4, 8–10). This stochastic activation may be particularly relevant in the context of DURTs and γδ T cells, which preferentially reside in nonlymphoid tissues (2, 4). Upon infection, a proportion of these tissue-resident T cells may expand either in response to cognate antigen or through a variety of TCR-independent mechanisms (bystander activation; ref. 11). In the case of stochastic activation of tissue-resident T cells, enrichment of a given T cell subset or clone would not necessarily indicate pathogen reactivity (Figure 1B).
Figure 1. Models of local enrichment of T cell clones.
(A) Antigen-specific T cell clones undergo antigen-driven clonal expansion and are specifically recruited to the infected tissue. (B) A subset of the circulating T cell repertoire stochastically seeds to the tissue and locally present T cell clones expand in response to infection in an antigen-driven or nonspecific manner. LN, lymph node.
Previous studies have shown that both MAITs and iNKTs are reduced in individuals with active Mycobacterium tuberculosis (Mtb) infection, which has led to the hypothesis that these cells may get recruited to the lung upon infection (12–16). Alternatively, these patients may have fewer circulating DURTs to start with, which could predispose them to infection (15). However, these studies have mostly analyzed T cells in the peripheral blood rather than at the site of infection using flow cytometry (14–16). As pointed out by Ogongo et al. in this issue, these types of experiments are confounded by the dynamic expression of cell-surface markers, which are widely used for the identification of T cell subsets but can be downregulated upon activation and thus affect the analysis (17–19). In the present study, the authors addressed both these limitations by analyzing T cells directly from diseased lungs using TCR sequencing (19, 20). This approach offers a unique opportunity to assess the DURT and γδ T cell immune response to Mtb, as the somatically rearranged genomic sequence encoding the TCR is not affected by the activation status of the cell. Access to lung resections further enabled the authors to analyze the TCR repertoire within the affected tissue and compare it with both matched blood samples and blood from healthy controls.
Two unexpected results
When Ogongo et al. analyzed the total TCR repertoire, they found increased clonality in the diseased lung tissue compared with healthy blood (19). This indicates enrichment of certain T cell clones at the site of infection and could be mediated by specific recruitment of antigen-specific cells or expansion of stochastically seeded tissue-resident clones (Figure 1). Next, the authors specifically focused on different unconventional T cell subsets. They found no enrichment of iNKT or GEM TCR sequences in the lungs of patients with tuberculosis compared with matched blood samples (19). Surprisingly, although the new data confirmed the loss of MAITs from the circulation, there was no concurrent enrichment of the MAIT TCR in the lung relative to the blood (19). This finding argues against a specific recruitment of circulating MAITs to the site of infection and instead points to a potential correlation between circulating MAITs and disease susceptibility. Interestingly, deconvoluting the analysis by disease state revealed a specific enrichment of MAITs in the lungs of HIV+ individuals previously infected with Mtb, indicating that MAITs are not depleted from the tissue upon HIV infection despite their loss in the peripheral blood (15, 17, 19, 21). Of note, a similar study has recently shown that MAIT-consistent TCRα chains are enriched in the bronchoalveolar (BAL) fluid compared with blood in patients with active tuberculosis (22). While the TCR repertoire in BAL fluid may differ from that in the lung, it should be noted that clinical indications necessitating surgical lung resections suggest a severely diseased host. Thus, Mtb-infected lung samples from surgical resections may not represent a model of Mtb control. Furthermore, antigen-driven, selective expansions of MAIT clones expressing specific TCRβ chains have been reported in the context of infection (23, 24), and hence determining paired TCR sequences by single-cell TCR sequencing may reveal clonal expansions not detectable by the analysis of TCRα chains only.
Ogongo et al. subsequently analyzed the γδ T cell compartment and showed that certain TCRδ clonotypes were highly expanded in Mtb-infected lungs and thus may play a role in the immune response to this pathogen. Although Vδ1+ γδ T cells are known to dominate in the tissue (2, 6), perhaps the most surprising finding of this study was a skewing of the pulmonary γδ TCR repertoire toward T cells that did not express Vγ9Vδ2. Vγ9Vδ2 γδ T cells are known to recognize mycobacterial phosphoantigens in a butyrophilin 3A1–dependent manner (2, 3, 6) but were found to be depleted in the infected lungs (19). Instead, there was significant heterogeneity among the predominant γδ TCRs even within the same lung, suggesting that the clonal expansions were highly localized. Similarly, Hunter et al. recently reported that distinct Vδ2–γδ clonotypes are overrepresented in the liver, further supporting the notion that tissue-resident γδ TCR repertoires differ from their circulating counterparts (10).
Strikingly, Ogongo et al. identified a number of TCRδ clonotypes that were highly expanded in the lungs of multiple individuals, albeit at lower frequencies than the previously characterized bona fide invariant TCRs expressed by MAITs or GEMs (19). Since enrichment for Vδ1+ γδ T cells was also seen in the absence of infection, one explanation for this unexpected result may be the stochastic, local expansion of tissue-resident clones by virtue of their relative abundance (Figure 1B). Whereas the proinflammatory microenvironment may cause unspecific bystander activation of these cells, it is tempting to speculate that novel mycobacterial antigens could drive this local expansion. These antigens may have remained undiscovered because of the dominance of phosphoantigen-responsive Vγ9Vδ2 cells in the blood. To distinguish between these two scenarios, defining the antigen specificity of this unique T cell subset will be of pivotal importance.
Concluding remarks
Here, Ogongo and colleagues describe a previously unappreciated population of unconventional T cells in Mtb-infected human lungs (19). Future studies should further characterize these pulmonary Vδ2– γδ T cells and include them in the development of novel therapeutic and vaccination strategies. Moreover, the robust detection of DURTs in the lungs of Mtb-infected individuals encourages further research into how we can harness these specialized T cell subsets in the fight against tuberculosis.
Acknowledgments
We apologize to those colleagues whose work we could not cite due to space constraints. This work was supported by VA Merit Award I01BX000533 (to DML); NIH grant R01 AI134790 (to DML and DAL); and NIH grant R01 AI129980 (to DML and DAL). The contents do not represent the views of the US Department of Veterans Affairs or the United States Government.
Version 1. 11/25/2019
Electronic publication
Version 2. 01/02/2020
Print issue publication
Footnotes
Conflict of interest: DAL and DML are co-inventors on patents that protect the development of tuberculosis antigens recognized by human CD8+ T cells for diagnostic and vaccine use. These are: US8961989B2, Methods for producing an immune response to tuberculosis; US8053181B2, Methods for detecting a Mycobacterium tuberculosis infection; US20150273039A1, Compositions comprising soluble HLA/M. Tuberculosis-specific ligand complexes and methods of production and use thereof; and US20180273536A1, Small molecules that bind mr1.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):68–70. https://doi.org/10.1172/JCI133119.
See the related article at Differential skewing of donor-unrestricted and γδ T cell repertoires in tuberculosis-infected human lungs.
Contributor Information
Corinna A. Kulicke, Email: kulicke@ohsu.edu.
Deborah A. Lewinsohn, Email: lewinsde@ohsu.edu.
David M. Lewinsohn, Email: lewinsod@ohsu.edu.
References
- 1.Van Rhijn I, Moody DB. Donor unrestricted T cells: a shared human T cell Response. J Immunol. 2015;195(5):1927–1932. doi: 10.4049/jimmunol.1500943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16(11):1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y, et al. A structural change in butyrophilin upon phosphoantigen binding underlies phosphoantigen-mediated Vγ9Vδ2 T cell activation. Immunity. 2019;50(4):1043–1053.e5. doi: 10.1016/j.immuni.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Fan X, Rudensky AY. Hallmarks of tissue-resident lymphocytes. Cell. 2016;164(6):1198–1211. doi: 10.1016/j.cell.2016.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joosten SA, et al. Harnessing donor unrestricted T-cells for new vaccines against tuberculosis. Vaccine. 2019;37(23):3022–3030. doi: 10.1016/j.vaccine.2019.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dantzler KW, de la Parte L, Jagannathan P. Emerging role of γδ T cells in vaccine-mediated protection from infectious diseases. Clin Transl Immunology. 2019;8(8):e1072. doi: 10.1002/cti2.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2(4):251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 8.Gebhardt T, Mueller SN, Heath WR, Carbone FR. Peripheral tissue surveillance and residency by memory T cells. Trends Immunol. 2013;34(1):27–32. doi: 10.1016/j.it.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Wakim LM, Waithman J, van Rooijen N, Heath WR, Carbone FR. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 2008;319(5860):198–202. doi: 10.1126/science.1151869. [DOI] [PubMed] [Google Scholar]
- 10.Hunter S, et al. Human liver infiltrating γδ T cells are composed of clonally expanded circulating and tissue-resident populations. J Hepatol. 2018;69(3):654–665. doi: 10.1016/j.jhep.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whiteside SK, Snook JP, Williams MA, Weis JJ. Bystander T cells: a balancing act of friends and foes. Trends Immunol. 2018;39(12):1021–1035. doi: 10.1016/j.it.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salou M, Franciszkiewicz K, Lantz O. MAIT cells in infectious diseases. Curr Opin Immunol. 2017;48:7–14. doi: 10.1016/j.coi.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Van Kaer L, Parekh VV, Wu L. The response of CD1d-restricted invariant NKT cells to microbial pathogens and their products. Front Immunol. 2015;6:226. doi: 10.3389/fimmu.2015.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11(8):701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 15.Wong EB, et al. Low levels of peripheral CD161++CD8+ mucosal associated invariant T (MAIT) cells are found in HIV and HIV/TB co-infection. PLoS ONE. 2013;8(12):e83474. doi: 10.1371/journal.pone.0083474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kee SJ, et al. Dysfunction of natural killer T cells in patients with active Mycobacterium tuberculosis infection. Infect Immun. 2012;80(6):2100–2108. doi: 10.1128/IAI.06018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leeansyah E, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 2013;121(7):1124–1135. doi: 10.1182/blood-2012-07-445429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson MT, et al. The response of natural killer T cells to glycolipid antigens is characterized by surface receptor down-modulation and expansion. Proc Natl Acad Sci U S A. 2003;100(19):10913–10918. doi: 10.1073/pnas.1833166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogongo PS, et al. Differential skewing of donor-unrestricted and γδ T cell repertoires in tuberculosis-infected human lungs. J Clin Invest. 2020;130(1):214–230. doi: 10.1172/JCI130711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matos TR, de Rie MA, Teunissen MBM. Research techniques made simple: high-throughput sequencing of the T-cell receptor. J Invest Dermatol. 2017;137(6):e131–e138. doi: 10.1016/j.jid.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cosgrove C, et al. Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood. 2013;121(6):951–961. doi: 10.1182/blood-2012-06-436436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong EB, et al. TRAV1-2+ CD8+ T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis. Commun Biol. 2019;2:203. doi: 10.1038/s42003-019-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold MC, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. 2014;211(8):1601–1610. doi: 10.1084/jem.20140507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howson LJ, et al. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with Salmonella paratyphi A. Nat Commun. 2018;9(1):253. doi: 10.1038/s41467-017-02540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]